Significance
Invariant natural killer T (iNKT) cells are innate-like T cells that recognize lipid antigens and play important roles in antimicrobial and tumor immunity. Functionally, iNKT cells have been classified in three effector subsets based on expression of specific transcription factors (TFs) and cytokine genes. We previously demonstrated that the TF Bcl11b controls glycolipid processing and presentation by double-positive thymocytes to iNKT precursors and thus their formation. Using a model that allows bypassing those defects, here we provide evidence that the TF Bcl11b is critical for effector iNKT1 and INKT2 subsets and overall survival of iNKT cells. Additionally we provide evidence that Bcl11b sustains cytokine production by iNKT1 and iNKT2 cells and restricts expression of the iNKT17 cell program in all effector subsets.
Keywords: iNKT cell program, transcription factor Bcl11b, iNKT1 effector cells, iNKT2 effector cells, iNKT17 effector cells
Abstract
Invariant natural killer T (iNKT) cells are innate-like T cells that recognize glycolipid antigens and play critical roles in regulation of immune responses. Based on expression of the transcription factors (TFs) Tbet, Plzf, and Rorγt, iNKT cells have been classified in effector subsets that emerge in the thymus, namely, iNKT1, iNKT2, and iNKT17. Deficiency in the TF Bcl11b in double-positive (DP) thymocytes has been shown to cause absence of iNKT cells in the thymus and periphery due to defective self glycolipid processing and presentation by DP thymocytes and undefined intrinsic alterations in iNKT precursors. We used a model of cre-mediated postselection deletion of Bcl11b in iNKT cells to determine its intrinsic role in these cells. We found that Bcl11b is expressed equivalently in all three effector iNKT subsets, and its removal caused a reduction in the numbers of iNKT1 and iNKT2 cells, but not in the numbers of iNKT17 cells. Additionally, we show that Bcl11b sustains subset-specific cytokine production by iNKT1 and iNKT2 cells and restricts expression of iNKT17 genes in iNKT1 and iNKT2 subsets, overall restraining the iNKT17 program in iNKT cells. The total numbers of iNKT cells were reduced in the absence of Bcl11b both in the thymus and periphery, associated with the decrease in iNKT1 and iNKT2 cell numbers and decrease in survival, related to changes in survival/apoptosis genes. Thus, these results extend our understanding of the role of Bcl11b in iNKT cells beyond their selection and demonstrate that Bcl11b is a key regulator of iNKT effector subsets, their function, identity, and survival.
Invariant natural killer T (iNKT) cells recognize glycolipid antigens presented by the MHC class I-like molecule CD1d and have been shown to play an important role not only in the immune response to bacterial pathogens, but also in antitumor immune responses (1, 2). iNKT cells bear a T-cell receptor (TCR) composed of Vα14–Jα18 chain paired with Vβ7, β8, and β2 in mice, and Vα24 and Vβ11 in humans (3). Following stimulation with glycolipid antigens or cytokines, iNKT cells quickly respond by producing cytokines, including IFNγ, IL-4, IL-13, IL-17, IL-10, and GM-CSF (4–9). This quick response gives them the innate-like attribute. Thymic iNKT precursors are selected on double-positive (DP) thymocytes, which present self glycolipids on CD1d molecules (10–12). Following selection, iNKT precursors go through four developmental stages: 0 (NK1.1−HSAhiCD44lo), 1 (NK1.1−HSAloCD44lo), 2 (NK1.1−HSAloCD44hi), and 3 (NK1.1+HSAloCD44hi) (13). iNKT cell migration out of the thymus occurs at stages 2 and 3 (13, 14). Similar to T helper cells and innate lymphoid cells (ILCs), iNKT cells have been classified into three distinct effector subsets, based on the expression of the TFs Tbet, PLZF, and Rorγt, namely, iNKT1 (TbethiPLZFlo), iNKT2 (TbetloPLZFhi), and iNKT17 (TbetloPLZFloRorγt+) (15). In B6 mice, the iNKT2 and iNKT17 subsets are found predominantly within developmental stage 2, whereas the iNKT1 subset is confined to stage 3 (15). Several transcription factors (TFs) have been found essential for iNKT cell progression through developmental stages, as well as for their effector functions. Tbet is critical for iNKT1 cell function and for terminal maturation and homeostasis (15, 16). Rorγt not only controls the iNKT17 pathway, but together with Runx1, regulates iNKT cell development (12, 15, 17). PLZF is expressed postselection and directs the development and effector program of iNKT cells (18, 19). E and Id proteins are important for both lineage choice between iNKT and T cells during selection and differentiation into iNKT1 and iNKT2 subsets (20–22). c-myb regulates CD1d levels on DP thymocytes, as well as Slamf1, Slamf6, and SAP on iNKT cells (23). Hobit controls maintenance of mature iNKT cells and their effector functions (24). Recently Lef1 was found to be essential for iNKT2 subset formation and function, and to regulate Gata3 and Thpok (25), both known to control CD4+ iNKT cells (26). TF Bcl11b plays a crucial role in T-cell lineage commitment (27, 28), selection, differentiation, and survival of thymocytes (29, 30), clonal expansion and effector function of CD8+ T cells (31), as well as suppression function of Treg cells (32). Additionally, Bcl11b restricts expression of Th2 lineage genes in Th17 cells in experimental autoimmune encephalomyelitis (EAE) (33). Bcl11b was recently found to sustain innate lymphoid type 2 cell (ILC2) program (34, 35, 36) and to suppress ILC3 program in ILC2s (36). Bcl11b’s deficiency in DP thymocytes resulted in lack of iNKT cells in the thymus and periphery (37, 38), despite the fact that the Vα14Jα18 TCR was normally rearranged (38). The defect was caused by the inability of Bcl11b−/− DP thymocytes to support the selection of iNKT precursors, due to defective glycolipid self-antigen processing/presentation. Additionally, Bcl11b−/− iNKT precursors, even when normally presented with glycolipid self-antigens, failed to generate iNKT cells, due to unidentified intrinsic defects (38). Here we set up a system to study the defects caused by the absence of Bcl11b in iNKT cells, using the PLZF-Cre mouse strain, which promotes removal of floxed alleles postselection of iNKT cells, namely starting with developmental stage 1 (18, 39, 40). Our study demonstrates that PLZF-Cre–mediated iNKT cell deletion of Bcl11b resulted in significantly reduced iNKT cells in the thymus and periphery, associated with reduction in survival in relation to changes in survival/apoptosis genes. iNKT1 and iNKT2 effector subsets were numerically reduced both in thymus and spleen, suggesting that these two subsets need Bcl11b. Additionally, levels of IFNγ and IL-4 within Bcl11b−/− iNKT1 and iNKT2 subsets, respectively, were reduced, demonstrating that these cells also have functional alterations. Although numbers of Bcl11b−/− iNKT17 cells were normal, IL-17 production was up-regulated together with increased levels of iNKT17 subset molecules, namely, IL23R and Nrp1. IL23R and Nrp1 were up-regulated not only in total iNKT cells and in iNKT17 subset, but also in the other two effector subsets, suggesting an additional role for Bcl11b in restricting the iNKT17 program in iNKT1 and iNKT2 subsets. Additionally, Bcl11b suppressed expression of some NK genes in iNKT cells. Thus, our results demonstrate a major role of Bcl11b in the development, function, identity and survival of iNKT cells.
Results
Bcl11b’s Removal Is Specific for iNKT Cells in Bcl11bF/F/PLZF-Cre Mice.
Given previous results in which absence of Bcl11b caused a developmental block in iNKT cells due to defective iNKT precursors (38), as well as defective glycolipid processing and presentation by DP thymocytes, we used the PLZF-Cre mouse strain to remove targeted alleles restrictively in iNKT cells, postselection (40). The PLZF-Cre deleter efficiently removed Bcl11b in splenic and thymic iNKT cells, starting with developmental stage 1 (Fig. S1 A–C). Given that ∼25% of T cells were previously reported to be positive for the Rosa 26-tdTomato reporter in PLZF-Cre mice (40), we evaluated removal of Bcl11b in PBS-57/CD1d− thymic CD4+ single positive (SP) and splenic CD4+ T cells and found no removal of Bcl11b in these cells (Fig. S1 A and B). Moreover, thymic SP and peripheral CD4+ and CD8+ T-cell populations remained numerically unaffected (Fig. S1 D and E). Considering that PLZF was found to be expressed in a precursor population of helper ILCs (41), and removal of Bcl11b in ILC2s caused down-regulation of St2 and derepression of Rorγt (36), we investigated whether Bcl11b is removed from ILC2s in Bcl11bF/F/PLZF-Cre mice. Our results show no removal of Bcl11b in these cells (Fig. S1F). Additionally, ILC2s of Bcl11bF/F/PLZF-Cre mice did not down-regulate St2 or up-regulate Rorγt (Fig. S1G). These results taken together demonstrate that Bcl11b’s removal is specific for iNKT cells in Bcl11bF/F/PLZF-Cre mice.
Fig. S1.
Bcl11b is specifically removed from iNKT cells in Bcl11bF/F/PLZF-Cre mice and not from CD4+ T cells or innate lymphoid type 2 cells (ILC2s). (A and B) Frequencies of Bcl11b+ PBS-57/CD1d+ T cells and PBS-57/CD1d− CD4+ T cells in thymus (A) and spleen (B) of Bcl11bF/F/PLZF-Cre and Bcl11bF/F mice evaluated by FACS. (C) Frequencies of Bcl11b+ iNKT cells in the four thymic developmental stages from Bcl11bF/F/PLZF-Cre and Bcl11bF/F mice. (D and E) Frequencies of DP, CD4, and CD8 SP thymocytes (D), and CD4+ and CD8+ T cells in spleen (E) of Bcl11bF/F/PLZF-Cre and WT control mice. (F) Bcl11b in ILC2s from mesenteric lymph nodes and lung of Bcl11bF/F/PLZF-Cre and WT mice. (G) St2 and Rorγt in lung Lin− CD90+CD127+CD25+ innate lymphoid cells of Bcl11bF/F/PLZF-Cre and WT mice. Data are representative of five pairs of mice (A and B), four pairs of mice (C), and three pairs of mice (D–G).
Bcl11bF/F/PLZF-Cre Mice Have Reduced Numbers of Thymic and Peripheral iNKT Cells and Developmental Alterations of iNKT Cells.
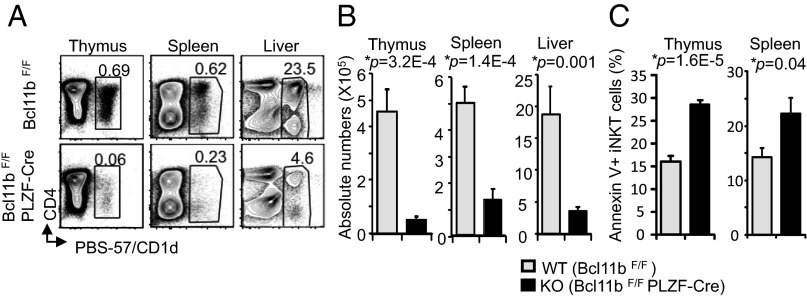
Ablation of Bcl11b with the PLZF-Cre deleter caused a severe reduction of the percentages and absolute numbers of iNKT cells in the thymus, spleen, and liver (Fig. 1 A and B and Fig. S2A), although the mean fluorescence intensity (MFI) of PBS-57–loaded CD1d tetramer bound by TCR remained similar between Bcl11b−/− and wild-type iNKT cells (Fig. S2B). We further asked the question whether the reduced numbers of iNKT cells in the absence of Bcl11b is caused by their reduced survival. Annexin V staining of thymic and splenic iNKT cells showed that more Bcl11b−/− iNKT cells stained for Annexin V compared with control, both in thymus and spleen (Fig. 1C and Fig. S2C). These results suggest that Bcl11b−/− iNKT cells have an increased tendency to die, which is likely to contribute to their numerical decrease.
Fig. 1.
Bcl11b's deficiency in iNKT cells causes a major reduction in thymic and peripheral iNKT cells associated with increased apoptosis. (A and B) Frequencies (A) and absolute numbers (B) of iNKT cells from Bcl11bF/F/PLZF-Cre (KO) and WT mice in thymus, spleen, and liver. The gated population in A shows percentages of CD1d-PBS-57+cells. (C) Frequencies of Annexin V+ iNKT cells from Bcl11bF/F/PLZF-Cre (KO) and Bcl11bF/F (WT) mice in thymus and spleen. Data are representative of several independent experiments with 10 pairs of mice (A and B) and 5 pairs of mice (C). P values determined by unpaired two-tailed Student’s t test are indicated. Means ± SEM.
Fig. S2.
Frequencies of iNKT cells in thymus, spleen, and liver of Bcl11bF/F/PLZF-Cre mice, survival of iNKT cells and levels of CD1d-PBS-57 bound by Bcl11b−/− iNKT cells. (A) Quantification of percentages of iNKT cells from Bcl11bF/F/PLZF-Cre and WT mice in thymus, spleen, and liver. (B) Mean fluorescent intensity (MFI) of CD1d-PBS-57 bound to iNKT cells from thymus, spleen, and liver of Bcl11bF/F/PLZF-Cre and Bcl11bF/F mice. Data are representative of eight pairs of mice (A) and five pairs of mice (B). P values are indicated. Means ± SEM. (C) Frequencies of Annexin V+ iNKT cells from Bcl11bF/F/PLZF-Cre (KO) and Bcl11bF/F (WT) mice in thymus and spleen. Data are representative of five pairs of mice.
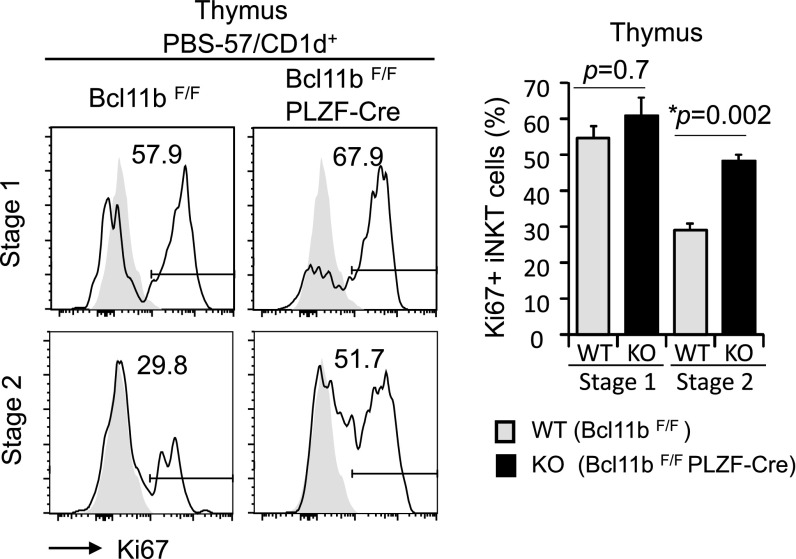
We further investigated the thymic iNKT developmental stages and found that Bcl11bF/F/PLZF-Cre mice had higher percentages of iNKT cells in stages 0–2, whereas the percentages of stage 3 iNKT cells were reduced (Fig. 2 A and B). The absolute numbers of stages 0 and 1 iNKT cells were similar in Bcl11bF/F/PLZF-Cre and wild-type mice, however, there was a substantial reduction of absolute numbers of iNKT cells in stages 2 and 3 (Fig. 2C). Given that iNKT cells go through a massive expansion as they transition between stages 1 and 2, we evaluated the Ki67, indicative of cells entering the cell cycle. Our results show that similar percentages of Bcl11b−/− and wild-type iNKT cells were positive for Ki67 in stage 1, whereas during stage 2, the percentages were increased for Bcl11b−/− iNKT cells (Fig. S3). Therefore, it is unlikely that the reduction in the numbers of iNKT cells in the absence of Bcl11b is caused by defective cell cycle entering, but rather by reduced survival and alterations in development.
Fig. 2.
Bcl11b's removal in iNKT cells causes developmental alterations. (A) Frequencies of iNKT cells from Bcl11bF/F/PLZF-Cre and WT mice in the four developmental stages (Left) based on surface CD24, CD44, and NK1.1 within the CD1d-PBS-57+ cells. (B and C) Quantification of percentages (B) and absolute numbers (C) of iNKT cells within the four developmental stages. Data are representative of several independent experiments with 10 pairs of mice. P values are indicated. Means ± SEM.
Fig. S3.
Ki67 levels in developmental stages 1 and 2. Frequencies of Ki67+ iNKT cells in thymic developmental stages 1 and 2 of Bcl11bF/F/PLZF-Cre (KO) and Bcl11bF/F (WT) mice. Data are representative of five pairs of mice. Means ± SEM.
Absence of Bcl11b Causes Up-Regulation of iNKT17 Genes, Down-Regulation of iNKT1 and iNKT2 Genes, and Alterations in Survival Genes.
Comparison of the mRNAs of Bcl11b−/− and wild-type iNKT cells showed that the prosurvival gene Bcl2 was down-regulated and the proapoptotic gene Bag2 was up-regulated in Bcl11b−/− iNKT cells (Fig. 3), thus supporting the observation that Bcl11b−/− iNKT cells have a reduction in survival. The mRNAs for the Tbx21 (Tbet), critical for the iNKT1 subset and its signature cytokine IFNγ, were reduced (Fig. 3). The mRNAs for the TFs Zbtb16 (PLZF) and Lef1, important for iNKT2 subset (15, 25), were also diminished, together with the mRNAs for the iNKT2 subset cytokines IL-4 and IL-13, and the receptor IL-4Rα. However, there was no change in the mRNAs for Gata3 and Zbtb7b (Thpok) (Fig. 3), known to be downstream of Lef1 (25). The mRNAs for the TFs Rorγt (Rorc) and Sox4, known to be important for the iNKT17 subset, were up-regulated (Fig. 3), together with the iNKT17 subset mRNAs for IL-17, IL-22, IL-23R, and Nrp1 (Fig. 3). In addition, the mRNAs for Ncr1 (Nkp46), Klrb1, Gzmb, and Gzmc were elevated (Fig. 3). The results of this analysis support the conclusion that Bcl11b controls survival of iNKT cells and effector iNKT subset genes.
Fig. 3.
Bcl11b-deficient iNKT cells show altered expression of genes critical for effector iNKT subsets and survival. Scatterplot of log2 expression values for mRNAs isolated from sorted iNKT cells of Bcl11bF/F/PLZF-CreVα14 Tg (KO) versus Vα14 Tg control mice (WT) treated with α-GalCer. Data are representative of two independent arrays. Relevant genes are listed. The heat map shows relevant genes off the scale in scatterplot.
iNKT1 and iNKT2 Effector Subsets Require Bcl11b.
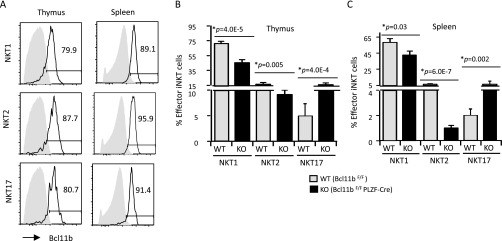
Given the up-regulation of iNKT17 genes and the down-regulation of iNKT1 and iNKT2 genes, we further evaluated the effector iNKT subsets in the absence of Bcl11b. We first assessed the expression of Bcl11b in these subsets and found that Bcl11b was expressed in thymic and splenic effector iNKT subsets at similar levels (Fig. S4A). We further evaluated the thymic and splenic effector iNKT subsets in the absence of Bcl11b, and found that the percentages and absolute numbers of iNKT1 and iNKT2 cells were reduced both in the thymus and periphery (Fig. 4 A–D and Fig. S4 B and C). Whereas there was an increase in the percentages of iNKT17 cells, the absolute numbers of iNKT17 cells remained unchanged (Fig. 4 A–D and Fig. S4 B and C). These data suggest that iNKT1 and iNKT2 subsets, but not the iNKT17 subset, require Bcl11b.
Fig. S4.
Bcl11b is expressed in all effector iNKT subsets and its depletion alters the frequencies of iNKT subsets. (A) Frequencies of Bcl11b+ cells in the thymic and splenic iNKT subsets defined as in Fig. 4. (B and C) Percentages of effector iNKT subsets in thymus (B) and spleen (C) of Bcl11bF/F/PLZF-Cre (KO) and Bcl11bF/F (WT) mice. Data are representative of five pairs of mice. P values are indicated. Means ± SEM.
Fig. 4.
iNKT1 and iNKT2 subset cell numbers are reduced in thymus and spleen of Bcl11bF/F/PLZF-Cre mice. (A–D) Frequencies (A and B) and absolute numbers (C and D) of effector iNKT subsets in thymus and spleen, identified as iNKT1 (TbethiPLZFlo), iNKT2 (TbetloPLZFhi), and iNKT17 (RorγthiPLZFlo) on gated CD1d-PBS-57+ cells from Bcl11bF/F/PLZF-Cre (KO) and Bcl11bF/F (WT) mice. Data are representative of 8–10 pairs of mice. P values are indicated. Means ± SEM.
Bcl11b−/− iNKT Effector Subsets Have Functional Alterations.
We further evaluated the cytokine production by total Bcl11b−/− iNKT cells, as well as by the effector subsets. Our results indicate that overall frequencies of IL-17–producing Bcl11b−/− iNKT cells were increased, whereas frequencies of IL-4– and IFNγ-producing Bcl11b−/− iNKT cells were reduced compared to wild-type iNKT cells both in thymus and spleen (Fig. S5 A and B), in agreement with the changes in the proportion of effector subsets. Within the iNKT1 and iNKT2 subsets, the percentages of Bcl11b−/− IFNγ- and IL-4–producing iNKT cells, respectively, were reduced both in thymus and spleen (Fig. 5 A–D). Additionally, the absolute numbers of Bcl11b−/− IFNγ- and IL-4–producing iNKT1 and INKT2 cells, respectively, were reduced as well (Fig. 5 E and F). Within the iNKT17 subset, the percentages of Bcl11b−/− IL-17–producing iNKT cells were increased in the thymus and spleen and the absolute numbers were slightly increased, but not significantly (Fig. 5 A–F). These results suggest that within the Bcl11b−/− iNKT1 and iNKT2 subsets, fewer cells produced IFNγ and IL-4, respectively, thus presenting functional alterations, whereas within the Bcl11b−/− iNKT17 subset, a larger percentage produced IL-17.
Fig. S5.
Bcl11b-deficient iNKT cells show altered cytokine production. Frequencies of IL-4–, IFNγ- (Top), and IL-4–, and IL-17 (Bottom)-producing iNKT cells in thymus (A) and spleen (B) from Bcl11bF/F/PLZF-Cre (KO) and Bcl11bF/F (WT) mice. For evaluation of cytokine production in thymus, total thymocytes were collected and treated in vitro with α-galactosylceramide (α-GalCer) for 72 h (A). For evaluation of splenic iNKT cells, mice were injected i.p. with α-GalCer and killed 4 h later (B). Data are representative of six pairs of mice.
Fig. 5.
Bcl11b-deficient iNKT cells are functionally altered. (A–F) Frequencies (A–D) and absolute numbers (E and F) of IL-4+, IFNγ+, and IL-17+ iNKT cells within the indicated effector subsets, iNKT1, iNKT2, and iNKT17, defined as in Fig. 4 A and B. Data are representative of several experiments with eight pairs of mice. P values are indicated. Means ± SEM.
Bcl11b Restricts Expression of iNKT17 Program in iNKT1 and iNKT2 Effector Subsets.
We further investigated whether other genes, dysregulated in the absence of Bcl11b from iNKT cells, were altered within the effector subsets or their difference in expression was a consequence of ratio change between the effector subsets.
Several mRNAs for iNKT17 genes were up-regulated, including for IL-23R and Nrp1 (Fig. 3). IL-23R and Nrp1 were up-regulated not only in the total iNKT cells and within the iNKT17 subset, but even in the iNKT1 and iNKT2 effector subsets (Fig. 6). These results suggest that Bcl11b may be overall implicated in restraining the iNKT17 program, including in iNKT1 and iNKT2 effector subsets.
Fig. 6.
Bcl11b-deficient iNKT cells, including iNKT1 and iNKT2 effector subsets up-regulate iNKT17 cell markers. Frequencies of IL-23R+, Nrp1+, NKp46+, and Granzyme B+ in total iNKT cells and in effector iNKT subsets of Bcl11bF/F/PLZF-Cre and Bcl11bF/F mice in thymus (A) and spleen (B). iNKT1, iNKT2, and iNKT17 are defined as in Fig. 4 A and B. Data are representative of three independent experiments.
We further evaluated Nkp46 and Gzmb, two natural killer (NK) genes up-regulated at mRNA level in total Bcl11b−/− iNKT cells and known to be derepressed following Bcl11b’s removal in other T-cell populations and progenitors (reviewed in ref. 42). Both Nkp46 and Gzmb were very modestly expressed in the wild-type thymic and splenic iNKT cells at steady state (Fig. 6), similar to what has been recently reported (43). Removal of Bcl11b resulted in the up-regulation of Nkp46 and Gzmb in all of the three effector subsets both in thymus and spleen (Fig. 6). As shown above, the mRNA encoding Lef1, recently found to be essential for iNKT2 cells (25), was reduced (Fig. 3). There was also a modest reduction of Lef1 in the total Bcl11b−/− iNKT cells, however, no change in iNKT2 subset (Fig. S6A), suggesting that the reduced Lef1 mRNA level in total Bcl11b−/− iNKT cells was simply due to the iNKT2 subset numerical reduction. In agreement with these results, no up-regulation of CD8 was observed in the Bcl11b−/− iNKT cells above wild-type background (Fig. S6B) and Gata3 remained unchanged (Fig. S6A).
Fig. S6.
Lef1 and Gata3 are not reduced in Bcl11b−/− iNKT cells belonging to the iNKT2 subset and CD8 is not up-regulated on Bcl11b-deficient iNKT cells. (A) Frequencies of Lef1+ and Gata3+ total iNKT and iNKT2 cells in thymus of Bcl11bF/F/PLZF-Cre and Bcl11bF/F mice evaluated by FACS. (B) Frequencies of thymic CD8+ iNKT cells from Bcl11bF/F/PLZF-Cre (KO) and Bcl11bF/F (WT) mice. Data are representative of of three pairs of mice.
Thus these results taken together suggest that Bcl11b is required to restrict iNKT17 genes overall in iNKT cells, including in effector iNKT1 and iNKT2 subsets. Additionally, Bcl11b represses expression of some NK genes in iNKT cells.
Discussion
Previous reports demonstrate that absence of the TF Bcl11b at DP stage of T-cell development caused a defect in self glycolipid processing and presentation, which impacted the selection of iNKT precursors (38). Additionally, Bcl11b−/− iNKT precursors had unknown intrinsic defects that blocked their development even when selected on DP thymocytes able to present glycolipids (38). Here we set up a system to study the specific role of Bcl11b in iNKT cells postselection, at developmental stage 1. Importantly, our results demonstrate specific and restrictive removal of Bcl11b with the PLZF-Cre system in iNKT cells and no removal of Bcl11b in other T cells or ILC2s. Using this system we found that deficiency of Bcl11b caused a significant decrease in iNKT cells in the thymus and periphery, related to reduced iNKT1 and iNKT2 cell numbers and to an overall reduced survival of iNKT cells attributed to changes in survival genes, similar to what was previously reported for Bcl11b−/− thymocytes (29, 30). Although iNKT1 and iNKT2 cell numbers were reduced in the absence of Bcl11b, there was no change in the numbers of iNKT17 cells, suggesting an essential role of Bcl11b in the control of effector iNKT1 and iNKT2 subsets. The reduction in the iNKT1 and iNKT2 subsets in the absence of Bcl11b is also in agreement with the reduction in developmental stage 3, predominantly represented by the iNKT1 subset, and the reduction in developmental stage 2, represented by iNKT2 and iNKT17 subsets. One possibility is that development of iNKT1 and iNKT2 subsets is dependent on Bcl11b, whereas the iNKT17 subset is not. Another possibility is that Bcl11b−/− iNKT1 and iNKT2 cells have altered survival, however, Bcl11b−/− iNKT17 cells survive normally. Not only were the iNKT1 and iNKT2 subsets numerically reduced, but the percentages of IFNγ- and IL-4–producing iNKT cells were also reduced within the iNKT1 and iNKT2 subsets, respectively, suggesting that iNKT1 and iNKT2 cells have functional alterations in the absence of Bcl11b. The TF Lef1, recently demonstrated to control Gata3 and IL-4 expression in iNKT2 cells (25), was not reduced in Bcl11b−/− iNKT2 cells. There was no reduction in Gata3 expression, suggesting that Bcl11b might be involved in the control of IL-4, independent of these transcriptional regulators. Also, Bcl11b might be implicated in the control of IFNγ either directly or through Tbet. Interestingly, levels of the IL-17, IL-23R, and Nrp1, all part of the iNKT17 program, were elevated in total and iNKT17 cells, which suggests a role of Bcl11b in restraining the iNKT17 program. IL-23R and Nrp1 were also up-regulated in iNKT1 and iNKT2 subsets, further suggesting that Bcl11b restricts expression of the iNKT17 program in iNKT1 and iNKT2 subsets. This is similar to the role of Bcl11b in ILC2s, in which Bcl11b represses the ILC3 program (34). As mentioned above Bcl11b−/− iNKT17 cells were not affected numerically in the absence of Bcl11b, similar to what happened in the absence of Bcl11b in Th17 cells during EAE (33). However, different from Bcl11b−/− Th17 cells in EAE (33), Bcl11b−/− iNKT17 cells did not express IL-4. Thus, Bcl11b regulates some common themes in some immune populations, such as ILCs and iNKT cells, however, it acts also in a context-dependent manner in other populations (reviewed in ref. 42). It remains to be established whether Bcl11b plays other roles in iNKT17 cells except restricting expression of its signature genes. In addition, as in other T-cell populations, Bcl11b represseed NKp46 and Gzmb, but not NK1.1, CD244, or myeloid genes, that are derepressed in the absence of Bcl11b in early thymocytes (27, 28). This again suggests that Bcl11b governs not only common, but also context-specific programs. The suppression of NKp46 and Gzmb can be related to overall repression of NK program. NKp46 was expressed in wild-type iNKT subsets at very low level, with the highest level in the iNKT17 subset, at least in the thymus. Therefore, it is possible that NKp46 is part of the iNKT17 program, similar to a subset of ILC3s, and absence of Bcl11b causes its increase in relation to derepression of iNKT17 genes or as mentioned above as part of restricting expression of NK genes. This remains to be established in the future.
In conclusion, our results demonstrate a critical role of Bcl11b in the control of iNKT1 and iNKT2 subsets and in sustaining their function. Bcl11b also restricts expression of the iNKT17 program, including in iNKT1 and iNKT2 subsets. In addition, Bcl11b is required for overall survival of iNKT cells and repression of some NK genes.
Materials and Methods
Detailed materials and methods are given in SI Materials and Methods.
Mice.
Bcl11bF/F/PLZF-Cre mice were generated by breeding Bcl11bF/F and PLZF-Cre mice, previously described (29, 30, 40). Vα14 mice were previously described (44). Mice were kept under specific pathogen-free conditions. All of the experiments were conducted in accordance with animal protocols approved by the Institutional Animal Care and Use Committee of the University of Florida and Albany Medical College.
Statistical Analysis.
The statistical difference between experimental groups was determined by unpaired two-tailed Student t test. The P values ≤0.05 were considered significant.
SI Materials and Methods
Cell Preparation and Flow Cytometry.
Single-cell suspensions were prepared from the thymus, spleen, and liver. Liver lymphocytes were enriched using a 33% Percoll gradient. In all cases, cells were incubated with Fc blocking Abs before staining with specific antibodies and tetramer. iNKT cells were identified using PE or APC-conjugated CD1d tetramer loaded with PBS57 or unloaded. FACS was conducted on a Cytek-updated eight color FACSCalibur using FlowJo acquisition software or on a 4-laser 15-color LSRII, using FACSdiva software. Data were analyzed by using FlowJo analysis software. Thymic, splenic, and liver iNKT cells were identified as CD1d-PBS-57+ cells in the total lymphocyte population.
Antibodies.
Fluorophore-conjugated Abs include: anti-CD3ε (145-2c11), -CD4 (GK1.5), -CD8α (53-6.7), -CD24 (M1/69), -CD44(IM7), -NK1.1 (PK-136), -B220 (RA36B2), -ICOS (C3984A), -Slamf6 (eBio13G3-19D), -Slamf1 (9D1), -PLZF (Mag21F7), -T-bet (4B10), -ROR-γT (AFKJS-9), –IL-4 (11b11), -IFNγ (XMG1.2), –IL-17A (17B7), -Nrp1 (3DS304M), -NKp46 (29A1.4), -Gata3 (TWAJ), -ThPOK (2POK), -Granzyme B (GB11), and Ki67 (SolA15) from eBioscience or Biolegend; anti–IL-23R (3C9) from BD Bioscience; anti-Lef1 (C12A5) from Cell Signaling; and anti-Bcl11b from Bethyl Laboratories. CD1d tetramer loaded with PBS57 or unloaded was provided by the Tetramer core facility of the NIH.
In Vitro and in Vivo Cytokine Production.
For in vitro cytokine production, total thymocytes were placed in 24-well plates, at a concentration of 10 × 106 cells per milliliter in RPMI medium, and stimulated for 72 h with 125 ng/mL α-galactosylceramide (KRN7000). Phorbol 12-myristate 13-acetate (PMA) (50 ng/mL), ionomycin (500 ng/mL), and Golgiplug (1 ng/mL) were added to the cells for the final 5 h. For in vivo stimulation of iNKT cells, mice were injected intraperitoneally with 4 μg α-galactosylceramide. Four hours later, splenocytes were collected, stained intracellularly for cytokines, and analyzed by flow cytometry.
Microarray Analysis.
Mice were injected intraperitoneally with 4 μg α-galactosylceramide. Splenocytes were collected 4 h later and surface stained with CD1d-PBS57 and anti-B220 antibodies. iNKT cells were sorted as CD1d-PBS57+B220− population. RNA was extracted with RNeasy Protocol (Qiagen) and analyzed for gene expression profiling using Agilent Single color 26655 platform at MOgene (GSE73679).
Acknowledgments
We thank Drs. William C. Curtiss and Shaukat Rangwala (MOgene) for microarray analysis, Drs. Liang Zhou and Shahram Salek-Ardankani (University of Florida) for reviewing the manuscript and for exciting discussions, and the Tetramer Facility of the NIH for CD1dT-PBS57+ tetramers. This work was supported by NIH Grant R01AI067846, the University of Florida Gatorade Trust (D.A.), and National Institute of Allergy and Infectious Diseases Grant R01AI083988 (to D.B.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. E.V.R. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE73679).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1521846113/-/DCSupplemental.
References
- 1.Kinjo Y, Kitano N, Kronenberg M. The role of invariant natural killer T cells in microbial immunity. J Infect Chemother. 2013;19(4):560–570. doi: 10.1007/s10156-013-0638-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fujii S, et al. NKT cells as an ideal anti-tumor immunotherapeutic. Front Immunol. 2013;4:409. doi: 10.3389/fimmu.2013.00409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salio M, Silk JD, Jones EY, Cerundolo V. Biology of CD1- and MR1-restricted T cells. Annu Rev Immunol. 2014;32:323–366. doi: 10.1146/annurev-immunol-032713-120243. [DOI] [PubMed] [Google Scholar]
- 4.Yoshimoto T, Paul WE. CD4pos, NK1.1pos T cells promptly produce interleukin 4 in response to in vivo challenge with anti-CD3. J Exp Med. 1994;179(4):1285–1295. doi: 10.1084/jem.179.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawano T, et al. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science. 1997;278(5343):1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 6.Burdin N, et al. Selective ability of mouse CD1 to present glycolipids: Alpha-galactosylceramide specifically stimulates V alpha 14+ NK T lymphocytes. J Immunol. 1998;161(7):3271–3281. [PubMed] [Google Scholar]
- 7.Gumperz JE, Miyake S, Yamamura T, Brenner MB. Functionally distinct subsets of CD1d-restricted natural killer T cells revealed by CD1d tetramer staining. J Exp Med. 2002;195(5):625–636. doi: 10.1084/jem.20011786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stetson DB, et al. Constitutive cytokine mRNAs mark natural killer (NK) and NK T cells poised for rapid effector function. J Exp Med. 2003;198(7):1069–1076. doi: 10.1084/jem.20030630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coquet JM, et al. Diverse cytokine production by NKT cell subsets and identification of an IL-17-producing CD4-NK1.1-NKT cell population. Proc Natl Acad Sci USA. 2008;105(32):11287–11292. doi: 10.1073/pnas.0801631105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gapin L, Matsuda JL, Surh CD, Kronenberg M. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat Immunol. 2001;2(10):971–978. doi: 10.1038/ni710. [DOI] [PubMed] [Google Scholar]
- 11.Bendelac A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J Exp Med. 1995;182(6):2091–2096. doi: 10.1084/jem.182.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egawa T, et al. Genetic evidence supporting selection of the Valpha14i NKT cell lineage from double-positive thymocyte precursors. Immunity. 2005;22(6):705–716. doi: 10.1016/j.immuni.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 13.Pellicci DG, et al. A natural killer T (NKT) cell developmental pathway iInvolving a thymus-dependent NK1.1(-)CD4(+) CD1d-dependent precursor stage. J Exp Med. 2002;195(7):835–844. doi: 10.1084/jem.20011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benlagha K, Kyin T, Beavis A, Teyton L, Bendelac A. A thymic precursor to the NK T cell lineage. Science. 2002;296(5567):553–555. doi: 10.1126/science.1069017. [DOI] [PubMed] [Google Scholar]
- 15.Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat Immunol. 2013;14(11):1146–1154. doi: 10.1038/ni.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Townsend MJ, et al. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004;20(4):477–494. doi: 10.1016/s1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 17.Michel ML, et al. Critical role of ROR-γt in a new thymic pathway leading to IL-17-producing invariant NKT cell differentiation. Proc Natl Acad Sci USA. 2008;105(50):19845–19850. doi: 10.1073/pnas.0806472105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kovalovsky D, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat Immunol. 2008;9(9):1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Savage AK, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29(3):391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Wu D, Jiang N, Zhuang Y. Combined deletion of Id2 and Id3 genes reveals multiple roles for E proteins in invariant NKT cell development and expansion. J Immunol. 2013;191(10):5052–5064. doi: 10.4049/jimmunol.1301252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Verykokakis M, et al. Essential functions for ID proteins at multiple checkpoints in invariant NKT cell development. J Immunol. 2013;191(12):5973–5983. doi: 10.4049/jimmunol.1301521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.D’Cruz LM, Stradner MH, Yang CY, Goldrath AW. E and Id proteins influence invariant NKT cell sublineage differentiation and proliferation. J Immunol. 2014;192(5):2227–2236. doi: 10.4049/jimmunol.1302904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu T, Simmons A, Yuan J, Bender TP, Alberola-Ila J. The transcription factor c-Myb primes CD4+CD8+ immature thymocytes for selection into the iNKT lineage. Nat Immunol. 2010;11(5):435–441. doi: 10.1038/ni.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Gisbergen KP, et al. Mouse Hobit is a homolog of the transcriptional repressor Blimp-1 that regulates NKT cell effector differentiation. Nat Immunol. 2012;13(9):864–871. doi: 10.1038/ni.2393. [DOI] [PubMed] [Google Scholar]
- 25.Carr T, et al. The transcription factor lymphoid enhancer factor 1 controls invariant natural killer T cell expansion and Th2-type effector differentiation. J Exp Med. 2015;212(5):793–807. doi: 10.1084/jem.20141849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, et al. The sequential activity of Gata3 and Thpok is required for the differentiation of CD1d-restricted CD4+ NKT cells. Eur J Immunol. 2010;40(9):2385–2390. doi: 10.1002/eji.201040534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li L, Leid M, Rothenberg EV. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science. 2010;329(5987):89–93. doi: 10.1126/science.1188989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li P, et al. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science. 2010;329(5987):85–89. doi: 10.1126/science.1188063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wakabayashi Y, et al. Bcl11b is required for differentiation and survival of alphabeta T lymphocytes. Nat Immunol. 2003;4(6):533–539. doi: 10.1038/ni927. [DOI] [PubMed] [Google Scholar]
- 30.Albu DI, et al. BCL11B is required for positive selection and survival of double-positive thymocytes. J Exp Med. 2007;204(12):3003–3015. doi: 10.1084/jem.20070863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang S, et al. Antigen-specific clonal expansion and cytolytic effector function of CD8+ T lymphocytes depend on the transcription factor Bcl11b. J Exp Med. 2010;207(8):1687–1699. doi: 10.1084/jem.20092136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vanvalkenburgh J, et al. Critical role of Bcl11b in suppressor function of T regulatory cells and prevention of inflammatory bowel disease. J Exp Med. 2011;208(10):2069–2081. doi: 10.1084/jem.20102683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Califano D, et al. Diverting T helper cell trafficking through increased plasticity attenuates autoimmune encephalomyelitis. J Clin Invest. 2014;124(1):174–187. doi: 10.1172/JCI70103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker JA, et al. Bcl11b is essential for group 2 innate lymphoid cell development. J Exp Med. 2015;212(6):875–882. doi: 10.1084/jem.20142224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu Y, et al. The transcription factor Bcl11b is specifically expressed in group 2 innate lymphoid cells and is essential for their development. J Exp Med. 2015;212(6):865–874. doi: 10.1084/jem.20142318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Califano D, et al. Transcription factor Bcl11b controls identity and function of mature type 2 innate lymphoid cells. Immunity. 2015;43(2):354–368. doi: 10.1016/j.immuni.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kastner P, et al. Bcl11b represses a mature T-cell gene expression program in immature CD4(+)CD8(+) thymocytes. Eur J Immunol. 2010;40(8):2143–2154. doi: 10.1002/eji.200940258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Albu DI, et al. Transcription factor Bcl11b controls selection of invariant natural killer T-cells by regulating glycolipid presentation in double-positive thymocytes. Proc Natl Acad Sci USA. 2011;108(15):6211–6216. doi: 10.1073/pnas.1014304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alonzo ES, Sant’Angelo DB. Development of PLZF-expressing innate T cells. Curr Opin Immunol. 2011;23(2):220–227. doi: 10.1016/j.coi.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang S, Laouar A, Denzin LK, Sant’Angelo DB. Zbtb16 (PLZF) is stably suppressed and not inducible in non-innate T cells via T cell receptor-mediated signaling. Sci Rep. 2015;5:12113. doi: 10.1038/srep12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature. 2014;508(7496):397–401. doi: 10.1038/nature13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Avram D, Califano D. The multifaceted roles of Bcl11b in thymic and peripheral T cells: Impact on immune diseases. J Immunol. 2014;193(5):2059–2065. doi: 10.4049/jimmunol.1400930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Farr AR, Wu W, Choi B, Cavalcoli JD, Laouar Y. CD1d-unrestricted NKT cells are endowed with a hybrid function far superior than that of iNKT cells. Proc Natl Acad Sci USA. 2014;111(35):12841–12846. doi: 10.1073/pnas.1323405111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thapa P, et al. The transcriptional repressor NKAP is required for the development of iNKT cells. Nat Commun. 2013;4:1582. doi: 10.1038/ncomms2580. [DOI] [PMC free article] [PubMed] [Google Scholar]