Significance
The ribosome is the protein-synthesizing machine of the cell and is a major target for antibiotics. The increase in multidrug-resistant bacteria has limited the utility of our current arsenal of clinically used antibiotics, highlighting the need for further development of compounds that have distinct binding sites and do not display cross-resistance. Using cryo-electron microscopy, we have visualized the binding site of the orthosomycins evernimicin and avilamycin on the bacterial 70S ribosome. The binding site and mode of interaction of evernimicin and avilamycin are distinct from other ribosome-targeting antibiotics. Together with single-molecule studies, our structures reveal how the orthosomycin antibiotics inhibit protein synthesis by preventing accommodation of the aminoacyl-tRNA at the A site of the ribosome.
Keywords: antimicrobial, cryo-EM, everninomicin, rRNA, Ziracin
Abstract
The ribosome is one of the major targets for therapeutic antibiotics; however, the rise in multidrug resistance is a growing threat to the utility of our current arsenal. The orthosomycin antibiotics evernimicin (EVN) and avilamycin (AVI) target the ribosome and do not display cross-resistance with any other classes of antibiotics, suggesting that they bind to a unique site on the ribosome and may therefore represent an avenue for development of new antimicrobial agents. Here we present cryo-EM structures of EVN and AVI in complex with the Escherichia coli ribosome at 3.6- to 3.9-Å resolution. The structures reveal that EVN and AVI bind to a single site on the large subunit that is distinct from other known antibiotic binding sites on the ribosome. Both antibiotics adopt an extended conformation spanning the minor grooves of helices 89 and 91 of the 23S rRNA and interacting with arginine residues of ribosomal protein L16. This binding site overlaps with the elbow region of A-site bound tRNA. Consistent with this finding, single-molecule FRET (smFRET) experiments show that both antibiotics interfere with late steps in the accommodation process, wherein aminoacyl-tRNA enters the peptidyltransferase center of the large ribosomal subunit. These data provide a structural and mechanistic rationale for how these antibiotics inhibit the elongation phase of protein synthesis.
Many clinically used antibiotics target the ribosome to inhibit bacterial growth (1). X-ray crystallography structures have revealed that the majority of antibiotics that target the large ribosomal subunit bind at or near the peptidyl-transferase center (PTC), the active site for peptide bond formation (1, 2). The emergence of multidrug resistance in pathogenic bacteria, which has the potential to render our current arsenal of antibiotics obsolete, highlights the need for the development of new antibiotics that target distinct sites on the ribosome. Although structurally uncharacterized, biochemical and resistance studies indicate that one such class of antibiotics is the orthosomycins (3), which includes evernimicin (originally termed everninomicin, and hereafter referred to as EVN) and avilamycin (AVI) (2).
AVI is produced by Streptomyces virdochromogenes strain Tü57 (4), whereas EVN was identified and isolated from the producer Micromonospora carbonacea (5, 6). EVN and AVI display excellent antimicrobial activity against Gram-positive bacteria (3), including methicillin-resistant Staphylococcus aureus (7), as well as some Gram-negative bacteria, such as Borrelia burgdorferi (8). Importantly, strains resistant to EVN and AVI do not display cross-resistance to any other known antimicrobial agents, including ribosome-targeting antibiotics, such as chloramphenicol, tetracycline, or erythromycin (9, 10).
EVN/AVI resistance in Streptococcus pneumoniae and in the archaeon Halobacterium halobium arises via mutations within helix 89 (H89) and H91 of the 23S rRNA (10–12). Resistance to EVN and AVI also occurs via the action of methyltransferases that modify H89 and H91 (13, 14). Consistently, both EVN/AVI protect nucleotides within H89 and H91 from chemical modification (11, 12), suggesting that these two rRNA helices comprise at least part of the orthosomycin binding site. Additionally, EVN/AVI resistance has been associated with mutations in Arg-51, Ile-52, and Arg-56 of the ribosomal protein L16 in Enterococcus faecalis, E. faecium, S. pneumoniae, and S. aureus (15–18). However, it remains unclear whether these effects are direct consequences of EVN/AVI interacting with L16 or are mediated indirectly via changes in the 23S rRNA, as observed for other ribosomal protein-derived resistance mechanisms (2).
As expected based on the locations of the reported resistance mutations, both AVI and EVN bind to the ribosomal 50S subunit (19) and inhibit protein synthesis in vivo and in vitro (19, 20). Subsequent in vitro studies revealed that EVN inhibits IF2-dependent 70S initiation complex formation (11, 21); however, the inhibitory effect of EVN is not restricted to translation initiation because toe-printing assays indicate that EVN also inhibits translation elongation (22). EVN and AVI do not inhibit puromycin reaction (11, 12) and do not compete for binding with antibiotics that target PTC of the ribosome, such as chloramphenicol, linezolid, lincomycin, or clindamycin (19). Moreover, EVN has no inhibitory effect on the ribosome-dependent GTPase activity of EF-G (21). EVN and AVI are hypothesized to inhibit elongation by preventing tRNA binding to the A site (12, 20); however, this model remains to be conclusively demonstrated.
AVI has long been used in animal feed as a growth promoter (Surmax/Maxus; Elanco Animal Health), thereby limiting its clinical usefulness. However, EVN (SCH27899; Ziracin) underwent phase II/III clinical trials before being dropped in 2000 by Schering-Plough because of side effects and poor solubility. Nevertheless, the lack of cross-resistance between AVI/EVN and other clinically used ribosome-targeting antibiotics makes the orthosomycins attractive for further investigation (9, 10). The total chemical synthesis of EVN (23) and the biosynthesis of novel AVI derivatives with improved solubility (24) provide a good basis for further drug development; however, a structural understanding of how these antibiotics interact with the ribosome is necessary to facilitate rational design of improved orthosomycin derivatives.
Here we present two cryo-EM structures of EVN or AVI in complex with the Escherichia coli 70S ribosome at 3.6- to 3.9-Å resolution. These structures reveal that the conserved heptasaccharide core of both orthosomycins spans across the minor grooves of H89 and H91 of the 23S rRNA, whereas the terminal dichloro-ring interacts with the arginine residues of ribosomal protein L16. The binding positions of EVN and AVI overlap with the elbow region of a tRNA bound in the A site. Consistently, single-molecule FRET (smFRET) imaging of the tRNA selection process demonstrates that EVN and AVI allow initial binding of aminoacyl-tRNA (aa-tRNA) at the A site, but prevent complete accommodation of the incoming aa-tRNA, thus providing a structural explanation of how orthosomycin antibiotics inhibit translation elongation.
Results and Discussion
Cryo-EM Structures of EVN and AVI in Complex with the E. coli 70S Ribosome.
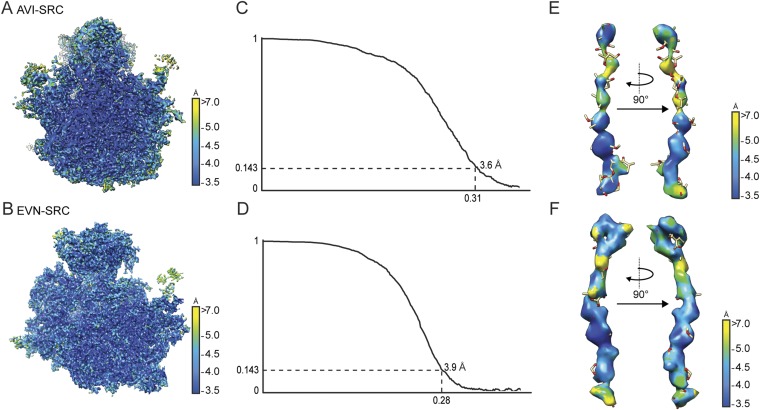
To determine the structures of EVN and AVI on the ribosome, we prepared Erm-stalled ribosome complexes (SRCs) as reported (25, 26). The SRCs were incubated with either 100 µM EVN or AVI, and the complexes were then subjected to single-particle cryo-EM analysis (Materials and Methods). The resulting cryo-EM reconstructions of the EVN- and AVI-SRC had an average resolution of 3.9 and 3.6 Å, respectively, with local resolution extending to 3.5 Å within the core of the ribosome (Fig. S1). Careful analysis of the cryo-EM maps revealed only a single binding site of EVN on the 50S subunit of the 70S ribosome, consistent with previous biochemical studies showing a 1:1 stoichiometry of EVN with the 50S subunit (19). In contrast to early reports that AVI binds to the 30S subunit (20), we observed only a single AVI binding site on the 50S subunit at the same location as EVN, a result that is consistent with the competition between these two antibiotics for ribosome binding (19).
Fig. S1.
Average and local resolution determination of AVI- and EVN-SRCs. (A and B) Transverse section of the cryo-EM reconstructions of the AVI-SRC (A) and the EVN-SRC (B) colored according to local resolution. (C and D) Average resolution of the AVI-SRC (C) and EVN-SRC (D) was 3.6 and 3.9 Å using the Fourier shell correlation (FSC) cutoff value of 0.143. Because of image processing with an absence of spatial frequencies >8 Å, the FSC value of 0.143 was used for average resolution determination (47). (E and F) Local resolution of the density for AVI (E) and EVN (F).
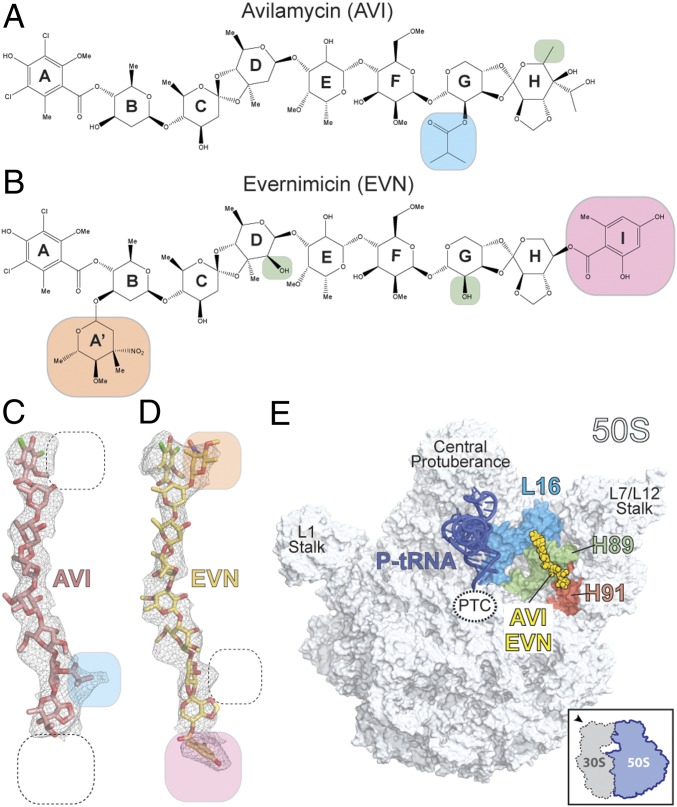
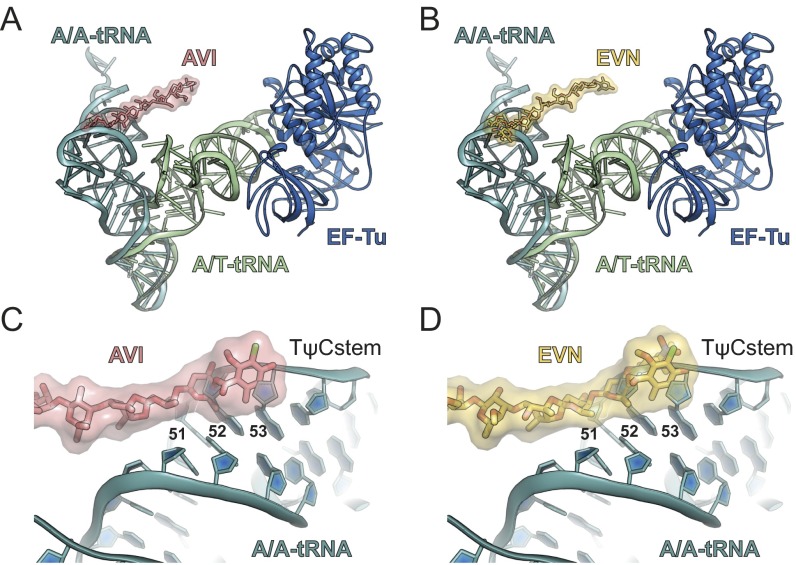
AVI has a terminal dichloroisoeverninic acid moiety (ring A) linked to a linear heptasaccharide chain consisting of d-olivose (rings B and C), 2-deoxy-d-evalose (ring D), 4-O-methyl-d-fucose (ring E), 2,6-di-O-methyl-d-mannose (ring F), the unusual pentose l-lyxose (ring G), and the bicyclic eurekanate (ring H) (ref. 27; Fig. 1A). Similar to AVI, EVN contains a nearly identical core heptasaccharide chain, but, unlike AVI, EVN is branched by a 2-deoxy-β-glycoside nitrosugar (ring A′) attached to ring B, and also contains an additional terminal benzyl moiety (ring I) attached to eurekanate ring H (28) (Fig. 1B). The presence of distinct electron density corresponding to the additional rings A′ and I of EVN in the cryo-EM map of the EVN-SRC, and absence of the same features in the AVI-SRC map, enabled us to unambiguously orient both AVI and EVN on the ribosome (Fig. 1 C and D). Despite the good fit of the refined molecular models to the cryo-EM electron density maps, higher resolution will be required to provide an unambiguous description of the hydrogen-bond interactions of the drugs with the ribosome. Nevertheless, these structures reveal that both drugs adopt elongated conformations on the ribosome, with the heptasaccharide rings B-H of both orthosomycins inserting into the minor grooves of H89 and H91 of the 23S rRNA (Fig. 1E and Movie S1) and the terminal ring A interacting with ribosomal protein L16 (Fig. 2A).
Fig. 1.
Cryo-EM reconstructions of EVN- and AVI-SRC. (A and B) Chemical structures of the orthosomycins AVI (A) and EVN (B), with compositional differences highlighted. (C and D) Cryo-EM electron densities (gray mesh) with fitted models for AVI (red; C) and EVN (yellow; D). (E) Overview of EVN/AVI binding site on the 70S ribosome (50S, gray, and 30S subunit omitted for clarity). Binding position of EVN/AVI (yellow) is shown relative to the P-site tRNA (blue), ribosomal protein L16 (cyan), H89 (green), and H91 (red).
Fig. 2.
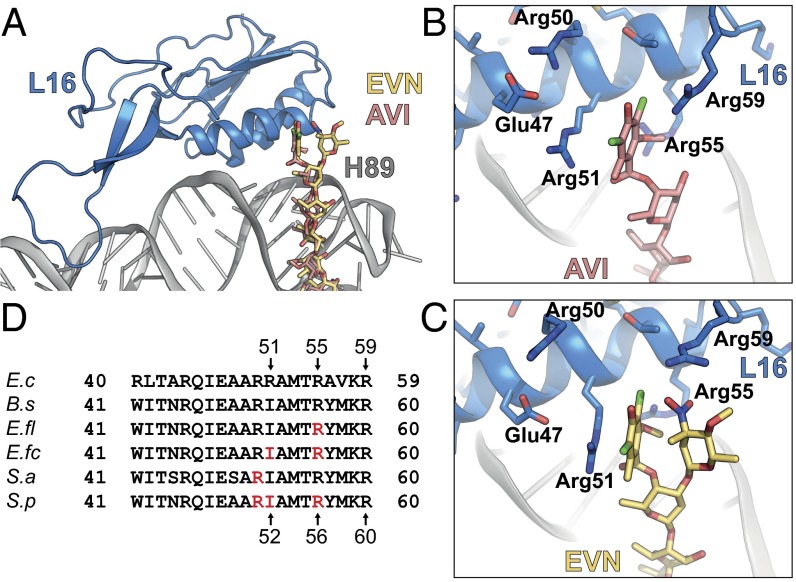
Interactions of EVN and AVI with the ribosomal protein L16. (A) Overview of L16 (blue) interactions with EVN (gold) and AVI (red). (B and C) Close-up views of showing interactions between Arg-51, -55, and -59 of L16 (blue) and ring A of AVI (B) and rings A and A′ of EVN (C). (D) Sequence alignment of the L16 from E. coli (E.c), B. subtilis (B.s), E. faecalis (E.fl), E. faecium (E.fc), S. aureus (S.a), and S. pneumoniae (S.p), with residues conferring resistance to EVN and AVI highlighted in red.
Interactions of EVN/AVI with Arginine Residues of L16.
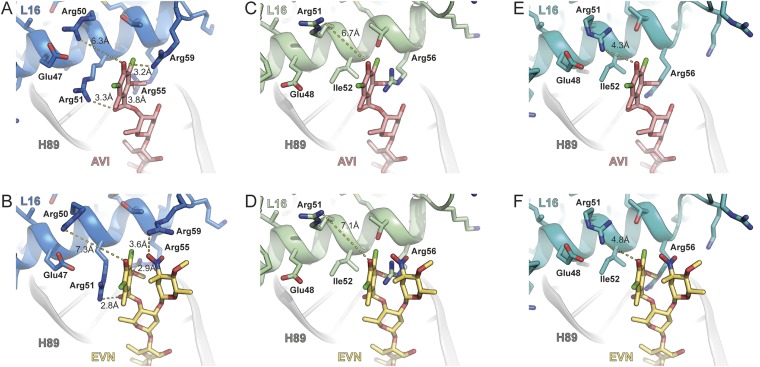
The terminal dichloroisoeverninic acid moiety (ring A) of AVI establishes stacking interactions with the side chain of Arg-51 of L16 (Fig. 2 A and B and Movie S1). In addition, the side chains of Arg-55 and -59 also approach ring A of AVI (Fig. 2B); however, the density for these side chains is less well defined. In contrast to AVI, the electron density for the terminal region of EVN is bifurcated (Fig. 1D), consistent with the presence of the additional 2-deoxy-β-glycoside nitrosugar (ring A′) (Fig. 1B). Unfortunately, the resolution does not allow unambiguous assignment of ring A and A′ to the bifurcated density. Therefore, our current model is based on the rationale that ring A of EVN occupies the same position as ring A of AVI, and the remaining density is then assigned to the ring A′ (Fig. 2C). Sequence alignments (Fig. 2D), as well as comparison with the structures of the B. subtilis 70S ribosome (29) and S. aureus 50S subunit (30) (Fig. S2), reveals that E. coli Arg-51 and -55 are equivalent to Ile-52 and Arg-56 in most Gram-positive bacteria. Consistently, mutations of Arg-56–His or Ile-52–Ser/Thr/Asn in L16 render E. faecalis, E. faecium, and S. pneumoniae isolates resistant to EVN and AVI (15–17). Chemical mutagenesis experiments in S. aureus led to the identification of strains with Arg-51–Cys or Arg-51–His mutations in L16 that conferred increased resistance to both compounds (18). In our structure, residue Arg-50 (E. coli), equivalent to residue Arg-51 in S. aureus, does not contact the drug (Fig. 2 B and C and Fig. S2). This finding suggests that the Arg-51–Cys/His mutations may indirectly confer EVN/AVI resistance in S. aureus, possibly by affecting the neighboring Ile-52 residue. Alternatively, EVN/AVI may interact with S. aureus ribosomes using a slightly different binding mode that enables direct interaction between Arg-51 and the drugs. Nevertheless, the finding that both EVN and AVI directly interact with L16 in the region where EVN/AVI resistance mutations occur illustrates the importance of this interaction for drug binding. Moreover, it also reveals that resistance occurs because the mutations directly perturb drug binding, rather than indirectly preventing drug binding by distorting the local rRNA conformation of H89/H91.
Fig. S2.
Comparison of AVI/EVN binding site with respect to E. coli, B. subtilis, and S. aureus L16. Interaction of AVI (red; A) and EVN (gold; B) with E. coli L16 (blue) compared with the relative position of B. subtilis L16 (green; C and D) (29) and S. aureus L16 (cyan; E and F) (30). In E. coli, Arg-50 is 6.3–7.3 Å from ring A of AVI/EVN, whereas the equivalent residue Arg-51 is 6.7–7.1 Å and 4.3–4.8 Å when superimposing AVI/EVN with B. subtilis L16 (green; C and D) (29) and S. aureus L16 (cyan; E and F) (30).
Interaction of EVN and AVI with H89 and H91 of the 23S rRNA.
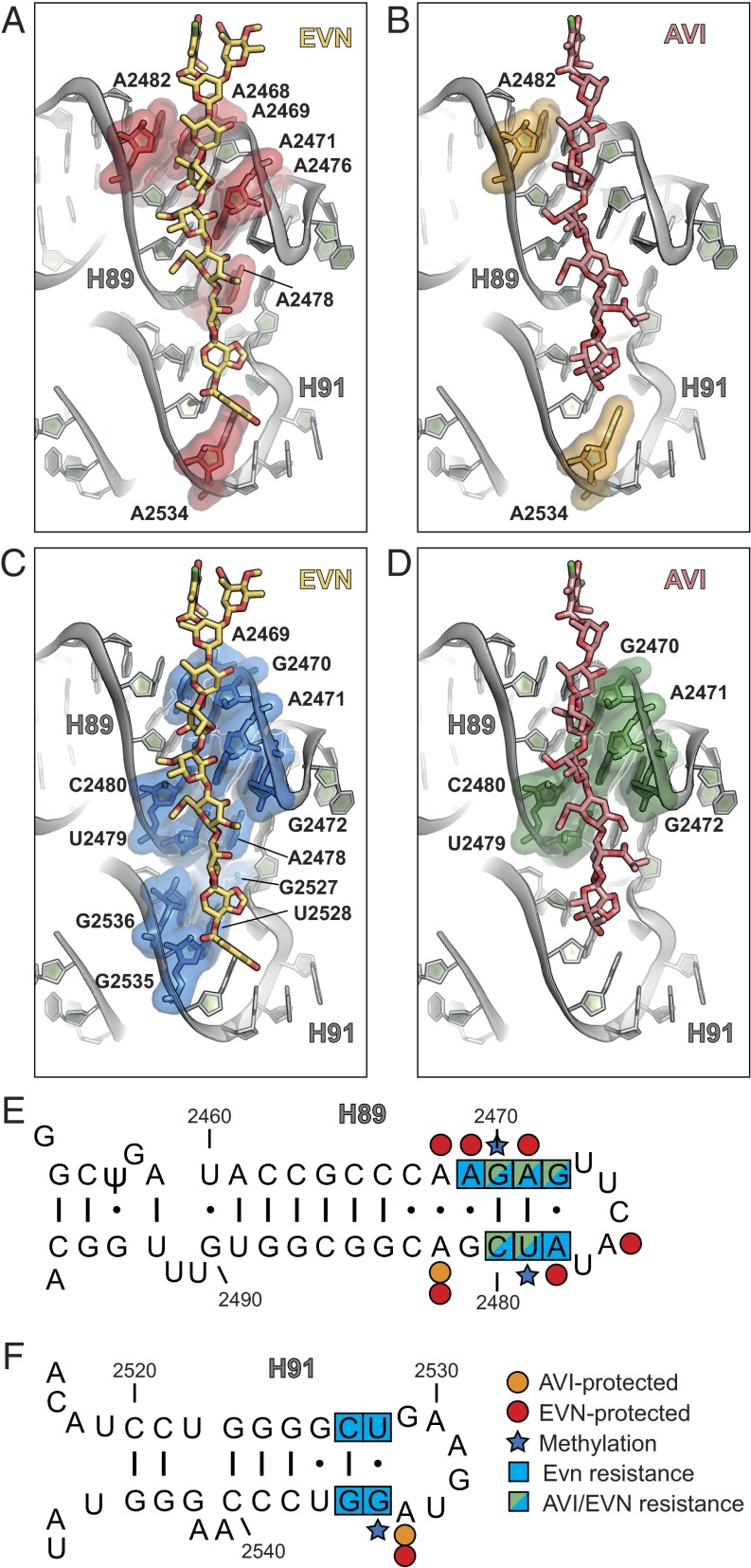
Within the limits of the present resolution, we observed no significant difference between the interaction of the conserved heptasaccharide cores of EVN and AVI with H89 and H91 of the 23S rRNA (Fig. 3 A–D). The largest interaction surface between the drugs and the ribosome encompasses rings B–F of EVN/AVI and the minor groove of H89, specifically, nucleotides A2468-G2472 and A2478-A2482 (Fig. 3 A–D), which base pair to form the stem of H89 (Fig. 3E). Additional interactions were observed between rings G and H of EVN/AVI with the minor groove of H91 (Fig. 3 A–D) formed by nucleotides G2527–U2528 and A2534–G2536 (Fig. 3F). This interaction pattern is consistent with footprinting data on E. coli, E. feacium, and H. halobium ribosomes showing that EVN protects multiple nucleotides within H89, including A2468, A2469, A2471, A2476, A2478, and A2482, as well as nucleotide A2534 in H91, from chemical modification by dimethyl sulfate (DMS) (11, 13) (Fig. 3 A, E, and F). Similarly, AVI protects A2482 in H89 and A2534 in H91 from chemical modification by DMS on E. coli 70S ribosomes (12) (Fig. 3 B, E, and F). The terminal benzyl moiety (ring I) of EVN establishes additional interactions with nucleotides within the loop of H91 (Fig. 3 A and C), which may contribute to the higher potency of EVN compared with AVI.
Fig. 3.
Interactions of EVN and AVI with H89 and H91 of the 23S rRNA. (A and B) Binding site of EVN (gold) (A) and AVI (red) (B), with nucleotides in H89 and H91 protected from DMS modification highlighted in red and orange, respectively. (C and D) Binding site of EVN (gold) (C) and AVI (red) (D), with resistance mutations in H89 and H91 highlighted in blue and green, respectively. (E and F) Secondary structure of 23S rRNA with zoom on H89 (E) and H91 (F), with nucleotides protected by EVN (red) and AVI (orange), EVN (blue) and AVI (green) resistance mutations and methylations (blue star) as indicated (10–18).
Resistance to EVN/AVI via 23S rRNA Mutations.
A striking correlation exists between the nucleotides that comprise the EVN/AVI binding site observed here and the reported mutations in 23S rRNA that confer resistance to these two drugs (Fig. 3 C–F). In S. pneumoniae, selection for EVN resistance led to the identification of 23S rRNA mutations A2469C, C2480U (in H89), G2535A, or G2536C (in H91) (ref. 10; Fig. 3 C, E, and F). The G2535A mutation was also subsequently reported to confer EVN resistance in E. faecalis (16). The archaeon H. halobium has also been used to select for EVN resistance, producing A2471G/C, A2478C, U2479C, and C2480A/U mutations in H89 and G2527A, U2528A, and G2535A mutations in H91 (11) (Fig. 3 C, E, and F). In contrast, selection for AVI using H. halobium only led to the identification of mutations within H89, namely, G2470U, A2471G, G2473U, U2479C, and C2480U (12) (Fig. 3 D and E). The increased frequency of resistance mutations located in H89, as well as the higher resistance conferred by these mutations compared with H91 mutations (11, 12), emphasizes the importance of the extensive interaction surface between rings B-F of EVN/AVI and nucleotides comprising the minor groove of H89. Because all of the reported mutations are expected to alter base-pairing potential, resistance is likely to arise from distortions of the helical geometry of H89 and H91, which thereby reduce the affinity of the drugs for their binding site.
Resistance to EVN/AVI via Methylation of the 23S rRNA.
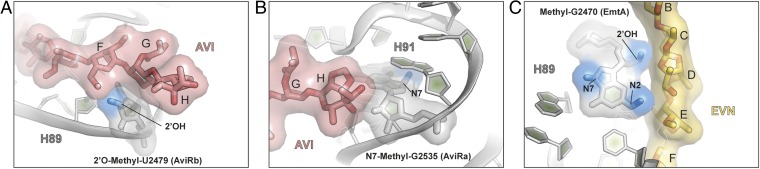
Analysis of the binding site of EVN and AVI reveals a structural basis for the resistance obtained via posttranslational modifications of nucleotides within H89 and H91 (13, 14) (Fig. 4). S. virdochromogenes Tü57, the producer of AVI, expresses two methyltransferases, AviRa and AviRb, which confer resistance to EVN/AVI. Whereas AviRb methylates the ribose 2′OH of U2479 within H89 to confer high-level AVI resistance, AviRa methylates the N7 position of G2535 within H91 to confer low-level resistance (14, 31). Inspection of the EVN/AVI binding site reveals that a 2′O-methylation of U2479 would lead to a direct clash with ring F of the drug (Fig. 4A). In contrast, N7-methylation of G2535 appears to neither interfere with the drug binding (Fig. 4B) nor disrupt base pairing with U2528, suggesting that methylation at this position indirectly confers resistance by inducing local conformational changes, possibly during ribosome assembly. We note that both AviRa and AviRb are required to obtain full protection against the AVI (31), suggesting that they function in a synergistic manner, similar to the methyltransferases that cause resistance to tylosin (32). The EVN methyltransferase EmtA, which was identified on a plasmid-borne insertion element in EVN-resistant E. faecium strains (isolated from animals given AVI as a growth promotant), was shown to methylate G2470 of H89 (13). Although the exact site of the modification has not been identified, we note that methylation of the N2 position of G2470 or the ribose 2′OH would lead to a direct clash with rings D and C, respectively, of EVN/AVI (Fig. 4C), whereas an N7-methylation would most likely confer resistance indirectly via conformational changes.
Fig. 4.
Structural basis for EVN/AVI resistance via methylation of the 23S rRNA residues. (A) The 2′O-methylation of U2479 in H89 by AviRb (14) clashes with the ring F of the drug. (B) N7 methylation of G2535 in H91 by AviRa (14) is located distal from the AVI binding site. (C) Methylation of the 2′OH of the ribose or N2 position in the nucleobase of G2470 by EmtA (13) clashes with EVN (gold), whereas the N7 position is distal to the drug-binding site.
Inhibition of IF2 and A-tRNA Accommodation by EVN and AVI.
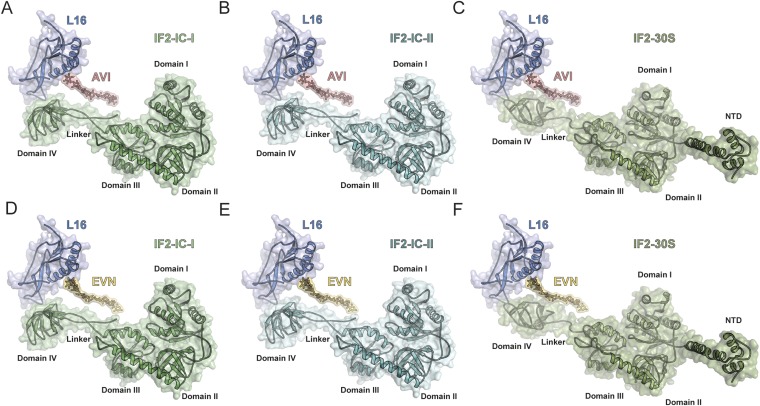
EVN has been reported to inhibit formation of the IF2-dependent 70S initiation complex (70S-IC) (11, 21). Therefore, we compared the binding sites of EVN/AVI relative to structures of IF2 on the 70S ribosome (33) and 30S subunit (34, 35). No overlap was observed between EVN/AVI and IF2 on the 70S, with the shortest distance between ring E of EVN/AVI being 2–3 Å away from the linker between domains III and IV of IF2 (Fig. S3). In contrast, alignment of IF2-30S complex to the AVI/EVN-SRC reveals a slight overlap between EVN/AVI and domain IV of IF2 (Fig. S3), suggesting that EVN/AVI may interfere with IF2-dependent 70S-IC formation by blocking a transient intermediate state of IF2 that arises upon subunit binding and transition from the 30S-IC to the 70S-IC.
Fig. S3.
Comparison of AVI/EVN binding site with respect IF2. Binding position of AVI (red; A–C) and EVN (D–F) on the E. coli 70S ribosome relative to IF2-IC conformation I (green; A and D) and IF2-IC conformation II (teal; B and E) on the 70S ribosome (33) and IF2-GTP conformation (olive; C and F) on the 30S subunit (34, 35).
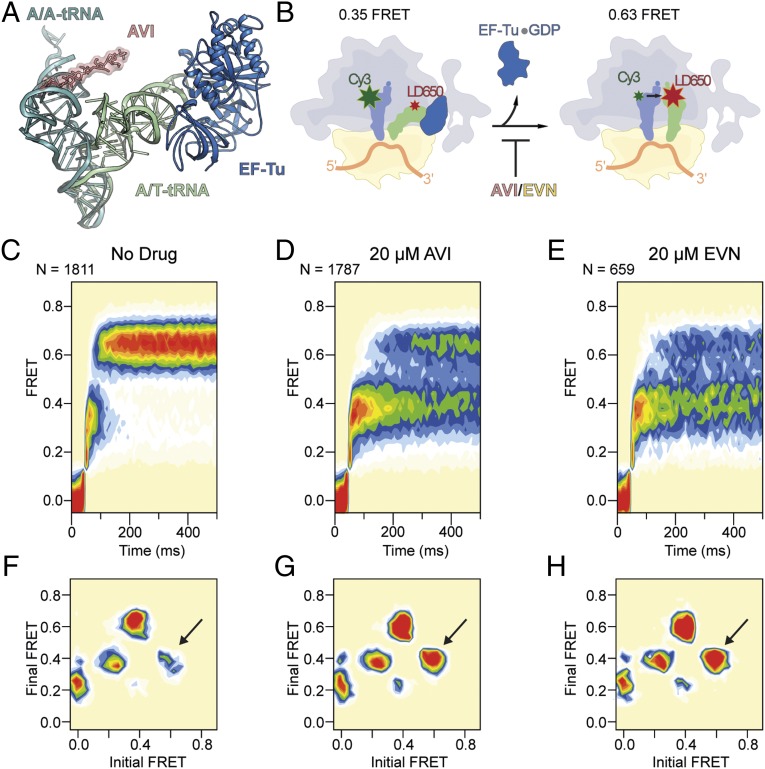
EVN and AVI have also been suggested to inhibit translation elongation by interfering with the tRNA binding to the A site of the ribosome (12, 20, 22). Therefore, we compared the binding position of EVN/AVI relative to the tRNA in the A/T state observed during decoding when the aa-tRNA is bound to the ribosome but still remains in complex with EF-Tu (36, 37), as well as with the tRNA in the classical A/A state in which the acceptor arm of the aa-tRNA is released from EF-Tu and has accommodated at the PTC on the large ribosomal subunit (38). These comparisons show that the EVN/AVI binding site does not overlap with aa-tRNA within the A/T state, whereas there is direct clash between rings A-C of EVN/AVI and nucleotides 51–53 within the stem region of the TΨC-loop (elbow region) of fully accommodated aa-tRNA (Fig. 5A, Fig. S4, and Movie S1).
Fig. 5.
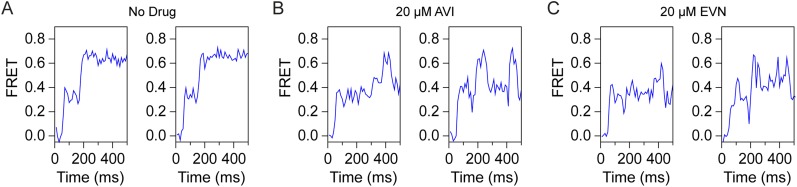
EVN/AVI inhibit accommodation of tRNA into the A site. (A) Comparison of the relative binding positions on the ribosome of AVI (red), EF-Tu (blue), A/T-tRNA (green) (36, 56), and A/A-tRNA (teal) (38). (B) Schematic diagram of smFRET measurements of tRNA selection. After delivery of EF-Tu⋅GTP⋅tRNA ternary complex containing cognate Phe-tRNAPhe(LD650) to the A site of E. coli 70S ribosomes containing tRNAiMet(Cy3) in the P site, tRNA motion can be tracked through the progression of FRET efficiencies from low (0.2) to intermediate (0.35) FRET during initial steps of selection to high (0.63) FRET upon A-site tRNA accommodation, which is inhibited by AVI/EVN. (C–E) Ensemble smFRET histograms showing the time course of aa-tRNA selection, imaged in the absence of drugs (C) or in the presence of 20 μM AVI (D) or 20 μM EVN (E). The histograms were postsynchronized by aligning each observed event to the first appearance of nonzero FRET states. (F–H) Transition density plots for the data shown in C–E, respectively. These 2D histograms juxtapose the FRET efficiencies immediately before and after FRET transitions. As indicated by arrows, EVN and AVI promote reversible transitions between high and intermediate FRET.
Fig. S4.
EVN and AVI specifically block accommodation of A-tRNA into the classical state. Representative smFRET traces exemplifying the processes shown in Fig. 5 C–E are shown. (A) In the absence of drug, aa-tRNA rapidly achieves the fully accommodated A/A state (0.63 FRET) via reversible forward-sampling from the 0.35 FRET A/T state. (B and C) AVI (B) and EVN (C) inhibit formation of the A/A state, leading to repeated, unsuccessful sampling events.
To investigate the impact of EVN and AVI on the selection and accommodation of aa-tRNA, we used pre-steady-state smFRET measurements that enable real-time visualization of tRNA motion during EF-Tu–catalyzed delivery of aa-tRNA to surface-immobilized ribosomes (39–41) (Fig. 5B). Here, the time evolution of FRET efficiency was monitored at 10 ms per frame time resolution within individual 70S ribosomes bound with (Cy3-s4U8)-labeled fMet-tRNAiMet in the P site upon stopped-flow injection of ternary complex containing EF-Tu, GTP and (LD650-acp3U47)-labeled Phe-tRNAPhe (Fig. 5B). As expected from previous studies (40, 41), in the absence of the drug, productive FRET events leading to the incorporation of aa-tRNA at the A site evolved from a low (∼0.2) to high (∼0.63) FRET state via the reversible transit of at least one intermediate (∼0.35) FRET configuration, which reflects the A/T state of the A-site tRNA (Fig. 5 B and C) (39, 40). Consistent with rapid aa-tRNA progression through the selection process, the time delay between the initial observation of low FRET and formation of the stable, high-FRET state, corresponding to the fully accommodated, classically configured A/A-tRNA position, was ∼60 ms (∼16 s−1) (Fig. 5 B and C and Fig. S5).
Fig. S5.
Relative position of EVN/AVI to A/T- and A/A-tRNA. (A and B) Comparison of the relative binding position on the ribosome of AVI (red; A) and EVN (B), with EF-Tu (blue) and A/T-tRNA (green) (36, 56), as well as with accommodated A/A-tRNA (teal) (38). (C and D) Zoom showing the overlap between AVI (red; C) and EVN (gold; D) with nucleotides 51–53 with the TΨC stem of the an accommodated A/A-tRNA (teal) (38).
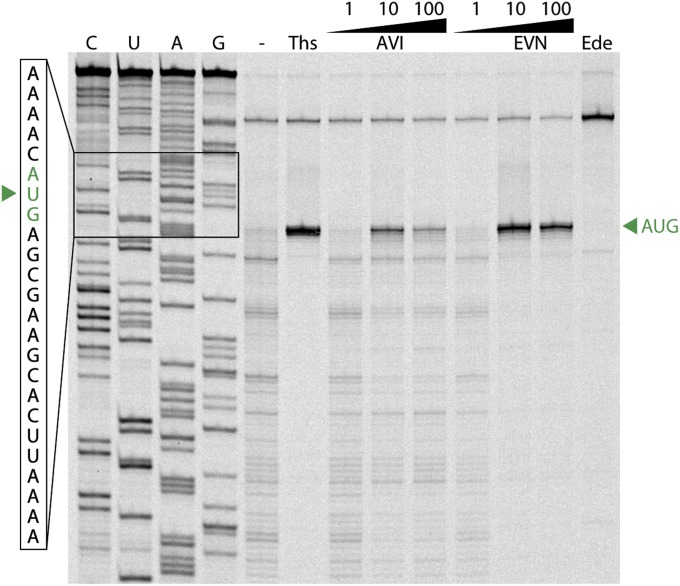
In the presence of saturating concentrations (20 µM) of EVN or AVI, aa-tRNA progression into the ribosome was strongly and specifically blocked during the transition between the A/T state (∼0.35 FRET) and the fully accommodated A/A state (∼0.63 FRET) (Fig. 5 D and E and Fig. S5). To examine the dynamics underlying this inhibition, we visualized the ensemble of observed molecular transitions using transition density plots (42). In this representation, observed transitions appear as peaks in a 2D histogram of initial and final FRET efficiencies (Fig. 5 F–H). The peak corresponding to reverse transitions from high to intermediate FRET is significantly enhanced in the presence of both EVN or AVI (arrows in Fig. 5 F–H), confirming that inhibition was characterized by an exacerbation of the reversible excursions between A/T and accommodated positions that normally accompany proofreading (40) (Fig. S4). These findings are in agreement with toe-printing experiments demonstrating that 70S ribosomes initiated on the AUG start codon of mRNA do not proceed into the elongation phase of translation when increasing concentrations of EVN or AVI are present (Fig. S6). Our findings contrast with a previous report (22) in which Evn did not appear to significantly affect the first elongation cycle, but, rather, allowed successive rounds of elongation before inhibition was observed. One possibility for this discrepancy is that the strength of the inhibition of the orthosomycins depends on the nature of the aa-tRNA that is being accommodated.
Fig. S6.
EVN and AVI block ribosomes at the start codon of the mRNA. Toe-printing assay monitoring translation in the presence of increasing concentrations (1, 10, and 100 μM) of EVN or AVI is shown. Additionally, control reactions without antibiotic (−) or including thiostrepton (Ths, 100 μM) or edeine (Ede, 50 μM) are shown. AUG designates location of the ribosomes stalled at the start codon. C, U, A, and G indicate the sequencing lanes.
Conclusion
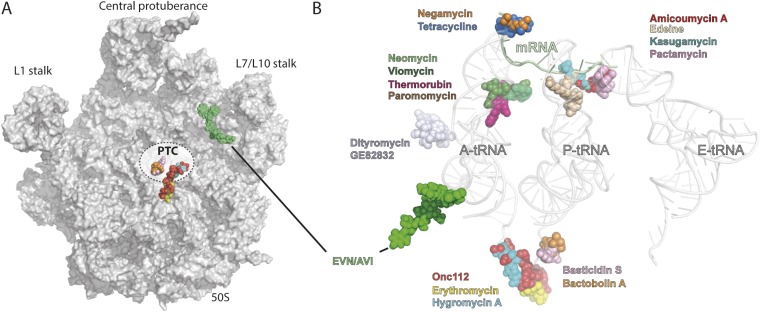
The cryo-EM structures of EVN- and AVI-SRC reported here reveal that both orthosomycins bind to a single site on the large subunit that is distinct from other known antibiotic binding sites on the ribosome (Fig. S7), explaining the lack of cross-resistance with other ribosome-targeting antibiotics (9, 10). The orthosomycin binding site comprises the minor grooves of H89 and H91 of the 23S rRNA, as well as arginine residues of L16 (Fig. 1E), consistent with available chemical protection and resistance data (Figs. 2–4) (10–18). The binding position for EVN and AVI provides a structural explanation for how the orthosomycins inhibit IF2-dependent 70S-IC formation (11, 21)—namely, by interfering with the transition from the IF2-30S conformation to the IF2-70S that occurs upon subunit joining (Fig. S3). Additionally, our smFRET data demonstrate that both EVN and AVI interfere with the accommodation of aa-tRNA at the A site of the ribosome (Fig. 5 C–H), consistent with the overlap between EVN/AVI and the elbow region of a fully accommodated A-tRNA (Fig. 5A). Overall, our study also demonstrates that cryo-EM can be used to determine de novo the binding site of antibiotics on the bacterial ribosome, as was also recently demonstrated for the antiprotozoan drug emetine in complex with the Plasmodium falciparum 80S ribosome (43).
Fig. S7.
Relative position of EVN/AVI to other ribosome-targeting antibiotics. (A) Overview of ribosomal 50S subunit (gray) with relative position of EVN/AVI (yellow) compared with known antibiotics that target the PTC. (B) Relative position of EVN/AVI (green) compared with known antibiotics that target the small and large ribosomal subunit, with A-, P-, and E-tRNAs shown for reference.
Materials and Methods
The SRCs were prepared essentially as described (25, 44). Cryo-EM data collection was performed on the Titan Krios (FEI) 300-kV TEM equipped with a Falcon II direct electron detector. Images of individual ribosome particles were aligned by using Motion Correction software (45), and then particles were selected automatically by using SIGNATURE (46). All images were processed by using a frequency-limited refinement protocol that prevents overfitting (47) using the SPIDER software package (48), as described (25, 44). The final maps were subjected to the program EM-BFACTOR (49) to apply an automatically determined negative B factor for sharpening of the map, and local resolution was calculated by using ResMap (50). Molecular models were fitted and adjusted by using COOT (51) and refined in Phenix (52). Model validation was carried out by using the MolProbity server (53), and the final model statistics are presented in Table S1. All figures showing atomic models as well as Movie S1 were generated by using PyMOL (Schrödinger). Fig. S1 was generated by using Chimera (54). The smFRET experiments were performed as described (39–41, 55). Further details can be found in SI Materials and Methods. The cryo-EM maps and models for the EVN- and AVI-SRC have been deposited in the EMDatabank (accession nos. EMD-8238 and EMD-8237) and the Protein Data Bank (PDB ID codes 5KCS and 5KCR).
Table S1.
Data collection and refinement statistics
| Data collection and refinement | EVN-SRC | AVI-SRC |
| Particles | 78,186 | 61,651 |
| Pixel size, Å | 1.108 | 1.108 |
| Defocus range, µm | 1.0–2.4 | 0.7–2.4 |
| Voltage, kV | 300 | 300 |
| Electron dose, e−/Å-2 | 28 | 20 |
| Map sharpening B factor, Å2 | −234.20 | −126.14 |
| Resolution, Å (0.143 FSC) | 3.9 | 3.6 |
| Model composition | ||
| Nonhydrogen atoms | 149,026 | 145,176 |
| Protein residues | 6,664 | 5,626 |
| RNA bases | 4,645 | 4,696 |
| Validation (proteins) | ||
| Poor rotamers, % | 0.00 | 0.08 |
| Ramachandran outliers, % | 2.61 | 1.68 |
| Ramachandran favored, % | 88.98 | 92.36 |
| Bad backbone bonds, % | 0.00 | 0.12 |
| Bad backbone angles | 0.00 | 0.01 |
| MolProbity score | 2.02 (74th percentile) | 2.16 (100th percentile) |
| Validation (nucleic acids) | ||
| Correct sugar puckers, % | 97.71 | 99.46 |
| Bad backbone conformations, % | 12.89 | 13.86 |
| Bad bonds, % | 0.05 | 0.39 |
| Bad angles | 0.01 | 0.01 |
| Clashscore, all atoms | 8.29 (80th percentile) | 15.25 (97th percentile) |
SI Materials and Methods
Cryo-EM and Single Particle Reconstruction.
The SRCs were prepared essentially as described (25, 44). The AVI- and EVN-SRCs (4 A260/mL) were applied to 2-nm precoated Quantifoil R3/3 holey carbon supported grids and vitrified by using a Vitrobot Mark IV (FEI). Data collection was performed at NeCEN on a Titan Krios (FEI) transmission electron microscope equipped with a Falcon II direct electron detector at 300 kV with a magnification of 125,085×, a pixel size of 1.108 Å, and a defocus range of 0.8–2.4 µm. The data were provided as a series of seven frames (dose per frame is 4 e−/Å2), from which we summed frames 1–4 (AVI-SRC) or 1–6 (EVN-SRC) (accumulated dose, including the pre-exposure frame, of 20 and 28 e−/Å2, respectively, as described in Table S1) after alignment by using Motion Correction software (45). Images were processed by using a frequency-limited refinement protocol that helps prevent overfitting (47), specifically by truncation of high frequencies (in this case at 7–8 Å using Butterworth filter). Power spectra and defocus values were determined by using CTFFIND4 (57). Data were processed further by using the SPIDER software package (48), in combination with an automated workflow as described (58). After initial, automated particle selection based on the program SIGNATURE (46), initial alignments were performed (102,837 particles for AVI-SRC and 127,205 particles for EVN-SRC) by using E. coli 70S ribosome as a reference structure (25). After 3D classification using an incremental K-means-like method of unsupervised 3D sorting (59), the final AVI- and EVN-SRC maps [61,651 (60%) and 78,186 (75%) particles, respectively] could be refined to average resolutions of 3.6 Å (AVI-SRC) and 3.9 Å (EVN-SRC) (Fig. S1). The final refined maps were subjected to the program EM-BFACTOR (49) to apply an automatically determined negative B-factor for sharpening of the map. Local resolution calculations were performed by using ResMap (50), revealing that the majority of the cores of the 30S and 50S subunits extended to 3.5 Å (Fig. S1).
Model Building and Structure Refinement Procedures.
The initial atomic model for the E. coli 70S ribosome was generated by using the 50S subunit from Protein Data Bank (PDB) ID code 4YBB (60) and the 30S subunit from PDB ID code 4TP9 (61). The initial model for the P-tRNA was based on PDB ID code 3TVF (62), and the model for the P-site mRNA was built de novo. Atomic models for AVI and EVN were generated from their known chemical structures by using PRODRG online software (63), which was also used to generate CIF restraints for the subsequent fitting and refinement. The initial models of the entire 70S ribosome with bound mRNA, P-site tRNA, and either AVI or EVN were fitted into the cryo-EM electron-density maps by real-space rigid-body refinement in PHENIX (52) with the ribosome split into multiple rigid-body domains. Resulting molecular models were further adjusted by using COOT (51) and refined in PHENIX (52). Model validation was carried out by using the MolProbity server (53). The final model statistics are presented in Table S1. All figures showing atomic models, as well as Movie S1, were generated by using PyMOL (Schrödinger). Fig. S1 was generated by using Chimera (54).
smFRET Imaging.
Single-molecule experiments were performed in Tris-polymix buffer containing 50 mM Tris-acetate (pH 7.5), 5 mM MgCl2, 100 mM KCl, 5 mM NH4(CH3COO), 0.5 mM CaCl2, 0.1 mM EDTA, 5 mM putrescine, 1 mM spermidine, 1.5 mM β-mercaptoethanol, and 1 mM GTP, in the presence of an oxygen-scavenging system consisting of 2 mM protocatechuic acid, 50 nM protocatechuate 3,4-dioxygenase, and photostabilizing compounds (1 mM Trolox, 1 mM cyclooctatetraene, and 1 mM nitrobenzyl-alcohol) (64). EF-Tu⋅GTP⋅Phe-tRNAPhe(LD650-acp3U47) ternary complex was prepared as described (39, 65). LD650 (Lumidyne Technologies) is an intramolecularly photostabilized derivative of Cy5 (66–68). Surface immobilization of ribosome complexes (0.5 nM) programmed with biotinylated mRNA was achieved by brief incubation in PEG-passivated, streptavidin-coated quartz microfluidic devices (55). Complexes lacking ribosomal protein L1 were used to minimize the contribution of hybrid state tRNA configurations after peptide bond formation (55).
smFRET data were acquired on a home-built prism-based total internal reflection microscope as described (41). Briefly, a 532-nm laser (Opus 532; Laser Quantum) was used to excite Cy3 fluorophores attached to tRNAfMet. Emitted fluorescence from Cy3 donor and LD650 acceptor fluorophores was collected by using a 1.27-NA 60× water immersion objective (Nikon), spectrally separated by using a MultiCam-LS device (Cairn) equipped with a dichroic mirror (T635lpxr-UF2; Chroma), and imaged onto two scientific complementary metal-oxide semiconductor cameras (Orca-Flash 4.0 V2; Hamamatsu Photonics) acquiring at 10-ms integration time. FRET efficiency time courses for individual ribosomes were calculated, selected, and analyzed by using the freely available MATLAB-based software package SPARTAN as described (41).
Toe-Printing Assay.
The position of the ribosome on the mRNA was monitored by using a toe-printing assay based on the PURExpress (New England Biolabs) in vitro coupled transcription-translation system, as described (69, 70). Briefly, each translation reaction consisted of 2 μL of solution A, 1.5 μL of solution B, and 1 μL (0.6 pmol) of DNA template (hns40aa): (5′- ATTAATACGACTCACTATAGGGATATAAGGAGGAAAACATATGAGCGAAGCACTTAAAATTCTGAACAACATCCGTACTCTTCGTGCGCAGGCAAGAGAATGTACACTTGAAACGCTGGAAGAAATGCTGGAAAAATTAGAAGTTGTTGTTAACGAACGTTGGATTTTGTAAGTGATAGAATTCTATCGTTAATAAGCAAAATTCATTATAACC-3′, with the ATG start and TAA stop codon highlighted in bold and the primer-binding site underlined) and 0.5 μL of additional agents (nuclease-free water, Ths, Ede, AVI, or EVN). The template was synthesized via PCR from gDNA (E. coli MG1655). Translation was performed at 37 °C for 15 min in 1.5-mL reaction tubes with constant shaking at 500 rpm. After translation, 2 pmol of Alexa 647-labeled NV-1 toe-printing primer (5′-GGTTATAATGAATTTTGCTTATTAAC-3′) was added to each reaction and incubated at 37 °C for 5 min without shaking. Reverse transcription (RT) was performed with 0.5 μL of AMV reverse transcriptase (NEB), 0.1 μL of dNTP mix (10 mM), and 0.4 μL of Pure System Buffer and incubated at 37 °C for 20 min. The RT reaction was quenched and RNA degraded by addition of 1 μL of 10 M NaOH and incubation for at least 15 min at 37 °C and then was neutralized with 0.7 μL of 25% HCl. Then, 20 μL of toe-printing resuspension buffer and 200 μL of PN1 buffer were added to each reaction before treatment with a QIAquick Nucleotide Removal Kit (Qiagen). The Alexa 647-labeled DNA was then eluted from the QIAquick columns with 80 μL of nuclease-free water. A vacuum concentrator was used to vaporize the solvent, and the Alexa 647-labeled DNA was then dissolved into 3.5 μL of formamide dye. The samples were heated to 95 °C for 5 min before being applied onto a denaturing 6% (vol/vol) polyacrylamide (19:1) sequencing gel containing 7 M urea. Gel electrophoresis was performed at 40 W and 2,000 V for 2 h. The GE Typhoon FLA9500 imaging system was subsequently used to scan the polyacrylamide gel.
Supplementary Material
Acknowledgments
We thank Rishi Matadeen and Sacha DeCarlo for data collection at the NeCEN facility, Leiden, The Netherlands; Charlotte Ungewickell for expert technical assistance; and Shura Mankin and Birte Vester for kindly providing copious amounts of EVN and AVI, respectively. This research was supported by Deutsche Forschungsgemeinschaft Grants FOR1805, WI3285/3-2, and GRK1721 (to D.N.W.); National Institutes of Health Grants GM079238 and GM098859 (to S.C.B.); the Human Frontiers of Science Program (D.N.W. and S.C.B.); and the Illinois State startup funds (Y.S.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The cryo-EM maps and models for the EVN- and AVI-SRC have been deposited in the EMDatabank (accession nos. EMD-8238 and EMD-8237) and the Protein Data Bank, www.pdb.org (PDB ID codes 5KCS and 5KCR).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1604790113/-/DCSupplemental.
References
- 1.Wilson DN. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol. 2014;12(1):35–48. doi: 10.1038/nrmicro3155. [DOI] [PubMed] [Google Scholar]
- 2.Wilson DN. The A-Z of bacterial translation inhibitors. Crit Rev Biochem Mol Biol. 2009;44(6):393–433. doi: 10.3109/10409230903307311. [DOI] [PubMed] [Google Scholar]
- 3.Wright D. The orthosomycins, a new family of antibiotics. Tetrahedron. 1979;35(10):1209–1327. [Google Scholar]
- 4.Buzzetti F, et al. [Avilamycin] Experientia. 1968;24(4):320–323. doi: 10.1007/BF02140794. [DOI] [PubMed] [Google Scholar]
- 5.Wagman GH, Luedemann GM, Weinstein MJ. Fermentation and isolation of everninomicin. Antimicrob Agents Chemother (Bethesda) 1964;10:33–37. [PubMed] [Google Scholar]
- 6.Weinstein MJ, Luedemann GM, Oden EM, Wagman GH. Everninomicin, a new antibiotic complex from Micromonospora Carbonacea. Antimicrob Agents Chemother (Bethesda) 1964;10:24–32. [PubMed] [Google Scholar]
- 7.Boucher HW, Thauvin-Eliopoulos C, Loebenberg D, Eliopoulos GM. In vivo activity of evernimicin (SCH 27899) against methicillin-resistant Staphylococcus aureus in experimental infective endocarditis. Antimicrob Agents Chemother. 2001;45(1):208–211. doi: 10.1128/AAC.45.1.208-211.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dever LL, Torigian CV, Barbour AG. In vitro activities of the everninomicin SCH 27899 and other newer antimicrobial agents against Borrelia burgdorferi. Antimicrob Agents Chemother. 1999;43(7):1773–1775. doi: 10.1128/aac.43.7.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanders WE, Jr, Sanders CC. Microbiological characterization of everninomicins B and D. Antimicrob Agents Chemother. 1974;6(3):232–238. doi: 10.1128/aac.6.3.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adrian PV, et al. Evernimicin (SCH27899) inhibits a novel ribosome target site: analysis of 23S ribosomal DNA mutants. Antimicrob Agents Chemother. 2000;44(11):3101–3106. doi: 10.1128/aac.44.11.3101-3106.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belova L, Tenson T, Xiong L, McNicholas PM, Mankin AS. A novel site of antibiotic action in the ribosome: Interaction of evernimicin with the large ribosomal subunit. Proc Natl Acad Sci USA. 2001;98(7):3726–3731. doi: 10.1073/pnas.071527498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kofoed CB, Vester B. Interaction of avilamycin with ribosomes and resistance caused by mutations in 23S rRNA. Antimicrob Agents Chemother. 2002;46(11):3339–3342. doi: 10.1128/AAC.46.11.3339-3342.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mann PA, et al. EmtA, a rRNA methyltransferase conferring high-level evernimicin resistance. Mol Microbiol. 2001;41(6):1349–1356. doi: 10.1046/j.1365-2958.2001.02602.x. [DOI] [PubMed] [Google Scholar]
- 14.Treede I, et al. The avilamycin resistance determinants AviRa and AviRb methylate 23S rRNA at the guanosine 2535 base and the uridine 2479 ribose. Mol Microbiol. 2003;49(2):309–318. doi: 10.1046/j.1365-2958.2003.03558.x. [DOI] [PubMed] [Google Scholar]
- 15.Aarestrup FM, Jensen LB. Presence of variations in ribosomal protein L16 corresponding to susceptibility of enterococci to oligosaccharides (avilamycin and evernimicin) Antimicrob Agents Chemother. 2000;44(12):3425–3427. doi: 10.1128/aac.44.12.3425-3427.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zarazaga M, Tenorio C, Del Campo R, Ruiz-Larrea F, Torres C. Mutations in ribosomal protein L16 and in 23S rRNA in Enterococcus strains for which evernimicin MICs differ. Antimicrob Agents Chemother. 2002;46(11):3657–3659. doi: 10.1128/AAC.46.11.3657-3659.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adrian PV, et al. Mutations in ribosomal protein L16 conferring reduced susceptibility to evernimicin (SCH27899): Implications for mechanism of action. Antimicrob Agents Chemother. 2000;44(3):732–738. doi: 10.1128/aac.44.3.732-738.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McNicholas PM, et al. Effects of mutations in ribosomal protein L16 on susceptibility and accumulation of evernimicin. Antimicrob Agents Chemother. 2001;45(1):79–83. doi: 10.1128/AAC.45.1.79-83.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McNicholas PM, et al. Evernimicin binds exclusively to the 50S ribosomal subunit and inhibits translation in cell-free systems derived from both gram-positive and gram-negative bacteria. Antimicrob Agents Chemother. 2000;44(5):1121–1126. doi: 10.1128/aac.44.5.1121-1126.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolf H. Avilamycin, an inhibitor of the 30 S ribosomal subunits function. FEBS Lett. 1973;36(2):181–186. doi: 10.1016/0014-5793(73)80364-3. [DOI] [PubMed] [Google Scholar]
- 21.Mikolajka A, et al. Differential effects of thiopeptide and orthosomycin antibiotics on translational GTPases. Chem Biol. 2011;18(5):589–600. doi: 10.1016/j.chembiol.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orelle C, et al. Tools for characterizing bacterial protein synthesis inhibitors. Antimicrob Agents Chemother. 2013;57(12):5994–6004. doi: 10.1128/AAC.01673-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nicolaou KC, et al. Total synthesis of everninomicin 13,384-1--Part 4: Explorations of methodology; stereocontrolled synthesis of 1,1′-disaccharides, 1,2-seleno migrations in carbohydrates, and solution- and solid-phase synthesis of 2-deoxy glycosides and orthoesters. Chemistry. 2000;6(17):3166–3185. doi: 10.1002/1521-3765(20000901)6:17<3166::aid-chem3166>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 24.Weitnauer G, et al. Novel avilamycin derivatives with improved polarity generated by targeted gene disruption. Chem Biol. 2004;11(10):1403–1411. doi: 10.1016/j.chembiol.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 25.Arenz S, et al. Drug sensing by the ribosome induces translational arrest via active site perturbation. Mol Cell. 2014;56(3):446–452. doi: 10.1016/j.molcel.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arenz S, Nguyen F, Beckmann R, Wilson DN. Cryo-EM structure of the tetracycline resistance protein TetM in complex with a translating ribosome at 3.9-Å resolution. Proc Natl Acad Sci USA. 2015;112(17):5401–5406. doi: 10.1073/pnas.1501775112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boll R, et al. The active conformation of avilamycin A is conferred by AviX12, a radical AdoMet enzyme. J Biol Chem. 2006;281(21):14756–14763. doi: 10.1074/jbc.M601508200. [DOI] [PubMed] [Google Scholar]
- 28.Ganguly AK. Ziracin, a novel oligosaccharide antibiotic. J Antibiot (Tokyo) 2000;53(10):1038–1044. doi: 10.7164/antibiotics.53.1038. [DOI] [PubMed] [Google Scholar]
- 29.Sohmen D, et al. Structure of the Bacillus subtilis 70S ribosome reveals the basis for species-specific stalling. Nat Commun. 2015;6:6941. doi: 10.1038/ncomms7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eyal Z, et al. Structural insights into species-specific features of the ribosome from the pathogen Staphylococcus aureus. Proc Natl Acad Sci USA. 2015;112(43):E5805–E5814. doi: 10.1073/pnas.1517952112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weitnauer G, et al. An ATP-binding cassette transporter and two rRNA methyltransferases are involved in resistance to avilamycin in the producer organism Streptomyces viridochromogenes Tü57. Antimicrob Agents Chemother. 2001;45(3):690–695. doi: 10.1128/AAC.45.3.690-695.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu M, Douthwaite S. Resistance to the macrolide antibiotic tylosin is conferred by single methylations at 23S rRNA nucleotides G748 and A2058 acting in synergy. Proc Natl Acad Sci USA. 2002;99(23):14658–14663. doi: 10.1073/pnas.232580599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sprink T, et al. Structures of ribosome-bound initiation factor 2 reveal the mechanism of subunit association. Sci Adv. 2016;2(3):e1501502. doi: 10.1126/sciadv.1501502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simonetti A, et al. Structure of the 30S translation initiation complex. Nature. 2008;455(7211):416–420. doi: 10.1038/nature07192. [DOI] [PubMed] [Google Scholar]
- 35.Simonetti A, et al. Involvement of protein IF2 N domain in ribosomal subunit joining revealed from architecture and function of the full-length initiation factor. Proc Natl Acad Sci USA. 2013;110(39):15656–15661. doi: 10.1073/pnas.1309578110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmeing TM, et al. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326(5953):688–694. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fischer N, et al. Structure of the E. coli ribosome-EF-Tu complex at <3 A resolution by C-corrected cryo-EM. Nature. 2015;520:567–570. doi: 10.1038/nature14275. [DOI] [PubMed] [Google Scholar]
- 38.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313(5795):1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 39.Blanchard SC, Gonzalez RL, Kim HD, Chu S, Puglisi JD. tRNA selection and kinetic proofreading in translation. Nat Struct Mol Biol. 2004;11(10):1008–1014. doi: 10.1038/nsmb831. [DOI] [PubMed] [Google Scholar]
- 40.Geggier P, et al. Conformational sampling of aminoacyl-tRNA during selection on the bacterial ribosome. J Mol Biol. 2010;399(4):576–595. doi: 10.1016/j.jmb.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Juette MF, et al. Single-molecule imaging of non-equilibrium molecular ensembles on the millisecond timescale. Nat Methods. 2016;13(4):341–344. doi: 10.1038/nmeth.3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McKinney SA, Joo C, Ha T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys J. 2006;91(5):1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong W, et al. Cryo-EM structure of the Plasmodium falciparum 80S ribosome bound to the anti-protozoan drug emetine. eLife. 2014;3:3. doi: 10.7554/eLife.03080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arenz S, et al. Molecular basis for erythromycin-dependent ribosome stalling during translation of the ErmBL leader peptide. Nat Commun. 2014;5:3501. doi: 10.1038/ncomms4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X, et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat Methods. 2013;10(6):584–590. doi: 10.1038/nmeth.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen JZ, Grigorieff N. SIGNATURE: A single-particle selection system for molecular electron microscopy. J Struct Biol. 2007;157(1):168–173. doi: 10.1016/j.jsb.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 47.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9(9):853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frank J, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116(1):190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 49.Fernández JJ, Luque D, Castón JR, Carrascosa JL. Sharpening high resolution information in single particle electron cryomicroscopy. J Struct Biol. 2008;164(1):170–175. doi: 10.1016/j.jsb.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 50.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11(1):63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 52.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 1):12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 55.Munro JB, Altman RB, O’Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell. 2007;25(4):505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schuette JC, et al. GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO J. 2009;28(6):755–765. doi: 10.1038/emboj.2009.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192(2):216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Becker T, et al. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature. 2012;482(7386):501–506. doi: 10.1038/nature10829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Loerke J, Giesebrecht J, Spahn CM. Multiparticle cryo-EM of ribosomes. Methods Enzymol. 2010;483:161–177. doi: 10.1016/S0076-6879(10)83008-3. [DOI] [PubMed] [Google Scholar]
- 60.Noeske J, et al. High-resolution structure of the Escherichia coli ribosome. Nat Struct Mol Biol. 2015;22(4):336–341. doi: 10.1038/nsmb.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Noeske J, et al. Synergy of streptogramin antibiotics occurs independently of their effects on translation. Antimicrob Agents Chemother. 2014;58(9):5269–5279. doi: 10.1128/AAC.03389-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G. A new understanding of the decoding principle on the ribosome. Nature. 2012;484(7393):256–259. doi: 10.1038/nature10913. [DOI] [PubMed] [Google Scholar]
- 63.Schüttelkopf AW, van Aalten DM. PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 8):1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 64.Dave R, Terry DS, Munro JB, Blanchard SC. Mitigating unwanted photophysical processes for improved single-molecule fluorescence imaging. Biophys J. 2009;96(6):2371–2381. doi: 10.1016/j.bpj.2008.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Munro JB, Altman RB, Tung CS, Sanbonmatsu KY, Blanchard SC. A fast dynamic mode of the EF-G-bound ribosome. EMBO J. 2010;29(4):770–781. doi: 10.1038/emboj.2009.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Juette MF, et al. The bright future of single-molecule fluorescence imaging. Curr Opin Chem Biol. 2014;20:103–111. doi: 10.1016/j.cbpa.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zheng Q, et al. Ultra-stable organic fluorophores for single-molecule research. Chem Soc Rev. 2014;43(4):1044–1056. doi: 10.1039/c3cs60237k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Akyuz N, et al. Transport domain unlocking sets the uptake rate of an aspartate transporter. Nature. 2015;518(7537):68–73. doi: 10.1038/nature14158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Starosta AL, et al. Translational stalling at polyproline stretches is modulated by the sequence context upstream of the stall site. Nucleic Acids Res. 2014;42(16):10711–10719. doi: 10.1093/nar/gku768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Seefeldt AC, et al. Structure of the mammalian antimicrobial peptide Bac7(1-16) bound within the exit tunnel of a bacterial ribosome. Nucleic Acids Res. 2016;44(5):2429–2438. doi: 10.1093/nar/gkv1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.