Significance
Overapplication of nitrogen (N) fertilizer causes delayed flowering and negatively impacts the function and composition of natural ecosystems and climate. In this study, we demonstrate that flowering time variations regulated by altered nitrogen levels are mediated by two key factors: ferredoxin-NADP+-oxidoreductase (FNR1) and the blue-light receptor cryptochrome 1 (CRY1). Nitrogen regulates FNR1 expression, thereby contributing to changes in NADPH/NADP+ and ATP/AMP ratios, which in turn activates adenosine monophosphate-activated protein kinase to modulate nuclear CRY1 abundance, which further acts in the N signal input pathway to affect central clock function and flowering time. A better understanding of N-regulated floral transition will offer biotechnological solutions to improve sustainable agriculture.
Keywords: adenosine monophosphate-activated protein kinase, circadian clock, cryptochrome 1, ferredoxin-NADP+-oxidoreductase 1, nitrogen-regulated flowering
Abstract
The phenomenon of delayed flowering after the application of nitrogen (N) fertilizer has long been known in agriculture, but the detailed molecular basis for this phenomenon is largely unclear. Here we used a modified method of suppression-subtractive hybridization to identify two key factors involved in N-regulated flowering time control in Arabidopsis thaliana, namely ferredoxin-NADP+-oxidoreductase and the blue-light receptor cryptochrome 1 (CRY1). The expression of both genes is induced by low N levels, and their loss-of-function mutants are insensitive to altered N concentration. Low-N conditions increase both NADPH/NADP+ and ATP/AMP ratios, which in turn affect adenosine monophosphate-activated protein kinase (AMPK) activity. Moreover, our results show that the AMPK activity and nuclear localization are rhythmic and inversely correlated with nuclear CRY1 protein abundance. Low-N conditions increase but high-N conditions decrease the expression of several key components of the central oscillator (e.g., CCA1, LHY, and TOC1) and the flowering output genes (e.g., GI and CO). Taken together, our results suggest that N signaling functions as a modulator of nuclear CRY1 protein abundance, as well as the input signal for the central circadian clock to interfere with the normal flowering process.
The transition from vegetative to reproductive development is a central event in the plant life cycle, which is coordinately regulated by various endogenous and external cues. In the model dicotyledonous plant species Arabidopsis thaliana, five distinct genetic pathways regulating flowering time have been established: the vernalization pathway, photoperiod pathway, gibberellin acid (GA) pathway, autonomous pathway, and endogenous (age) pathway (1). These pathways ultimately converge to regulate a set of floral integrator genes, FLOWERING LOCUS T (FT) and SUPPRESSOR OF CONSTANS 1 (SOC1), which in turn activate the expression of floral meristem identity genes to trigger the formation of flowers (2–4).
Plants use the circadian clock as the timekeeping mechanism to measure day length and to ensure flowering at the proper season (5, 6). As a facultative long-day (LD) plant, Arabidopsis flowers earlier under LD conditions than under short-day (SD) conditions. Forward genetics in A. thaliana have identified the GI-CO-FT hierarchy as the canonical genetic pathway promoting flowering specifically under LD conditions (5, 7, 8). In this pathway, GI (GIGANTEA) can be considered the output point of the circadian clock to control flowering by regulating CONSTANS (CO) expression in the right phase, which activates expression of FT and TSF (TWIN SISTER OF FT) in the companion cells of the phloem within the vascular tissue (2, 9). FT and TSF proteins act as the long-sought florigens that move from leaves to the apical meristem to induce genes required for reproductive development (2–4). Both GI and CO are regulated by the circadian clock and by light signaling simultaneously and at both transcriptional and posttranscriptional levels, to ensure the transcription of FT under LD conditions, but not under SD conditions (10).
Nitrogen (N) availability is one of the key factors controlling developmental and growth to ensure plant survival and reproduction (11). Arabidopsis, like other plants, flowers earlier under low-nitrate conditions (11). Previous studies have reported that the flowering activators CO, FT, LEAFY (LFY), and APETALA1 (AP1) are induced, but the flowering repressor FLC (FLOWERING LOCUS C) is repressed in low-nitrate conditions (12, 13). There are also reports that nitrate availability controls the GA pathway at different levels (GA biosynthesis, perception, and signaling) to influence the timing of vegetative to reproductive phase change (13, 14). However, Castro Marín et al. (15) showed that low nitrate induced flowering by a pathway downstream of the floral integrators and independent of photoperiod, GA, and autonomous pathways. Thus, the detailed molecular mechanisms, especially the key factors to sense and transmit the N signal to regulate flowering remain elusive.
In this study, we used a modified method of suppression-subtractive hybridization (SSH) to identify two key factors involved in N-regulated flowering: ferredoxin-NADP+-oxidoreductase (FNR1) (16) and the blue-light photoreceptor cryptochrome 1 (CRY1) (17). Their loss-of-function mutants are insensitive to altered N levels. We show that N regulates FNR1 at the transcription level, thus affecting ratios of NADPH/NADP+ and ATP/AMP, which in turn affect adenosine monophosphate-activated protein kinase (AMPK) activity and nuclear CRY1 protein abundance. Our data imply that the nuclear level of CRY1 functions as an input cue to regulate the amplitude of circadian clock transcripts, thereby controlling flowering time.
Results
Identification of FNR1 and CRY1 as N-Responsive Genes.
Earlier microarray studies have shown that thousands of Arabidopsis genes (∼7% of the Arabidopsis transcriptome) are N-responsive (18). To search for key factors involved in N-regulated flowering, we performed a modified SSH screen with Arabidopsis seedlings grown on media containing different levels of N. We first determined the appropriate N treatment levels and Arabidopsis floral transition times. When grown at reduced N levels (1/20 N, NH4NO3, and KNO3 equally reduced), the flowering time was shortened from to 21 d from 25 d when grown on normal-N (NN) medium (1/2 MS medium containing 10 mM NH4NO3 and 9.4 mM KNO3). When grown on high-N (HN; 2×N) MS medium (40 mM NH4NO3 and 37.6 mM KNO3), the flowering time was delayed to 32 d. A further reduction of N levels to 1/50 N resulted in a severely stressed phenotype with increased anthocyanin accumulation (SI Appendix, Fig. S1).
Consistent with the observed alteration in flowering time, quantitative RT-PCR analysis revealed that expression of LFY and FT began to increase at day 11 for plants grown in low-N (LN; 1/20 N) medium, day 13 for plants grown in NN medium, and day 15 for plants grown in HN medium (SI Appendix, Fig. S2). This finding validates the suitability of our experimental conditions for screening N-responsive genes.
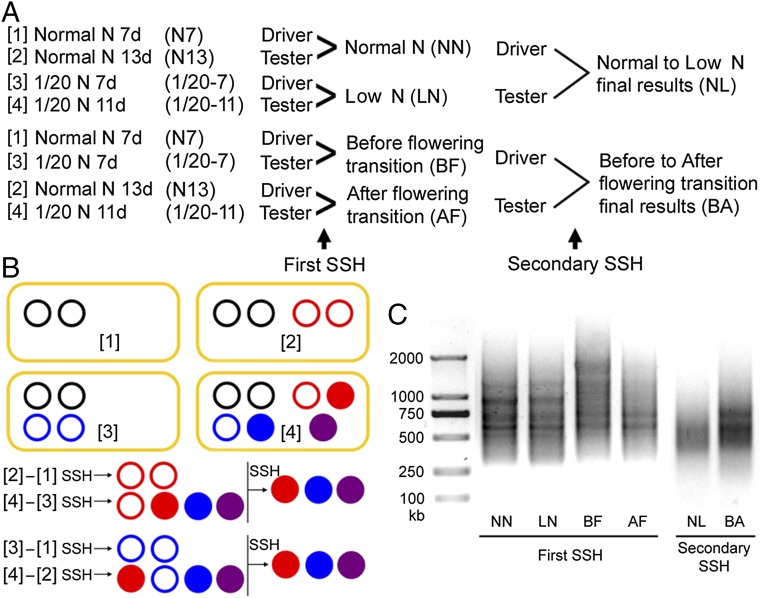
To identify the key players involved in N regulation of flowering, we performed a modified SSH screen with RNA samples collected before and after the floral transition. Four total RNA samples were collected: in NN before floral transition at the seventh day, in NN after floral transition at the 13th day, in LN before floral transition at the seventh day, and in LN after floral transition at the 11th day. Pairwise comparison of these samples through two rounds of SSH screens (Fig. 1) revealed that genes encoding FNR1 and CRY1 are prominently enriched in both final results (SI Appendix, Tables S1 and S2). Induction of FNR1 and CRY1 gene expression by LN (increased by 5.2-fold for FNR1 and by 4.8-fold for CRY1) was further confirmed by quantitative RT-PCR (SI Appendix, Fig. S3). Statistical analysis revealed that the abundance of FNR1 and CRY1 transcript is negatively correlated with flowering time (P < 0.05), implying that FNR1 and CRY1 act as two positive regulators of N-regulated flowering.
Fig. 1.
Procedures, schematic diagram of the SSH screens, and results. (A) Experimental procedure for SSH. (B) Schematic diagram of two-round SSH. Red circles indicate floral transition-induced genes, which can be further induced by 1/20 LN conditions (red disk); blue circles indicate LN-induced genes, which can be further induced after floral transition (blue disk). Purple disks indicate genes expressed only at 1/20 LN conditions and after floral transition (two-condition synergistic genes). (C) PCR verification of first-round and second-round SSH.
Furthermore, the FNR1 expression can be induced by high-Fe and S conditions (0.4 mM FeSO4·7H2O in the MS medium) and CRY1 expression can be induced by blue-light treatment (SI Appendix, Fig. S3), in disagreement with previous reports (17, 19) but consistent with reported transcriptome analyses (20, 21).
Arabidopsis fnr1 and cry1 Mutants Are Insensitive to N Changes.
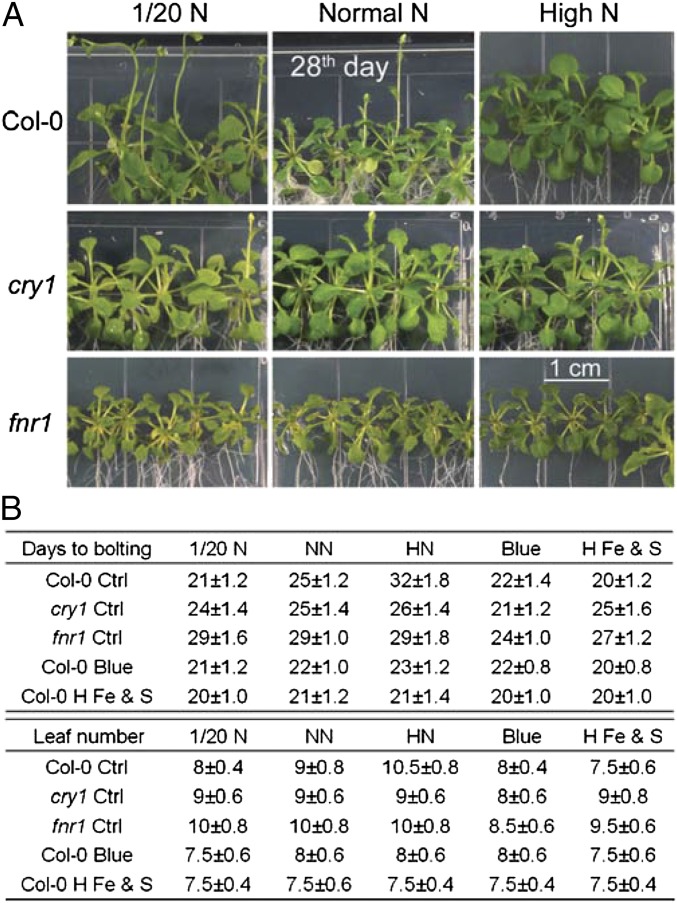
To test whether FNR1 and CRY1 play roles in N-regulated flowering, we examined the responsiveness of Arabidopsis fnr1 and cry1 mutants to different levels of N under LD conditions. Both medium-grown and soil-grown fnr1 mutants exhibited a late-flowering phenotype (29 d in NN medium), that could be altered by changing the N levels in the growth medium (P > 0.05). Although the cry1 mutant exhibited a normal flowering phenotype under regular N supply (25 d in NN medium), the flowering time of the mutant also was not altered by changing N levels (P > 0.05; Fig. 2 and SI Appendix, Fig. S4). Thus, both the Arabidopsis fnr1 and cry1 mutants displayed insensitivity to N level changes in term of flowering time. In addition, blue-light treatment (presumably to induce CRY1 expression; Fig. 1) led wild type (WT), fnr1, and cry1 plants to flower earlier than under NN conditions (P < 0.05); whereas high-Fe and S growth conditions (presumably to induce FNR1 expression; Fig. 1) did not promote early flowering in the cry1 mutant (P > 0.05; Fig. 2B). These results suggest that CRY1 may work downstream of FNR1 in the N-signaling pathway. Although previous studies reported that both the fnr2 mutant (16, 22) and the cry2 mutant (19, 23) had a late-flowering phenotype, here these mutants exhibited normal responses to N changes (P < 0.05; SI Appendix, Table S3), suggesting that neither FNR2 nor CRY2 plays an essential role in N-regulated flowering time.
Fig. 2.
fnr1 and cry1 mutants are insensitive to N levels. (A) 28-d-old WT Columbia (Col-0) plants and the fnr1 and cry1 mutant derivatives grown in MS medium under different N conditions. (B) Flowering times of WT, fnr1, and cry1 plants grown in MS medium. Days to flowering and rosette leaf number were scored (mean ± SD; n ≥ 25 plants). blue, blue-light treatment; H Fe & S, high-Fe and S (0.4 mM FeSO4) treatment.
We next checked the responsiveness of WT, cry1, and fnr1 plants to two different N sources to assess for a preference for ammonium or nitrate. WT plants flowered earlier when grown in medium supplemented with 1/20 ammonium (2.94 mM NH4Cl; at 21 d, as in the 1/20 LN condition), but flowered later when grown in medium supplemented with high levels of NH4+ nitrogen (117.6 mM NH4Cl; at 32 d, as in the HN condition). However, no difference in flowering time was observed for either the cry1 or fnr1 mutant when grown on 1/20 ammonium or high-NH4+ conditions. WT plants showed severely stressed phenotypes in either the 1/20 NO3− (2.94 mM NaNO3) or high-NO3− (117.6 mM NaNO3) condition, implying a preference for ammonium as the N source; however, the cry1 and fnr1 mutants showed less-stressed phenotypes in both the 1/20 NO3− and high-NO3− conditions (SI Appendix, Fig. S5), indicating that the cry1 and fnr1 mutants are less sensitive to nitrate changes.
We next questioned whether the LN-promoted flowering is FNR1-specific. A previous study reported that the chl1-5 (nitrate transporter AtNRT1.1) mutant exhibits a delayed flowering phenotype (24), whereas the double mutant of nitrate reductase, nia1/nia2, shows an early flowering phenotype (25). Interestingly, the flowering time in these mutants exhibited normal responses to altered N levels (SI Appendix, Fig. S6), demonstrating that the altered flowering phenotype of the cry1 or fnr1 mutant is not a general phenotype of N deficiency and is presumably related to the cooperation between N signaling and the flowering pathway.
LN Conditions Increase NADPH/NADP+ and ATP/AMP Ratios.
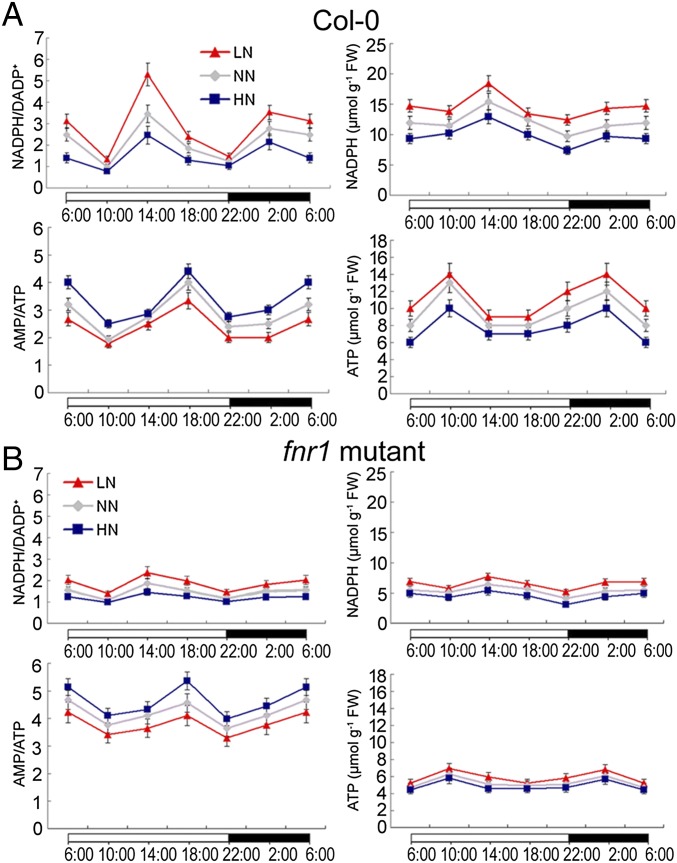
FNR is a ubiquitous flavoenzyme that oxidizes the final reduced product of the photosynthetic electron transport chain, ferredoxin (Fd), to reduce NADP+, resulting in ATP production. Therefore, FNR levels (activity) regulate cellular NADPH/NADP+ and ATP/AMP ratios (16, 22). Consistent with LN induction of FNR1 expression, we detected higher NADPH/NADP+ and lower AMP/ATP ratios in WT plants in LN conditions than in NN conditions (P < 0.05). Conversely, lower NADPH/NADP+ and higher AMP/ATP ratios were detected in HN conditions (P < 0.05; Fig. 3A). As expected, the fnr1 mutant always had a lower NADPH/NADP+ ratio and a higher AMP/ATP ratio independent of N level compared with the WT plants (Fig. 3B).
Fig. 3.
NADPH levels, ATP levels, and NADPH/NADP+ and AMP/ATP ratios in WT (A) and fnr1 mutant (B) plants over a 24-h period. Error bars show SD (n = 5).
LN Conditions Decrease Nuclear AMPK Activity and Nuclear CRY1 Phosphorylation.
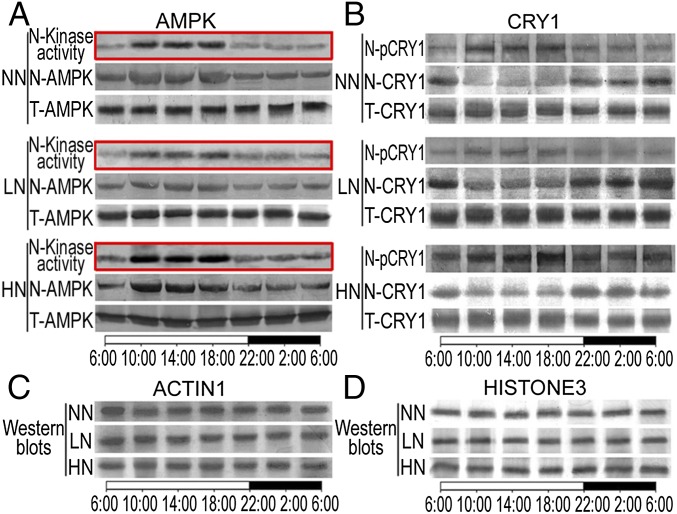
In mammalian cells, the AMP/ATP ratio affects AMPK activity, which in turn affects nuclear cryptochrome phosphorylation and peripheral clock phase (26). The Arabidopsis SnRK superfamily is homologous to mammalian AMPK and is composed of three distinct subfamilies, SnRK1, SnRK2, and SnRK3 (27). Members of the Arabidopsis SnRK family have been reported to play roles in diverse stress and metabolic signaling (27). To test whether Arabidopsis AMPK is regulated by N levels and involved in the regulation of nuclear cryptochrome phosphorylation, we examined the activity of Arabidopsis AMPKα1 (the catalytic subunit of AMPK) under different N levels. AMPKα1 is highly conserved in eukaryotes (27). Arabidopsis AMPKα1 homologs KIN10 (AT3G01090) and KIN11 (AT3G29160) proteins share 79.3% similarity with the human AMPKα1 (SI Appendix, Fig. S7). Western blot analysis showed that the anti-human AMPKα1 antibody raised against conserved peptides of the catalytic subunit can also recognize the Arabidopsis AMPKα1 homologs KIN10 and KIN11 specifically (SI Appendix, Figs. S7 and S8). An immunoprecipitation kinase assay showed that Arabidopsis AMPKα1 is activated by high levels of AMP under HN conditions (Fig. 4A). Similar to the reported robust circadian rhythm of nuclear localization for the mouse AMPKα1 subunit (26), nuclear AMPKα1 protein levels in Arabidopsis also exhibited a robust circadian rhythm, although the total cellular AMPKα1 content remained relative stable. In addition, nuclear AMPKα1 activity was much higher during the day than at night (Fig. 4A). Strikingly, we found that HN conditions increased the nuclear AMPKα1 protein level and its oscillation amplitude, whereas LN conditions decreased them (the phase was not changed; Fig. 4A). These observations suggest that N levels affect nuclear AMPKα1 level (activity) via the circadian clock, which in turn regulates flowering time in Arabidopsis. Consistent with this hypothesis, simultaneous loss of KIN10 and KIN11 function, which encode two homologs closely related to the human AMPKα1, caused reduced sensitivity to N level alteration, despite the normal phenotype of the kin10 or kin11 single mutant (SI Appendix, Fig. S9).
Fig. 4.
N enhances nuclear AMPK activity and phosphorylates and destabilizes nuclear CRY1. (A) Nuclear AMPK activity (N-kinase activity), nuclear AMPK protein level (N-AMPK), and total cellular AMPK protein level (T-AMPK) of WT plants grown under 1/20 LN, NN, or HN conditions over a 24-h period. Autoradiographs are marked with red boxes. (B) Nuclear phosphorylated CRY1 (N-pCRY1), nuclear CRY1 protein level (N-CRY1), and total cellular CRY1 protein level (T-CRY1) of WT plants grown under LN, NN, and HN conditions over a 24-h period. (C) ACTIN1 served as a loading control for total cellular proteins. (D) HISTONE3 served as a loading control for nuclear proteins.
It was previously shown that in mammalian cells, AMPK-mediated phosphorylation of CRY1 promotes ubiquitin-dependent CRY1 degradation (28), and thus the peak time of AMPKα1 nuclear localization coincides with minimal nuclear CRY1 (26). Similarly, we found that nuclear AMPKα1 peaked synchronously with nuclear CRY1 phosphorylation in Arabidopsis, and that the peak time of nuclear AMPKα1 level and its activity coincided with maximal nuclear CRY1 phosphorylation and minimal nuclear CRY1 protein level (Fig. 4B). This result suggests that Arabidopsis AMPKα1 also mediates nuclear CRY1 phosphorylation and its subsequent degradation. In support of this notion, we found that the half-life of nuclear CRY1 protein levels decreased in response to increased N levels (SI Appendix, Fig. S10). Moreover, the AMPK agonist aminoimidazole carboxamide ribonucleotide (AICAR) (26) mimicked the effect of HN conditions by causing late flowering, whereas the effect of AMPK inhibitor dorsomorphin (29) resembled the effect of LN conditions by causing early flowering (SI Appendix, Figs. S11 and S12 and Table S3). Furthermore, nuclear AMPKα1 level (activity) and nuclear CRY1 phosphorylation were always higher and nuclear CRY1 protein level was always lower in the fnr1 mutant than in WT plants (SI Appendix, Figs. S11 and S12). Taken together, these results support the hypothesis that AMPKα1 plays a key role in nuclear CRY1 phosphorylation and degradation, and that this process is regulated by FNR1 activity.
N Regulates the Circadian Clock Through the CRY1 Input Pathway.
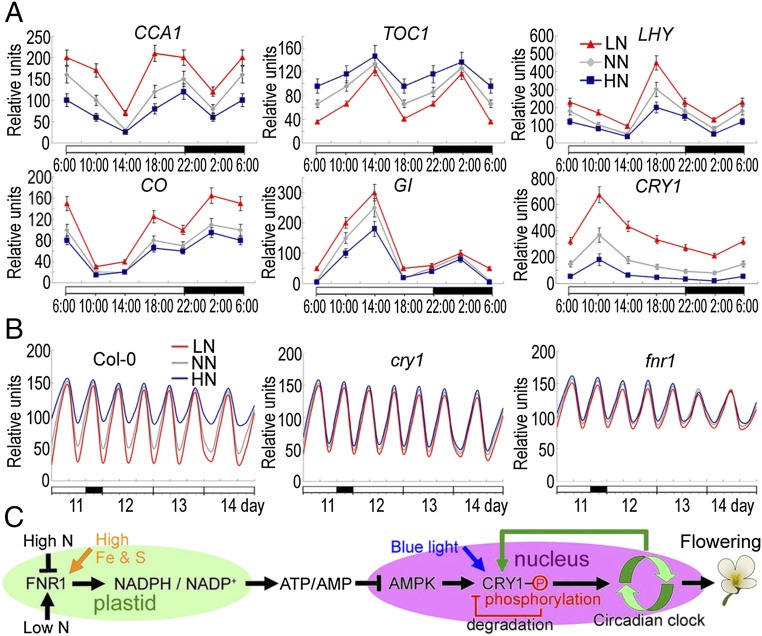
In Arabidopsis, cryptochromes act as part of the input pathways to the circadian clock, which subsequently affects the expression of key components of the central oscillator, such as LHY (LATE ELONGATED HYPOCOTYL), CCA1 (CIRCADIAN CLOCK ASSOCIATED 1), and TOC1 (TIMING OF CAB EXPRESSION 1) (30–32). As shown in Fig. 5A, the oscillation phase and abundance of CCA1 and LHY transcripts were inversely correlated with the expression of TOC1. Transcripts of two output genes, CO and GI, also fluctuated inversely with each other (Fig. 5A). To further test the role of N signaling in regulating the circadian clock, we compared the effects of altered N levels on the phase and amplitudes of CCA1, LHY, and TOC1. We found that LN conditions increased the amplitudes of all circadian transcripts throughout the circadian cycle, whereas HN conditions decreased the amplitudes of these genes in WT plants (P < 0.05); however, no phase shift was observed for these circadian clock genes (Fig. 5 A and B). Furthermore, the AMPK agonist AICAR affected the expression pattern of these genes similarly to HN conditions, whereas the AMPK inhibitor dorsomorphin affected the expression pattern of these circadian transcripts in a manner similar to LN conditions (S SI Appendix, Fig. S13). These results support the notion that altered N levels serve as a signal to regulate circadian clock function, and that AMPK plays an essential role in this regulation.
Fig. 5.
N suppresses the photoperiod pathway by inhibiting circadian transcript oscillation. (A) mRNA abundances of CCA1, TOC1, LHY, CO, GI, and CRY1 in WT plants grown in LN, NN, and HN conditions over a 24-h period. Specific gene expression levels are represented as the percentage relative to ACTIN7 expression level. Error bars show SDs (n = 5). (B) Circadian accumulation of TOC1 mRNA in WT (Col-0), fnr1, and cry1 plants under 3-d continuous light. Samples were collected from seedlings grown in 16-h light/8-h dark for 11 d, and from seedlings that were then transferred to continuous white light for 3 d. (C) Diagram of the putative N-regulated flowering pathway. Phosphorylation of CRY1 triggers its degradation. The CRY1 transcription is feedback-regulated by the circadian clock.
Interestingly, we found that the expression levels and amplitudes of these circadian transcripts were insensitive to N changes in the cry1 mutant, but were always lower in the fnr1 mutant, independent of N level (P > 0.05; SI Appendix, Fig. S14). These results suggest that FNR1 and CRY1 may play key roles in sensing and transmitting the N signal into the central clock to regulate the clock function and thus flowering.
Discussion
In this study, we have identified FNR1 and CRY1 as two critical players in N-regulated flowering time in Arabidopsis. We found that AMPK activity and nuclear localization are rhythmic and inversely correlated with nuclear CRY1 protein abundance. Correlation coefficients (R2) between parameters of NADPH/NADP+, AMP/ATP, nuclear AMPK activity, nuclear CRY1 phosphorylation level, and nuclear CRY1 protein level range from 0.83 to 0.99 (SI Appendix, Table S4). Our collective results lead us to propose a working model in which the nuclear level of CRY1 acts as a modulator in the N-signaling pathway to regulate the amplitudes of the circadian clock, thereby controlling flowering time (Fig. 5C). In Arabidopsis sugar signaling, photosynthesis also has a profound effect on the entrainment and maintenance of robust circadian rhythms, and thus regulates floral transition (33).
Our model is analogous to the earlier finding that AMPKα1 plays an important role in peripheral circadian clock entrainment in mammals in response to nutritional status through regulation of nuclear CRY1 phosphorylation and degradation (26, 34–36). Thus, the nutritional status-AMPK-CRY1 pathway may represent a conserved mechanism in higher eukaryotes. Our model is also consistent with a previous finding that the central clock gene CCA1 acts as a “master regulator” of the organic N response network (37). Expression of CCA1 is repressed by organic N, and this in turn affects the expression of glutamine synthetase 1.3 (GLN1.3), glutamate dehydrogenase 1 (GDH1), bZIP1, and asparagine synthetase 1 (ASN1) (37). Furthermore, chromatin immunoprecipitation assays have shown that CCA1 directly binds to the promoter regions of GLN1.3, GDH1, and bZIP1. These results support the hypothesis that CCA1 is a key regulator of N assimilation, and that in turn N metabolites modulate CCA1 expression, allowing N assimilation to regulate the Arabidopsis circadian clock (37).
It has been shown that the major function of CRY1 is to mediate the blue light-dependent de-etiolation process, whereas CRY2 mediates primarily the photoperiod regulation of floral initiation (19, 23). However, in the present study, the cry2 mutant exhibited normal responses to N, suggesting that CRY2 does not play an essential role in N-regulated flowering. Previous studies showed that CRY2 may mediate photoperiodic control of floral initiation by mediating light suppression of the COP1-dependent degradation of CONSTANS (10, 38), regulating light entrainment of the circadian clock to affect the expression of CO (39), or directly modulating the transcription of FT through interaction with CIBs, a group of basic helix-loop-helix transcription factors (40). In the present study, we have shown that in response to HN conditions, AMPK-mediated nuclear CRY1 phosphorylation triggers nuclear CRY1 degradation when total cellular CRY1 protein levels remain relatively stable and then inhibit circadian clock oscillations to interfere with flowering control. Consistently, it has been reported that hy4/cry1 alleles cause late flowering in SD conditions (41), and that a gain-of-function mutation in CRY1 promotes flowering in Arabidopsis (42). It also has been reported that the cry1, cry2 double mutation delays flowering in monochromatic blue light, whereas neither monogenic cry1 nor cry2 single mutant exhibits late flowering in blue light, suggesting that CRY1 and CRY2 play a somewhat redundant role in regulation of flowering (43).
Of note, some previous studies have indicated that Arabidopsis CRY1 is a light-stable protein, and that blue light-induced CRY1 phosphorylation is not accompanied by a decrease in its steady-state protein level (44). The discrepancies in CRY1 stability and degradation in response to blue light and N signal might be related to CRY1’s differing subcellular localizations under different light conditions (45). It is also interesting to note that the blue light-cryptochrome pathways show epistatic effects on plant flowering control, as N-delayed flowering can be overcome by blue light. Although our data suggest that posttranslational regulation of nuclear CRY1 protein abundance represents a major regulatory mechanism, transcriptional regulation of CRY1 also may play a role in this process. Alternatively, the induction of CRY1 expression by LN conditions might be due to a regulatory feedback mechanism (green arrow in Fig. 5C).
Like HN conditions, nitric oxide (NO) also represses floral transition by inhibiting circadian output of CO and GI expression in Arabidopsis (46), rather than key components of the central oscillator (LHY, CCA1, or TOC1). Therefore, N and NO may regulate plant flowering through different molecular signaling pathways.
In agricultural practice, N shortage causes early flowering, whereas excessive application of N fertilizers usually results in undesirable late flowering and delayed maturation (11). Other environmental stresses, such as salt, drought, heat, cold, and UV stress, also promote flowering, and this has been interpreted as a strategy to ensure seed production for plants grown under unfavorable conditions (47, 48). Designing strategies to control the timing of flowering is of pivotal importance for crop production. Our results reported here suggest that the N-regulated flowering pathway is adjusted by treatment with blue light or by regulation of CRY1 expression in Arabidopsis, which may offer a new testable solution to managing flowering time in crops.
Materials and Methods
Plant materials and growth conditions, N level adjustments, flowering time analysis, and details of SSH, quantitative RT-PCR, NADPH/NADP+ and ATP/AMP determination, protein extraction and immunoblotting (including the nuclear CRY1 phosphorylation assay), and the immunoprecipitation nuclear AMPK assay are described in detail in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Michelle Hanna (University of Saskatchewan) for proofreading the manuscript. This work was supported by the Ministry of Science and Technology of China (Grant 2013CB967300, to Y. He), the National Natural Science Foundation of China (Grants 31300207, to Z.-W.Z.; 31400242, to F.X.; and 31530006, to Y.H.), and the Preeminent Youth Fund of Sichuan Province (Grant 2015JQO045, to S.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1602004113/-/DCSupplemental.
References
- 1.Srikanth A, Schmid M. Regulation of flowering time: All roads lead to Rome. Cell Mol Life Sci. 2011;68(12):2013–2037. doi: 10.1007/s00018-011-0673-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T. TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol. 2005;46(8):1175–1189. doi: 10.1093/pcp/pci151. [DOI] [PubMed] [Google Scholar]
- 3.Corbesier L, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316(5827):1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- 4.Jang S, Torti S, Coupland G. Genetic and spatial interactions between FT, TSF and SVP during the early stages of floral induction in Arabidopsis. Plant J. 2009;60(4):614–625. doi: 10.1111/j.1365-313X.2009.03986.x. [DOI] [PubMed] [Google Scholar]
- 5.Samach A, Coupland G. Time measurement and the control of flowering in plants. BioEssays. 2000;22(1):38–47. doi: 10.1002/(SICI)1521-1878(200001)22:1<38::AID-BIES8>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 6.Yanovsky MJ, Kay SA. Living by the calendar: How plants know when to flower. Nat Rev Mol Cell Biol. 2003;4(4):265–275. doi: 10.1038/nrm1077. [DOI] [PubMed] [Google Scholar]
- 7.Suárez-López P, et al. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001;410(6832):1116–1120. doi: 10.1038/35074138. [DOI] [PubMed] [Google Scholar]
- 8.Searle I, Coupland G. Induction of flowering by seasonal changes in photoperiod. EMBO J. 2004;23(6):1217–1222. doi: 10.1038/sj.emboj.7600117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wigge PA, et al. Integration of spatial and temporal information during floral induction in Arabidopsis. Science. 2005;309(5737):1056–1059. doi: 10.1126/science.1114358. [DOI] [PubMed] [Google Scholar]
- 10.Valverde F, et al. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303(5660):1003–1006. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- 11.Vidal EA, Moyano TC, Canales J, Gutiérrez RA. Nitrogen control of developmental phase transitions in Arabidopsis thaliana. J Exp Bot. 2014;65(19):5611–5618. doi: 10.1093/jxb/eru326. [DOI] [PubMed] [Google Scholar]
- 12.Kant S, Peng M, Rothstein SJ. Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in Arabidopsis. PLoS Genet. 2011;7(3):e1002021. doi: 10.1371/journal.pgen.1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu T, et al. Nitrate or NaCl regulates floral induction in Arabidopsis thaliana. Biologia. 2013;68:215–222. [Google Scholar]
- 14.Osnato M, Castillejo C, Matías-Hernández L, Pelaz S. TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nat Commun. 2012;3:808. doi: 10.1038/ncomms1810. [DOI] [PubMed] [Google Scholar]
- 15.Castro Marín I, et al. Nitrate regulates floral induction in Arabidopsis, acting independently of light, gibberellin, and autonomous pathways. Planta. 2011;233(3):539–552. doi: 10.1007/s00425-010-1316-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lintala M, et al. Structural and functional characterization of ferredoxin-NADP+-oxidoreductase using knock-out mutants of Arabidopsis. Plant J. 2007;49(6):1041–1052. doi: 10.1111/j.1365-313X.2006.03014.x. [DOI] [PubMed] [Google Scholar]
- 17.Ahmad M, Cashmore AR. HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature. 1993;366(6451):162–166. doi: 10.1038/366162a0. [DOI] [PubMed] [Google Scholar]
- 18.Yang XS, et al. Gene expression biomarkers provide sensitive indicators of in planta nitrogen status in maize. Plant Physiol. 2011;157(4):1841–1852. doi: 10.1104/pp.111.187898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mockler T, et al. Regulation of photoperiodic flowering by Arabidopsis photoreceptors. Proc Natl Acad Sci USA. 2003;100(4):2140–2145. doi: 10.1073/pnas.0437826100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang XN, et al. HFR1 is crucial for transcriptome regulation in the cryptochrome 1-mediated early response to blue light in Arabidopsis thaliana. PLoS One. 2008;3(10):e3563. doi: 10.1371/journal.pone.0003563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai HL, et al. HUA ENHANCER1 is involved in posttranscriptional regulation of positive and negative regulators in Arabidopsis photomorphogenesis. Plant Cell. 2014;26(7):2858–2872. doi: 10.1105/tpc.114.126722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemaire SD, et al. The complex regulation of ferredoxin/thioredoxin-related genes by light and the circadian clock. Planta. 1999;209(2):221–229. doi: 10.1007/s004250050626. [DOI] [PubMed] [Google Scholar]
- 23.Guo H, Yang H, Mockler TC, Lin C. Regulation of flowering time by Arabidopsis photoreceptors. Science. 1998;279(5355):1360–1363. doi: 10.1126/science.279.5355.1360. [DOI] [PubMed] [Google Scholar]
- 24.Guo FQ, Wang R, Chen M, Crawford NM. The Arabidopsis dual-affinity nitrate transporter gene AtNRT1.1 (CHL1) is activated and functions in nascent organ development during vegetative and reproductive growth. Plant Cell. 2001;13(8):1761–1777. doi: 10.11054/TPC.010126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seligman K, Saviani EE, Oliveira HC, Pinto-Maglio CA, Salgado I. Floral transition and nitric oxide emission during flower development in Arabidopsis thaliana is affected in nitrate reductase-deficient plants. Plant Cell Physiol. 2008;49(7):1112–1121. doi: 10.1093/pcp/pcn089. [DOI] [PubMed] [Google Scholar]
- 26.Lamia KA, et al. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326(5951):437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crozet P, et al. Mechanisms of regulation of SNF1/AMPK/SnRK1 protein kinases. Front Plant Sci. 2014;5:190. doi: 10.3389/fpls.2014.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee Y, Kim EK. AMP-activated protein kinase as a key molecular link between metabolism and clockwork. Exp Mol Med. 2013;45:e33. doi: 10.1038/emm.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hong CC, Yu PB. Applications of small molecule BMP inhibitors in physiology and disease. Cytokine Growth Factor Rev. 2009;20(5-6):409–418. doi: 10.1016/j.cytogfr.2009.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Somers DE, Devlin PF, Kay SA. Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science. 1998;282(5393):1488–1490. doi: 10.1126/science.282.5393.1488. [DOI] [PubMed] [Google Scholar]
- 31.Barak S, Tobin EM, Andronis C, Sugano S, Green RM. All in good time: The Arabidopsis circadian clock. Trends Plant Sci. 2000;5(12):517–522. doi: 10.1016/s1360-1385(00)01785-4. [DOI] [PubMed] [Google Scholar]
- 32.Ni Z, et al. Altered circadian rhythms regulate growth vigour in hybrids and allopolyploids. Nature. 2009;457(7227):327–331. doi: 10.1038/nature07523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haydon MJ, Mielczarek O, Robertson FC, Hubbard KE, Webb AA. Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature. 2013;502(7473):689–692. doi: 10.1038/nature12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davies SP, Carling D, Munday MR, Hardie DG. Diurnal rhythm of phosphorylation of rat liver acetyl-CoA carboxylase by the AMP-activated protein kinase, demonstrated using freeze-clamping: Effects of high-fat diets. Eur J Biochem. 1992;203(3):615–623. doi: 10.1111/j.1432-1033.1992.tb16591.x. [DOI] [PubMed] [Google Scholar]
- 35.Witters LA, Kemp BE, Means AR. Chutes and ladders: The search for protein kinases that act on AMPK. Trends Biochem Sci. 2006;31(1):13–16. doi: 10.1016/j.tibs.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 36.Jeon SM, Chandel NS, Hay N. AMPK regulates NADPH homeostasis to promote tumour cell survival during energy stress. Nature. 2012;485(7400):661–665. doi: 10.1038/nature11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gutiérrez RA, et al. Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc Natl Acad Sci USA. 2008;105(12):4939–4944. doi: 10.1073/pnas.0800211105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H, Ma LG, Li JM, Zhao HY, Deng XW. Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science. 2001;294(5540):154–158. doi: 10.1126/science.1063630. [DOI] [PubMed] [Google Scholar]
- 39.Zhu H, et al. Cryptochromes define a novel circadian clock mechanism in monarch butterflies that may underlie sun compass navigation. PLoS Biol. 2008;6(1):e4. doi: 10.1371/journal.pbio.0060004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu H, et al. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science. 2008;322(5907):1535–1539. doi: 10.1126/science.1163927. [DOI] [PubMed] [Google Scholar]
- 41.Bagnall DJ, King RW, Hangarter RP. Blue-light promotion of flowering is absent in hy4 mutants of Arabidopsis. Planta. 1996;200(2):278–280. doi: 10.1007/BF00208319. [DOI] [PubMed] [Google Scholar]
- 42.Exner V, et al. A gain-of-function mutation of Arabidopsis cryptochrome1 promotes flowering. Plant Physiol. 2010;154(4):1633–1645. doi: 10.1104/pp.110.160895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mockler TC, Guo H, Yang H, Duong H, Lin C. Antagonistic actions of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development. 1999;126(10):2073–2082. doi: 10.1242/dev.126.10.2073. [DOI] [PubMed] [Google Scholar]
- 44.Shalitin D, Yu X, Maymon M, Mockler T, Lin C. Blue light-dependent in vivo and in vitro phosphorylation of Arabidopsis cryptochrome 1. Plant Cell. 2003;15(10):2421–2429. doi: 10.1105/tpc.013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang HQ, et al. The C termini of Arabidopsis cryptochromes mediate a constitutive light response. Cell. 2000;103(5):815–827. doi: 10.1016/s0092-8674(00)00184-7. [DOI] [PubMed] [Google Scholar]
- 46.He Y, et al. Nitric oxide represses the Arabidopsis floral transition. Science. 2004;305(5692):1968–1971. doi: 10.1126/science.1098837. [DOI] [PubMed] [Google Scholar]
- 47.Martínez C, Pons E, Prats G, León J. Salicylic acid regulates flowering time and links defence responses and reproductive development. Plant J. 2004;37(2):209–217. doi: 10.1046/j.1365-313x.2003.01954.x. [DOI] [PubMed] [Google Scholar]
- 48.Achard P, et al. Integration of plant responses to environmentally activated phytohormonal signals. Science. 2006;311(5757):91–94. doi: 10.1126/science.1118642. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.