Significance
The tricarboxylic acid (TCA) cycle plays a central role in the metabolism of many pathogens, but few inhibitors of this pathway currently exist. Our study describes the first small molecule inhibitor, to our knowledge, of the fumarate hydratase enzyme in the TCA cycle of Mycobacterium tuberculosis. We also report the discovery of an allosteric regulatory site that confers selectivity of the inhibitor’s activity over the homologous human enzyme. Our findings illustrate the potential to target vulnerable metabolic enzymes effectively and selectively, despite their high degree of evolutionary conservation.
Keywords: allosteric regulation, selective inhibition, fumarate hydratase, Mycobacterium tuberculosis, TCA cycle
Abstract
Enzymes in essential metabolic pathways are attractive targets for the treatment of bacterial diseases, but in many cases, the presence of homologous human enzymes makes them impractical candidates for drug development. Fumarate hydratase, an essential enzyme in the tricarboxylic acid (TCA) cycle, has been identified as one such potential therapeutic target in tuberculosis. We report the discovery of the first small molecule inhibitor, to our knowledge, of the Mycobacterium tuberculosis fumarate hydratase. A crystal structure at 2.0-Å resolution of the compound in complex with the protein establishes the existence of a previously unidentified allosteric regulatory site. This allosteric site allows for selective inhibition with respect to the homologous human enzyme. We observe a unique binding mode in which two inhibitor molecules interact within the allosteric site, driving significant conformational changes that preclude simultaneous substrate and inhibitor binding. Our results demonstrate the selective inhibition of a highly conserved metabolic enzyme that contains identical active site residues in both the host and the pathogen.
Because the tricarboxylic acid (TCA) cycle connects many pathways of cellular metabolism, preventing the function of this cycle through enzyme inhibition is an attractive strategy for targeting infectious agents (1). In Mycobacterium tuberculosis, experimental evidence (2) has suggested that fumarate hydratase, the essential enzyme responsible for the reversible conversion of fumarate to (l)-malate, is a vulnerable target. This vulnerability is in part due to the fact that, unlike other bacteria (such as Escherichia coli), M. tuberculosis expresses only one enzyme that performs this function (3). In addition to its role in metabolism under aerobic conditions, fumarate hydratase has also garnered interest because of the discovery of a flux toward the reverse TCA cycle under hypoxic conditions in nonreplicating M. tuberculosis (2, 4, 5). However, despite these discoveries, no small molecule inhibitor of the M. tuberculosis fumarate hydratase has been reported. The discovery of such an inhibitor would provide an important tool to begin probing the role of the TCA cycle in both actively replicating and nonreplicating bacteria.
From the standpoint of drug development, however, targeting the M. tuberculosis fumarate hydratase poses a significant challenge, because the protein is highly evolutionarily conserved. In particular, the human and M. tuberculosis homologs share identical active site residues as well as 53% overall sequence identity (6, 7). Both homologs form a stable homotetramer containing four active sites, and every active site is composed of residues from three enzyme subunits. Each dumbbell-shaped subunit within the tetramer contains three domains: an N-terminal domain, a central domain, and a C-terminal domain (8–10). The N- and C-terminal domains are predominantly α-helical and linked by the central domain that consists of five tightly packed helices. The central domains of the four subunits pack together into a 20-helix bundle to form the tetrameric structure. Each subunit organizes in a head to head fashion with one subunit and a head to tail fashion with the remaining two subunits. These structural similarities further increase the challenge of selective inhibition.
Here, we report the discovery of the first selective small molecule inhibitor, to our knowledge, of the M. tuberculosis fumarate hydratase. The selectivity results from the binding of the inhibitor to a previously unidentified allosteric site composed of residues that are not conserved between the human and the M. tuberculosis homologs. Using X-ray crystallography and steady-state kinetics, we define the location of this binding pocket and assess the effect of the inhibitor on both enzyme structure and function. These results illustrate the potential for fumarate hydratase to be a tractable target for drug development against M. tuberculosis.
Results
Fluorescence-Based Assays to Monitor Fumarate Hydratase Enzyme Activity.
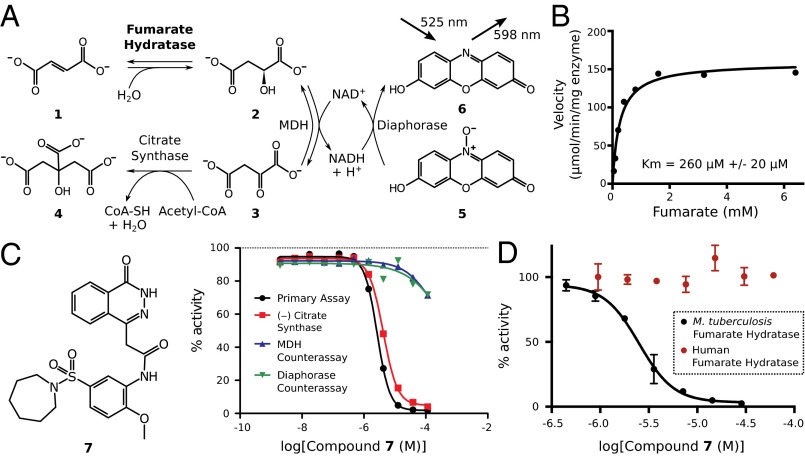
To identify inhibitors of the M. tuberculosis fumarate hydratase, we developed a fluorescence-based assay to monitor the enzyme’s activity. Inspired by others who have used enzyme kinetics to investigate the TCA cycle (11), we modeled the assay after the natural progression of the cycle under aerobic conditions. In this assay design, fumarate hydratase requires coupling to a minimum of two enzymes. The first coupled enzyme, malate dehydrogenase (MDH), generates the fluorescent molecule NADH on oxidizing (l)-malate to oxaloacetate; however, the equilibrium of this reaction favors (l)-malate. Therefore, a second coupled enzyme is required to obtain a reliable readout. We chose to include the enzyme diaphorase, which consumes the NADH from the MDH reaction and regenerates NAD+. The use of diaphorase and its substrate resazurin (7-hydroxy-3H-phenoxazin-3-one) has the additional benefit of forming the fluorescent molecule resorufin (7-hydroxy-3H-phenoxazin-3-one) as a product. Thus, we were able to monitor resorufin fluorescence at 598 nm as a means of measuring fumarate hydratase activity. The detection at a wavelength of 598 nm is preferable to commonly used shorter wavelengths, such as 340 nm, where the intrinsic fluorescence of compounds in small molecule libraries results in assay interference (12). Finally, we also incorporated the enzyme citrate synthase into the assay design, because the thermodynamically favorable cleavage of the thioester bond of acetyl-CoA in this reaction significantly speeds the assay progression by shifting the equilibrium state toward the products, thereby making a high-throughput screen feasible (Fig. 1A).
Fig. 1.
A high-throughput screen identifies inhibitors of the M. tuberculosis fumarate hydratase. (A) Schematic of the primary fluorescence-based high-throughput screening assay used to monitor fumarate hydratase activity. Fumarate (1) is used to initiate the reaction, and (l)-malate (2), oxaloacetate (3), and citrate (4) are produced. The assay is monitored by measuring the conversion of resazurin (5) to resorufin (6). (B) The saturation curve, fit using the Michaelis–Menten model, for the fumarate hydratase enzyme as measured by the assay shown in A. Data are reported as an average of replicates (n = 3), and error bars indicate SEMs. (C) The structure and dose–response profiles of the most potent compound (7) from the screen. Inhibition is observed in both the primary assay (black) and a variation of the assay in which citrate synthase is removed (red). (D) The dose–response curve obtained for 7 (black; n = 3). The compound shows no inhibitory effect on the human fumarate hydratase (red; n = 2). Data are reported as an average of replicates, and error bars indicate SEMs.
A titration of the fumarate hydratase enzyme in our final assay conditions showed proportional changes in the initial rate of the fluorescence output, indicating that we were accurately monitoring the enzyme’s activity (SI Appendix, Fig. S1). We further tested the reliability of the assay by measuring the initial reaction rate at varying substrate concentrations. The resulting highly reproducible saturation curve was fit using the Michaelis–Menten model (13, 14), yielding a Km of 260 ± 20 μM for fumarate (Fig. 1B). To evaluate enzyme activity in the presence of small molecules, we elected to measure fluorescence immediately on initiation of the reaction and again after 5 min; 5 min was found to be a suitable time point, because the reaction displayed a wide signal window of 1,104 ± 13 relative fluorescence units (RFU) between positive and negative (no fumarate hydratase enzyme) controls with a factor (15) of 0.71.
Identification of Fumarate Hydratase Inhibitors Using High-Throughput Screening.
We conducted a screen of 479,984 small molecules in a 1,536-well format (16, 17). The compound library contains a diverse set of molecules with drug-like physicochemical properties and includes compounds with known biological activity (18). Each compound was screened at four concentrations ranging from 115 to 2.3 μM. We selected a group of 1,856 compounds that showed potential activity as defined by a dose–response curve class (17) of either 1 or 2, and evaluated their reproducibility by rescreening them in the assay at 11 concentrations ranging from 115 μM to 2.0 nM. This group of compounds was also subjected to rigorous counterscreening to eliminate those molecules affecting resorufin production through mechanisms other than fumarate hydratase inhibition. Compounds affecting MDH, citrate synthase, and diaphorase activity were identified and discarded using truncated versions of the primary assay (SI Appendix, Fig. S2). In addition to eliminating inhibitors of the coupled enzymes, the counterassays allowed for removal of molecules that affected fluorescence because of inherent properties of the compound or nonspecific effects in the assay conditions.
We identified two structurally similar candidate molecules that bear no overt resemblance to fumarate and (l)-malate (SI Appendix, Fig. S3). This observation prompted us to examine the more potent of the two compounds [N-(5-(azepan-1-ylsulfonyl)-2-methoxyphenyl)-2-(4-oxo-3,4-dihydrophthalazin-1-yl)acetamide (7)] in detail (Fig. 1C). The compound was resynthesized to confirm its identity and purity (SI Appendix, Fig. S4). Subsequent revalidation of 7 revealed a highly reproducible dose–response curve with an IC50 of 2.5 ± 1.1 μM, a maximum inhibition of 97% ± 3%, and notably, a Hill slope of 2.4 ± 0.5 (Fig. 1D). This Hill slope suggests that a minimum of two inhibitor molecules must be functionally important for the inhibition of catalysis at a given active site (19).
Validation of Binding Using X-Ray Crystallography.
To validate the binding of 7 to fumarate hydratase, we performed X-ray crystallographic studies of the enzyme. We crystallized the protein in the absence of 7 in a solution containing 200 mM magnesium formate and determined the structure at 1.86-Å resolution (SI Appendix, Table S1) [Protein Data Bank (PDB) ID code 5F92]. Formate ions from the solution are bound at two of the enzyme active sites in a fashion that mimics substrate binding, with the formate carboxylate ions making identical hydrogen bonding interactions as the carboxylates of (l)-malate described in a previously published structure (PDB ID code 4ADL) (SI Appendix, Fig. S5) (7). The binding of substrate-mimicking formate ions in two of four available active sites is also consistent with the structure of fumarate hydratase in complex with (l)-malate. We illustrate the relative positions of the formate-bound and unoccupied active sites in SI Appendix, Fig. S6.
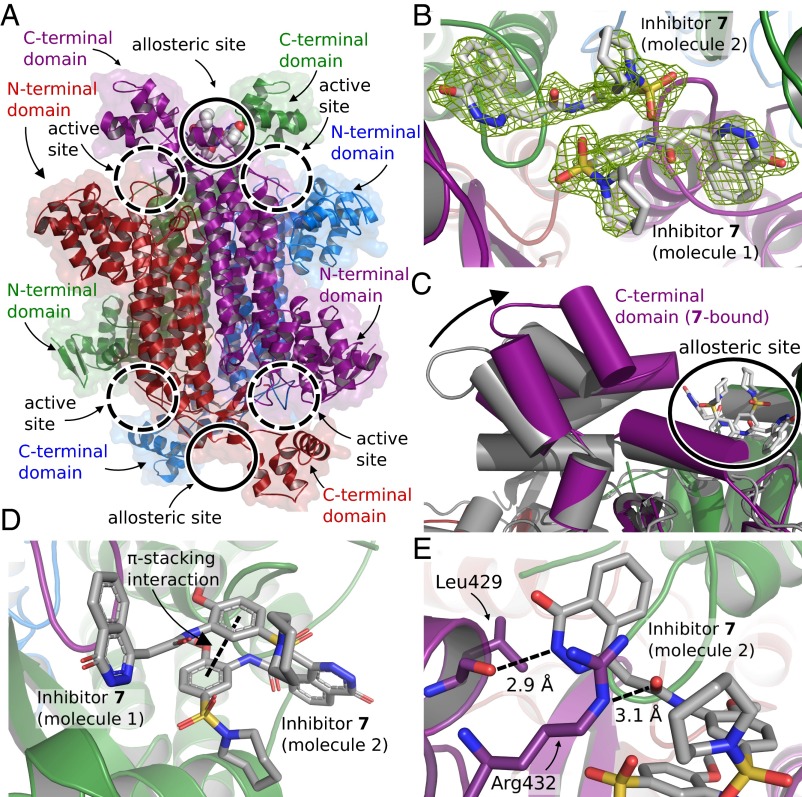
To obtain a protein–inhibitor complex, we soaked the formate-bound crystals with a saturated solution of 7, resulting in a structure at 2.0-Å resolution (SI Appendix, Table S1) (PDB ID code 5F91). Analysis of the structure reveals Fourier difference density that unambiguously identifies a binding site of two molecules of 7 ∼20 Å away from each of the two nearest active sites (Fig. 2 A and B). This binding site is located at the interface of the two C-terminal domains of subunits A and C. Because of the enzyme’s internal symmetry, a second identical binding site should also exist between the C-terminal domains of subunits B and D; however, no inhibitor is observed at this location, most likely because the site is engaged in a crystal contact (SI Appendix, Figs. S7 and S8).
Fig. 2.
A crystal structure of the M. tuberculosis fumarate hydratase at 2.0-Å resolution reveals the existence of an allosteric site. (A) A ribbon/surface representation of the overall structure of the homotetrameric M. tuberculosis fumarate hydratase with 7 bound to an allosteric site. The dashed circles indicate the location of the four active sites, and the solid circles indicate the location of the two allosteric sites. Colors represent the subunits of the tetramer: green, subunit A; blue, subunit B; purple, subunit C; red, subunit D. (B) Fourier difference density (green mesh; contoured at 3σ) calculated before 7 was added to the model. (C) Superimposition of the fumarate hydratase enzyme bound with formate (gray) or 7 (colors). An arrow indicates the conformational change in the C-terminal domain of subunit C on binding of 7. The solid circle indicates the location of the bound inhibitor at the allosteric site. (D) The stabilizing stacking interaction (indicated by a dashed line) of the central phenyl rings of 7 within the allosteric site. (E) Snapshot of several key interactions between 7 and surrounding amino acid residues. A hydrogen bond between the backbone oxygen atom of Leu429 and the 2-position of the quinazolinone ring of 7 is shown with a dashed line. Arg432 hydrogen bonds to the amide oxygen atom of 7 and also forms a stacking interaction with the quinazolinone ring.
The binding of 7 results in a dramatic conformational change, in which the C-terminal domain of subunit C rotates to interact with the inhibitor molecules (Fig. 2C). Consistent with this observation of a significant structural rearrangement, the length of the crystallographic c axis decreases by ∼5 Å (SI Appendix, Table S1). The structural change results in the loss of electron density of both the two formate ions and the active site loop 316A–323A in the nearest active site. This effect on the substrate-mimicking formate ions and the active site assembly suggests that the binding site of 7 is allosteric in nature.
One of the key features of the two bound inhibitor molecules is an unusual π-stacking interaction between their two core phenyl rings within the allosteric site (Fig. 2D). This interaction between the two molecules of 7 is likely to be an important contributor to binding. Furthermore, each molecule of 7 makes several key interactions with surrounding amino acids. Each inhibitor molecule interacts with the nearest Arg432 (of either subunit A or C), with the residue forming a stacking interaction with the inhibitor’s quinazolinone ring and a hydrogen bond to its amide oxygen atom. In addition, the backbone oxygen atom of each Leu429 hydrogen bonds to the nitrogen atom at the 2-position of the nearest inhibitor molecule’s quinazolinone ring (Fig. 2E and SI Appendix, Fig. S9). A stereoimage of the two molecules of 7 in their electron density, with a depiction of additional interactions with the surrounding amino acids and solvent molecules, can be found in SI Appendix, Fig. S10.
Examination of the Mode of Inhibition.
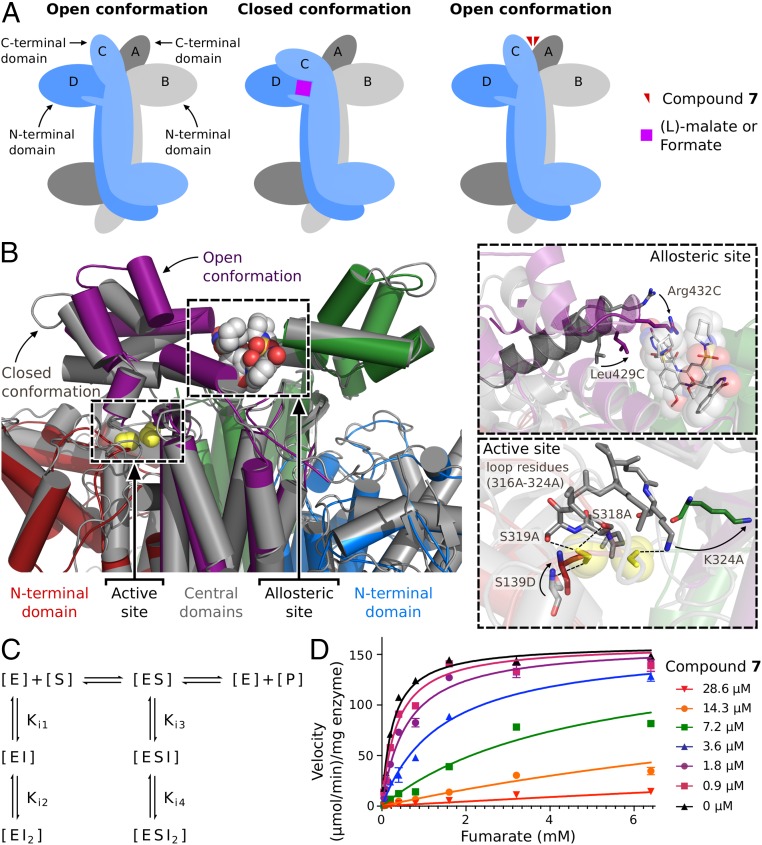
The loss of the two formate ions from the active site adjacent to 7 suggests that the binding of the two inhibitor molecules to the allosteric site competitively precludes substrate binding in the neighboring active sites. To investigate this observation further, we assessed the nature of the protein’s conformational change by examining the subunit that exhibits the greatest movement on inhibitor binding (subunit C). It has been previously reported that the C-terminal domain of each subunit of the M. tuberculosis fumarate hydratase enzyme possesses both an “open” and a “closed” conformational state. The closed conformational state is induced on substrate binding at the nearest active site and involves a 34° inward rotation of the C-terminal domain from the open unbound form (9). In the formate-bound structure, subunit C aligns with the closed conformation of a subunit with (l)-malate bound to the nearest active site (subunit rmsd = 0.8 Å; PDB ID code 4ADL), because the formate ions mimic the binding mode of the carboxylates of (l)-malate (SI Appendix, Fig. S11A). On binding of 7, however, the C-terminal domain rotates outward to align with the substrate-free enzyme in the open conformation (subunit rmsd = 1.3 Å; PDB ID code 3NO9) (SI Appendix, Fig. S11B). This movement suggests that inhibitor binding stabilizes the subunit’s C-terminal domain in an open-like form, thereby preventing substrate binding and catalysis in a competitive fashion (Fig. 3A). Notably, the C-terminal domain of subunit A, which also contributes to the allosteric site, remains in the open conformation in both the formate- and the 7-bound structures, because its adjacent active site is always unoccupied.
Fig. 3.
Structural data and enzyme kinetics support a competitive mode of inhibition. (A) Cartoon representation of fumarate hydratase in the presence of substrate and the allosteric inhibitor, 7. (Left) In the absence of substrate or substrate mimic, the C-terminal domain of subunit C adopts the open conformation. (Center) When (l)-malate or formate is bound to the active site, the C-terminal domain of subunit C adopts the closed conformation. (Right) In the presence of the allosteric inhibitor, subunit C shifts to the open conformation. The electron density for the formate ions is lost, suggesting that substrate and inhibitor cannot be simultaneously bound. (B) A depiction of the fumarate hydratase active site and allosteric site when formate (gray structure; formate is in yellow) or 7 is bound (colored structure). On binding of 7, the C-terminal domain of subunit C (purple) rotates into the open conformation. (Upper Inset) At the allosteric site, Leu429C and Arg432C shift toward the inhibitor. (Lower Inset) At the active site, the binding of 7 results in the loss of electron density of the loop residues 316A–323A as well as the movement of active site residues Lys324A and Ser139D. These active site residues form critical hydrogen bonds (dashed lines) to the substrate-mimicking formate ions in the formate-bound structure. (C) The equilibrium binding model that accounts for two molecules of inhibitor (I) binding to the enzyme (E) or enzyme–substrate complex (ES) in the presence of substrate (S) and product (P). (D) The saturation curves of the fumarate hydratase enzyme in the presence of different concentrations of 7, fit using the model shown in C. Data are reported as an average of replicates (n = 2), and error bars indicate SEMs.
The effect of the inhibitor on the residues of the allosteric site and the adjacent active site is illustrated in Fig. 3B. On binding of 7, Leu429C and Arg432C shift to form interactions with the inhibitor within the allosteric site (Fig. 3B, Upper Inset). In the adjacent active site, the electron density of the two formate ions is lost. Notably, the conformational change affects active site assembly by increasing the flexibility of the loop residues 316A–323A, such that these residues can no longer be modeled in the 7-bound structure (SI Appendix, Fig. S12). The loss of the electron density of this loop is significant, because two of the residues, Ser318A and Ser319A, form critical hydrogen bonds with formate ions in the formate-bound structure. This change in the active site is likely an effect of the concerted motion of the C-terminal domain of subunit C induced by the binding of 7. The movement of the C-terminal domain of subunit C from the closed conformation to the open conformation eliminates the stabilizing interaction between Asn413C and the backbone oxygen atom of the loop residue Ile320A (SI Appendix, Fig. S13). The loss of this interaction likely contributes to the increased flexibility of the critical loop residues 316A–323A at the active site. Additional active site residues are also affected in the inhibitor-bound structure. Lys324A and Ser139D, which both form hydrogen bonds with the formate ions in the formate-bound structure, are displaced in the 7-bound structure. As a result of the movement, the γ-oxygen of Ser139D occupies the former location of a formate ion (Fig. 3B, Lower Inset). Together, these changes in the active site on binding of 7 suggest that simultaneous binding of inhibitor and substrate cannot occur and that 7 is a competitive inhibitor of enzyme activity.
In addition to the structural evidence, we also assessed the mechanism of inhibition of 7 in solution using steady-state kinetic experiments. We generated a matrix of reaction progress curves for fumarate hydratase in the presence of varying concentrations of substrate and 7, and then plotted the enzymatic saturation curves at each inhibitor concentration. Given that substrate cooperativity was not observed at the fumarate concentrations used in the assay, the kinetic data were analyzed with respect to an independent catalytic site. Furthermore, because the crystal structure suggests that the binding of two inhibitor molecules disrupts active site assembly, the model for determining mode of inhibition was modified to incorporate the equilibrium binding of two inhibitor molecules (Fig. 3C). The rate equation derived from this model represents general mixed inhibition (Eq. 1). [All standard models of inhibition (competitive, noncompetitive, uncompetitive, and mixed) are incorporated within this model, and on fitting of a dataset, the model may reduce to a specific case depending on the resultant Ki values (20).]
We fit the rate equation for the model shown in Fig. 3C to our kinetic data (Fig. 3D), and the resulting Ki values support a competitive mode of inhibition. The values for Ki1 and Ki2, which represent the affinities of the inhibitor for the enzyme without substrate bound, were found to be 6.3 ± 4.4 and 3.2 ± 0.3 μM, respectively. By contrast, Ki3 and Ki4, which represent the affinities of the inhibitor for the enzyme–substrate complex, were found to contribute negligibly to the fit [(1/Ki3) 10−14 and (1/Ki4) 10−16]. This result suggests that the binding of the inhibitor to the enzyme alone results in inhibition in a purely competitive fashion. The values of apparent Vmax and apparent Km for the fits shown in Fig. 3D have been calculated for each inhibitor concentration and are presented in SI Appendix, Table S2. Consistent with competitive inhibition, apparent Vmax does not change as inhibitor concentration increases, whereas the apparent Km is observed to increase.
Allosteric Site Binding Confers Selectivity.
After the identification of the allosteric site, we aligned the amino acid sequences of the M. tuberculosis and the human fumarate hydratase enzymes (SI Appendix, Fig. S14A). The alignment reveals that, despite a 53% sequence identity and identical active site residues, the allosteric site residues making direct interactions with the inhibitor molecules are not conserved in the human homolog (SI Appendix, Fig. S14B). The difference in these amino acid residues suggests the potential for selectivity of 7 over the human enzyme.
To investigate this hypothesis, we modified the enzymatic assay shown in Fig. 1A to report on human fumarate hydratase activity (SI Appendix, Fig. S14 C and D). We then treated the human enzyme with 7 at concentrations up to 122 μM. We observed no inhibition of the human enzyme, indicating that 7 is highly selective (SI Appendix, Fig. S14E). The striking difference in the effect of 7 on the activity of the two enzymes is illustrated in Fig. 1D.
Effect of the Inhibitor on M. tuberculosis H37Rv.
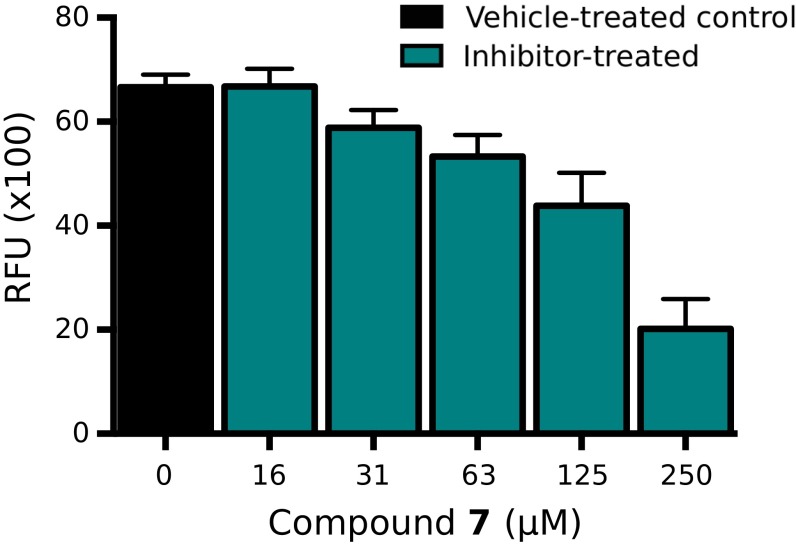
To evaluate the potential of 7 in a biological context, we examined the effect of our hit compound on the survival and growth of M. tuberculosis H37Rv. When evaluating survival by detection of colony-forming units, we did not observe any bactericidal effect when cells were treated with 7 under either aerobic or anaerobic conditions (SI Appendix, SI Methods and Fig. S15). However, treatment of H37Rv with 7 under aerobic conditions resulted in a dose-dependent effect on bacterial growth rate (Fig. 4). Cells constitutively expressing GFP (21) were used to monitor the growth rate of the bacteria. Twelve days after treatment with 7 at a concentration of 250 μM, bacterial cells exhibited 35% ± 6% growth relative to a vehicle-treated control culture. The growth rate could not be monitored in a similar fashion under anaerobic conditions, because these cells are nonreplicating.
Fig. 4.
Compound 7 shows inhibition of bacterial growth rate under aerobic conditions. Treatment of M. tuberculosis H37Rv with the fumarate hydratase inhibitor 7 under aerobic conditions results in a decrease in bacterial growth. The fluorescence of cells constitutively expressing GFP was measured 12 d after dosing. Cultures were grown in 7H9 medium supplemented with glucose and casitone. The control cultures were treated with DMSO (black column), and 7 was dosed at concentrations up to 250 μM (blue columns). The relative fluorescence units (RFU) are represented as an average of replicates (n = 4), and error bars indicate SEMs.
Although we were able to measure a dose-dependent change in bacterial growth rate under aerobic conditions in the presence of 7, the magnitude of this effect is small. The observed effect could be limited by the hit compound’s potency and cell permeability, or the effect could be the result of an off-target mechanism within the cell. To investigate these possibilities, a medicinal chemistry campaign to optimize the aforementioned properties of the hit compound will be required.
Discussion
Although the properties of fumarate hydratase enzymes have been studied extensively since the 1950s (22–25), reports of targeted small molecule modulators of their activity have been scarce. Pyromellitic acid (benzene-1,2,4,5-tetracarboxylic acid) has been previously reported as a weak inhibitor of some fumarate hydratase homologs (26), but it shows no inhibitory activity with respect to the M. tuberculosis enzyme. We have identified the first, to our knowledge, effective and selective small molecule inhibitor of the M. tuberculosis fumarate hydratase. The structural features of our hit compound immediately suggest a potential allosteric effect. The compound lacks acidic functional groups to mimic substrate binding at the active site, and its molecular weight is more than triple that of the enzyme’s natural substrates. A crystal structure of our hit bound to fumarate hydratase has identified the location of an allosteric site as well as provided a rationale for the observed selectivity over the homologous human enzyme.
In addition to the location of an allosteric site, the crystal structure has also revealed an unusual compound binding mode, with two molecules bound at the interface of two protein subunits. Although this structural information provides a guide for the rational design of additional inhibitors, modifications to the compound structure that preserve potency may be challenging to identify. Compound analogs must not only preserve interactions with the enzyme but also maintain the stacking interactions between the two inhibitor molecules. One potential design strategy would be to explore an array of linkers to join the two inhibitor molecules. Importantly, design strategies should take advantage of the knowledge that the allosteric inhibitor likely prevents catalysis by locking the enzyme subunits in an open conformation, in which the substrate cannot bind. With this information in mind, any small molecule or peptidomimetic that stabilizes the open conformation of the enzyme could potentially be an effective inhibitor.
The discovery of an allosteric regulatory site raises the possibility that a natural biomolecule binds to fumarate hydratase at this location in vivo. Although we were unable to identify such a biomolecule, it is certainly possible that the allosteric site serves a yet unknown biological function. This function could be limited to simple regulation of enzymatic activity, or it could involve a second unrelated process. Dual functions of several metabolic enzymes have recently been reported (27–30), and in particular, the human fumarate hydratase has been implicated in the DNA damage response (31–33). The existence of these dual functions of metabolic enzymes suggests that potential biological functions of this allosteric site deserve additional exploration.
Although many inhibitors exist for M. tuberculosis enzymes in metabolic pathways ranging from glycolysis (34) to sulfur metabolism (35), there are, to our knowledge, no reported inhibitors that target the TCA cycle in this organism. This dearth of inhibitors is likely a result of both the redundancy and the evolutionary conservation of many enzymes in the pathway. Our work, however, exposes a vulnerability in this pathway in the form of an allosteric site that could be exploited to target these bacteria. When treating H37Rv with the allosteric inhibitor under aerobic conditions, we observe a decrease in bacterial growth rate. Although we cannot rule out the possibility that this result is caused by an off-target mechanism, the presence of a dose-dependent effect on growth in a biological context supports additional investigation to optimize the potency, solubility, and cell permeability of this molecule for greater biological activity. This optimization would facilitate studies to verify the mechanism of the compound’s biological effect and assess the role of fumarate hydratase in various environments. In describing a small molecule inhibitor of the M. tuberculosis fumarate hydratase and characterizing its binding to a previously unidentified allosteric site, we illustrate the feasibility of selectively inhibiting this highly conserved metabolic enzyme, while providing a starting point for the development of these more potent probes.
Materials and Methods
High-Throughput Screen and Counterscreens.
The M. tuberculosis fumarate hydratase was expressed and purified as described in SI Appendix. The high-throughput screens were run in 1,536-well plates with a final well volume of 4 μL. All measurements were performed in 50 mM Tris (pH 8), 5 mM MgCl2, and 0.01% Brij 35. Final concentrations of reagents were as follows: 10 nM M. tuberculosis fumarate hydratase, 10 U/mL MDH, 1 U/mL citrate synthase, 150 μM NAD+, 200 μM acetyl-CoA, 0.05 mg/mL diaphorase, 0.05 mM resazurin, and 400 μM fumaric acid. All reagents, with the exception of the purified fumarate hydratase, were purchased from Sigma-Aldrich. Detailed protocols for the high-throughput screen and counterscreens can be found in SI Appendix.
Synthesis and Characterization of 7.
The synthetic scheme for the preparation of 7 (SI Appendix, Fig. S4) and the detailed compound characterization (SI Appendix, SI Methods) can be found in SI Appendix.
Crystallization, Data Processing, and Structure Determination.
For the formate-bound structure, crystals were grown by sitting drop vapor diffusion at 20 °C in 17.0% (wt/vol) PEG 3350, 5% (vol/vol) DMSO, and 200 mM magnesium formate by combining 2 μL 14 mg/mL protein solution [10 mM Tris (pH 8.0), 150 mM NaCl, 0.5 mM Tris(2-carboxyethyl)phosphine hydrochloride] with 1 μL reservoir.
The crystals with 7 bound were obtained by soaking of the formate-bound crystals. Crystals [reservoir: 17.0% (wt/vol) PEG 3350, 5% (vol/vol) DMSO, 200 mM magnesium formate] were soaked for 16 h by adding 2 μL containing a saturated solution of 7 in 19% (wt/vol) PEG 3350, 7.5% (vol/vol) DMSO, and 200 mM magnesium formate to the sitting drop. Before data collection, crystals were cryocooled in liquid nitrogen.
Detailed methods on crystallization trials, data processing, and structure determination can be found in SI Appendix. Coordinates and structure factors for the formate-bound structure and the complex with 7 have been deposited in the PDB under ID codes 5F92 and 5F91, respectively.
Mode of Inhibition Steady-State Kinetic Assay.
The mode of inhibition assay was run in 1,536-well plates under the same conditions as the primary high-throughput screen described above, with the following modifications: (i) a new batch of enzyme was purified for this study, resulting in the fumarate hydratase concentration being lowered to 1.25 nM, and (ii) to prevent pH changes at high substrate concentrations, sodium fumarate dibasic was used in place of fumaric acid to initiate the reaction. The inhibitor was tested in a seven-point dilution ranging from 28.6 μM to 447 nM. Nine concentrations of sodium fumarate dibasic, ranging from 0 to 6.4 mM, were tested. Each plate was read in kinetic mode on a ViewLux High-Throughput CCD Imager (Perkin-Elmer) equipped with standard UV fluorescence optics (525-nm excitation and 598-nm emission) for 10 min. The change in fluorescence intensity over the 10-min reaction period was normalized against no substrate controls, and the resulting initial reaction rates were plotted. The enzymatic saturation curves were fit using the following rate equation derived using the steady-state approximation from the model of general mixed inhibition pictured in Fig. 3C:
| [1] |
Human Fumarate Hydratase Selectivity.
The selectivity assay was conducted in 384-well black Greiner plates, with a final well volume of 40 μL. All measurements were performed in 50 mM Tris (pH 8), 5 mM MgCl2, and 0.01% Brij 35. Final concentrations of reagents were as follows: 10 nM human fumarate hydratase (Sigma-Aldrich), 10 U/mL MDH, 1 U/mL citrate synthase, 150 μM NAD+, 200 μM acetyl-CoA, 0.05 mg/mL diaphorase, 0.05 mM resazurin, and 400 μM fumaric acid. A detailed assay protocol can be found in SI Appendix.
Growth Rate of H37Rv Under Aerobic Conditions.
An M. tuberculosis strain H37Rv containing a plasmid with the gfp gene encoding GFP under control of the constitutive msp12 promoter of Mycobacterium marinum was used to monitor growth. Because the plasmid contains a selectable kanamycin marker, cultures were maintained in 25 μg/mL kanamycin in 7H9 medium (Difco) supplemented with 4 g/L glucose and 0.3 g/L casitone (Difco). Experiments were conducted in black, clear-bottomed 96-well plates (Greiner). Early log phase cultures were diluted 125-fold, and 50 μL were plated into each well; 50 μL of a 2× compound solution were added to the cells. Compound 7 was dosed at concentrations ranging from 16 to 250 μM. Each plate contained an isoniazid control dosed at concentrations ranging from 0.07 to 19 μM. Plates were incubated at 37 °C and read every 2 d for 12 d on an OPTIMA Microplate Reader (BMG Labtech) using the bottom read setting with excitation at 485 nm and emission at 520 nm.
Supplementary Material
Acknowledgments
We thank Dr. Christina Spry, Dr. David Garboczi, Dr. Apostolos Gittis, Dr. Paresma Patel, and Dr. Tathagata Mukherjee for helpful advice and discussions. We also thank Sam Michael, Charles Bonney, Dr. Rajarshi Guha, and Paul Shinn for help with the high-throughput screen and compound management. Some experiments were performed in the Crystallographic X-Ray Facility at the Department of Biochemistry, University of Cambridge. We thank Dr. Dimitri Chirgadze for his assistance in using these facilities. We also thank Diamond Light Source and staff for the provision of beam time at beamlines I04 and I24. M.K. is supported by the National Institutes of Health–Oxford–Cambridge Scholars Program and the Washington University in St. Louis Medical Scientist Training Program. This work was supported by the Division of Preclinical Innovation, National Center for Advancing Translational Sciences and the Intramural Research Program of the National Institute of Allergy and Infectious Diseases.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5F92 and 5F91).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1600630113/-/DCSupplemental.
References
- 1.Boshoff HI, Barry CE., 3rd Tuberculosis - metabolism and respiration in the absence of growth. Nat Rev Microbiol. 2005;3(1):70–80. doi: 10.1038/nrmicro1065. [DOI] [PubMed] [Google Scholar]
- 2.Watanabe S, et al. Fumarate reductase activity maintains an energized membrane in anaerobic Mycobacterium tuberculosis. PLoS Pathog. 2011;7(10):e1002287. doi: 10.1371/journal.ppat.1002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guest JR, Miles JS, Roberts RE, Woods SA. The fumarase genes of Escherichia coli: Location of the fumB gene and discovery of a new gene (fumC) J Gen Microbiol. 1985;131(11):2971–2984. doi: 10.1099/00221287-131-11-2971. [DOI] [PubMed] [Google Scholar]
- 4.Park HD, et al. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol Microbiol. 2003;48(3):833–843. doi: 10.1046/j.1365-2958.2003.03474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eoh H, Rhee KY. Multifunctional essentiality of succinate metabolism in adaptation to hypoxia in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2013;110(16):6554–6559. doi: 10.1073/pnas.1219375110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Estévez M, Skarda J, Spencer J, Banaszak L, Weaver TM. X-ray crystallographic and kinetic correlation of a clinically observed human fumarase mutation. Protein Sci. 2002;11(6):1552–1557. doi: 10.1110/ps.0201502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takeuchi T, Schumacker PT, Kozmin SA. Identification of fumarate hydratase inhibitors with nutrient-dependent cytotoxicity. J Am Chem Soc. 2015;137(2):564–567. doi: 10.1021/ja5101257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weaver TM, Levitt DG, Donnelly MI, Stevens PP, Banaszak LJ. The multisubunit active site of fumarase C from Escherichia coli. Nat Struct Biol. 1995;2(8):654–662. doi: 10.1038/nsb0895-654. [DOI] [PubMed] [Google Scholar]
- 9.Mechaly AE, et al. Conformational changes upon ligand binding in the essential class II fumarase Rv1098c from Mycobacterium tuberculosis. FEBS Lett. 2012;586(11):1606–1611. doi: 10.1016/j.febslet.2012.04.034. [DOI] [PubMed] [Google Scholar]
- 10.Weaver T, Banaszak L. Crystallographic studies of the catalytic and a second site in fumarase C from Escherichia coli. Biochemistry. 1996;35(44):13955–13965. doi: 10.1021/bi9614702. [DOI] [PubMed] [Google Scholar]
- 11.Gibon Y, et al. A Robot-based platform to measure multiple enzyme activities in Arabidopsis using a set of cycling assays: Comparison of changes of enzyme activities and transcript levels during diurnal cycles and in prolonged darkness. Plant Cell. 2004;16(12):3304–3325. doi: 10.1105/tpc.104.025973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simeonov A, et al. Fluorescence spectroscopic profiling of compound libraries. J Med Chem. 2008;51(8):2363–2371. doi: 10.1021/jm701301m. [DOI] [PubMed] [Google Scholar]
- 13.Michaelis L, Menten ML. Die kinetik der invertinwirkung. Biochem Z. 1913;49:333–369. [Google Scholar]
- 14.Michaelis L, Menten ML, Johnson KA, Goody RS. The original Michaelis constant: Translation of the 1913 Michaelis-Menten paper. Biochemistry. 2011;50(39):8264–8269. doi: 10.1021/bi201284u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 16.Yasgar A, et al. Compound management for quantitative high-throughput screening. JALA Charlottesv Va. 2008;13(2):79–89. doi: 10.1016/j.jala.2007.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inglese J, et al. Quantitative high-throughput screening: A titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci USA. 2006;103(31):11473–11478. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCarthy A. The NIH Molecular Libraries Program: Identifying chemical probes for new medicines. Chem Biol. 2010;17(6):549–550. doi: 10.1016/j.chembiol.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 19.Weiss JN. The Hill equation revisited: Uses and misuses. FASEB J. 1997;11(11):835–841. [PubMed] [Google Scholar]
- 20.Strelow J, et al. Mechanism of action assays for enzymes. In: Sittampalam GS, et al., editors. Assay Guidance Manual. Eli Lily & Company and the National Center for Advancing Translational Sciences; Bethesda: 2012. [PubMed] [Google Scholar]
- 21.Abrahams GL, et al. Pathway-selective sensitization of Mycobacterium tuberculosis for target-based whole-cell screening. Chem Biol. 2012;19(7):844–854. doi: 10.1016/j.chembiol.2012.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alberty RA, Massey V, Frieden C, Fuhlbrigge AR. Studies of the enzyme fumarase. III. The dependence of the kinetic constants at 25° upon the concentration and pH of phosphate buffers. J Am Chem Soc. 1954;76(9):2485–2493. [Google Scholar]
- 23.Massey V. Studies on fumarase. II. The effects of inorganic anions on fumarase activity. Biochem J. 1953;53(1):67–71. doi: 10.1042/bj0530067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massey V. Studies on fumarase. III. The effect of temperature. Biochem J. 1953;53(1):72–79. doi: 10.1042/bj0530072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Massey V. Studies on fumarase. 4. The effects of inhibitors on fumarase activity. Biochem J. 1953;55(1):172–177. doi: 10.1042/bj0550172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beeckmans S, Van Driessche E. Pig heart fumarase contains two distinct substrate-binding sites differing in affinity. J Biol Chem. 1998;273(48):31661–31669. doi: 10.1074/jbc.273.48.31661. [DOI] [PubMed] [Google Scholar]
- 27.Hall DA, et al. Regulation of gene expression by a metabolic enzyme. Science. 2004;306(5695):482–484. doi: 10.1126/science.1096773. [DOI] [PubMed] [Google Scholar]
- 28.Kim JW, Dang CV. Multifaceted roles of glycolytic enzymes. Trends Biochem Sci. 2005;30(3):142–150. doi: 10.1016/j.tibs.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 29.Huberts DH, van der Klei IJ. Moonlighting proteins: An intriguing mode of multitasking. BBA-Mol. Cell Res. 2010;1803(4):520–525. doi: 10.1016/j.bbamcr.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 30.Copley SD. Moonlighting is mainstream: Paradigm adjustment required. BioEssays. 2012;34(7):578–588. doi: 10.1002/bies.201100191. [DOI] [PubMed] [Google Scholar]
- 31.Yogev O, Naamati A, Pines O. Fumarase: A paradigm of dual targeting and dual localized functions. FEBS J. 2011;278(22):4230–4242. doi: 10.1111/j.1742-4658.2011.08359.x. [DOI] [PubMed] [Google Scholar]
- 32.Yogev O, et al. Fumarase: A mitochondrial metabolic enzyme and a cytosolic/nuclear component of the DNA damage response. PLoS Biol. 2010;8(3):e1000328. doi: 10.1371/journal.pbio.1000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeffery CJ. Moonlighting proteins--an update. Mol Biosyst. 2009;5(4):345–350. doi: 10.1039/b900658n. [DOI] [PubMed] [Google Scholar]
- 34.Capodagli GC, Sedhom WG, Jackson M, Ahrendt KA, Pegan SD. A noncompetitive inhibitor for Mycobacterium tuberculosis’s class IIa fructose 1,6-bisphosphate aldolase. Biochemistry. 2014;53(1):202–213. doi: 10.1021/bi401022b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palde PB, et al. First-in-class inhibitors of sulfur metabolism with bactericidal activity against non-replicating M. tuberculosis. ACS Chem Biol. 2016;11(1):172–184. doi: 10.1021/acschembio.5b00517. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.