Significance
Neurofibromatosis type 1 (NF1) and Legius syndrome (LS) are disorders characterized by deregulation of the Ras/MAPK pathway, which is central to a variety of cellular processes, including growth and differentiation. Both neurofibromin protein and Sprouty-related, EVH1 domain-containing protein 1 (Spred1) inhibit the Ras/MAPK pathway, and loss-of-function alterations in their encoding genes cause NF1 or LS, respectively. Such alterations are also often found in sporadic tumors unrelated to NF1 or LS. An earlier report described a cellular complex containing neurofibromin and Spred1; the details of these interactions remained unknown. Here we show that the EVH1 domain of Spred1 binds to the GTPase activating protein-related domain of neurofibromin without interfering with its catalytic activity. We further describe how a disease associated mutation in neurofibromin prevents Spred1 binding.
Keywords: cancer, signal transduction, protein–protein interaction, RASopathy, nontruncating mutations
Abstract
Neurofibromatosis type 1 (NF1) and Legius syndrome are related diseases with partially overlapping symptoms caused by alterations of the tumor suppressor genes NF1 (encoding the protein neurofibromin) and SPRED1 (encoding sprouty-related, EVH1 domain-containing protein 1, Spred1), respectively. Both proteins are negative regulators of Ras/MAPK signaling with neurofibromin functioning as a Ras-specific GTPase activating protein (GAP) and Spred1 acting on hitherto undefined components of the pathway. Importantly, neurofibromin has been identified as a key protein in the development of cancer, as it is genetically altered in a large number of sporadic human malignancies unrelated to NF1. Spred1 has previously been demonstrated to interact with neurofibromin via its N-terminal Ena/VASP Homology 1 (EVH1) domain and to mediate membrane translocation of its target dependent on its C-terminal Sprouty domain. However, the region of neurofibromin required for the interaction with Spred1 has remained unclear. Here we show that the EVH1 domain of Spred1 binds to the noncatalytic (GAPex) portion of the GAP-related domain (GRD) of neurofibromin. Binding is compatible with simultaneous binding of Ras and does not interfere with GAP activity. Our study points to a potential targeting function of the GAPex subdomain of neurofibromin that is present in all known canonical RasGAPs.
Neurofibromatosis type 1 (NF1) is an autosomal dominant genetic disorder with an incidence of 1 in 3,500 newborns and full penetrance. Patients with NF1 have an increased risk of developing the disease-typical neurofibromas consisting of benign and malignant tumors of the nervous system. Additional symptoms include characteristic pigmentation anomalies (café au lait macules, axillary freckling), melanocytic hamartomas of the iris (Lisch nodules), bone deformations, and learning disabilities, altogether defining a clinical picture that is far from being well understood and for which no cure is available (1). The tumor suppressor gene NF1 encodes the giant signal regulatory protein neurofibromin and is mutated in NF1 patients. The reported alterations (currently >2,000 mutations are listed in the Human Gene Mutation Database) include splice variants, nonsense and missense mutations, insertions and deletions, duplications and rearrangements, as well as repeat variations (www.hgmd.cf.ac.uk/ac/index.php). About 10% of the mutations comprise nontruncating (in-frame) deletions or missense mutations that are in principle compatible with the translation of the protein product (2).
Neurofibromin is a 320-kDa Ras-specific GTPase activating protein (RasGAP) that uses the central GAP-related domain (GRD) to enhance the GTPase activity of the small guanine nucleotide binding protein Ras, thereby down-regulating its biological activity (3–5). The NF1 gene has gained considerable impact as it is frequently mutated in a variety of very aggressive human malignancies, including glioblastoma, lung adenocarcinoma, acute myeloid leukemia, and ovarian and breast cancers (6–12). Using structural biology approaches, we previously discovered a bipartite module located C-terminal to the GRD that is composed of a glycerophospholipid binding Sec14-like domain associated with a previously undetected C-terminal pleckstrin homology (PH)-like domain (13–15). Although a number of interaction partners of neurofibromin have been reported, the significance of the underlying protein–protein interaction is not clearly established in many cases, as reviewed in refs. 2 and 16.
The recently described Legius syndrome (LS) shares the milder features with NF1, including café au lait macules, axillary freckling, and sometimes macrocephaly, but does not appear to include the more severe clinical manifestations of NF1 such as neurofibromas, optic pathway gliomas, or osseous lesions (17). LS is caused by germ-line mutations in the SPRED1 gene (17, 18) encoding the Sprouty-related, EVH1 domain-containing protein 1 (Spred1), which has been demonstrated to specifically inhibit the Ras/MAPK pathway in response to growth factor-, cytokine-, and chemokine-induced ERK activation (19–22). In pediatric acute myeloblastic leukemia, Spred1 acts as a tumor suppressor (23). The 50-kDa protein comprises an N-terminal Ena/VASP Homology 1 (EVH1) domain (24) and a C-terminal Sprouty-related domain (SPR) separated by a central c-Kit binding domain (19). EVH1 domains are PH-like protein–protein interaction modules (25, 26) that bind to proline rich sequence stretches of the type FPPPP (27, 28). Most of the reported interaction partners of Spred1 involve binding the C-terminal SPR domain with unclear function or significance (29). Most of the LS-associated mutations found in the SPRED1 gene result in premature stop codons, presumably associated with truncated translation products, and the majority of the identified nontruncating missense mutations are located within the EVH1 domain (18).
Using MS combined with a tandem affinity purification (TAP) approach, neurofibromin was identified as a Spred1-interacting protein (30, 31). Stowe and coworkers performed a detailed analysis using overexpressed as well as endogenous proteins to suggest a model for the role of Spred1 in Ras/MAPK inhibition, in which the EVH1 domain serves as the binding site for neurofibromin; the complex is then translocated to the plasma membrane via the Sprouty domain of Spred1. However, it was not clear from that study which region of the 2,818 residues comprising neurofibromin is necessary for Spred1 recognition and whether the interaction was direct or mediated by another cellular factor (31).
In this study, we tested binding of purified recombinant Spred1(EVH1) to different segments of neurofibromin that were prepared following recombinant expression in Escherichia coli or in insect cells. We find that the EVH1 domain binds to the GRD of neurofibromin with nanomolar affinity and that the interaction is fully compatible with binding of activated Ras without interfering with RasGAP activity. Furthermore, our study identifies the extra domain (GAPex) of the GRD as the binding site of the Spred1(EVH1) domain. GAPex is present in known canonical RasGAPs but its function has thus far remained unclear. A mutant neurofibromin harboring a disease-associated single residue deletion in the identified Spred1-binding site fails to interact with Spred1 and does not translocate to the plasma membrane but retains GAP activity, underscoring the physiological importance of this interaction.
Results
Spred1 Binds to the GRD of Neurofibromin and Localizes the Domain at the Membrane.
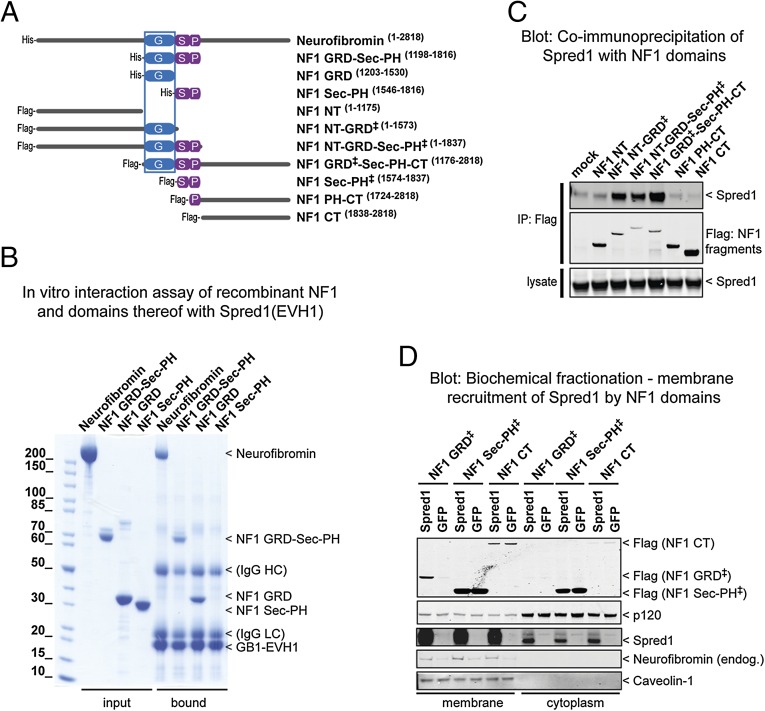
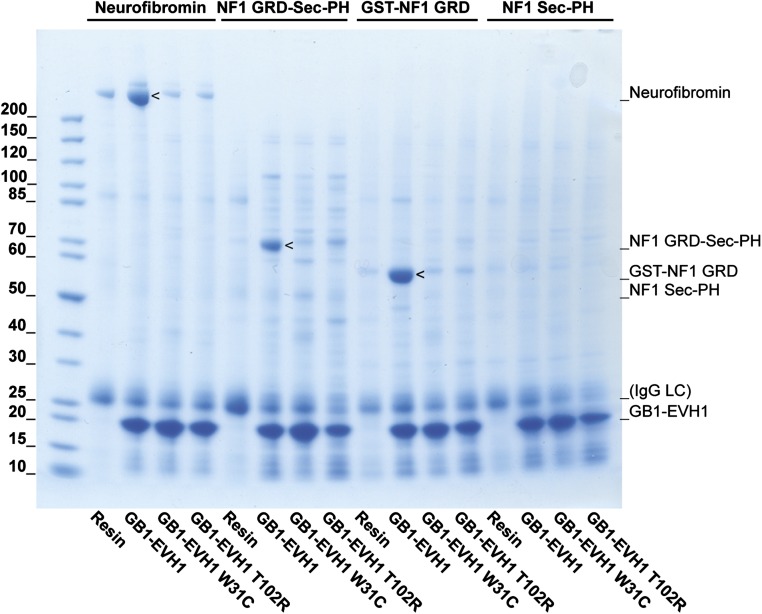
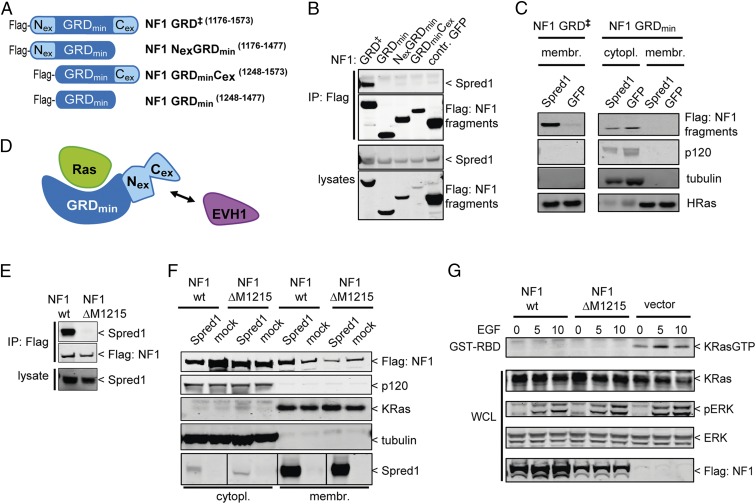
To define the Spred1(EVH1) binding site on neurofibromin, we purified recombinant neurofibromin and soluble segments thereof (schematically shown in Fig. 1A) to homogeneity and analyzed their interaction with the EVH1 domain of Spred1 using an in vitro affinity purification approach. Immobilized recombinant Spred1(EVH1) captured all neurofibromin protein variants containing the GRD including the isolated GRD (Fig. 1B). Two LS patient-derived pathogenic missense mutations T102→R and W31→C (18) located on the surface of the EVH1 domain prevented the interaction with GRD-containing fragments (24) (Fig. S1). This result was supported and confirmed by analysis of several Flag-tagged neurofibromin constructs (Fig. 1A) for their ability to coimmunoprecipitate with Spred1 from human embryonic kidney cell lysates overexpressing both proteins (Fig. 1C). The observed failure of the constructs containing the N- and C-terminal portions of neurofibromin to bind but not of those containing the GRD strongly suggests that no other Spred1 binding site is present on neurofibromin. Membrane localization, which is a requirement to inactivate membrane-anchored Ras, is mediated by Spred1/2 (30). To determine whether membrane recruitment of neurofibromin by Spred1 depends on the GRD, we performed biochemical fractionation of human embryonic kidney cells in which either Spred1 or GFP (as a negative control) was expressed together with the various Flag-tagged neurofibromin constructs (Fig. 1D). As expected, Spred1 expression increased the membrane localization of endogenous neurofibromin. For the Flag-tagged neurofibromin fragments, Spred1 expression specifically localized the Flag-tagged GRD-containing neurofibromin fragments to the membrane, but not those fragments lacking the GRD. Thus, our experiments define the GRD of neurofibromin as the binding site for the Spred1(EVH1) domain.
Fig. 1.
The GRD of neurofibromin binds to Spred1 and localizes to the membrane. (A) Schematic presentation of the human neurofibromin constructs used to map the binding site of Spred1 on neurofibromin as shown in B and C. (B) In vitro binding study: purified recombinant neurofibromin proteins were incubated with GB1-tagged Spred1(EVH1) fusion proteins immobilized on IgG resin. Bound proteins were eluted and analyzed by SDS/PAGE, and protein bands were visualized by Coomassie staining. Neurofibromin constructs containing the GRD did bind to Spred1(EVH1). (C) Immunoprecipitation of Flag-tagged neurofibromin proteins overexpressed together with Spred1 in 293T cells. Proteins were separated by SDS/PAGE and identified by Western blotting. Only neurofibromin constructs containing the GRD were able to pull down Spred1. (D) Biochemical fractionation of 293T cells coexpressing the Flag-tagged neurofibromin constructs indicated on top of the gel and either Spred1 or GFP as a negative control. Proteins were analyzed as in C. Spred1 expression specifically localized the GRD to the membrane but not the other neurofibromin fragments.
Fig. S1.
Patient-derived mutations in the EVH1 domain prevent neurofibromin binding. Purified recombinant neurofibromin protein or lysates from bacteria expressing the neurofibromin domains GRD-Sec-PH, GRD, or Sec-PH were incubated with GB1-tagged EVH1 fusion proteins immobilized on IgG resin. Bound proteins were eluted and analyzed by SDS/PAGE, and protein bands were visualized by Coomassie staining. Only neurofibromin constructs containing the GRD did bind to EVH1. LS patient-derived mutations (W31→C or T102→R) located in the Spred1(EVH1) domain prevented the interaction with neurofibromin. Note that, in this experiment, the GRD is fused to the 25-kDa GST protein, altering its apparent molecular weight.
The GRD of Neurofibromin Forms a Ternary Complex with EVH1 and Activated Ras.
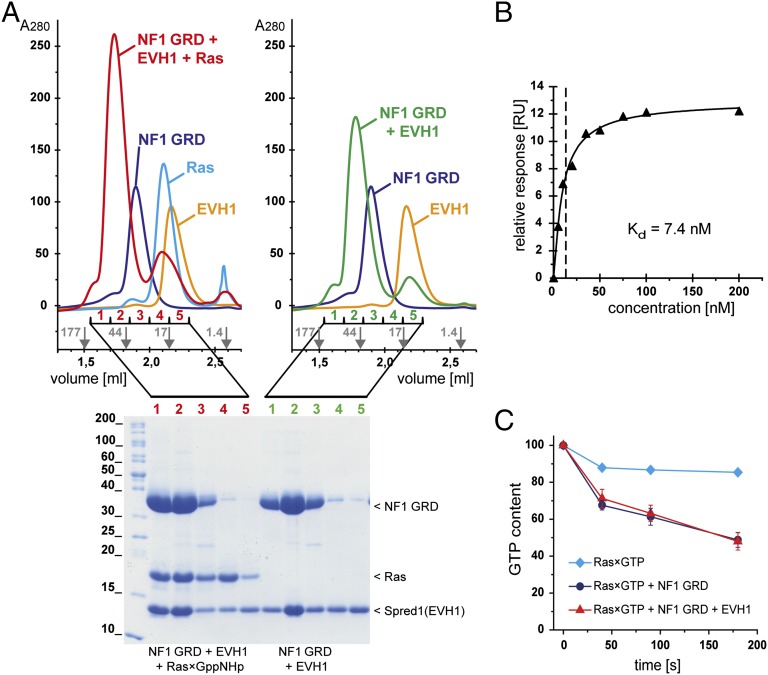
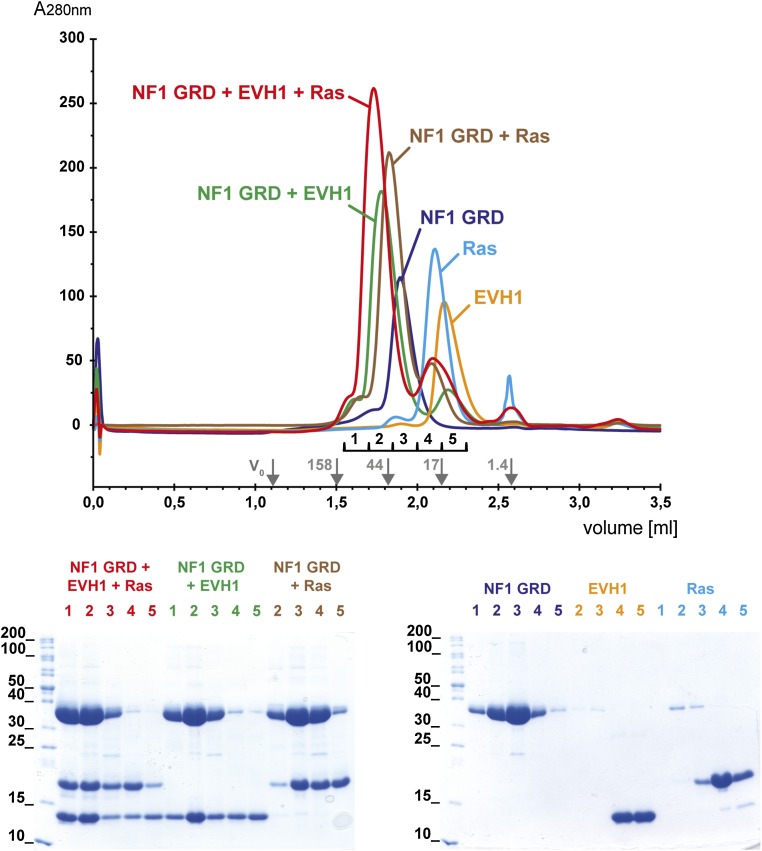
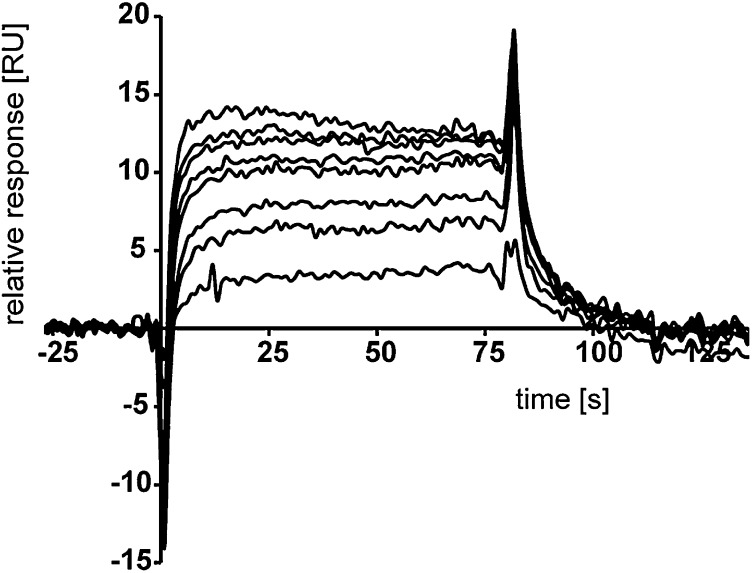
The GRD is probably the best investigated part of the neurofibromin protein, especially its interaction with and enzymatic activity toward activated Ras (4, 32–36). We were interested in whether the neurofibromin–Spred interaction competes with Ras recognition or whether the GRD harbors an additional yet unknown protein binding site. To answer this question, we prepared recombinant Spred1(EVH1), neurofibromin(GRD), and Ras bound to the nonhydrolysable GTP analog GppNHp (referred to as Ras×GppNHp). The integrity of the purified recombinant proteins was analyzed by MS, revealing masses of 40,126 (GRD, calculated Mr = 40,125), 13,363 [Spred1(EVH1), calculated Mr = 13,494], and 18,852 (H-Ras, calculated Mr = 18,853) confirming the expected masses for the GRD and for H-Ras and indicating cleavage of the N-terminal methionine (Δm = 132) of Spred1(EVH1). Using these proteins, we performed analytical gel filtration experiments, and we were able to isolate a stable ternary complex consisting of the GRD, Ras×GppNHp, and the EVH1 domain of Spred1 [termed GRD•Ras×GppNHp•Spred1(EVH1)], as well as a binary complex of the GRD and the EVH1 domain of Spred1 [termed GRD•Spred1(EVH1)] (Fig. 2A and Fig. S2). The formation of the previously characterized GRD•Ras×GppNHp complex was used as a positive control (4, 33). Thus, both activated Ras and the Spred1(EVH1) domain can simultaneously bind to the GRD. Using a multiple angle light scattering (MALS) detector, we determined the molecular masses for the GRD, Spred1(EVH1), and the binary GRD•Spred1(EVH1) complex eluting in one peak from a size exclusion column, suggesting a 1:1 stoichiometry of the GRD to Spred1(EVH1).
Fig. 2.
GRD, Spred1(EVH1), and activated Ras form stable complexes. (A) Separation of a ternary GRD•Spred1(EVH1)•Ras×GppNHp complex (Upper Left; chromatogram in red) and of a binary GRD•Spred1(EVH1) complex (Upper Right; chromatogram in green). Equimolar amounts of the indicated purified recombinant proteins were mixed and analyzed by analytical gel filtration chromatography. Positions of size marker proteins are indicated by gray arrows. The indicated fractions from the gel filtration column were analyzed by SDS/PAGE [15% (wt/vol) acrylamide], and protein bands were visualized by Coomassie staining. (B) Surface plasmon resonance supported quantification of the GRD-Spred1(EVH1) affinity. Spred1(EVH1) (at concentrations between 5 and 200 nM) was injected over a surface loaded with 25 nM GRD. Relative responses at saturation were plotted against the Spred1(EVH1) concentration. The Kd corresponds to the Spred1(EVH1) concentration at 50% of the maximum intensity (saturation). (C) Enzymatic GAP activity of the GRD toward Ras×GTP in the presence and absence of Spred1(EVH1). Ras×GTP was incubated at room temperature with (blue circles) or without GRD (cyan diamonds); the former experiment was repeated in the presence of an excess of Spred1(EVH1) (red triangles). Samples from the reactions were taken at the indicated time points, and the nucleotides were qualified and quantified by HPLC. SEM values were calculated for the average of three independent experiments and displayed in the diagram (SEM < 0.01 is not visible).
Fig. S2.
NF1 GRD, Spred1(EVH1), and activated Ras form stable complexes. Analysis of complex formation between NF1 GRD, activated Ras, and Spred1(EVH1) using analytical gel filtration. Indicated purified recombinant proteins were either analyzed separately [NF1 GRD blue, Ras×GppNHp cyan, and Spred1(EVH1) orange chromatogram] or in an equimolar mixture. Five fractions of 150 µL each were collected between 1.55- and 2.30-mL elution volume. Gel filtration standards (gray arrows indicate respective bands) were run under the same conditions to allow the estimation of the molecular weight of the isolated species. The individual proteins elute as monomers. The chromatograms are presented in an overlay to display changes of the peak intensities and shifts of elution volumes due to formation of the complexes NF1 GRD•Ras×GppNHp•Spred1(EVH1) (red), NF1 GRD•Spred1(EVH1) (green), and NF1 GRD•Ras×GppNHp (brown chromatogram, positive control). The protein content of the peaks was analyzed by SDS PAGE, fraction numbers are indicated on top of the gels using the same color as in the corresponding chromatogram.
To quantify the Spred1–neurofibromin affinity, we performed surface plasmon resonance experiments injecting Spred1(EVH1), activated Ras (Ras×GppNHp), or inactive Ras [Ras bound to GDP (referred to as Ras×GDP), as a negative control] over the GRD immobilized to surfaces (Fig. 2B and Fig. S3). Fitting the response curve for a 1:1 interaction model, as determined by the light scattering analysis, allowed the calculation of a dissociation constant (Kd) of 7.4 nM for the GRD•Spred1(EVH1) complex and of 6.2 µM for the GRD•Ras×GppNHp complex.
Fig. S3.
Surface plasmon resonance raw data of one representative titration experiment. Spred1(EVH1) (at 5, 10, 20, 35, 50, 75, 100, and 200 nM) was injected over a Ni-NTA chip surface loaded with 25 nM His-GRD. Values from the second channel (background from unloaded Ni-NTA chip) were withdrawn. Relative response data were plotted against the time.
Spred1(EVH1) Binding to the GRD Does Not Affect RasGAP Activity.
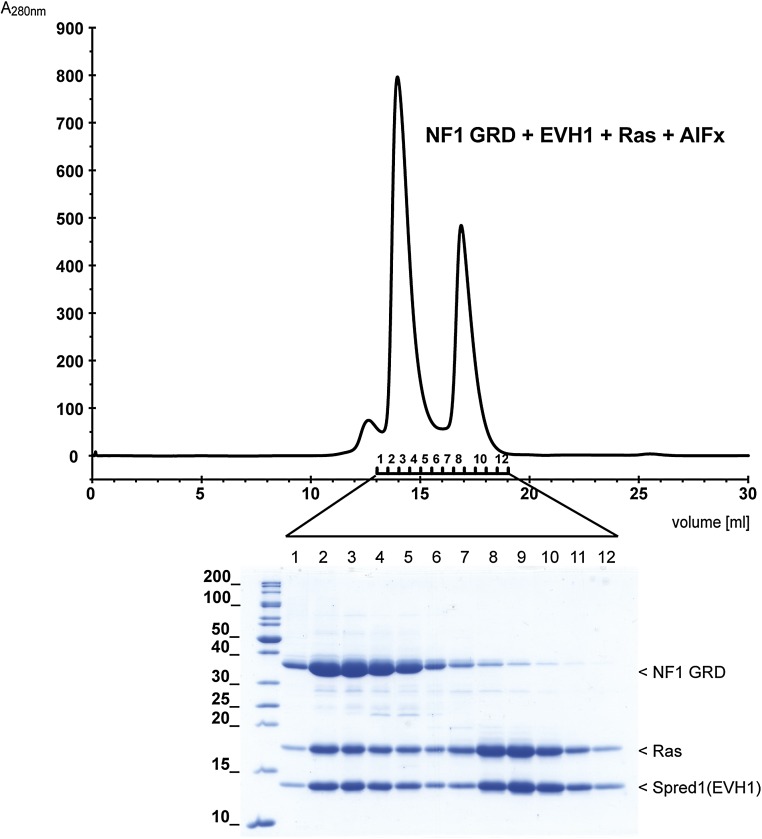
The GTPase reaction of Ras has been widely studied in vivo and in vitro. Because we observed binding of EVH1 to the GRD•Ras complex, we aimed to analyze potential influences on the catalytic property of the GRD. We set up an in vitro GAP reaction and monitored the GRD-catalyzed Ras×GTP hydrolysis by HPLC analysis. Comparison of the reaction rates in the absence and presence of Spred1(EVH1) did not reveal significant differences (Fig. 2C), suggesting that Spred does not modulate the catalytic activity of the GRD. Consistently, we could isolate a stable transition state mimicking complex of the GRD and Ras×GDP in the presence of aluminum fluoride (32) binding to Spred1(EVH1) (Fig. S4). These data suggest that the interaction site of Spred on the GRD is distant to and independent of the Ras binding site.
Fig. S4.
Spred1(EVH1) binds to a transition state complex of the GAP reaction. Spred1(EVH1) and Ras×GDP were mixed in a twofold molar excess with NF1 GRD in the presence of aluminum and fluoride ions and injected to a size exclusion column. The indicated fractions were analyzed by SDS/PAGE (15% acrylamide), and protein bands were visualized by Coomassie staining. The proteins elute in two main fractions, where the one at higher molecular weight represents NF1 GRD•Ras×GDP×AlFx•Spred1(EVH1) demonstrating that the formation of a stable transition state mimicking complex (32) is compatible with Spred1(EVH1) binding. The excess of the smaller Ras and Spred1(EVH1) proteins elute in the second fraction due to similar molecular weight.
The Extra Domain of the GRD Contains the Binding Site of Spred1(EVH1).
The simultaneous binding of Spred1(EVH1) and activated Ras to the GRD raises the question of where the EVH1 binding region is located on the GRD. Proteolysis experiments and structural analysis of a 333-residue neurofibromin GRD (NF1-333) revealed a bipartite module consisting of a 230-residue central domain exhibiting full GAP activity (termed NF230, GAPc, or GRDmin) and an extra domain (GAPex), formed by residues located N- and C-terminally to GRDmin (Nex and Cex, respectively) as schematically outlined in Fig. 3A (33, 37). Because the Spred1 interaction with the GRD neither influences Ras binding nor its catalytic GAP activity, we suspected that the location of the EVH1 binding site may be on the extra domain. Overexpression of deletion mutants of the GRD either lacking the N-terminal or the C-terminal part or the entire GAPex in an embryonic kidney cell line and immunoprecipitation, as well as biochemical fractionation experiments, revealed that the extra domain of the GRD is necessary for interaction with Spred1 and for membrane recruitment of neurofibromin (Fig. 3 B and C). Thus, EVH1 binds to the extra domain of the GRD as schematically shown in Fig. 3D. In addition, a neurofibromin protein carrying a patient-derived single residue deletion in the N-terminal part of GAPex (ΔM1215) (38) failed to coimmunoprecipitate with Spred1 and to localize to the membrane upon Spred1 overexpression (Fig. 3 E and F). Although the NF1 ΔM1215 mutant could not be localized to the membrane by Spred, sufficient overexpression of this mutant, such that it was localized throughout the cell, showed that NF1 ΔM1215 retains GAP activity in human embryonic kidney cells by suppressing basal and EGF-induced Ras×GTP loading (Fig. 3G).
Fig. 3.
An intact GAPex is required for Spred1(EVH1) binding. (A) Schematic presentation of the constructs used in this experiment outlining the location of the N- and C-terminal parts forming the extra domain of the GRD. (B) Untagged Spred1 and Flag-tagged GRD constructs or GFP as a negative control were expressed in 293T cells. Immunoprecipitation with anti-Flag beads using the indicated lysates shows that Spred1 does not associate with GRD proteins lacking the N-terminal, C-terminal, or both N- and C-terminal portions forming GAPex. (C) Spred1 or GFP (negative control) was expressed along with Flag-tagged GRD or Flag-tagged GRDmin in 293T cells. Cells were subject to biochemical fractionation of membrane and cytoplasmic compartments. Proteins in the fractions were separated by SDS/PAGE and identified by subsequent Western blotting. Expression of Spred1 moved the GRD to the membrane but did not alter the localization of the GRD lacking the extra domain (GRDmin). (D) Cartoon of the presumed ternary complex formed by the GRD, Ras, and Spred1(EVH1) according to our mapping data. (E) Same experiment as in B but using full-length Flag-tagged neurofibromin, either as WT protein or harboring a single residue deletion in the N-terminal part of the extra domain (NF1 ΔM1215). WT neurofibromin (NF1 wt) coimmunoprecipitated with Spred1, whereas the ΔM1215 mutation in the extra domain prevented the interaction. (F) Similar fractionation experiment as in C but following the transfection of plasmids encoding full-length Flag-tagged neurofibromin, either as WT protein (NF1 wt) or carrying the ΔM1215 mutation or an empty vector as a negative control together with Spred1. Overexpression of Spred1 enhanced the localization of WT neurofibromin to the membrane but not of the ΔM1215 mutant. (G) Overexpression of the neurofibromin ΔM1215 mutant in 293T cells suppressed EGF-induced GTP loading of Ras.
Discussion
In this study, we show that neurofibromin interacts with Spred1 using its GAP-related domain. In detail, the interaction occurs via the noncatalytic extra domain of the GRD, is compatible with Ras binding, and does not affect GAP catalysis. Our study defines GAPex, the function of which has been unclear, as a protein-interaction module. The observation that the Spred1 binding region is close to but apparently not overlapping with the Ras-binding site may point to a Ras-targeting function of the Spred1–neurofibromin interaction, which is in agreement with our previous studies, where we showed that Spred1 binding enables neurofibromin to act on Ras in cells, presumably by bringing neurofibromin into proximity to Ras at the plasma membrane. Indeed, we showed that loss of function mutations in Spred1’s C-terminal Sprouty domain could be rescued by targeting the mutant Spred1 protein to the plasma membrane with sequences derived from the Ras CAAX box (30).
We were surprised to discover that Spred1 binds at the GRD region of neurofibromin because another RasGAP, p120GAP (RASA1) is recruited to the plasma membrane through a very different mechanism: SH2 domains bind to phospho-tyrosines on receptor tyrosine kinases, and so position p120GAP in proximity to Ras×GTP (39). We therefore might have expected that Spred1 would bind to a domain elsewhere in the neurofibromin protein to recruit it to the plasma membrane. However, Spred proteins are thought to be intimately related to regulating signal transduction from receptor tyrosine kinases, such as the FGF receptor (40, 41). In contrast, neurofibromin-related proteins are well known in organisms such as Saccharomyces cerevisiae, which, of course, lack tyrosine kinase signaling altogether. The yeast IRA1 and IRA2 proteins are the best studied neurofibromin related proteins in distantly related organisms (42). Therefore, neurofibromin is likely to play important and highly conserved roles in aspects of cell physiology that are unrelated to tyrosine kinase signaling, and, in this sense, it would be surprising if neurofibromin and IRA proteins shared a domain devoted to Spred1 binding, as S. cerevisiae lack Spred proteins.
Sequence comparisons and structural studies of canonical RasGAPs, including p120GAP, synGAP, and neurofibromin, identify the presence of a GAPex domain that is located next to the central catalytic GAP domain and forms a conserved structure (33, 43, 44). Secondary structure prediction analyses of the IRA1/2 proteins suggest that the extra domain might be conserved in yeast. We speculate that GAPs may interact with proteins that perform functions similar to that of Spred1, and we are actively attempting to identify such novel partners and to determine their role in GAP and Ras regulation. Spred1 appears to play a role in positioning neurofibromin at the membrane, where it can interact with Ras. The localization of Ras to specific types of membranes is likely cell type dependent, and it is certainly possible that the regulation of Spred1 recruitment to different types of membranes has an effect on the regulation of Ras signaling, which will be addressed in future studies. However, the mechanism by which Spred1 associates with the membrane is not clear. The Sprouty domain is thought to be palmitoylated and necessary for membrane anchoring (21). The kit-binding domain may direct the Spred•neurofibromin complex to specific receptors within the membrane, and, perhaps, binding of Spred1 to the GAPex domain brings neurofibromin to Ras in a much more specific manner than by plain membrane recruitment. Our data represent a step toward a better understanding of the precise molecular mechanism by which Ras proteins are regulated. Defining the mechanistic details of the underlying protein–protein interactions will have to await the molecular structures of the complexes identified in this study.
Materials and Methods
Expression Plasmids.
The expression constructs encoding recombinant human NF1 GRD-Sec-PH and NF1 Sec-PH have been previously described (45). A synthetic gene encoding human neurofibromin (45) was inserted into pACEBac1 finally encoding an N-terminally His-tagged protein. NF1 GRD used for in vitro experiments was expressed from a pET21 derivative in which a synthetic gene encoding residues D1203–H1530 of human neurofibromin preceded by a (His)6 tag was inserted. Flag-tagged neurofibromin constructs used for transfection experiments in HEK293T cells encoded residues M1-T1175 (NF1 NT), M1-Y1573 (NF1 NT-GRD‡), M1-A1837 (NF1 NT-GRD-Sec-PH‡), Q1574-A1837 (NF1 Sec-PH‡), R1176-V2818 (NF1 GRD‡-Sec-PH-CT), K1724-V2818 (NF1 PH-CT), L1838-V2818 (NF1 CT), R1176-Y1573 (NF1 GRD‡), R1176-F1477 (NF1 NexGRDmin), D1248-Y1573 (NF1 GRDminCex), and D1248-F1477 (NF1 GRDmin) in a pcDNA3.1 vector. Numbering corresponds to neurofibromin isoform 2 (NP_000258.1). Note: the domain boundaries of the GRD and the Sec-PH used for transfections differs slightly from the recombinant proteins (NF1 GRD and NF1 Sec-PH) described below and were therefore labeled NF1 GRD‡ and NF1 Sec-PH‡. Human Spred1 and GFP were expressed from a pcDNA3.1 vector as described previously (30). A plasmid encoding untagged Spred1(EVH1) (residues S13–S130) was generated by inserting the corresponding synthetic gene into pET21d. The same cDNA was also cloned in pET21d downstream of a gene encoding the IgG binding protein G B1 domain (46, 47). Point mutations were introduced to generate the pathogenic mutated (T102→R, W31→C) EVH1 variants of the GB1-fusion proteins. H-Ras (M1-H166) was expressed from a construct previously described (48).
Preparation of Recombinant Proteins.
The expression and purification of NF1 GRD-Sec-PH and NF1 Sec-PH have been previously described (45). Human neurofibromin was expressed in insect cells (SF21). NF1 GRD used for in vitro experiments was expressed in E. coli strain BL21 Star (DE3). All recombinant neurofibromin proteins were purified by metal chelate affinity and preparative size exclusion chromatography. Spred1(EVH1) and mutated forms (T102→R, W31→C) were expressed in E. coli strain BL21 Star (DE3). The introduction of the point mutation was verified by MS. Untagged Spred1(EVH1) was purified by cation exchange and preparative size exclusion chromatography. H-Ras (M1-H166) was expressed as previously described (49) and purified by anion exchange and preparative size exclusion chromatography, yielding Ras in the GDP bound state (named Ras×GDP). The integrity of the recombinant proteins used for the in vitro experiments described below was analyzed by electrospray ionization MS.
Ras loaded with guanosine 5′-[βγ-imido]triphosphate (named Ras×GppNHp) was prepared following the concept described previously (50) but with the following modifications: Mg2+ was withdrawn from recombinant Ras×GDP (0.5 mM) by treatment with EDTA (1 mM) to enhance the nucleotide exchange reaction (49), and released GDP was hydrolyzed by the addition of alkaline phosphatase in the presence of 200 mM (NH4)2SO4 while GppNHp (added in a 1.5-fold molar excess) was incorporated. After GDP degradation was completed, as monitored by HPLC analysis, Ras×GppNHp was purified from the reaction mixture by size exclusion chromatography. Ras in its activated state (Ras×GTP) was prepared as follows: Ras×GDP was incubated with a molar excess of EDTA in the presence of GTP (100-fold molar excess) at 273 K for 1 h. The Ras protein was purified from the excess of nucleotide by size exclusion chromatography, yielding between 50% and 74% Ras×GTP.
In Vitro Interaction Assay.
Bacterial lysates generated from GB1-Spred1(EVH1) expression cultures were incubated with IgG-sepharose. The resins were washed in 20 mM Tris buffer (pH 8.0) containing 300 mM NaCl, 2 mM MgCl2, 2 mM DTT, and 2% (vol/vol) glycerol and purified recombinant neurofibromin proteins were added. After a 30-min incubation at 273 K, the resins were washed again using the same buffer. Bound proteins were eluted in SDS sample buffer and analyzed by SDS/PAGE [4–12% (wt/vol) acryl amide gradient].
Immunochemical Analysis and Biochemical Fractionation.
HEK293T cells were grown in DMEM with 10% (wt/vol) bovine calf serum and grown to 70% confluency at the time of transfection. Cells were transfected with Lipofectamine 2000 and cultivated for another 24–48 h. For immunoprecipitation experiments, cells were lysed in a buffer containing 20 mM Tris (pH 7.5), 100 mM NaCl, 20 mM MgCl2, 1% (vol/vol) Triton-X100, 1 mM DTT, and protease and phosphatase inhibitors. For biochemical fractionation, cells were resuspended in hypotonic lysis buffer [10 mM Tris (pH 7.5), 1 mM EDTA, 1 mM DTT, supplemented with protease and phosphatase inhibitors], kept on ice for 20 min, and then passaged through a 25-G needle 15 times. Nuclei and unbroken cells were then removed by centrifugation at 100 × g for 3 min; the NaCl concentration was adjusted to 100 mM, and the supernatant was then centrifuged at 10,000 × g for 15 min to pellet the membrane fraction. The resulting supernatant was then centrifuged at 100,000 × g for 30 min to clear the cytoplasmic fraction. Membrane pellets were resuspended in SDS/PAGE sample buffer. The generated fractions were analyzed by Western blotting. The Spred1 antibody was from Abcam; the neurofibromin, p120GAP, H-Ras, and tubulin antibodies were from Santa Cruz Biotechnology; and the FLAG, ERK, and K-Ras antibodies were from Sigma.
Analytical Gel Filtration.
GRD, Ras×GppNHp, and Spred1(EVH1) were each diluted to 100 µM and injected into a Superdex 200 increase 5/150 analytical SEC column (GE Healthcare) equilibrated in 20 mM Tris buffer (pH 8.0) containing 150 mM NaCl and 2 mM MgCl2. Proteins were UV detected at 280 nm, and additionally, the molecular weight of the eluted species was determined by multiple angle light scattering (MALS) using a miniDAWN TREOS detector (WYATT). For analysis of complex formation, proteins were mixed at 100 µM each in the same total volume and analyzed under identical conditions. A similar experiment was performed using Ras×GDP in the presence of an equimolar amount of aluminum ions (Al3+) and a 330-fold excess of fluoride together with the GRD and Spred1(EVH1). The protein content of the collected fractions was analyzed by SDS/PAGE [15% (wt/vol) acrylamide] and subsequent Coomassie staining. Blue dextran and a protein standard mixture were run under the same conditions to determine the void volume v0 of the size exclusion column and the elution volume of the size reference proteins.
Surface Plasmon Resonance Experiments.
A Ni2+-loaded NTA sensor chip in a BiacoreX100 system (GE Healthcare) was coated with the His-tagged GRD in 10 mM Hepes buffer (pH 7.4) containing 150 mM NaCl and 0.05% (vol/vol) polysorbate 20. For experiments including Ras, the buffer was supplemented with 1 mM MgCl2. The second flow cell with immobilized Ni2+ ions was used as a reference surface. Dilution series in the range between 5 and 200 nM of Spred1(EVH1) and 700 nM and 50 µM of Ras×GppNHp (positive control) or Ras×GDP (negative control) were prepared and injected. The dissociation constant Kd was determined by nonlinear fitting of the sensorgrams to a 1:1 interaction model using Biacore X100 software (version 2.0.1). At least two independent titration experiments were performed, and results were averaged.
GAP Assays.
Ras×GTP samples (20–100 µM Ras×GTP in 50 mM Tris⋅HCl, pH 7.4, 150 mM NaCl, 10 mM MgCl2, and 2 mM DTT) were incubated at 293 K in the absence or presence of the GRD (molar ratio of GRD to Ras×GTP 1:2,000) or the GRD together with an 100-fold excess of Spred1(EVH1) relative to the GRD. Samples were taken after 40, 90, and 180 s and heat denatured for 60 s at 95 °C. The released nucleotide was qualified by reversed phase HPLC as previously described (51). For HPLC quantification, standards with GDP and GTP concentrations between 1 and 100 µM and protein samples from the GAP reactions were analyzed in triplicates. GAP activity was determined in three independent experiments for each of the experimental setups. The initial loading state of Ras×GTP was set to 100%. The SEM was calculated using the formula SEM = with n = 3. EGF stimulation and measurement of GAP activity from 293T lysates was performed as described previously (30).
Acknowledgments
We thank Jeanette Seiler, Julianna Leuenberger, Angela Ausserbichler, and Jacqueline Galeas for excellent technical assistance; Herbert Lindner and Taras Stasyk for performing MS analysis; Andreas Naschberger and Stefan Lechner for providing purified neurofibromin proteins for mapping experiments; and Peter E. Wright (The Scripps Research Institute) for providing a plasmid encoding the GB1 tag. This work was funded by the Austrian Science Fund (FWF) Grant SFB021 (to K.S.) and by a grant from the Children’s Tumor Foundation (to E.L.M.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1607298113/-/DCSupplemental.
References
- 1.Riccardi VM. Neurofibromatosis: Phenotype Natural History, and Pathogenesis. 2nd Ed Johns Hopkins Univ Press; Baltimore: 1992. [Google Scholar]
- 2.Upadhyaya M, Cooper DN, editors. Neurofibromatosis Type 1: Molecular and Cellular Biology. Springer; New York: 2012. [Google Scholar]
- 3.Ballester R, et al. The NF1 locus encodes a protein functionally related to mammalian GAP and yeast IRA proteins. Cell. 1990;63(4):851–859. doi: 10.1016/0092-8674(90)90151-4. [DOI] [PubMed] [Google Scholar]
- 4.Martin GA, et al. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell. 1990;63(4):843–849. doi: 10.1016/0092-8674(90)90150-d. [DOI] [PubMed] [Google Scholar]
- 5.Xu GF, et al. The catalytic domain of the neurofibromatosis type 1 gene product stimulates ras GTPase and complements ira mutants of S. cerevisiae. Cell. 1990;63(4):835–841. doi: 10.1016/0092-8674(90)90149-9. [DOI] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding L, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455(7216):1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sangha N, et al. Neurofibromin 1 (NF1) defects are common in human ovarian serous carcinomas and co-occur with TP53 mutations. Neoplasia (New York, N.Y.) 2008;10(12):1362–1372. doi: 10.1593/neo.08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kan Z, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466(7308):869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 10.Boudry-Labis E, et al. French ALFA group Neurofibromatosis-1 gene deletions and mutations in de novo adult acute myeloid leukemia. Am J Hematol. 2013;88(4):306–311. doi: 10.1002/ajh.23403. [DOI] [PubMed] [Google Scholar]
- 11.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hölzel M, et al. NF1 is a tumor suppressor in neuroblastoma that determines retinoic acid response and disease outcome. Cell. 2010;142(2):218–229. doi: 10.1016/j.cell.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aravind L, Neuwald AF, Ponting CP. Sec14p-like domains in NF1 and Dbl-like proteins indicate lipid regulation of Ras and Rho signaling. Curr Biol. 1999;9(6):R195–R197. doi: 10.1016/s0960-9822(99)80127-4. [DOI] [PubMed] [Google Scholar]
- 14.D’Angelo I, Welti S, Bonneau F, Scheffzek K. A novel bipartite phospholipid-binding module in the neurofibromatosis type 1 protein. EMBO Rep. 2006;7(2):174–179. doi: 10.1038/sj.embor.7400602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Welti S, D’Angelo I, Scheffzek K. Neurofibromatoses. Karger; Würzburg, Germany: 2008. p. 192. [Google Scholar]
- 16.Ratner N, Miller SJ. A RASopathy gene commonly mutated in cancer: The neurofibromatosis type 1 tumour suppressor. Nat Rev Cancer. 2015;15(5):290–301. doi: 10.1038/nrc3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brems H, et al. Germline loss-of-function mutations in SPRED1 cause a neurofibromatosis 1-like phenotype. Nat Genet. 2007;39(9):1120–1126. doi: 10.1038/ng2113. [DOI] [PubMed] [Google Scholar]
- 18.Brems H, et al. Review and update of SPRED1 mutations causing Legius syndrome. Hum Mutat. 2012;33(11):1538–1546. doi: 10.1002/humu.22152. [DOI] [PubMed] [Google Scholar]
- 19.Wakioka T, et al. Spred is a Sprouty-related suppressor of Ras signalling. Nature. 2001;412(6847):647–651. doi: 10.1038/35088082. [DOI] [PubMed] [Google Scholar]
- 20.Miyoshi K, et al. The Sprouty-related protein, Spred, inhibits cell motility, metastasis, and Rho-mediated actin reorganization. Oncogene. 2004;23(33):5567–5576. doi: 10.1038/sj.onc.1207759. [DOI] [PubMed] [Google Scholar]
- 21.Nonami A, et al. The Sprouty-related protein, Spred-1, localizes in a lipid raft/caveola and inhibits ERK activation in collaboration with caveolin-1. Genes Cells. 2005;10(9):887–895. doi: 10.1111/j.1365-2443.2005.00886.x. [DOI] [PubMed] [Google Scholar]
- 22.Kim HJ, Bar-Sagi D. Modulation of signalling by Sprouty: A developing story. Nat Rev Mol Cell Biol. 2004;5(6):441–450. doi: 10.1038/nrm1400. [DOI] [PubMed] [Google Scholar]
- 23.Pasmant E, et al. SPRED1, a RAS MAPK pathway inhibitor that causes Legius syndrome, is a tumour suppressor downregulated in paediatric acute myeloblastic leukaemia. Oncogene. 2015;34(5):631–638. doi: 10.1038/onc.2013.587. [DOI] [PubMed] [Google Scholar]
- 24.Harmer NJ, Sivak JM, Amaya E, Blundell TL. 1.15 A crystal structure of the X. tropicalis Spred1 EVH1 domain suggests a fourth distinct peptide-binding mechanism within the EVH1 family. FEBS Lett. 2005;579(5):1161–1166. doi: 10.1016/j.febslet.2004.11.114. [DOI] [PubMed] [Google Scholar]
- 25.Scheffzek K, Welti S. Pleckstrin homology (PH) like domains - versatile modules in protein-protein interaction platforms. FEBS Lett. 2012;586(17):2662–2673. doi: 10.1016/j.febslet.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 26.Ball LJ, Jarchau T, Oschkinat H, Walter U. EVH1 domains: Structure, function and interactions. FEBS Lett. 2002;513(1):45–52. doi: 10.1016/s0014-5793(01)03291-4. [DOI] [PubMed] [Google Scholar]
- 27.Peterson FC, Volkman BF. Diversity of polyproline recognition by EVH1 domains. Front Biosci (Landmark Ed) 2009;14:833–846. doi: 10.2741/3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prehoda KE, Lee DJ, Lim WA. Structure of the enabled/VASP homology 1 domain-peptide complex: A key component in the spatial control of actin assembly. Cell. 1999;97(4):471–480. doi: 10.1016/s0092-8674(00)80757-6. [DOI] [PubMed] [Google Scholar]
- 29.Bundschu K, Walter U, Schuh K. Getting a first clue about SPRED functions. BioEssays. 2007;29(9):897–907. doi: 10.1002/bies.20632. [DOI] [PubMed] [Google Scholar]
- 30.Stowe IB, et al. A shared molecular mechanism underlies the human rasopathies Legius syndrome and Neurofibromatosis-1. Genes Dev. 2012;26(13):1421–1426. doi: 10.1101/gad.190876.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McClatchey AI, Cichowski K. SPRED proteins provide a NF-ty link to Ras suppression. Genes Dev. 2012;26(14):1515–1519. doi: 10.1101/gad.197434.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mittal R, Ahmadian MR, Goody RS, Wittinghofer A. Formation of a transition-state analog of the Ras GTPase reaction by Ras-GDP, tetrafluoroaluminate, and GTPase-activating proteins. Science. 1996;273(5271):115–117. doi: 10.1126/science.273.5271.115. [DOI] [PubMed] [Google Scholar]
- 33.Scheffzek K, et al. Structural analysis of the GAP-related domain from neurofibromin and its implications. EMBO J. 1998;17(15):4313–4327. doi: 10.1093/emboj/17.15.4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahmadian MR, Hoffmann U, Goody RS, Wittinghofer A. Individual rate constants for the interaction of Ras proteins with GTPase-activating proteins determined by fluorescence spectroscopy. Biochemistry. 1997;36(15):4535–4541. doi: 10.1021/bi962556y. [DOI] [PubMed] [Google Scholar]
- 35.Cichowski K, Jacks T. NF1 tumor suppressor gene function: Narrowing the GAP. Cell. 2001;104(4):593–604. doi: 10.1016/s0092-8674(01)00245-8. [DOI] [PubMed] [Google Scholar]
- 36.Zhu Y, Parada LF. Neurofibromin, a tumor suppressor in the nervous system. Exp Cell Res. 2001;264(1):19–28. doi: 10.1006/excr.2000.5138. [DOI] [PubMed] [Google Scholar]
- 37.Ahmadian MR, Wiesmüller L, Lautwein A, Bischoff FR, Wittinghofer A. Structural differences in the minimal catalytic domains of the GTPase-activating proteins p120GAP and neurofibromin. J Biol Chem. 1996;271(27):16409–16415. doi: 10.1074/jbc.271.27.16409. [DOI] [PubMed] [Google Scholar]
- 38.Fahsold R, et al. Minor lesion mutational spectrum of the entire NF1 gene does not explain its high mutability but points to a functional domain upstream of the GAP-related domain. Am J Hum Genet. 2000;66(3):790–818. doi: 10.1086/302809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaplan DR, Morrison DK, Wong G, McCormick F, Williams LT. PDGF beta-receptor stimulates tyrosine phosphorylation of GAP and association of GAP with a signaling complex. Cell. 1990;61(1):125–133. doi: 10.1016/0092-8674(90)90220-9. [DOI] [PubMed] [Google Scholar]
- 40.Sivak JM, Petersen LF, Amaya E. FGF signal interpretation is directed by Sprouty and Spred proteins during mesoderm formation. Dev Cell. 2005;8(5):689–701. doi: 10.1016/j.devcel.2005.02.011. [DOI] [PubMed] [Google Scholar]
- 41.Sasaki A, Taketomi T, Wakioka T, Kato R, Yoshimura A. Identification of a dominant negative mutant of Sprouty that potentiates fibroblast growth factor- but not epidermal growth factor-induced ERK activation. J Biol Chem. 2001;276(39):36804–36808. doi: 10.1074/jbc.C100386200. [DOI] [PubMed] [Google Scholar]
- 42.Broach JR. Ras-regulated signaling processes in Saccharomyces cerevisiae. Curr Opin Genet Dev. 1991;1(3):370–377. doi: 10.1016/s0959-437x(05)80302-8. [DOI] [PubMed] [Google Scholar]
- 43.Pena V, et al. The C2 domain of SynGAP is essential for stimulation of the Rap GTPase reaction. EMBO Rep. 2008;9(4):350–355. doi: 10.1038/embor.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scheffzek K, Lautwein A, Kabsch W, Ahmadian MR, Wittinghofer A. Crystal structure of the GTPase-activating domain of human p120GAP and implications for the interaction with Ras. Nature. 1996;384(6609):591–596. doi: 10.1038/384591a0. [DOI] [PubMed] [Google Scholar]
- 45.Bonneau F, Lenherr ED, Pena V, Hart DJ, Scheffzek K. Solubility survey of fragments of the neurofibromatosis type 1 protein neurofibromin. Protein Expr Purif. 2009;65(1):30–37. doi: 10.1016/j.pep.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 46.Sugase K, Landes MA, Wright PE, Martinez-Yamout M. Overexpression of post-translationally modified peptides in Escherichia coli by co-expression with modifying enzymes. Protein Expr Purif. 2008;57(2):108–115. doi: 10.1016/j.pep.2007.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou P, Lugovskoy AA, Wagner G. A solubility-enhancement tag (SET) for NMR studies of poorly behaving proteins. J Biomol NMR. 2001;20(1):11–14. doi: 10.1023/a:1011258906244. [DOI] [PubMed] [Google Scholar]
- 48.John J, Schlichting I, Schiltz E, Rösch P, Wittinghofer A. C-terminal truncation of p21H preserves crucial kinetic and structural properties. J Biol Chem. 1989;264(22):13086–13092. [PubMed] [Google Scholar]
- 49.Tucker J, et al. Expression of p21 proteins in Escherichia coli and stereochemistry of the nucleotide-binding site. EMBO J. 1986;5(6):1351–1358. doi: 10.1002/j.1460-2075.1986.tb04366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.John J, et al. Kinetics of interaction of nucleotides with nucleotide-free H-ras p21. Biochemistry. 1990;29(25):6058–6065. doi: 10.1021/bi00477a025. [DOI] [PubMed] [Google Scholar]
- 51.Ahmadian MR, et al. Guanosine triphosphatase stimulation of oncogenic Ras mutants. Proc Natl Acad Sci USA. 1999;96(12):7065–7070. doi: 10.1073/pnas.96.12.7065. [DOI] [PMC free article] [PubMed] [Google Scholar]