Significance
The process of trabeculation is central to heart development and maturation, as it allows the increase in muscle mass before the formation of coronaries. This complex process involves a number of morphological changes in a subset of cardiomyocytes, resulting in their delamination from the compact layer. As cardiomyocytes delaminate, they must also remain attached to the compact layer. We identified Erb-b2 receptor tyrosine kinase 2-mediated relocalization of N-cadherin (Cdh2) as a mechanism underlying the formation of cell–cell junctions between trabecular and compact layer cardiomyocytes. Interestingly, we found that blood flow and heart contraction are also essential for the localization of Cdh2 molecules. These studies further our understanding of the complex cell biological processes underlying the maturation of the vertebrate heart.
Keywords: N-cadherin, trabeculation, heart development, Erbb2 signaling, Cdh2-EGFP
Abstract
During cardiac trabeculation, cardiomyocytes delaminate from the outermost (compact) layer to form complex muscular structures known as trabeculae. As these cardiomyocytes delaminate, the remodeling of adhesion junctions must be tightly coordinated so cells can extrude from the compact layer while remaining in tight contact with their neighbors. In this study, we examined the distribution of N-cadherin (Cdh2) during cardiac trabeculation in zebrafish. By analyzing the localization of a Cdh2-EGFP fusion protein expressed under the control of the zebrafish cdh2 promoter, we initially observed Cdh2-EGFP expression along the lateral sides of embryonic cardiomyocytes, in an evenly distributed pattern, and with the occasional appearance of punctae. Within a few hours, Cdh2-EGFP distribution on the lateral sides of cardiomyocytes evolves into a clear punctate pattern as Cdh2-EGFP molecules outside the punctae cluster to increase the size of these aggregates. In addition, Cdh2-EGFP molecules also appear on the basal side of cardiomyocytes that remain in the compact layer. Delaminating cardiomyocytes accumulate Cdh2-EGFP on the surface facing the basal side of compact layer cardiomyocytes, thereby allowing tight adhesion between these layers. Importantly, we find that blood flow/cardiac contractility is required for the transition from an even distribution of Cdh2-EGFP to the formation of punctae. Furthermore, using time-lapse imaging of beating hearts in conjunction with a Cdh2 tandem fluorescent protein timer transgenic line, we observed that Cdh2-EGFP molecules appear to move from the lateral to the basal side of cardiomyocytes along the cell membrane, and that Erb-b2 receptor tyrosine kinase 2 (Erbb2) function is required for this relocalization.
To maximize its function, the heart undergoes a series of morphological changes during development, with trabeculation being one of the main processes (1–5). Trabeculae initially appear as myocardial ridges in the outer curvature of the ventricle, and they are important for cardiac function, as evidenced by studies of hypo- and hypertrabeculation models (3, 6–11). Previous studies have shown that Erbb2 signaling is essential for trabeculation (3, 6, 7, 10, 11). Moreover, disturbing blood flow or cardiac contractility in the ventricle perturbs trabeculation (4, 12, 13), suggesting an important role for mechanical forces in this process. Our understanding of the cellular mechanisms regulating trabeculation remains fairly limited. Cardiomyocytes in the early heart tube show an epithelium-like morphology (3, 14). During trabeculation, some cardiomyocytes delaminate and enter the trabecular layer, where they join other trabecular cardiomyocytes to form ridge-like structures (3, 15). Previous studies have shown that the most proximal cardiomyocytes in the trabecular layer remain tightly attached to the basal side of compact layer cardiomyocytes (3, 15). Some of the key molecules involved in cell–cell adhesion belong to the cadherin family, and they also play crucial roles in epithelial cell morphology and behavior, including cell migration (16–18). Studies of epithelial cells in culture have shown that changes in cell morphology are accompanied by the extensive remodeling of cell–cell junctions; for example, cell–cell adhesion can be remodeled by regulating the expression and/or endocytic recycling of cadherin (19–23). Mechanical tension plays a crucial role in regulating the size of cell–cell junctions, and local tension generated by the actomyosin network may also modulate cell–cell junction remodeling (24–26). These forces can activate vinculin and stabilize E-cadherin/VE-cadherin-mediated cell–cell adhesion (27–29). Additional studies have shown that cadherin punctae formed by the clustering of E-cadherin along cell–cell boundaries can increase in size during the maturation of cell–cell junctions, and that they are important for cell–cell adhesion and force transmission in vivo (26, 30). How E-cadherin molecules come together and form these punctae has been under intense investigation (31): Some studies have suggested that E-cadherin molecules are able to move along lateral membranes (32, 33), and one of the common themes emerging is the importance of the cortical actin cytoskeleton in their formation (26, 31).
Cdh2 (N-cadherin) adhesive junctions play an important role in mechanical coupling between cardiomyocytes (34, 35). Despite its potential importance, no detailed analysis has been carried out on the organization of Cdh2 during heart development. As it is amenable to high-resolution imaging during its formation and maturation, the zebrafish heart is a good model to study the reorganization of Cdh2 under the influence of mechanical forces and during cardiomyocyte delamination (15, 36). Here we show that Cdh2 molecules first appear on the lateral sides of cardiomyocytes, where they cluster to form punctae. As trabeculation is initiated, Cdh2 molecules appear on the basal side of compact layer cardiomyocytes, possibly by moving from the lateral sides along the cell membrane. We also show that blood flow and/or cardiac contractility play an essential role in the distribution of Cdh2, although one distinct from the role played by Erbb2 signaling, a critical regulator of cardiac trabeculation.
Results
Dynamic Distribution of Cdh2-EGFP During Cardiac Trabeculation.
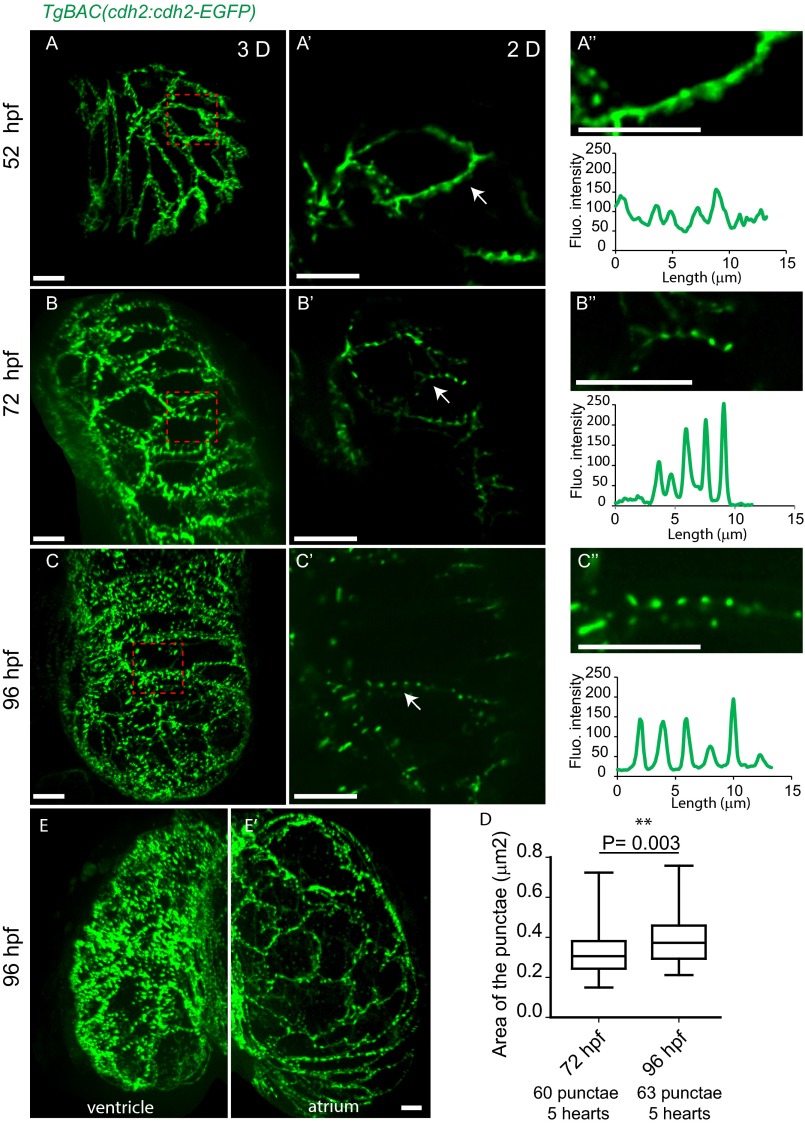
To investigate the question of cardiomyocyte adhesion during cardiac trabeculation in zebrafish, we focused on Cdh2, a highly expressed adhesion protein in cardiomyocytes. To analyze the localization of Cdh2, we used a BAC transgenic line expressing a Cdh2-EGFP fusion protein under the control of the zebrafish cdh2 promoter (37). Cdh2 localization in the heart was analyzed at three different developmental stages: 52 hours post fertilization (hpf) (before trabeculation has begun), 72 hpf (after the onset of trabeculation), and 96 hpf (when pronounced trabeculae have formed) (Fig. 1 A–C). At 52 hpf, Cdh2-EGFP molecules appear to be fairly evenly distributed along the lateral sides of cardiomyocytes with the occasional appearance of punctae (Fig. 1 A–A′′ and D and Movie S1). At 72 and 96 hpf, Cdh2-EGFP punctae have become more pronounced and are present all along the lateral sides (Fig. 1 B–B′′ and C–C′′ and Movies S2 and S3). At 96 hpf, Cdh2-EGFP molecules have also started to appear on the basal side of compact layer cardiomyocytes (Fig. 1 C–C′′ and Movie S3). The cartoons in Fig. 1 depict the localization of Cdh2-EGFP at different stages of heart development. We next examined in more detail the transition from even to punctate distribution of Cdh2-EGFP along the lateral sides of cardiomyocytes (Fig. S1 A–A′′ and C–C′′), and measurements of fluorescence intensity show the progression into a clear punctate pattern (Fig. S1 A′′–C′′). Cross-sectional area measurements of Cdh2-EGFP punctae also revealed that their size at 96 hpf is higher compared with their size at 72 hpf (Fig. S1D and Fig. S2), suggesting that Cdh2-EGFP molecules outside the punctae have joined them, causing an increase in size. A similar punctate distribution of Cdh2-EGFP molecules could be observed in atrial cardiomyocytes at 96 hpf, whereas at this stage, no Cdh2-EGFP signal could be observed on their basal surface (Fig. S1 E and E′ and Movie S4).
Fig. 1.
Dynamic distribution of Cdh2-EGFP during cardiac trabeculation. (A–C) 2D and (A′–C′) 3D views of TgBAC(cdh2:cdh2-EGFP) expression during zebrafish heart development, focusing on the ventricular chamber. (A′′–C′′) 3D views of single cardiomyocytes (see also Movies S1–S3); area outlined by yellow dashed lines represents the basal surface of cardiomyocytes and area between yellow and red dashed lines represents their lateral surface; illustration of Cdh2-EGFP distribution in cardiomyocytes. At 52 hpf, Cdh2-EGFP expression appears evenly distributed; by 72 hpf, punctae appear; and by 96 hpf, Cdh2-EGFP expression appears on the basal side of cardiomyocytes. (D–D′′ and E–E′′) TgBAC(cdh2:cdh2-EGFP);Tg(myl7:MKATE-CAAX) expression in developing hearts. Cdh2-EGFP is initially localized at the lateral side of cardiomyocytes (D); it is then also observed on the basal side of cardiomyocytes (D′); and after cardiomyocytes delaminate, it can be observed between compact layer cardiomyocytes and adjacent trabecular cardiomyocytes (D′′); white arrows point to Cdh2-EGFP expression (cartoon representation is shown below). (E–E′′) Redistribution of Cdh2-EGFP to the surface of compact layer cardiomyocytes directly adjacent to trabecular cardiomyocytes. Arrows indicate the apparent movement of Cdh2-EGFP (Movie S5). (Scale bars, 10 μm in A–C, A′–C′, A′′–C′′, and D–D′′; 5 μm in E′′.)
Fig. S1.
Localization of Cdh2-EGFP expression during heart development. (A–A′′) At 52 hpf, Cdh2-EGFP expression is evenly distributed along the lateral sides of cardiomyocytes with the occasional appearance of punctae. (B–B′′ and C–C′′) punctate pattern of Cdh2-EGFP expression at 72 and 96 hpf (lateral views). (A′–C′) Zoomed images of the regions outlined in the red dashed boxes in A–C. (A′′–C′′) Zoomed images of the regions indicated by the white arrows in A′–C′. (D) Increase in the area of Cdh2-EGFP punctae. (E and E′) As in the ventricle (E), cardiomyocytes in the atrium (E′) show lateral and punctate distribution of Cdh2-EGFP; however, Cdh2-EGFP is not observed on the basal surface of atrial cardiomyocytes at this stage (Movie S4). (Scale bars: 10 µm.)
Fig. S2.
Cdh2-EGFP puncta area measurements. (A) Single Cdh2-EGFP puncta, with a and b depicting major and minor axes. (B) Illustration of Cdh2-EGFP puncta. (C) 3D intensity distribution of the Cdh2-EGFP puncta from A. (D) Full-width and half-maximum major and minor axes of the Cdh2-EGFP puncta shown in A (using Fiji software). From these measurements, we calculated the cross-sectional area of Cdh2-EGFP punctae (Area = πab/4).
We also examined in more detail the localization of Cdh2-EGFP molecules as cardiomyocytes start to delaminate to form the trabecular layer. With the help of high-resolution confocal imaging, we observed that although Cdh2-EGFP molecules are initially localized at cell–cell junctions of cardiomyocytes (Fig. 1D, white arrows), by 78 hpf, they start to appear on the basal side of compact layer cardiomyocytes (Fig. 1D′, white arrows). As cardiomyocyte delamination proceeds, Cdh2-EGFP molecules begin to appear all around the surface of trabeculating cardiomyocytes, including along the surface adjacent to the basal side of compact layer cardiomyocytes (Fig. 1 D′′ and E–E′′, Fig. S3, and Movie S5). To confirm these observations, we generated mosaic hearts by transplanting Tg(myl7:cdh2-tdTomato) cells at midblastula stages into TgBAC(cdh2:cdh2-EGFP) host embryos of the same age. Analyzing such chimeric heart revealed the close apposition of Cdh2-tdTomato molecules on trabecular cardiomyocytes and Cdh2-EGFP molecules on adjacent compact layer cardiomyocytes (Fig. S4).
Fig. S3.
Localization of Cdh2-EGFP during trabeculation. (A–A′′) 2D picture of three different hearts, showing the localization of Cdh2-EGFP molecules during trabeculation. (B) 3D reconstructed images of cardiomyocytes from A. Red, blue, and green colors are used to represent compact cardiomyocytes, trabecular cardiomyocyte (pseudocoloring), and Cdh2-EGFP molecules, respectively. (Scale bars: 10 µm.)
Fig. S4.
Localization of Cdh2-EGFP during trabeculation. (A–A′′) 3D projection views of Tg(cmlc2:cdh2-tdTomato) donor-derived cardiomyocytes in a TgBAC(cdh2:cdh2-EGFP) host heart. (C–C′′) Zoomed images of the regions outlined by the white box (B–B′′); white arrow points to Cdh2-EGFP between the trabecular and compact cardiomyocytes (C′′). (Scale bars: 10 µm.)
Potential Movement of Cdh2-EGFP During Cardiac Trabeculation.
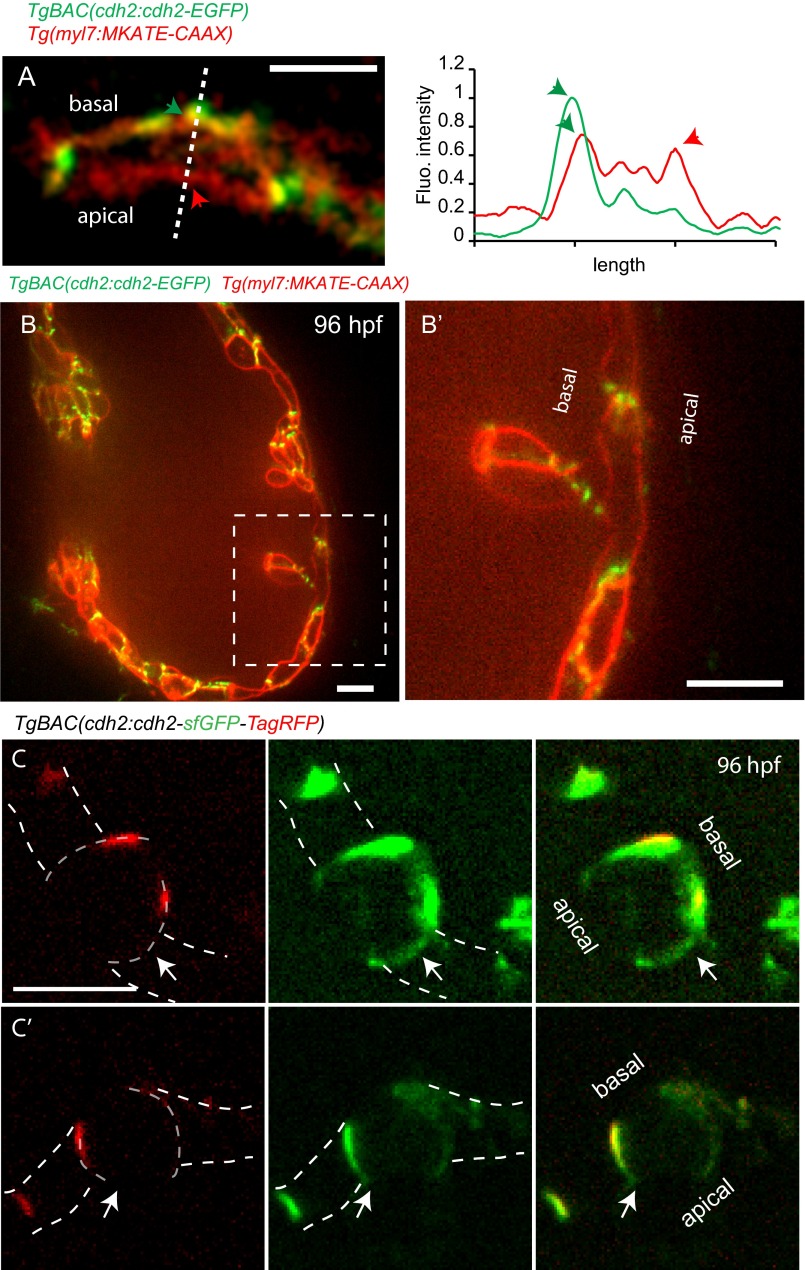
To investigate how Cdh2 becomes localized to the basal side of compact layer cardiomyocytes at the onset of trabeculation, we considered two different models: Cdh2 recruitment directly to the basal side of cardiomyocytes through endocytic vesicles or from newly generated molecules, and Cdh2 movement from the lateral to the basal side along the cell membrane. High-resolution imaging of cardiomyocytes in vivo did not reveal a significant amount of Cdh2-EGFP in their cytoplasm, suggesting an endocytic vesicle-independent recruitment of Cdh2-EGFP to their basal side (Fig. 2 A–A′′ and Fig. S5 B and B′). The first signs of cardiac trabeculation appear around 60 hpf and become more pronounced by 80 hpf (3, 4). To capture trabeculation processes in real time, we acquired time-lapse movies of beating hearts starting at 76 hpf. For this analysis, we imaged Tg(myl7:MKATE-CAAX);TgBAC(cdh2:cdh2-EGFP) embryos in which the cardiomyocyte membrane is labeled by MKATE, and Cdh2 by EGFP (Movie S6). From these data, it appears that Cdh2-EGFP may be moving from the lateral to the basal side of cardiomyocytes, along the cell membrane (Fig. 2 A–A′′, Top). Line scan quantification of Cdh2-EGFP signal in the cytoplasm and basal membranes of cardiomyocytes at several points indicated that the Cdh2-EGFP signal was present only in cell membranes (Fig. 2 A–A′′, Bottom and Fig. S5A), further suggesting that Cdh2-EGFP molecules move from the lateral to the basal side of cardiomyocytes along cell membranes, and not through endocytic vesicles. To examine the movement of Cdh2 molecules during trabeculation more closely, we used a transgenic line in which Cdh2 is tagged with a tandem fluorescent timer [tFT; TgBAC(cdh2:cdh2-sfGFP-TagRFP)] (37); this tFT consists of superfolder GFP (sfGFP), which takes 5 min to fold, and tag red fluorescent protein (TagRFP), which takes 100 min to fold (38). Use of this transgenic line allowed us to calculate the intensity ratio of sfGFP and tagRFP (37, 39, 40) to determine the age of Cdh2 molecules present in various parts of the cardiomyocytes. We analyzed the expression of the tFT reporter in cardiomyocytes in the compact layer either in the process of delaminating or after delamination. sfGFP and tagRFP expression could be observed simultaneously in all these cardiomyocytes (Fig. 2 B–B′′ and D–D′′, white arrows), indicating the presence of aged Cdh2 and not a rapid turnover. Interestingly, in delaminating cardiomyocytes, tagRFP expression was observed mostly at the basolateral side (Fig. 2 C′ and E and Fig. S5), whereas sfGFP was observed all along the lateral sides (Fig. 2 C and E and Fig. S5), suggesting that Cdh2 molecules move toward the basal side. The tFT intensity ratio in the basolateral side of delaminating cardiomyocytes was higher than that on the apical–lateral side (Fig. 2 E and F), further suggesting that Cdh2 molecules in the basolateral regions are older than those in the apical–lateral regions. Taken together, these data suggest that at these stages, Cdh2 molecules are first delivered to the apical–lateral regions and then move to the basal side of cardiomyocytes.
Fig. 2.
Potential movement of Cdh2-EGFP during cardiac trabeculation. (A–A′′) Spinning disk confocal images of TgBAC(cdh2:cdh2-EGFP);Tg(myl7:MKATE-CAAX) hearts. Cdh2-EGFP appears to relocalize from cell–cell junctions to the basal region of cardiomyocytes, as seen in the single-plane images at different points and depicted in the illustrations here (single-plane images taken from Movie S6). (Bottom) Line scan quantification of Cdh2-EGFP signal in the cytoplasm (red) and basal membrane (green) of cardiomyocytes. (B–D′′) Spinning disk confocal images of TgBAC(cdh2:cdh2-sfGFP-TagRFP);Tg(myl7:nlDsRedExpress) hearts. (B–B′′) Compact layer cardiomyocytes, (C–C′′) delaminating cardiomyocyte, and (D–D′′): delaminated cardiomyocyte; dotted line outlines cardiomyocytes, and the intense red signal in the cytoplasm marks the nucleus. cdh2:Cdh2-tFT expression analysis suggests Cdh2 recruitment to the apical–lateral region of compact layer cardiomyocytes; red arrows in C–C′′ point to an apical–lateral region with newly recruited Cdh2 displaying sfGFP signal without TagRFP signal; white arrows in B–B′′ and D–D′′ point to a region with aged Cdh2 displaying both sfGFP and TagRFP simultaneously. (E) Representative fluorescence intensity distribution of TagRFP and sfGFP in the region depicted by the dotted line in C′′. (F) Scatterplot showing quantification of Cdh2-tFT intensity ratio on the basolateral (yellow arrow in C′′ and G) and apical–lateral (red arrow in C′′ and G) sides of compact layer cardiomyocytes. (G) Diagram depicting hypothetical Cdh2 recruitment to the basal side of the cardiomyocyte plasma membrane from the apical end of cell–cell junctions; the two colors are physically separated for easier visualization. (Scale bars, 5 μm.)
Fig. S5.
Cdh2-EGFP molecules appear to move to the basal region of compact layer cardiomyocytes along the cell membrane. Spinning disk confocal images of a TgBAC(cdh2:cdh2-EGFP);Tg(myl7:MKATE-CAAX) heart. (A) Picture taken from time-lapse Movie S6; line scan quantification of Cdh2-EGFP and MKATE-CAAX; green and red arrows point to the basal and apical sides, respectively. (B′) Zoomed image of the region outlined by white dashed box. (B) Cdh2-EGFP is observed mainly on the cardiomyocyte’s cell membrane, and not in the cytoplasm. (C and C′) Spinning disk confocal images of TgBAC(cdh2:cdh2-sfGFP-TagRFP) cardiomyocytes. cdh2:Cdh2-tFT analysis indicates Cdh2 recruitment to the apical–lateral region of cardiomyocytes; white arrows point to a region with newly recruited Cdh2 displaying sfGFP signal without TagRFP signal. (Scale bar: 10 µm.)
Cardiac Contraction Is Required for Cdh2-EGFP Clustering.
Another factor involved in the organization of cadherin molecules is mechanical force (32, 41). Our initial studies revealed the progressive clustering of evenly distributed Cdh2-EGFP along the lateral sides of cardiomyocytes during larval stages. We hypothesized that the increasing mechanical force of the developing heart stimulates the clustering of evenly distributed Cdh2-EGFP into punctae. To test this hypothesis, we inhibited heart contractions by using a tnnt2 morpholino (42) and then imaged Cdh2-EGFP localization in cardiomyocytes. Surprisingly, in tnnt2 morphant hearts, Cdh2-EGFP punctae failed to form, and Cdh2-EGFP molecules remained evenly distributed at cell–cell junctions (Fig. 3B). To examine whether mechanical forces are needed to maintain Cdh2-EGFP localization in punctae, we stopped the beating hearts between 72 and 96 hpf (by 72 hpf, a clear punctate pattern is formed), using 20 mM 2,3-butanedione monoxime (BDM), a myosin inhibitor. Interestingly, in the absence of mechanical forces, Cdh2-EGFP punctae that were already formed at 72 hpf turned into evenly distributed Cdh2-EGFP by 96 hpf (Fig. 3C). These observations suggest that mechanical forces are essential for both the formation and maintenance of Cdh2-EGFP punctae. To further analyze the involvement of mechanical forces in the clustering of Cdh2-EGFP molecules, we carried out time-lapse imaging in the presence and after removal of BDM. After BDM treatment for 100 min, Cdh2-EGFP punctae turned into evenly distributed Cdh2-EGFP (Fig. 3 D–D′′, Fig. S6 A–A′′, and Movies S7 and S8). To determine whether the effect of BDM on Cdh2-EGFP distribution was transient or permanent, we treated the larvae with BDM for 5 h and then washed out the BDM to restart the heart. Interestingly, as the larval heart started beating again, Cdh2-EGFP molecules began reclustering (Fig. 3 E–E′′, Fig. S6 B–B′′, and Movie S9). One of the main cytoskeletal components attached to cadherin molecules is actin. With spinning disk confocal imaging of Tg(myl7:LIFEACT-GFP);Tg(myl7:cdh2-tdTomato) hearts, we found that actin bundles were strongly attached to Cdh2-tdTomato punctae (Fig. 3 F–H and Fig. S7). We hypothesized that the strong interactions between the actin bundles and Cdh2 molecules limited their dynamic motion inside the punctae. To test this hypothesis, we used fluorescence recovery after photobleaching (FRAP) experiments to analyze the dynamics of Cdh2-EGFP in punctae, as well as in regions of even distribution. Evenly distributed Cdh2-EGFP molecules at cardiomyocyte cell–cell junctions at 52 hpf were able to recover after photobleaching (Fig. 3I′′ and Fig. S8). However, Cdh2-EGFP punctae at 96 hpf did not reform after photobleaching, even though some of the evenly distributed Cdh2-EGFP molecules appeared to diffuse into the photobleached region (Fig. 3J′′ and Fig. S8). In contrast, in 96 hpf BDM-treated larvae, Cdh2-EGFP molecules were clearly able to diffuse after photobleaching (Fig. 3K′′). The data from these FRAP experiments suggest that evenly distributed Cdh2-EGFP molecules are able to move along cell–cell junctions. However, once clustered in punctae, their movements may become more restricted, possibly because of their tight connections to actin fibers.
Fig. 3.
Cardiac contraction is required for Cdh2-EGFP clustering. Spinning disk confocal images of TgBAC(cdh2:cdh2-EGFP) hearts. Localization of Cdh2-EGFP at 96 hpf in wild-type (A) and stopped hearts using tnnt2a morpholino (B) or 20 mM BDM (C). (D–D′′ and E–E′′) Single plane images at different points. (D–D′′) Cdh2-EGFP in punctae became evenly distributed upon treatment with BDM. (E–E′′) Removal of BDM resulted in the reclustering of Cdh2-EGFP into a punctate pattern. (F–H) Spinning disk confocal images of Tg(myl7:LIFEACT-GFP);Tg(myl7:cdh2-tdTomato) hearts focusing on the basal side of compact layer cardiomyocytes. Red arrows point to colocalization of actin bundle and Cdh2 punctae. On BDM treatment, the association between Cdh2-GFP punctae and actin bundles appears disrupted. (I–K) Dynamics of Cdh2-EGFP localization revealed by fluorescence recovery after photobleaching. (I′′) Recovery of Cdh2-EGFP signal in a 52 hpf heart after photobleaching. (J′′) After photobleaching of Cdh2-EGFP puncta at 96 hpf, it was not able to reform even though a diffuse Cdh2-EGFP signal is observed in the bleached region; zoomed images from bleached region. (K′′) Recovery of Cdh2-EGFP signal in a nonbeating heart at 96 hpf after photobleaching. (Scale bars, 10 μm in A–C, D′′–E′′, and F–H; 5 μm in I′′–K′′.)
Fig. S6.
Cardiac contraction is essential for Cdh2 clustering. 2D views of TgBAC(cdh2:cdh2-EGFP) hearts. Line scan quantification of Cdh2-EGFP signal in the region depicted by the red dotted line. (A–A′′) After treatment with BDM, Cdh2-EGFP expression became evenly distributed. (B–B′′) Removal of BDM resulted in the reclustering of Cdh2-EGFP expression into a punctate pattern. (Scale bars: 10 µm.)
Fig. S7.
Cardiac contractions are essential for localization of Cdh2 molecules and organization of actin fibers. 3D views of Tg(myl7:LIFEACT-GFP);Tg(myl7:cdh2-tdTomato) hearts. (A and B) Localization of Cdh2 molecules and organization of actin fibers at 52 and 96 hpf; white arrows point to the thick actin bundles. (C) Relocalization of Cdh2 molecules and reorganization of the actin fibers after BDM treatment. (D) Illustration of actin cytoskeleton reorganization under high mechanical force. (Scale bars: 10 μm.)
Fig. S8.
Recovery of Cdh2-EGFP localization after photobleaching. (A) Kymograph showing the recovery of Cdh2-EGFP molecules after FRAP at 52 hpf and 96 hpf, and BDM-treated larvae at 96 hpf (see Fig. 3 I–I′′ and K–K′′). (B) Line scan quantification of Cdh2-EGFP recovery from the kymograph, region depicted by the dotted line in A, green (quick and partial recovery), red (slow and partial recovery), and blue (slow and substantial recovery).
We also analyzed the organization of α-catenin in stopped hearts and found that α-catenin molecules became evenly distributed after 20 mM BDM treatment and remained colocalized with Cdh2 molecules (Fig. S9), suggesting the cadherin complex does not dissociate when the heart stops beating. At 96 hpf, the organization of the actin cytoskeleton in 20 mM BDM-treated cardiomyocytes resembled that seen in 52 hpf untreated cardiomyocytes (Fig. 3H). During larval development, the increase in cardiac contraction leads to increasing forces across cardiomyocyte junctions and may help the clustering of Cdh2 molecules and the bundling of actin fibers (Fig. 3 F–H and Fig. S7). Altogether, these data indicate that the distribution of Cdh2 molecules at cardiomyocyte cell–cell junctions is regulated by mechanical force.
Fig. S9.
Integrity of the cadherin–catenin complex is not affected when cardiac contractions are stopped using BDM. 3D views of Gt(ctnna-citrine);Tg(myl7:cdh2-tdTomato) hearts. Localization of Cdh2 tdTomato and Ctnna1-citrine, (A) control, and (B) after 20 mM BDM treatment. (Scale bar: 10 µm.)
Erbb2 Signaling Is Essential for Cdh2-EGFP Localization on the Basal Side of Cardiomyocytes.
Previous studies have shown that Erbb2 signaling is essential for cardiac trabeculation in mouse (6, 7) and zebrafish (3, 4). To determine whether Erbb2 signaling was necessary for the Cdh2 localization patterns described thus far, we used an Erbb2 inhibitor as well as erbb2 mutant zebrafish. In both erbb2 mutant and Erbb2 inhibitor-treated zebrafish, Cdh2-EGFP molecules failed to appear on the basal side of cardiomyocytes (Fig. 4 A′–A′′ and B′–B′′), suggesting Erbb2 signaling also plays a role in Cdh2 reorganization during heart development. Furthermore, using the tFT [TgBAC(cdh2:cdh2-sfGFP-TagRFP)] fish in combination with the Erbb2 inhibitor, we observed a higher value of Cdh2-tFT intensity ratio on the lateral sides of cardiomyocytes, suggesting that the hypothesized movements of Cdh2-EGFP from cell–cell junctions stop when Erbb2 signaling is inhibited (Fig. 4 D, E, and E′ and Fig. S10). Last, we measured the cross-sectional area of Cdh2-EGFP punctae in Erbb2 inhibitor-treated zebrafish and found no significant changes, suggesting Erbb2 signaling is not involved in the size regulation of Cdh2-EGFP punctae (Fig. 4G). Taken together, these data suggest Erbb2 signaling is essential for the appearance of Cdh2 molecules to the basal side of cardiomyocytes (Fig. 4H).
Fig. 4.
Erbb2 signaling is essential for Cdh2-EGFP localization on the basal side of cardiomyocytes. (A–A′′ and B–B′′) Spinning disk confocal images of TgBAC(cdh2:cdh2-EGFP) hearts. Localization of Cdh2-EGFP in 96 hpf control (wild-type), Erbb2 inhibitor-treated, and erbb2 mutant hearts is shown in the single 2D planes (A–A′′) and 3D reconstructed images (B–B′′). Illustrations of Cdh2-EGFP distribution are shown on the right. (C–C′′) Red, blue, and yellow rectangles shown in the zoomed images are from B–B′′. (D and E) tFT intensity ratio of Cdh2 molecules measured in the regions indicated by the white arrows (E–E′). (F) Schematic diagram depicting Cdh2 localization only at the cell–cell junctions in Erbb2 inhibitor-treated or erbb2 mutant larvae at 96 hpf. (G) Cross-sectional area of Cdh2-EGFP punctae at 96 hpf in DMSO control and Erbb2 inhibitor-treated larvae. (H) Illustration comparing Cdh2 organization in wild-type, stopped, and Erbb2 inhibitor-treated/mutant hearts. (Scale bar: 10 μm in A–A′′, B–B′′, and C–C′′; 5 μm in E and E′.)
Fig. S10.
Erbb2 signaling is required for Cdh2-EGFP distribution. (A) Red and green fluorescence sparsely distributed on the basal side of cardiomyocytes (control larva, left), indicating the dynamics of Cdh2-tFT. (B) Cdh2-tFT molecules are colocalized in cell–cell junctions, and thus show a yellow color in Erbb2 inhibitor-treated larva, suggesting the dynamics of Cdh2 are blocked. (Scale bars: 10 µm.)
Discussion
Several studies have shown that cadherin molecules play a central role in epithelial integrity and tissue morphogenesis (16, 17), and moreover, that dysregulation of cadherin is linked to a number of diseases, including cancer (23, 43, 44). The classical cadherins represent the main cadherin family present in cell–cell junctions and include E-cadherin, VE cadherin, and N-cadherin. Within this family, E-cadherin has been most widely studied. In epithelial cells, E-cadherin exhibits various patterns of localization that are mainly categorized into two forms: even distribution and punctae. These categories are based on the association of adherens junctions with actin filaments (17), and each pattern of E-cadherin localization has distinct functions in epithelial cells. Our studies show that during cardiac development in zebrafish, another cadherin, N-cadherin, also has distinct localization patterns. At embryonic stages, N-cadherin molecules are distributed mostly evenly along the lateral sides of cardiomyocytes, with the occasional appearance of punctae. During larval stages, these punctae become more distinct. Presumably, evenly distributed N-cadherin joins nascent clusters, thereby increasing the size of N-cadherin punctae. Studies with E-cadherin have revealed that the size, distribution, and dynamics of E-cadherin clusters play an essential role in local tensile force transmission and the strength of adhesive junctions (30, 45–47). As cadherin molecules mostly interact with the actin cytoskeleton, the clustering of N-cadherin molecules at larval stages likely recruits more actin fibers and helps organize the actin cytoskeleton in maturing cardiomyocytes. We have previously shown that the actin cytoskeleton in cardiomyocytes is substantially increased in larval stages compared with in embryonic stages (48). Taken together with the present data, these observations suggest that the clustering of N-cadherin molecules may function to increase the mechanical stability of cardiomyocytes and enable them to withstand the increasing mechanical force. However, how this process is controlled molecularly remains to be investigated.
EMT (epithelial to mesenchymal transition) is one of the main cellular events during development, as well as cancer progression, and the exact mechanisms of EMT in vivo remain unclear. Studies in epithelial cells have shown that during EMT, they lose apicobasal polarity, and E-cadherin disassociates and is subsequently down-regulated (23, 43). To prevent the complete extrusion of cells during their delamination from an epithelium, they need to make strong cell–cell attachments with the epithelial sheet. Our study reveals that N-cadherin molecules are able to localize on the basal side of compact layer cardiomyocytes, where they can interact with cadherins on the surface of delaminating cardiomyocytes, and thereby prevent their complete extrusion. We show that during trabeculation, N-cadherin appears to move to the basal side of cardiomyocytes. This localization of N-cadherin requires Erbb2 signaling, and our observations with the protein timer indicate that newly synthesized N-cadherin molecules are moving from the lateral to the basal surface of cardiomyocytes. It will be important to investigate exactly how N-cadherin molecules undergo this movement, as well as how they are excluded from the apical side.
Another factor involved in the organization of cadherin molecules is mechanical force (24). Abnormal heart beats, a common symptom of cardiac disease, lead to changes in the contractile forces of the heart. Other studies have also shown that cadherin clustering is essential to maintaining the mechanical integrity of cells (30, 45, 46). However, whether mechanical forces also play a role in clustering cadherin molecules through interaction with alpha-catenin and vinculin remains unclear. A previous study has shown that the size of adherens junctions is regulated by endothelial cell–cell tugging forces (41), suggesting these forces can regulate VE-cadherin and lead to an increase in its clustering. During heart development, cell–cell tugging forces between cardiomyocytes also increase as the heart contracts more vigorously. Presumably, tugging forces may help N-cadherin to cluster in cardiomyocytes, similar to what has been described for VE-cadherin in endothelial cells. Also, high mechanical forces allow vinculin to bind alpha-catenin, and vinculin is able to recruit more actin bundles (27). Thus, mechanical forces mediated clustering of N-cadherin, and interaction of alpha-catenin and vinculin may help the reorganization of the actin cytoskeleton during heart development (Fig. S7). Attenuation of actomyosin-mediated mechanical forces using Latrunculin A and Rock inhibitors may not affect the size of E-cadherin punctae (46). However, our studies indicate that actomyosin-mediated mechanical forces play an essential role in both the formation and maintenance of N-cadherin punctae in cardiomyocytes, revealing a potential difference between N-cadherin and E-cadherin. It is well known that reorganization of cadherin molecules in cell–cell junctions plays a key role in modulating cell morphology. Thus, mechanical forces, via their effect on N-cadherin distribution, might influence cardiomyocyte hypertrophy and hyperplasia. How mechanical forces modulate the reorganization of N-cadherin molecules is another important area for future studies.
Materials and Methods
Zebrafish.
All husbandry and experiments involving zebrafish (Danio rerio) were carried out according to protocols approved by local authorities. Transgenic zebrafish lines used in this study include TgBAC(cdh2:cdh2-EGFP,crybb1:ECFP)zf517 (37), Tg(myl7:MKATE-CAAX)Sd11 (49), TgBAC(cdh2:cdh2-sfGFP-TagRFP, crybb1:ECFP)zf516 (37), Tg(-0.8myl7:nlsDsRedExpress)hsc4 (50), Tg(myl7:cdh2-tdTomato)bns78, Gt(Ctnna-citrine)ct3a (51), and Tg(myl7:LIFEACT-GFP)s974 (48).
In Vivo Fluorescence Imaging and Image Processing of Stopped Hearts.
Zebrafish embryos and larvae were mounted in 1% low-melting agarose containing 0.2% (wt/vol) tricaine and imaged using a spinning disk confocal microscope (CSU-X1 Yokogawa; Zeiss) or confocal microscope (LSM 780; Zeiss) equipped with microscope objectives 40× (1.1 NA), 40× (1 NA), and 63× (1.3 NA), and two Hamamatsu ORCA-Flash4.0 V2 Digital CMOS cameras. Imaris (Bitplane) was used for 3D image processing, Fiji was used for 2D image processing, and images were prepared with the help of Adobe Illustrator and Photoshop.
Time-Lapse Imaging of Beating Hearts.
Zebrafish embryos and larvae were mounted in 1% low-melting agarose containing 0.01% (wt/vol) tricaine. The fish were maintained at 28 °C in a temperature-controlled chamber. Time-lapse movies of the larval hearts were taken using a spinning disk confocal microscope (CSU-X1 Yokogawa; Zeiss) using a 63× (1.3 NA) objective. Movies were collected at 4 Z-position 0.2 µm apart at each developmental point, and the time between these developmental points was 5–10 min for 4–5 h. The data were aligned and analyzed in Fiji.
Morpholino Oligonucleotides.
tnnt2a morpholino (5′-CATGTTTGCTCTGATCTGACACGCA-3′; 0.25 ng) was injected into early-stage embryos to prevent cardiac contractility (42).
Chemical Treatments.
BDM (20 mM; Sigma) was used to stop the zebrafish larval heart. We added 5–10 µM of the Erbb2 inhibitor PD168393 (Calbiochem) at 72 hpf and imaged the larvae at 96 hpf.
FRAP Experiments.
FRAP on Cdh2-EGFP within the heart was performed on a confocal microscope (LSM 780; Zeiss) equipped with a 40× 1.00 NA objective, using a UV (405 nm) laser.
Cell Transplantation.
Cells were taken from Tg(myl7:cdh2-tdTomato) donor embryos at midblastula stages and transplanted into TgBAC(cdh2:cdh2-EGFP,crybb1:ECFP) host embryos of the same age. Hosts were then imaged at 96 hpf.
Supplementary Material
Acknowledgments
We thank Darren Gilmour for providing the cadherin transgenic lines; Arica Beisaw, Yu Hsuan Carol Yang, Michelle Collins, and Rashmi Priya for discussions and/or critical reading of the manuscript; Radhan Ramadass for help with confocal microscopy, as well as feedback on the manuscript; and Sharon Meaney-Gardian, Sabine Fischer, Carmen Büttner, Nana Fukuda, Sophie Mucenieks, and Marianne Ploch for excellent assistance.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1606385113/-/DCSupplemental.
References
- 1.Sedmera D, et al. Functional and morphological evidence for a ventricular conduction system in zebrafish and Xenopus hearts. Am J Physiol Heart Circ Physiol. 2003;284(4):H1152–H1160. doi: 10.1152/ajpheart.00870.2002. [DOI] [PubMed] [Google Scholar]
- 2.Stankunas K, et al. Endocardial Brg1 represses ADAMTS1 to maintain the microenvironment for myocardial morphogenesis. Dev Cell. 2008;14(2):298–311. doi: 10.1016/j.devcel.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu J, et al. A dual role for ErbB2 signaling in cardiac trabeculation. Development. 2010;137(22):3867–3875. doi: 10.1242/dev.053736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peshkovsky C, Totong R, Yelon D. Dependence of cardiac trabeculation on neuregulin signaling and blood flow in zebrafish. Dev Dyn. 2011;240(2):446–456. doi: 10.1002/dvdy.22526. [DOI] [PubMed] [Google Scholar]
- 5.Staudt D, Stainier D. Uncovering the molecular and cellular mechanisms of heart development using the zebrafish. Annu Rev Genet. 2012;46:397–418. doi: 10.1146/annurev-genet-110711-155646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gassmann M, et al. Aberrant neural and cardiac development in mice lacking the ErbB4 neuregulin receptor. Nature. 1995;378(6555):390–394. doi: 10.1038/378390a0. [DOI] [PubMed] [Google Scholar]
- 7.Lee KF, et al. Requirement for neuregulin receptor erbB2 in neural and cardiac development. Nature. 1995;378(6555):394–398. doi: 10.1038/378394a0. [DOI] [PubMed] [Google Scholar]
- 8.Meyer D, Birchmeier C. Multiple essential functions of neuregulin in development. Nature. 1995;378(6555):386–390. doi: 10.1038/378386a0. [DOI] [PubMed] [Google Scholar]
- 9.Suri C, et al. Requisite role of angiopoietin-1, a ligand for the TIE2 receptor, during embryonic angiogenesis. Cell. 1996;87(7):1171–1180. doi: 10.1016/s0092-8674(00)81813-9. [DOI] [PubMed] [Google Scholar]
- 10.Morris JK, et al. Rescue of the cardiac defect in ErbB2 mutant mice reveals essential roles of ErbB2 in peripheral nervous system development. Neuron. 1999;23(2):273–283. doi: 10.1016/s0896-6273(00)80779-5. [DOI] [PubMed] [Google Scholar]
- 11.Tidcombe H, et al. Neural and mammary gland defects in ErbB4 knockout mice genetically rescued from embryonic lethality. Proc Natl Acad Sci USA. 2003;100(14):8281–8286. doi: 10.1073/pnas.1436402100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chi NC, et al. Genetic and physiologic dissection of the vertebrate cardiac conduction system. PLoS Biol. 2008;6(5):e109. doi: 10.1371/journal.pbio.0060109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samsa LA, et al. Cardiac contraction activates endocardial Notch signaling to modulate chamber maturation in zebrafish. Development. 2015;142(23):4080–4091. doi: 10.1242/dev.125724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trinh LA, Stainier DY. Fibronectin regulates epithelial organization during myocardial migration in zebrafish. Dev Cell. 2004;6(3):371–382. doi: 10.1016/s1534-5807(04)00063-2. [DOI] [PubMed] [Google Scholar]
- 15.Staudt DW, et al. High-resolution imaging of cardiomyocyte behavior reveals two distinct steps in ventricular trabeculation. Development. 2014;141(3):585–593. doi: 10.1242/dev.098632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris TJ, Tepass U. Adherens junctions: From molecules to morphogenesis. Nat Rev Mol Cell Biol. 2010;11(7):502–514. doi: 10.1038/nrm2927. [DOI] [PubMed] [Google Scholar]
- 17.Takeichi M. Dynamic contacts: Rearranging adherens junctions to drive epithelial remodelling. Nat Rev Mol Cell Biol. 2014;15(6):397–410. doi: 10.1038/nrm3802. [DOI] [PubMed] [Google Scholar]
- 18.Niessen CM, Leckband D, Yap AS. Tissue organization by cadherin adhesion molecules: Dynamic molecular and cellular mechanisms of morphogenetic regulation. Physiol Rev. 2011;91(2):691–731. doi: 10.1152/physrev.00004.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lochter A, et al. Matrix metalloproteinase stromelysin-1 triggers a cascade of molecular alterations that leads to stable epithelial-to-mesenchymal conversion and a premalignant phenotype in mammary epithelial cells. J Cell Biol. 1997;139(7):1861–1872. doi: 10.1083/jcb.139.7.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marambaud P, et al. A presenilin-1/gamma-secretase cleavage releases the E-cadherin intracellular domain and regulates disassembly of adherens junctions. EMBO J. 2002;21(8):1948–1956. doi: 10.1093/emboj/21.8.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maretzky T, et al. ADAM10 mediates E-cadherin shedding and regulates epithelial cell-cell adhesion, migration, and beta-catenin translocation. Proc Natl Acad Sci USA. 2005;102(26):9182–9187. doi: 10.1073/pnas.0500918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steinhusen U, et al. Cleavage and shedding of E-cadherin after induction of apoptosis. J Biol Chem. 2001;276(7):4972–4980. doi: 10.1074/jbc.M006102200. [DOI] [PubMed] [Google Scholar]
- 23.Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009;28(1-2):15–33. doi: 10.1007/s10555-008-9169-0. [DOI] [PubMed] [Google Scholar]
- 24.Lecuit T, Yap AS. E-cadherin junctions as active mechanical integrators in tissue dynamics. Nat Cell Biol. 2015;17(5):533–539. doi: 10.1038/ncb3136. [DOI] [PubMed] [Google Scholar]
- 25.Lecuit T, Lenne PF, Munro E. Force generation, transmission, and integration during cell and tissue morphogenesis. Annu Rev Cell Dev Biol. 2011;27:157–184. doi: 10.1146/annurev-cellbio-100109-104027. [DOI] [PubMed] [Google Scholar]
- 26.Maître JL, et al. Adhesion functions in cell sorting by mechanically coupling the cortices of adhering cells. Science. 2012;338(6104):253–256. doi: 10.1126/science.1225399. [DOI] [PubMed] [Google Scholar]
- 27.Yonemura S, Wada Y, Watanabe T, Nagafuchi A, Shibata M. alpha-Catenin as a tension transducer that induces adherens junction development. Nat Cell Biol. 2010;12(6):533–542. doi: 10.1038/ncb2055. [DOI] [PubMed] [Google Scholar]
- 28.Huveneers S, et al. Vinculin associates with endothelial VE-cadherin junctions to control force-dependent remodeling. J Cell Biol. 2012;196(5):641–652. doi: 10.1083/jcb.201108120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.le Duc Q, et al. Vinculin potentiates E-cadherin mechanosensing and is recruited to actin-anchored sites within adherens junctions in a myosin II-dependent manner. J Cell Biol. 2010;189(7):1107–1115. doi: 10.1083/jcb.201001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Truong Quang BA, Mani M, Markova O, Lecuit T, Lenne PF. Principles of E-cadherin supramolecular organization in vivo. Curr Biol. 2013;23(22):2197–2207. doi: 10.1016/j.cub.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 31.Yap AS, Gomez GA, Parton RG. Adherens Junctions Revisualized: Organizing Cadherins as Nanoassemblies. Dev Cell. 2015;35(1):12–20. doi: 10.1016/j.devcel.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 32.Kametani Y, Takeichi M. Basal-to-apical cadherin flow at cell junctions. Nat Cell Biol. 2007;9(1):92–98. doi: 10.1038/ncb1520. [DOI] [PubMed] [Google Scholar]
- 33.Wang YC, Khan Z, Kaschube M, Wieschaus EF. Differential positioning of adherens junctions is associated with initiation of epithelial folding. Nature. 2012;484(7394):390–393. doi: 10.1038/nature10938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferreira-Cornwell MC, et al. Remodeling the intercalated disc leads to cardiomyopathy in mice misexpressing cadherins in the heart. J Cell Sci. 2002;115(Pt 8):1623–1634. doi: 10.1242/jcs.115.8.1623. [DOI] [PubMed] [Google Scholar]
- 35.Bagatto B, Francl J, Liu B, Liu Q. Cadherin2 (N-cadherin) plays an essential role in zebrafish cardiovascular development. BMC Dev Biol. 2006;6:23. doi: 10.1186/1471-213X-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beis D, Stainier DY. In vivo cell biology: Following the zebrafish trend. Trends Cell Biol. 2006;16(2):105–112. doi: 10.1016/j.tcb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 37.Revenu C, et al. Quantitative cell polarity imaging defines leader-to-follower transitions during collective migration and the key role of microtubule-dependent adherens junction formation. Development. 2014;141(6):1282–1291. doi: 10.1242/dev.101675. [DOI] [PubMed] [Google Scholar]
- 38.Merzlyak EM, et al. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat Methods. 2007;4(7):555–557. doi: 10.1038/nmeth1062. [DOI] [PubMed] [Google Scholar]
- 39.Donà E, et al. Directional tissue migration through a self-generated chemokine gradient. Nature. 2013;503(7475):285–289. doi: 10.1038/nature12635. [DOI] [PubMed] [Google Scholar]
- 40.Khmelinskii A, et al. Tandem fluorescent protein timers for in vivo analysis of protein dynamics. Nat Biotechnol. 2012;30(7):708–714. doi: 10.1038/nbt.2281. [DOI] [PubMed] [Google Scholar]
- 41.Liu Z, et al. Mechanical tugging force regulates the size of cell-cell junctions. Proc Natl Acad Sci USA. 2010;107(22):9944–9949. doi: 10.1073/pnas.0914547107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sehnert AJ, et al. Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nat Genet. 2002;31(1):106–110. doi: 10.1038/ng875. [DOI] [PubMed] [Google Scholar]
- 43.Wheelock MJ, Shintani Y, Maeda M, Fukumoto Y, Johnson KR. Cadherin switching. J Cell Sci. 2008;121(Pt 6):727–735. doi: 10.1242/jcs.000455. [DOI] [PubMed] [Google Scholar]
- 44.El-Amraoui A, Petit C. Cadherins as targets for genetic diseases. Cold Spring Harb Perspect Biol. 2010;2(1):a003095. doi: 10.1101/cshperspect.a003095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin AC. Pulsation and stabilization: Contractile forces that underlie morphogenesis. Dev Biol. 2010;341(1):114–125. doi: 10.1016/j.ydbio.2009.10.031. [DOI] [PubMed] [Google Scholar]
- 46.Cavey M, Rauzi M, Lenne PF, Lecuit T. A two-tiered mechanism for stabilization and immobilization of E-cadherin. Nature. 2008;453(7196):751–756. doi: 10.1038/nature06953. [DOI] [PubMed] [Google Scholar]
- 47.Wu SK, et al. Cortical F-actin stabilization generates apical-lateral patterns of junctional contractility that integrate cells into epithelia. Nat Cell Biol. 2014;16(2):167–178. doi: 10.1038/ncb2900. [DOI] [PubMed] [Google Scholar]
- 48.Reischauer S, Arnaout R, Ramadass R, Stainier DY. Actin binding GFP allows 4D in vivo imaging of myofilament dynamics in the zebrafish heart and the identification of Erbb2 signaling as a remodeling factor of myofibril architecture. Circ Res. 2014;115(10):845–856. doi: 10.1161/CIRCRESAHA.115.304356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin YF, Swinburne I, Yelon D. Multiple influences of blood flow on cardiomyocyte hypertrophy in the embryonic zebrafish heart. Dev Biol. 2012;362(2):242–253. doi: 10.1016/j.ydbio.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takeuchi JK, et al. Chromatin remodelling complex dosage modulates transcription factor function in heart development. Nat Commun. 2011;2:187. doi: 10.1038/ncomms1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trinh A, et al. A versatile gene trap to visualize and interrogate the function of the vertebrate proteome. Genes Dev. 2011;25(21):2306–2320. doi: 10.1101/gad.174037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.