Significance
Clustered regularly interspaced short palindromic repeat (CRISPR)-CRISPR associated (Cas) immunity relies on adaptive acquisition of spacers from foreign DNA into prokaryotes genomes. Efficient “primed” acquisition leads to selective intake of extra spacers from DNA molecules recognized by Cas proteins complexed with CRISPR RNAs partially matching foreign DNA. In contrast, fully matching targets are destroyed without new spacer acquisition. We show that when the rate of degradation of DNA with fully and partially matching targets is made equal, fully matching protospacers stimulate primed adaptation much more efficiently than partially matching ones. Thus, different outcomes of CRISPR-Cas response to two kinds of targets are caused by much more rapid destruction of fully matching targets rather than by structural differences in complexes formed on fully and partially matching targets.
Keywords: CRISPR-Cas, CRISPR interference, primed CRISPR adaptation
Abstract
Prokaryotic clustered regularly interspaced short palindromic repeat (CRISPR)-CRISPR associated (Cas) immunity relies on adaptive acquisition of spacers—short fragments of foreign DNA. For the type I-E CRISPR-Cas system from Escherichia coli, efficient “primed” adaptation requires Cas effector proteins and a CRISPR RNA (crRNA) whose spacer partially matches a segment (protospacer) in target DNA. Primed adaptation leads to selective acquisition of additional spacers from DNA molecules recognized by the effector–crRNA complex. When the crRNA spacer fully matches a protospacer, CRISPR interference—that is, target destruction without acquisition of additional spacers—is observed. We show here that when the rate of degradation of DNA with fully and partially matching crRNA targets is made equal, fully matching protospacers stimulate primed adaptation much more efficiently than partially matching ones. The result indicates that different functional outcomes of CRISPR-Cas response to two kinds of protospacers are not caused by different structures formed by the effector–crRNA complex but are due to the more rapid destruction of targets with fully matching protospacers.
Clustered regularly interspaced short palindromic repeat (CRISPR)-CRISPR associated (Cas) systems provide their prokaryotic hosts with small RNA-based defense against mobile genetic elements such as viruses and plasmids (1–3). Although evolutionary and mechanistically diverse, all such systems comprise CRISPR DNA arrays of identical repeats separated by unique spacers and cas genes (4). Functionally, CRISPR-Cas systems can be divided into two modules. The acquisition module appropriates spacers from foreign DNA into CRISPR arrays and consists of proteins Cas1 and Cas2, homologous in all CRISPR-Cas systems (4). The Cas1 and Cas2 proteins from Escherichia coli alone are able to perform the spacer acquisition reaction in vitro (5) and are also sufficient for spacer acquisition in vivo in the absence of other Cas proteins (6, 7). Whenever a new spacer is acquired, a new copy of CRISPR repeat is also generated (1, 6).
Acquired spacers become a source of small CRISPR RNAs (crRNAs) programmed against DNA from which they originated. Individual crRNAs are bound by Cas effector proteins from the interference module and recognize foreign nucleic acids through complementary interactions between the targeted sequence (protospacer) and matching crRNA spacer (2). Interference module proteins are diverse, and this diversity forms a basis of classification of CRISPR-Cas systems into two classes and several types (4). In DNA-targeting type I and type II CRISPR-Cas systems, target recognition requires, in addition to crRNA spacer–target protospacer complementarity, a protospacer adjacent motif (PAM) (8, 9) recognized by effector proteins (10–12). Upon target DNA recognition, a stable R-loop containing locally melted protospacer DNA and an RNA–DNA heteroduplex is formed (13, 14). R-loop formation is followed by target DNA destruction either by the effector complex alone (type II) or through recruitment of an additional endonuclease Cas3 (type I systems) (4). Cas1 and Cas2 are not required for interference in vivo (2) and in vitro (15, 16).
Point mutations in protospacer or associated PAM decrease the affinity of crRNA–effector complex binding to protospacer (17). Under pressure from CRISPR-Cas, mobile genetic elements accumulate such mutations, which allows them to escape CRISPR interference (8, 17). For several type I systems, spacer acquisition from DNA molecules containing “escape” protospacers is very strongly stimulated (18–22) compared with “naïve” adaptation, which requires just Cas1 and Cas2. In the case of the E. coli type I CRISPR-Cas system, this specific version of spacer acquisition, referred to as “primed adaptation” (18), requires not just Cas1 and Cas2 but also all other components of the effector Cascade complex (Cse1, Cse2, Cas7, Cas5, Cas6e, and crRNA) and the Cas3 nuclease. Primed adaptation is beneficial to the host, as it leads to specific acquisition of spacers from genetic parasites that “learned” to evade defenses provided by earlier acquired spacers.
Because primed adaptation relies on specific recognition of partially matching protospacers by crRNA, a question arises as to whether such a complex, which causes preferential acquisition of new spacers from a DNA strand opposite to the one recognized by crRNA (18), is different from a complex formed with a fully matching protospacer, which leads to CRISPR interference. Recently, biophysical methods were used to present evidence that effector complexes formed by the E. coli Cascade with partially matching protospacers are structurally different from complexes formed with fully matching protospacers (23) and may stimulate recruitment of adaptation module enzymes (24). Here, we asked whether interactions with fully matching protospacers can support primed adaptation. Unexpectedly, our data show that interaction of the effector complex with a fully matching protospacer in fact stimulates primed adaptation to a much greater extent than the interaction with a partially matching protospacer. The apparent absence of efficient adaptation from DNA with fully matching protospacers stems from the fact that such DNA molecules are destroyed rapidly, leaving no time for spacer acquisition to occur. Our results thus suggest that there may be no qualitative difference between effector complexes formed on fully and partially matching targets: Different biological outcomes are the consequences of different kinetics of degradation of and spacer adaptation from the same target. When viewed in this context, the priming phenomenon itself must be considered as a direct consequence of target interference, which provides substrates for adaptation and directs the adaptation machinery to foreign DNA, minimizing the probability of self-destruction.
Materials and Methods
Strains and Plasmids.
E. coli KD263 (K-12 F+, lacUV5-cas3 araBp8-cse1, CRISPR I: repeat-spacer g8-repeat, CRISPR II deleted) has been described (25). KD546 is a KD263 derivative containing araBp promoter for cas3 expression and a lacUV5 promoter inserted between the cas6e and cas1 genes by means of recombineering (26). Strain KD384 is equal to KD263 but contains a trap spacer (5′-CGCAGATACCAAATACTGTTCTTCTAGTGTAG-3′) instead of a g8 spacer. It was also obtained by recombineering.
The pT7Blue-based plasmids pG8 and pG8mut carrying a 209-bp M13 fragment with the g8 protospacer (genome positions 1311–1519) with or without an escape mutation C1T at the first position of the protospacer have been described in ref. 18. The pRSF plasmid derivatives containing g8 or g8 mutant protospacers (pRSF_G8 and pRSF_G8mut) were constructed by subcloning the 209-bp fragments into NcoI and NotI sites of the pRSF-1b vector (Novagen). The plasmids RwFm and RmFw were generated by cloning a synthetic oligonucleotide duplex containing g8 or g8 mutant protospacer and functional ATG PAM into HindIII and XbaI sites of the pT7Blue vector and subsequent cloning of a 1,895-bp M13 fragment containing the g8 protospacer (genome positions 6874–1519) with or without the C1T mutation into the XbaI and KpnI sites. The resulting plasmids RwFm and RmFw contain both g8 and g8 mutant protospacers located on different strands separated by a 1,741-bp fragment.
CRISPR Interference and Adaptation Assays.
E. coli KD263, KD546, or KD384 were transformed with individual plasmids or plasmid combinations, and transformants were selected on LB agar plates containing 50 μg/mL ampicillin (to select for pT7Blue-based plasmids) with or without 50 μg/mL kanamycin to select for pRSF-based plasmids. Individual colonies were inoculated for overnight growth at 37 °C in liquid LB with appropriate antibiotics. Aliquots of overnight cultures were diluted 100 times into fresh LB without antibiotic and allowed to grow until the culture OD600 reached ∼0.5. At this point, half of the culture was induced with 1 mM arabinose and 1 mM IPTG. Another half of the culture remained uninduced. The cultivation was continued, and aliquots were withdrawn at various times postinduction. Where indicated, induction at of cultures that reached a higher OD600 (OD600 ∼1) was used. Serial dilutions of withdrawn culture aliquots were plated on LB agar with or without 50 μg/mL ampicillin to obtain colony forming unit (CFU) numbers. To observe plasmid loss, the plasmid was purified from identical volumes of induced culture aliquots using Thermo Scientific GeneJET Plasmid Miniprep Kit, and purified plasmids were analyzed on 0.9% agarose gels. CRISPR array expansion was monitored by PCR. For KD263 and KD546 cultures, amplifications with Ec-LDR-F (5′-AAGGTTGGTGGGTTGTTTTTATGG-3′) and M13g8 (5′-GGATCGTCACCCTCAGCAGCG-3′) primers were performed. The latter primer anneals to the g8 spacer. Ec-LDR-F anneals to the leader sequence upstream of the CRISPR array. To analyze CRISPR array expansion in KD384 cultures induced at OD600 = 1, amplification with Ec-LDR-F and Ec_minR (5′-CGAAGGCGTCTTGATGGGTTTG-3′) was used. Ec_minR anneals to the region downstream of the CRISPR array.
New Spacer Composition Analysis.
To monitor the composition of acquired spacers, PCR products corresponding to expanded CRISPR arrays were gel purified with QIAquick Gel Extraction Kit (QIAGEN) and sequenced using Miseq Illumina in pair-end 250 bp-long reads mode according to the manufacturer’s protocols. Raw sequencing reads preprocessing was done using ShortRead and BioString packages (27, 28). After adapter and quality (score of >20) trimming, reads with two or more CRISPR repeats were filtered, and sequences between two repeats were extracted as spacers. For reads corresponding to multiple spacer acquisitions, only spacers that were acquired first were selected for further analysis to avoid biases in spacer composition caused by secondary priming. Spacers were mapped to the source plasmid with no mismatches tolerated. Graphical representation was done with the EasyVisio tool developed by E. Rubtsova.
Results
Experimental Setup.
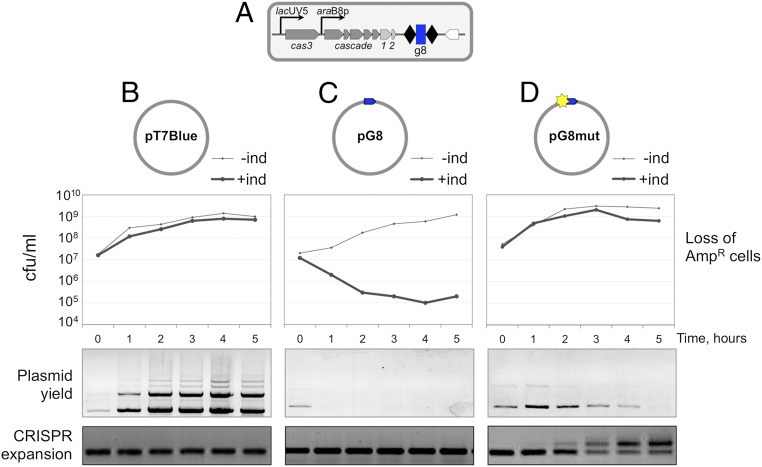
E. coli K12 KD263 cells (25) (Fig. 1A) express cas genes from inducible promoters and contain a minimized CRISPR array with a single g8 spacer derived from the M13 phage. The KD263 cells were transformed with ampicillin-resistant pT7Blue vector plasmid (Fig. 1B); pG8, a pT7Blue derivative containing a cloned g8 protospacer fully matching the g8 spacer with a functional ATG PAM (Fig. 1C); or pG8mut, which contains a C1T mutation in the seed region of the protospacer (Fig. 1D). Ampicillin-resistant transformants were grown in the absence of ampicillin until OD600 = 0.5, after which cas gene expression was induced. Induced cultures grew at the same rate as uninduced controls. However, cells transformed with pG8 rapidly lost ampicillin resistance upon induction (Fig. 1C), whereas control cells transformed with pT7Blue remained antibiotic-resistant (Fig. 1B). Induced cultures of cells transformed with pG8mut remained antibiotic-resistant for most of the experiment, with some loss becoming apparent at later times (Fig. 1D). Thus, the C1T mutation in the protospacer renders CRISPR-Cas interference inefficient, as expected. Plasmid loss was also monitored directly, by purifying plasmids from aliquots of induced cultures and electrophoretic analysis. The results showed that the yield of pT7Blue increased along with culture density (Fig. 1B). The pG8 plasmid was effectively purged from the culture 1 h postinduction (Fig. 1C). The amount of pG8mut plasmid was close to that in control pT7Blue harboring cultures 1 h postinduction and then slowly decreased (Fig. 1D). Thus, a mismatch between the g8 crRNA spacer and the C1T protospacer does not completely inactivate CRISPR interference. The low yield of pG8mut is in apparent contrast with the high level of ampicillin resistance in induced cultures. Because the culture continued to grow at least between the first and second hour postinduction with no detectable loss of ampicillin-resistant cells, it follows that CRISPR interference decreased the copy number of pG8mut without having a visible effect on antibiotic resistance. We note that pT7Blue and its derivatives are high-copy number plasmids (∼500 plasmid copies per cell) (29), and so antibiotic resistance can be detected even when the number of plasmids inside cells is decreased by the action of the CRISPR-Cas system.
Fig. 1.
CRISPR interference and primed CRISPR adaptation against matching and partially matching protospacer targets located on plasmids. (A) The E. coli KD263 cells capable of inducible cas gene expression and carrying an engineered CRISPR array with a single g8 spacer are schematically shown. E. coli KD263 was transformed with ampicillin-resistant pT7Blue vector (B) or pT7Blue-based plasmid pG8 (C) with a fully matching g8 protospacer with a functional PAM or pG8mut (D), carrying, respectively, a protospacer with a functional PAM and a C1T mutation in the protospacer. Protospacers are shown as blue arrows with the arrowpoint directed away from the 5′-ATG-3′ PAM. The C1T mutation is shown as a yellow star. Graphs show the number of ampicillin-resistant CFUs in cultures transformed with each plasmid with or without induction of cas gene expression. Plasmid DNA was purified from cells collected at the times indicated and resolved by agarose gel electrophoresis. The products of PCR amplification with primers annealing at the g8 spacer and further upstream in the CRISPR array leader are shown at the bottom.
Aliquots of induced cultures were also subjected to PCR amplification using a pair of primers of which one annealed to the g8 spacer and another to the upstream leader region outside the array. In cultures transformed with pT7Blue or pG8, only a PCR product corresponding to the unexpanded CRISPR array was observed (Fig. 1 B and C). In contrast, in cultures of cells transformed with pG8mut, time-dependent accumulation of longer PCR product corresponding to an insertion of an additional repeat-spacer unit in the array was observed (Fig. 1D). Accumulation of new spacers was barely detectable 1 h postinduction, but by 5 h, most cells in the culture contained expanded arrays.
Primed Adaptation from Targets Containing Fully and Partially Matching Protospacers Located in Trans.
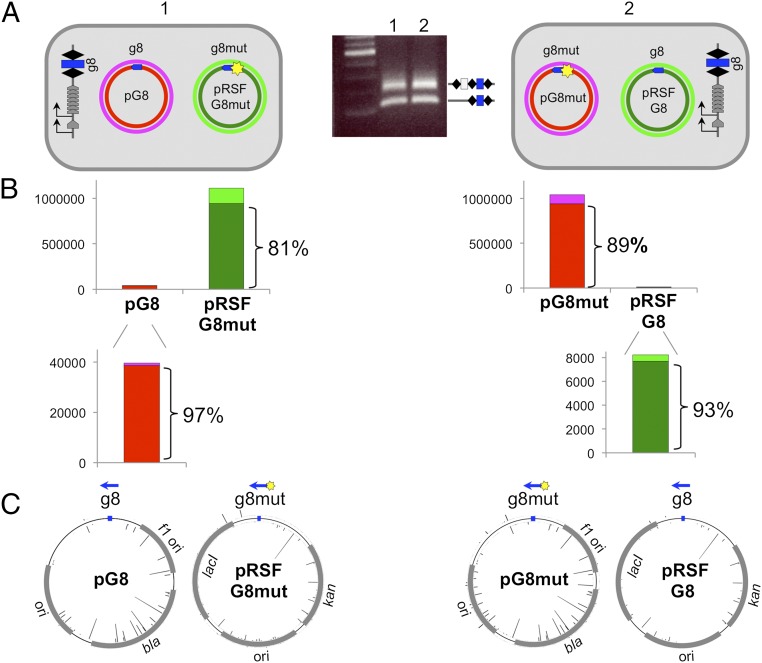
We considered that ongoing primed adaptation from a plasmid containing a mutant protospacer could act in trans and stimulate acquisition from a plasmid with a fully matching protospacer present in the same cell. Accordingly, pRSF-based plasmids with a wild-type or mutant g8 protospacer were created. These plasmids are compatible with pG8 and pG8mut plasmids. Plasmid pairs indicated in Fig. 2A were cotransformed in KD263. Upon induction, robust adaptation was observed in cultures containing either plasmid pair (Fig. 2A). PCR fragments corresponding to expanded CRISPR arrays were subjected to Illumina sequencing, and extracted sequences of acquired spacers were mapped. Because the two plasmids were different from each other, spacers could be unequivocally mapped to a plasmid from which they originated. For each plasmid pair analyzed, most (97% and 99%) spacers originated from a plasmid that carried the mutant protospacer, and a strong (81% and 89%) bias for selection of spacers from a nontargeted strand was observed (Fig. 2B, Top). When spacers originating from plasmids with fully matching protospacers were analyzed, an even higher bias of spacer selection from the nontargeted strand was also observed (97% and 93%; Fig. 2B, Bottom). Mapping of acquired spacers showed that their distribution (and therefore the use of donor protospacers) was very close for identical (pRSF- or pT7Blue-based) plasmids with g8 or g8mut protospacers (Fig. 2C).
Fig. 2.
CRISPR adaptation from matching and partially matching protospacer targets located in an in trans configuration. (A) The E. coli KD263 cells transformed with compatible pT7Blue or pRSF-based plasmids each carrying a wild-type or a mutant, C1T version of the g8 protospacer are schematically shown. Protospacers are indicated as blue arrows located on the nontargeted DNA strand of each plasmid, with the arrow pointing away from the ATG PAM. Results of a CRISPR adaptation experiment with the cells transformed with two plasmids (culture 1 and culture 2) are shown in the middle. (B) Results of overall mapping of spacers acquired by cells shown in A to separate strands of each plasmid are shown. Numbers on vertical axes show the number or reads. The color code used shows the origin of spacers and matches colors used to indicate the individual plasmid strands in A. (C) Mapping of individual acquired spacers on donor plasmids. Blue arrows indicate the position of the g8 protospacer. Bars protruding inside and outside the circles indicating plasmids represent spacers derived from different strands. Bar height indicates the relative efficiency of spacer acquisition from this position.
Trap Protospacer–Spacer Interaction Reveals Primed Adaptation from Fully Matching Targets.
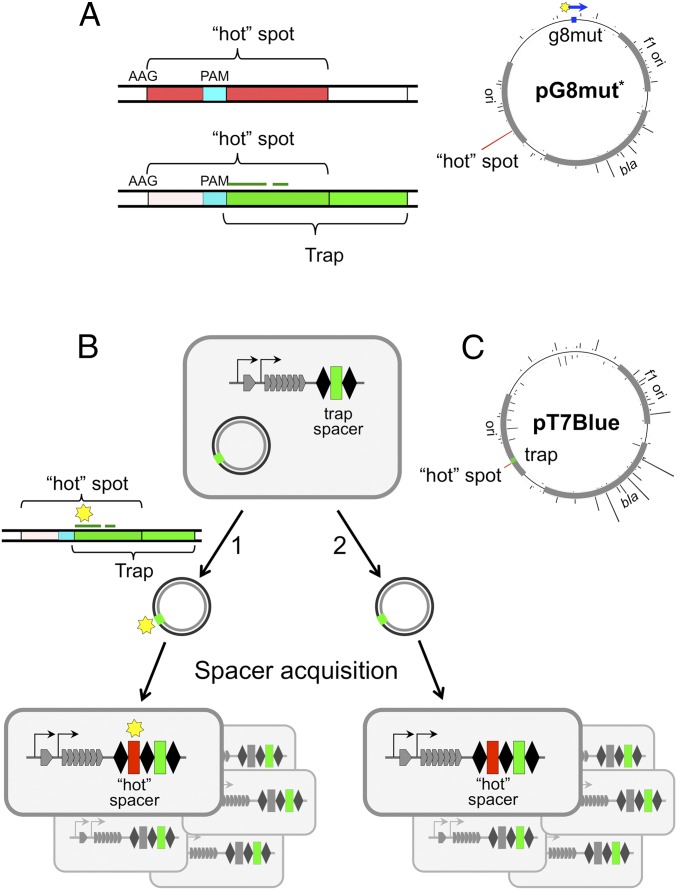
The results presented above suggest that primed adaptation may be initiated upon the interaction of Cascade–crRNA complexes with fully matching protospacers that are subject to CRISPR interference. However, it is possible that initially rare plasmids with escape mutations in protospacer or PAM accumulate in the course of CRISPR interference and are next used for primed spacer acquisition. To rule out such a possibility, the following approach was used. Some protospacers from the pT7Blue vector backbone reproducibly contribute a disproportionately large number of spacers to cells that undergo primed adaptation (30). We found one such “hot” protospacer that contained a centrally located internal GAG—an interference-proficient PAM (Fig. 3A). We reasoned that a 32-bp sequence located downstream of this trinucleotide could itself function as a priming protospacer in the presence of appropriate crRNA. For reasons outlined subsequently, we refer to this priming protospacer as “trap.” An E. coli strain KD384 containing a CRISPR array with a trap spacer was constructed (Fig. 3B) and used in a primed adaptation experiment with the pT7Blue plasmid. As shown in Fig. 3A, the entire seed region of the trap protospacer and its PAM are contained within the hot protospacer. If adaptation was indeed driven by priming on plasmid variants with mutations in the trap protospacer seed or PAM, spacers originating from the overlapping hot protospacer shall contain corresponding substitutions (Fig. 3B, pathway 1). In contrast, if adaptation was primed on wild-type trap, spacers acquired from an overlapping hot protospacer should contain an unchanged, wild-type trap segment (Fig. 3B, pathway 2).
Fig. 3.
Using a trap spacer to show adaptation from fully matching protospacers. (A) Mapping of spacers acquired by KD263 cells transformed with pG8mut* are shown [note that the orientation of the protospacer in pG8mut (Figs 1. and 2) and in pG8mut* is different]. The height of bars emanating from a circle representing pG8mut* shows the relative amounts of spacers acquired from this site. The sequence of a hot protospacer (indicated by a red bar) is expanded. It contains an internal PAM sequence (cyan) defining a trap protospacer shown in green. The position of the trap protospacer seed with respect to the partially overlapping hot protospacer is shown by a thin green line. (B) The E. coli KD384 cells containing a trap spacer in their CRISPR array and transformed with the pT7Blue plasmid are schematically shown to the left. At the right, structures of CRISPR arrays of cells that acquired a hot spacer (red) from a protospacer overlapping with protospacer recognized by trap crRNA are schematically shown. (C) Mapping of spacers acquired by KD384 cells transformed with pT7Blue. The position of the priming trap protospacer is shown as a green rectangle. Red bar shows observed efficiency of spacer acquisition from the overlapping hot protospacer.
The pT7Blue plasmid was rapidly lost from KD384 cultures upon induction of cas genes expression, and no CRISPR array expansion was detected by PCR, an expected result for a fully matching target with an interference-competent PAM. However, when cas gene expression was induced in late KD384 cultures, a low amount of PCR fragments corresponding to expanded CRISPR arrays was detected. Induction of cas genes after cessation of cell growth increases the proportion of cells with expanded arrays, as at standard conditions (inducing cells and allowing them to grow in the absence of ampicillin over extended periods of time) the proportion of cells that have lost the plasmid due to CRISPR interference increases steadily over the course of the experiment. Plasmid-free cells do not acquire spacers, but their nonexpanded CRISPR arrays contribute a strong background signal during PCR amplification, making it difficult to detect rare cells with expanded arrays. PCR fragments corresponding to expanded arrays were Illumina sequenced, and sequences corresponding to newly acquired spacers were retrieved and mapped on the parent plasmid (Fig. 3C). A strand bias similar to that observed during adaptation primed by g8 crRNA from the pG8mut plasmid was observed (compare Fig. 3C and Fig. 3A). A total of 2,844 reads of hot spacer were obtained. All of them contained a wild-type trap sequence. Therefore, we conclude that primed adaptation can be initiated by effector complex interaction with a protospacer preceded by an interference-proficient PAM and fully matching the crRNA spacer.
Primed Adaptation from Targets Containing Fully and Partially Matching Protospacers Located in Cis.
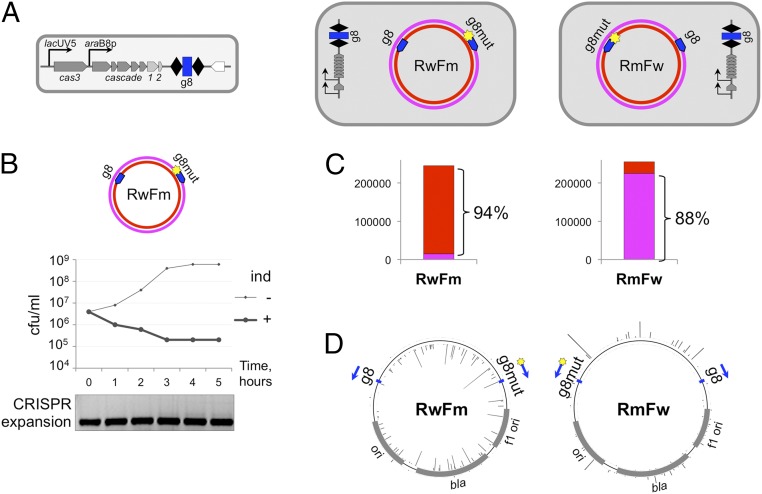
No conclusions about the relative efficiency of primed adaptation from DNA containing fully matching and partially matching targets can be made from experiments with a trap protospacer. To directly compare these efficiencies, pT7Blue-based plasmids carrying two g8 protospacers—one wild-type and another with the C1T substitution—were created. The protospacers were located in opposing orientations such that each one of them primed an acquisition of spacers from different DNA strands. If the two protospacers were equally efficient in priming, no overall strand bias of acquired spacers should be observed. Conversely, a bias toward one of the strands would indicate that priming from one of the protospacers is more efficient. The KD263 cells transformed with the double-protospacer RwFm plasmid were indistinguishable from cells transformed with a single fully matching protospacer pG8 plasmid: They rapidly lost ampicillin resistance upon induction and did not efficiently acquire new spacers (Fig. 4A). Thus, a fully matching protospacer is “dominant” over the partially matching one when present in cis. Trace amounts of PCR fragments with expanded CRISPR arrays were recovered from induced late KD263 cultures carrying RwFm or RmFw, a double-protospacer plasmid, which contains a reciprocal combination of protospacers (Fig. 4B). After high-throughput sequencing and mapping of acquired spacers, a strong strand bias was observed (Fig. 4C). Unexpectedly, the direction of the bias was consistent with more active primed adaptation initiated at the fully matching protospacer for both plasmids.
Fig. 4.
CRISPR adaptation from matching and partially matching protospacer targets located in an in cis configuration. (A) The E. coli KD263 cells transformed with pT7Blue-based RwFm or RmFw plasmids carrying a wild-type and a mutant, C1T, version of the g8 protospacer on different strands are schematically shown at the top. (B) A graph showing the number of CFUs on LB plates containing ampicillin in KD263 cultures transformed with one of the plasmids shown in A with or without induction of cas gene expression. Shown below are the results of agarose gel electrophoresis of PCR amplification products obtained from aliquots of induced culture with primers annealing at the g8 spacer and further upstream in the KD263 CRISPR array leader. (C) Bar graphs showing the overall statistics of acquired spacers in each cell culture shown in A with spacers mapped to either plasmid DNA strand shown in different colors (color coded as in plasmid schematics in A). (D) Mapping of spacers acquired by KD263 cells transformed with RwFm or RmFw. The positions of priming protospacers are shown by blue arrows.
Increased Cas1 and Cas2 Production Leads to Efficient Primed Adaptation from Targets Containing a Protospacer Fully Matching crRNA.
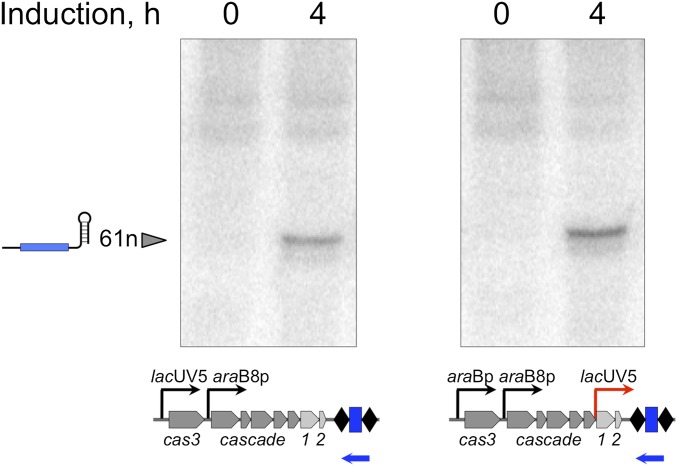
The result presented in the previous section argues that initiation of primed adaptation from protospacers with a functional PAM and fully matching crRNA spacer occurs very efficiently, but the overall yield is nevertheless low, because the targeted plasmid is rapidly destroyed by the interference machinery. Therefore, if the rate of spacer acquisition were increased, one could expect efficient primed adaptation even with fully matching targets. To test this prediction, KD456, a derivative of the KD263 strain with an additional inducible lacUV5 promoter inserted in front of cas1cas2 genes, was constructed (Fig. 5A). This strain was transformed with pG8, or pG8mut, and after induction of cas gene expression, spacer acquisition was monitored. Upon induction, the level of g8 crRNA was the same in both strains (Fig. S1), an expected result, as the crRNA precursor is transcribed from its own promoter (31). Spacers were efficiently acquired by exponentially growing KD456 cultures transformed with either plasmid (Fig. 5A, lanes 3 and 4). The control KD263 cultures behaved as expected—efficient CRISPR array expansion was only observed in the presence of pG8mut (lane 2). Because increased concentration of Cas1 and Cas2 could have stimulated nonprimed adaptation (6), acquired spacers were sequenced and analyzed. Mapping showed that the distribution of donor protospacers along the plasmid backbone, the strand bias, and the AAG PAM preference were typical for primed adaptation (Fig. 5B). Thus, increasing the intracellular concentration of Cas1 and Cas2 (or the ratio of these proteins to Cascade–crRNA and/or Cas3) is sufficient to allow primed adaptation from targets containing a protospacer with a functional PAM and an exact match with the crRNA spacer.
Fig. 5.
Increased expression of cas1 and cas2 leads to primed adaptation from fully matching protospacer targets. (A) An agarose gel showing the results of separation of products of PCR amplification of CRISPR arrays from aliquots of KD263 (lanes 1 and 2) and KD546 (lanes 3 and 4) transformed with indicated plasmids. (B) Results of mapping of spacers acquired by KD263 cultures transformed with pG8mut (2) and KD546 cultures transformed by pG8 and pG8mut (3, 4) on donor plasmids.
Fig. S1.
Northern blotting of RNA prepared from induced KD263 (Left) and KD546 (Right) cells. Total RNA was isolated from 5 mL of indicated E. coli cell cultures grown for 4 h after induction with 1 mM arabinose and 1 mM IPTG or in the absence of inducers. We separated 10 μg of total RNAs on a denaturing 8 M urea/15% (wt/vol) polyacrylamide gel and electrophoretically transferred it to the Hybond-XL membrane (GE Healthcare). ExpHyb hybridization solution (Clontech) was used for hybridization according to the manufacturer’s instructions for 1 h at 37 °C with 32P-end–labeled, g8 spacer-specific probe (5′-GCGGGATCGTCACCCTCAGCAGCGAAAGACAG-3′, indicated as a blue leftward arrow) as described previously (27).
Discussion
In this work, we demonstrate that primed adaptation by the type I-E CRISPR-Cas system from E. coli can occur after the Cascade–crRNA complex interacts with a fully matching protospacer that is subject to interference. In fact, when matched and mismatched protospacers are positioned on the same DNA molecule, primed adaptation initiated by recognition of a fully matching target is about 10 times more efficient than from a mismatched target. In contrast, when tested separately, mismatched rather than matched targets behave as preferred substrates for primed adaptation (18, 19). We propose that the apparent lack of adaptation from target DNA molecules with protospacers fully matching crRNA spacers is a consequence of rapid destruction of such targets. Target destruction occurs due to the activity of the Cas3 helicase–endonuclease that recognizes Cascade–crRNA complexes bound to protospacers (15, 16). We further propose that Cas3-mediated target destruction is a source of substrates for spacer acquisition machinery, the Cas1–Cas2 complex, during primed adaptation. Our data, as well as data from others, show that interference is initiated on either matching or partially matching protospacers (32), albeit with different efficiencies. The Cascade–crRNA interaction with fully matching protospacers leads to rapid degradation of DNA molecules that contain them. Apparently, intermediates of target DNA degradation are transient and disappear after an initial burst, leaving no substrates for Cas1–Cas2-mediated adaptation, which is either an intrinsically slower process or is limited by the insufficient concentrations of Cas1 and Cas2, which are encoded by promoter-distal genes of the cse1–cas2 operon. Increasing the concentration of Cas1 and Cas2 leads to robust spacer acquisition, presumably by allowing Cas1 and Cas2 to capture the transient intermediates of Cas3 action.
Cascade interaction with mutant, escape protospacers causes a much slower loss of targeted DNA molecules. This must be a combined effect of lowered binding affinity of Cascade–crRNA to mutated protospacers (and the consequent lower rate of their degradation) and the ability of plasmid copy number maintenance mechanisms (or phage genome replication during infection) to temporarily and/or partially compensate for the pressure exerted by the interference machinery. At these conditions of “prolongated interference,” a steady-state amount of intermediates of target destruction must exist that can be acted upon by the adaptation machinery, leading over time to apparently robust adaptation in the absence of strong interference.
When a fully matched and a partially matched protospacer are positioned on the same DNA molecule, adaptation directed by priming at the fully matching protospacer is much more efficient. Because at these conditions the rate of target plasmid degradation is equal for both protospacers, the relative efficiencies of spacer acquisition directed by either protospacer shall be directed, at least in part, by the efficiency of target protospacer recognition by Cascade–crRNA complexes. The reported binding affinities of Cascade charged with g8 crRNA for fully matching and C1T mutant protospacer differ about 50-fold in vitro (17). This compares well with the 10-fold difference in primed adaptation efficiency in two-protospacer plasmid experiments in vivo.
The proposed intimate link between target interference and primed adaptation can be extended to naïve adaptation, which depends only on Cas1 and Cas2. One can hypothesize that various cellular processes, unrelated to CRISPR interference, can also generate DNA fragments recognized by Cas1–Cas2, leading to a low level of spacer adaptation, particularly at elevated concentrations of these proteins. Recent reports indicate that there is some preference of the naïve adaptation to chi sites (33), implicating RecBCD machinery or rather intermediates produced by its action as potential substrates of Cas1–Cas2 action.
A strand bias of spacer acquisition specific for primed adaptation must be arising due to strand specificity of Cas3 action. The known preference of Cas3 for cleavage of the nontarget strand in the R-loop formed by the Cascade–crRNA interaction with a protospacer and the 3′–5′ helicase/nuclease activity of Cas3 are consistent with the experimentally observed direction of the bias and suggest that intermediates used by Cas1–Cas2 must be single-stranded, at least initially. On the other hand, partially double-stranded DNA fragments were observed bound to Cas1–Cas2 complexes in crystal structures (34, 35), and these fragments are incorporated by Cas1–Cas2 into CRISPR arrays in vitro (5). Thus, splayed double-stranded fragments are credible substrates for spacer integration. It was suggested that such spacer integration substrates might be generated after annealing of single-stranded fragments generated by Cas3 (36), which should allow the strand bias to be maintained. Identifying in vivo products of Cas3 interference and their mode of interaction with Cas1–Cas2 will bring important mechanistic insights into our understanding of a functional link between CRISPR interference and primed adaptation.
Acknowledgments
This study was supported by NIH Grant GM10407 (to K.S.); Russian Science Foundation Grant 14-14-00988 (to K.S.); Ministry of Education and Science of Russian Federation Project 14.B25.31.0004 (to K.S.); Russian Foundation for Basic Research Foundation Grant 14-04-00916 (to E. Savitskaya); and Skoltech for institutional support (K.S.). Funding for an open access charge was provided by Skoltech, Russian Science Foundation, and Ministry of Education and Science of Russian Federation grants.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1602639113/-/DCSupplemental.
References
- 1.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 2.Brouns SJ, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321(5891):960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322(5909):1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makarova KS, et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13(11):722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nuñez JK, Lee AS, Engelman A, Doudna JA. Integrase-mediated spacer acquisition during CRISPR-Cas adaptive immunity. Nature. 2015;519(7542):193–198. doi: 10.1038/nature14237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40(12):5569–5576. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arslan Z, Hermanns V, Wurm R, Wagner R, Pul Ü. Detection and characterization of spacer integration intermediates in type I-E CRISPR-Cas system. Nucleic Acids Res. 2014;42(12):7884–7893. doi: 10.1093/nar/gku510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deveau H, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190(4):1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mojica FJ, Díez-Villaseñor C, García-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155(Pt 3):733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 10.Sashital DG, Wiedenheft B, Doudna JA. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol Cell. 2012;46(5):606–615. doi: 10.1016/j.molcel.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anders C, Niewoehner O, Duerst A, Jinek M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513(7519):569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hayes RP, et al. Structural basis for promiscuous PAM recognition in type I-E Cascade from E. coli. Nature. 2016;530(7591):499–503. doi: 10.1038/nature16995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jore MM, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol. 2011;18(5):529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- 14.Szczelkun MD, et al. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc Natl Acad Sci USA. 2014;111(27):9798–9803. doi: 10.1073/pnas.1402597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Westra ER, et al. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell. 2012;46(5):595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mulepati S, Bailey S. In vitro reconstitution of an Escherichia coli RNA-guided immune system reveals unidirectional, ATP-dependent degradation of DNA target. J Biol Chem. 2013;288(31):22184–22192. doi: 10.1074/jbc.M113.472233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Semenova E, et al. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci USA. 2011;108(25):10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Datsenko KA, et al. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- 19.Fineran PC, et al. Degenerate target sites mediate rapid primed CRISPR adaptation. Proc Natl Acad Sci USA. 2014;111(16):E1629–E1638. doi: 10.1073/pnas.1400071111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li M, Wang R, Zhao D, Xiang H. Adaptation of the Haloarcula hispanica CRISPR-Cas system to a purified virus strictly requires a priming process. Nucleic Acids Res. 2014;42(4):2483–2492. doi: 10.1093/nar/gkt1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richter C, et al. Priming in the Type I-F CRISPR-Cas system triggers strand-independent spacer acquisition, bi-directionally from the primed protospacer. Nucleic Acids Res. 2014;42(13):8516–8526. doi: 10.1093/nar/gku527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Westra ER, et al. Parasite exposure drives selective evolution of constitutive versus inducible defense. Curr Biol. 2015;25(8):1043–1049. doi: 10.1016/j.cub.2015.01.065. [DOI] [PubMed] [Google Scholar]
- 23.Blosser TR, et al. Two distinct DNA binding modes guide dual roles of a CRISPR-Cas protein complex. Mol Cell. 2015;58(1):60–70. doi: 10.1016/j.molcel.2015.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Redding S, et al. Surveillance and processing of foreign DNA by the Escherichia coli CRISPR-Cas system. Cell. 2015;163(4):854–865. doi: 10.1016/j.cell.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shmakov S, et al. Pervasive generation of oppositely oriented spacers during CRISPR adaptation. Nucleic Acids Res. 2014;42(9):5907–5916. doi: 10.1093/nar/gku226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morgan M, et al. ShortRead: A bioconductor package for input, quality assessment and exploration of high-throughput sequence data. Bioinformatics. 2009;25(19):2607–2608. doi: 10.1093/bioinformatics/btp450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pages HP (2015) Biostrings: String Objects Representing Biological Sequences, and Matching Algorithms. R Package Version 2.38.0.
- 29.Sambrook JF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed Cold Spring Harbor Lab Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 30.Semenova E, et al. The Cas6e ribonuclease is not required for interference and adaptation by the E. coli type I-E CRISPR-Cas system. Nucleic Acids Res. 2015;43(12):6049–6061. doi: 10.1093/nar/gkv546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pougach K, et al. Transcription, processing and function of CRISPR cassettes in Escherichia coli. Mol Microbiol. 2010;77(6):1367–1379. doi: 10.1111/j.1365-2958.2010.07265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xue C, et al. CRISPR interference and priming varies with individual spacer sequences. Nucleic Acids Res. 2015;43(22):10831–10847. doi: 10.1093/nar/gkv1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levy A, et al. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature. 2015;520(7548):505–510. doi: 10.1038/nature14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nuñez JK, Harrington LB, Kranzusch PJ, Engelman AN, Doudna JA. Foreign DNA capture during CRISPR-Cas adaptive immunity. Nature. 2015;527(7579):535–538. doi: 10.1038/nature15760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, et al. Structural and mechanistic basis of PAM-dependent spacer acquisition in CRISPR-Cas systems. Cell. 2015;163(4):840–853. doi: 10.1016/j.cell.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 36.Amitai G, Sorek R. CRISPR-Cas adaptation: Insights into the mechanism of action. Nat Rev Microbiol. 2016;14(2):67–76. doi: 10.1038/nrmicro.2015.14. [DOI] [PubMed] [Google Scholar]