Significance
The mammalian target of rapamycin complex 1 (mTORC1) signaling pathway is an important facet of the immune system, including that it regulates T-cell differentiation and activation. Here, we report that type 2 phosphatidylinositol-5-phosphate 4-kinase gamma (protein, PI5P4Kγ; gene, PIP4K2C) plays a role in the regulation of the immune system by manipulating mTORC1 signaling. These results suggest that the SNP at the PIP4K2C locus (rs1678542) in human patients with autoimmunity might cause a decrease in PI5P4Kγ expression and thereby an increase in mTORC1 signaling. In addition, these results imply that inhibition of mTORC1 would be beneficial to these patients. These studies also suggest that agents that inhibit PIP4K2C function could be useful to enhance cancer immunotherapy.
Keywords: PIP4K2C, mTORC1, autoimmunity, PI5P4K, inflammation
Abstract
Type 2 phosphatidylinositol-5-phosphate 4-kinase (PI5P4K) converts phosphatidylinositol-5-phosphate to phosphatidylinositol-4,5-bisphosphate. Mammals have three enzymes PI5P4Kα, PI5P4Kβ, and PI5P4Kγ, and these enzymes have been implicated in metabolic control, growth control, and a variety of stress responses. Here, we show that mice with germline deletion of type 2 phosphatidylinositol-5-phosphate 4-kinase gamma (Pip4k2c), the gene encoding PI5P4Kγ, appear normal in regard to growth and viability but have increased inflammation and T-cell activation as they age. Immune cell infiltrates increased in Pip4k2c−/− mouse tissues. Also, there was an increase in proinflammatory cytokines, including IFNγ, interleukin 12, and interleukin 2 in plasma of Pip4k2c−/− mice. Pip4k2c−/− mice had an increase in T-helper-cell populations and a decrease in regulatory T-cell populations with increased proliferation of T cells. Interestingly, mammalian target of rapamycin complex 1 (mTORC1) signaling was hyperactivated in several tissues from Pip4k2c−/− mice and treating Pip4k2c−/− mice with rapamycin reduced the inflammatory phenotype, resulting in a decrease in mTORC1 signaling in tissues and a decrease in proinflammatory cytokines in plasma. These results indicate that PI5P4Kγ plays a role in the regulation of the immune system via mTORC1 signaling.
Type 2 phosphatidylinositol-5-phosphate 4-kinase (PI5P4K) converts phosphatidylinositol-5-phosphate to phosphatidylinositol-4,5-bisphosphate. Mammals have three genes, PIP4K2A, PIP4K2B, and PIP4K2C that encode the enzymes PI5P4Kα, PI5P4Kβ, and PI5P4Kγ, respectively.
All three isoforms are highly expressed in brain, whereas their relative expressions in other tissues vary. PI5P4Kα is highly expressed in spleen and the peripheral blood. PI5P4Kβ is highly expressed in muscle, whereas PI5P4Kγ is highly expressed in kidney (1). In kidney, PI5P4Kγ is mostly detected in the cortex and the outer medulla (1). All tissues examined appear to express at least one isoform of PI5P4K. At the cellular level, PI5P4Ks are found in several organelles, including plasma membrane, cytosol, nucleus, and vesicular compartments (2, 3). It is not simple to define unique compartmentalization of the individual enzymes because all three isoforms can homodimerize or heterodimerize with each other. At the sequence level, PI5P4Kα and PI5P4Kβ are more homologous to each other than either is to PI5P4Kγ. Also, PI5P4Kγ is only about 1% as active as the other isoforms, raising the possibility that its major role may be to localize or regulate the activities of PI5P4Kα and PI5P4Kβ.
Germline deletion of both alleles of Pip4k2a or Pip4k2b in mice results in healthy mice that live a normal life span. The Pip4k2b−/− mice have increased insulin sensitivity and are protected from obesity, insulin resistance, and type 2 diabetes when placed on a high-fat diet (4). Also, crossing the Pip4k2b−/− mice with Trp53−/− mice results in early embryonic lethality for the subset of embryos that are Pip4k2b−/−, Trp53−/−, indicating a synthetic lethality relationship between these genes. In contrast, Pip4k2a−/− mice do not exhibit any of the phenotypes observed in the Pip4k2b−/− mice (5). They are not protected from obesity or insulin resistance, they do not exhibit a synthetic lethality relationship with Trp53, and in all ways examined, they resemble wild-type mice (5). However, germline deletion of one allele of Pip4k2a in the context of germline Pip4k2b−/− causes enhancement of the phenotypes of the Pip4k2b−/− mice. Deletion of both alleles of Pip4k2a and Pip4k2b did not have any observable effect on embryonic development up until the time of birth, but resulted in perinatal lethality of all pups. These results indicate that these genes do not play a major role in embryonic development and that Pip4k2a provides a backup for Pip4k2b that becomes critical at the time of birth.
Whereas less is known about PI5P4Kγ, it has been linked to the mammalian target of rapamycin signaling complex (mTORC) and to cellular immunity. Mackey et al. argued that PI5P4Kγ was negatively regulated by mTORC1 through direct phosphorylation at serine 324 and serine 328 (6). On the other hand, it was reported that knocking out the only PI5P4K isoform in Drosophila resulted in lower body weight and that this correlated with decreased mTORC1 signaling (7). Of particular interest, multiple studies have shown a link between a SNP (rs1678542) in the human PIP4K2C locus and familial autoimmunity (8, 9). The immune system is a complex network that evolved to protect organisms from invasion of various microbes. Whereas an active immune system protects from microbes, overactivation of the immune system can result in autoimmunity. An understanding of the molecular underpinnings of an overactive or underactive immune system is important for developing therapies for immune-related diseases from immunodeficiency to autoimmunity and even cancers. mTORC1, a central regulator of cell survival, growth, and metabolism, also plays a critical role in regulation of immune cells. mTORC1 responds to intra- and extracellular signals such as growth factors, oxygen levels, energy status, and amino acid levels (10). The mTOR pathway is activated during various cellular processes, including T-cell activation, insulin resistance, and tumor formation. The immunosuppressive drug rapamycin that has been used clinically to prevent transplant rejection, directly binds to mTORC1 to suppress immune responses (11). One of the identified roles of mTORC1 in the immune system is to direct T-cell-fate decisions. mTORC1 positively regulates differentiation of the Th1 and Th17 subset of Th cells (12). On the other hand, mTORC1 is a negative regulator of Treg differentiation (13) and at the same time is required to maintain Treg function (14).
Here, we present the first characterization to our knowledge of mice with germline deletion of Pip4k2c. Surprisingly, Pip4k2c−/− mice exhibit a phenotype that is quite different from Pip4k2a−/− or Pip4k2b−/− mice. These mice develop normally and are not protected from obesity, insulin resistance, or diabetes, but rather develop enhanced immune responses, resulting in autoimmunity. In addition, they exhibit hyperactivation of mTORC1 signaling in multiple tissues, suggesting that Pip4k2c negatively regulates mTORC1. These results, along with a recent observation that the enzyme encoded by this gene is a substrate of mTORC1 (6), suggest a close relationship between mTORC1 and Pip4k2c. Moreover, the hyperimmune phenotype of the Pip4k2c−/− mice could be partially ameliorated by treatment with the mTORC1 inhibitor, rapamycin. Importantly, a SNP (rs1678542) located near the PIP4K2C locus has been correlated with familial autoimmunity (8) and the results presented here suggest that loss of PI5P4Kγ function could explain this disease.
Results
Generation of Pip4k2c−/− Mice.
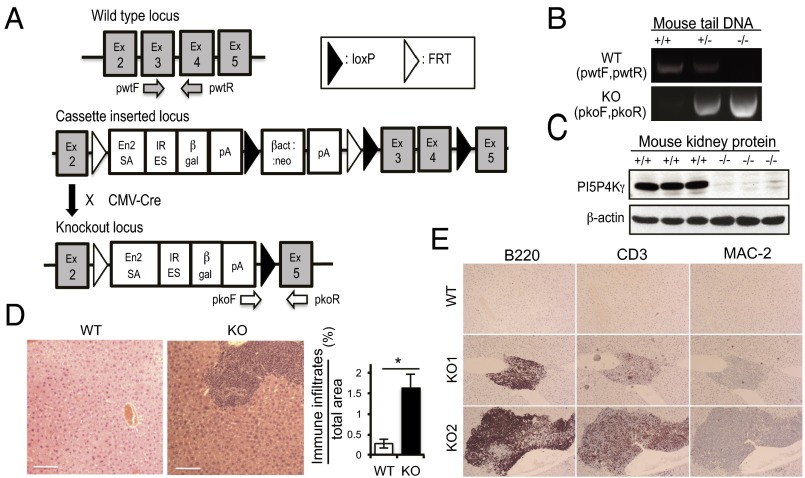
To investigate the role of PI5P4Kγ in mammals, PI5P4Kγ knockout (Pip4k2c−/−) mice were generated using Pip4k2c-targeted ES cell clones obtained from the knockout mouse project (KOMP) repository (www.komp.org). In the targeting construct, exons 3 and 4 of Pip4k2c were bracketed by loxP sequences, so deletion of these exons could remove much of the kinase domain and place the protein out of the correct reading frame (Fig. 1A). ES cell clones were karyotyped and selected for microinjection. Because the targeted clones were C57BL/6N (agouti)-derived JM8A3.N1, the cells were injected into C57BL/6J (black) blastocysts. Three chimeric mice were born and they were backcrossed with C57BL/6 mice for three generations. Then whole body knockout mice were obtained by crossing the transgenic mice with B6.C-Tg(CMV-cre)1Cgn/J. The Pip4k2c+/− mice were crossed with Pip4k2c+/− mice to get Pip4k2c+/+ (WT) mice and Pip4k2c−/− (KO) mice to use for the ensuing experiments. The wild-type allele and knockout allele of each mouse was confirmed by genomic DNA PCR (Fig. 1B) and Western blot for protein expression (Fig. 1C).
Fig. 1.
Generation of Pip4k2c−/− mice. (A) Schematic representation of the Pip4k2c locus before and after deletion of the critical exons (exons 3 and 4). The deletion of the critical exons was induced by Cre-lox recombination. β-gal, β-galactosidase; En2SA, engrailed 2 gene splice acceptor sequence; Ex, exon; neo, neomycin; pA, polyA signal. (B) PCR analysis of genomic DNA prepared from mouse tail. Primers used for genotyping by PCR are pwtF, pwtR, pkoF, and pkoR. (C) Western blotting of proteins prepared from mouse kidney. (D) Immune cell infiltration in the liver of Pip4k2c−/− mice. H&E staining readily identifies immune infiltrates in the liver tissue from Pip4k2c−/− mice. (Scale bars, 100 μm.) The area of immune infiltrates of H&E-stained liver tissues was quantified using ImageJ software. Twenty-five Pip4k2c−/− mice and 12 wild-type mice (four images per mouse) were examined. The results are presented as means ± SE of the means. (E) Immune infiltrates in liver tissue of Pip4k2c−/− mice are mostly T cells and B cells. The Pip4k2c−/− liver sections of two animals (KO1 and KO2) were incubated with anti-CD3, anti-B220, and anti-Mac2 antibodies, respectively.
Increased Inflammation in Pip4k2c−/− Mice.
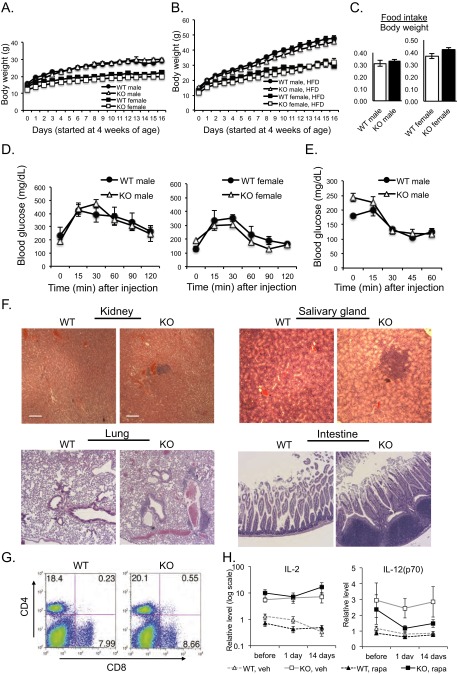
Pip4k2c−/− mice bred normally and grew into adulthood, displaying no obvious growth or behavioral abnormalities (Fig. S1 A–E). Unlike Pip4k2b−/− mice, these mice did not have enhanced insulin sensitivity and were not protected from obesity on a high-fat diet (Fig. S1 A–E).
Fig. S1.
Phenotypes of Pip4k2c−/− mice. (A and B) The growth curves of wild-type and Pip4k2c−/− mice. Mice (started at the age of 4 wk) were weighed weekly for 16 wk. (A) Mice placed on a normal diet. n > 9 per group. (B) Mice placed on a high-fat diet. n > 13 per group. The results are presented as means ± SE of the means. (C) Food intake of wild-type and Pip4k2c−/− mice. The food intake was measured by housing one or two mice of the same genotype per cage and weighing the food in the cages every morning at 11:00 AM every 3 d for 2 wk. The amount of food consumed was divided by the average body weight of the mice in each cage. n = 5 per group. The results are presented as means ± SE of the means. (D) Glucose tolerance test of wild-type and Pip4k2c−/− mice. After weaning, the 4-wk-old mice were placed on a high-fat diet for 5 mo. The mice were i.p. injected with 1 mg of glucose per gram of body weight from a 20 mg/mL solution of glucose in 0.9% NaCl. Blood glucose levels were measured with a One Touch Basic glucose meter before injection of glucose and at indicated time points. n > 5 per group. The results are presented as means ± SE of the means. (E) Insulin tolerance test of wild-type and Pip4k2c−/− mice. The mice were starved from 9:00 AM to 1:00 PM and at 1 PM, the mice were i.p. injected with Novolin-R at a dose of 0.5 units/kg of body weight. Blood glucose levels were measured with a One Touch Basic glucose meter before injection of glucose and at indicated time points, three WT mice and six KO mice (6 mo old). The results are presented as means ± SE of the means. (F) Immune cell infiltration in the kidney, salivary gland, lungs, and intestine of Pip4k2c−/− mice. H&E staining readily identifies immune infiltrates in the indicated tissue of 12- to 14-mo-old Pip4k2c−/− mice. (Scale bars, 100 μm.) (G) Flow cytometry of CD4+ and CD8+ T cells isolated from spleens of wild-type versus Pip4k2c−/− mice. The most representative data from three independent experiments are given, with n > 3 mice (12–14 mo old) from each group. (H) Effects of rapamycin treatment of wild-type mice or Pip4k2c−/− mice on plasma IL-2 and IL-12(p70) levels. Plasma cytokines were detected using multiplex cytokine ELISA. Plasma was collected before the treatment (“before”), 24 h after the first treatment (“1 day”), and a day after the final treatment (“14 day”) from the following four groups: wild type with vehicle, wild type with rapamycin (3 mg⋅kg−1⋅d−1), knockout with vehicle, and knockout with rapamycin (3 mg⋅kg−1⋅d−1), approximately four to eight mice (12–14 mo old) per group, daily i.p. injections for 2 wk. Data were normalized to the mean plasma level of IL-2 and IL-12(p70) in wild-type mice before therapy with 1 on the y axis indicating the mean of wild-type pretreatment (WT, vehicle, before): IL-2: 11.2115 RLU, IL-12(p70): 24.359 RLU. The results are presented as means ± SE of the means.
Because a SNP (rs1678542) near the human PIP4K2C locus has been linked to autoimmunity, we examined whether Pip4k2c−/− mice exhibit any inflammatory phenotype. We carried out a complete necropsy of Pip4k2c−/− mice at different ages. We found that immune cells formed clusters in the organs of mature Pip4k2c−/− mice (8–14 mo of age). The immune cell infiltration was observed in the liver, kidney, salivary glands, lungs, and intestine of the mice (Fig. 1D and Fig. S1F). To measure the surface area of infiltrating immune cells in the liver, we paraformaldehyde fixed and paraffin embedded liver tissues and performed H&E staining. The ratio of immune infiltrates per total area was significantly increased in the Pip4k2c−/− mice (Fig. 1D), which indicated that the Pip4k2c−/− mice developed chronic inflammation without a specific trigger such as infection or injuries. To identify which type of immune cells infiltrated the organs, we stained the liver tissue sections with anti-CD3, anti-B220, and anti-Mac2 antibodies. The infiltrating immune cells in the Pip4k2c−/− livers consisted of mostly CD3+ T cells and B220+ B cells (Fig. 1E).
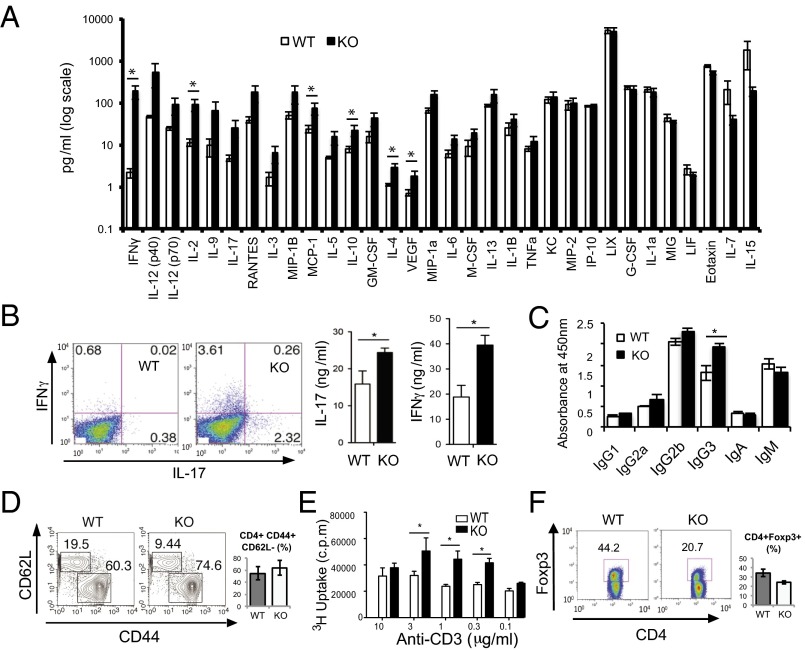
Moreover, the plasma levels of various proinflammatory cytokines increased in the Pip4k2c−/− mice, including the Th1-type cytokines IFNγ, IL-12, and IL-2 (Fig. 2A). Additionally, IL-17 and IFNγ secreted by CD4+ T cells isolated from spleen were higher in the Pip4k2c−/− mice (Fig. 2B). As T-cell cytokines can affect Ig class switching, we performed Ig isotyping. It is known that IL-17 drives B cells to undergo preferential isotype class switching to IgG3 and IgG2a (15). We found that the level of IgG3 increased in the plasma of Pip4k2c−/− mice, which agreed with the increase in IL-17 levels (Fig. 2C).
Fig. 2.
Proinflammatory cytokines are increased in Pip4k2c−/− mice. (A) Plasma cytokines were detected using multiplex cytokine ELISA. The experiments were performed on >10 mice (12–14 mo old) per group, with two measurements per mouse. The y axis is in logarithmic scale. The results are presented as means ± SE of the means. *P < 0.05. (B) T-cell–derived IFNγ and IL-17 levels are elevated in Pip4k2c−/− mice. Flow cytometry of IL-17 and IFNγ secretion by CD4+ T cells isolated from spleen-indicated groups. The data are representative of three independent experiments with n > 3 mice (12–14 mo old) per group. *P < 0.05 (Student’s t test, error bars represent SD). (C) Plasma IgG3 levels are elevated in Pip4k2c−/− mice. Plasma Ig levels were measured using ELISA. Three Pip4k2c−/− mice (12 mo old) and four age-matched wild-type mice were examined. The results are presented as means ± SE of the means. *P < 0.05. (D) Pip4k2c−/− mice exhibit an increase in CD44+ active T cells and a decrease in CD62L+ naïve T cells. Flow cytometry is shown of CD44+ and CD62L+ T cells isolated from spleen-indicated groups. The most representative data from three independent experiments are given, with n > 3 mice (12–14 mo old) from each group. (E) T cells from Pip4k2c−/− mouse have enhanced growth rates. CD4+ T cells were isolated from spleens of 12-mo-old mice and cultured in media containing 3H-thymidine. The level of radioactivity was measured by liquid scintillation. The data are presented as mean 3H-thymidine incorporation (cpm ± SEM, performed in triplicate). *P < 0.05. (F) Regulatory T cells are suppressed in Pip4k2c−/− mice. Flow cytometry of Foxp3+ and CD4+ T cells isolated from spleen-indicated groups. The most representative data from three independent experiments are given, with n > 3 mice (12–14 mo old) from each group.
T Cells Are Hyperactivated in Pip4k2c−/− Mice.
Flow cytometry was used to determine the activity of T cells by counting CD44+ and CD62L+ T cells. We found that Pip4k2c−/− mice have more CD44+-activated T cells and fewer CD62L+ naïve T cells compared with the wild-type mice (Fig. 2D). These data suggested that T cells were more activated in Pip4k2c−/− mice. Also, Pip4k2c−/− mice showed increased CD4+ and CD8+ populations, indicating an increase of Th cells and cytotoxic T cells (Fig. S1G).
To investigate proliferation of T cells, we isolated CD4+ T cells from the spleens from Pip4k2c−/− mice and wild-type mice. The cells were seeded in 96-well plates coated with anti-CD3 and anti-CD28. After 48 h of culture, 3H-thymidine was added for another 16 h before measuring the 3H-thymidine incorporation (Fig. 2E). The Pip4k2c−/− T cells showed significantly higher proliferation rates than T cells from wild-type mice. In accordance with increased T-cell activation in Pip4k2c−/− mice, the number of Foxp3+CD4+ Treg cells decreased in Pip4k2c−/− mice (Fig. 2F). Therefore, these results indicate that Pip4k2c−/− mice developed inflammation with an activation of T cells.
Increased mTORC1 Signaling in Pip4k2c−/− Mice.
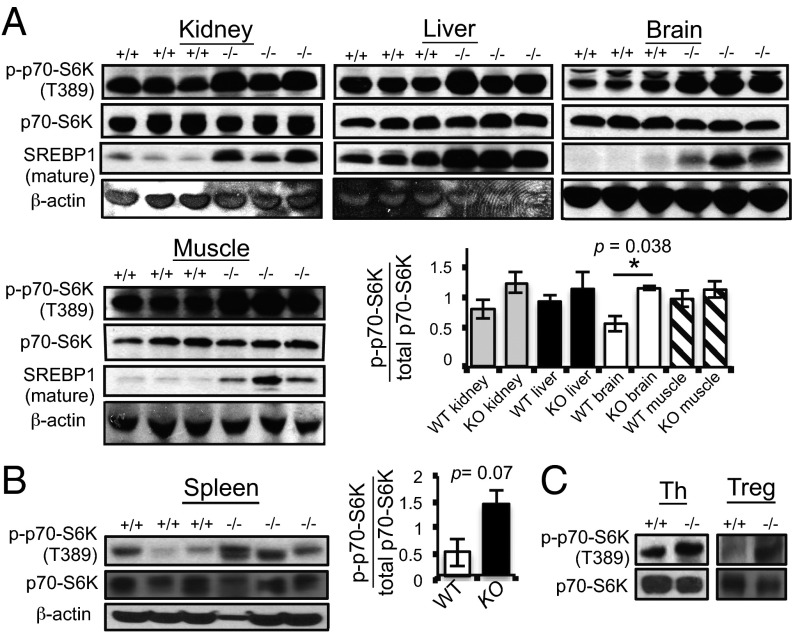
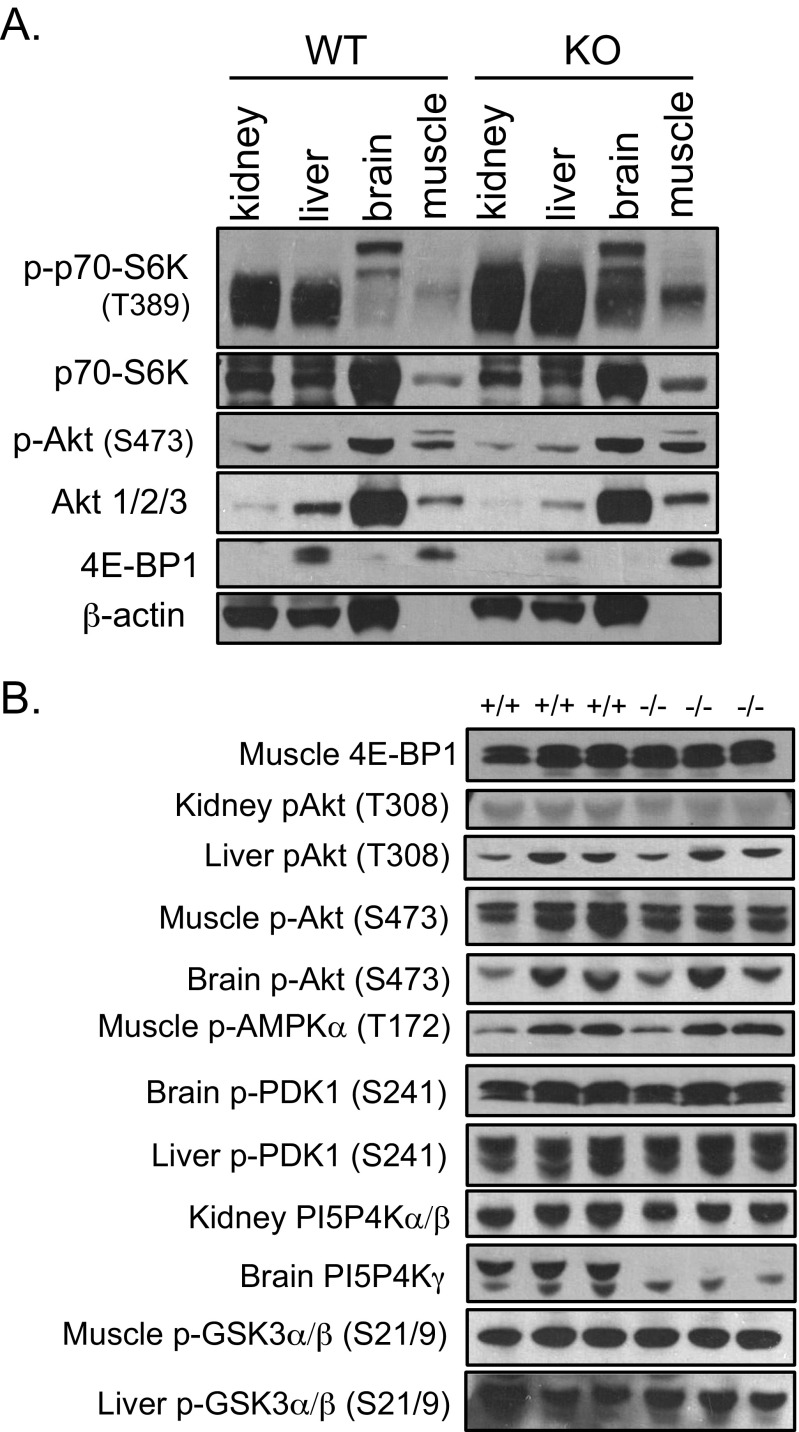
Because the immune system is hyperactivated in Pip4k2c−/− mice, as indicated by an increase in inflammation concomitantly with an increase in T-cell activation, an increase in Th-cell population and a decrease in Treg-cell population, we next investigated if mTORC1 signaling, which is known to regulate diverse immune cell types, (including T cells, macrophages, dendritic cells, neutrophils, and mast cells) was altered in Pip4k2c−/− mice. We found that phosphorylation of p70-S6K on threonine 389 (Thr389, a direct substrate of mTORC1) was increased in kidney, liver, brain, and muscle tissues from Pip4k2c−/− mice compared with wild-type mice (Fig. 3A). Phosphorylation of Thr389 of p70-S6K was also enhanced in spleen, the major immune system organ (Fig. 3B). Levels of mature SREBP1 have recently been shown to be a downstream reporter for mTORC1 activity (16). Consistent with the increased p70-S6K phosphorylation, we also found that levels of mature SREBP1 were significantly higher in various tissues from the Pip4k2c−/− mice (Fig. 3A). On the other hand, other upstream and downstream components of the mTORC1 pathway, including Akt, AMPK, PDK1, GSK3α/β, and 4E-BP1, did not exhibit significant changes in phosphorylation sites that regulate the activity of this pathway (Fig. S2). The failure to see significant changes in these other components probably reflects robust feedback control at each of these steps and further supports the concept that Pip4k2c regulates a step quite proximal to mTORC1. These results indicate that mTORC1 signaling is highly activated in Pip4k2c−/− mice, which suggests that increased mTORC1 signaling could explain the hyperactive immune system in Pip4k2c−/− mice.
Fig. 3.
Signaling downstream of mTORC1 is up-regulated in various tissues of Pip4k2c−/− mice. (A) p70-S6K Thr389 phosphorylation, total p70-S6K, and SREBP1 (cleaved mature form) were blotted for in kidney, liver, brain, and muscles from 12-mo-old wild-type and Pip4k2c−/− mice (three mice per group). The bar graph shows the ratio of p70-S6K Thr389 phosphorylation over total p70-S6K. The results are presented as means ± SE of the means. *P < 0.05. (B) p70-S6K Thr389 phosphorylation and total p70-S6K were blotted for spleen of 12-mo-old mice (three mice per group). The bar graph shows the ratio of p70-S6K Thr389 phosphorylation over total p70-S6K. The results are presented as means ± SE of the means. (C) Th cells and Treg cells isolated from the spleens of 12-mo-old mice (three mice per group). p70-S6K Thr389 phosphorylation and total p70-S6K were blotted.
Fig. S2.
Signaling pathway components of PI3K–Akt–mTORC1 in various tissues of Pip4k2c−/− mice. (A) p70-S6K Thr389 phosphorylation, total p70-S6K, Akt Ser473 phosphorylation, total Akt 1/2/3, 4E-BP1, and β-actin were blotted for kidney, liver, brain, and muscles from 12-mo-old wild-type (Left) and Pip4k2c−/− mice (Right). (B) Other components of PI3K–Akt–mTORC1 were blotted for indicated tissues of 12-mo-old mice (three mice per group).
Rapamycin Reduces mTORC1 Signaling and the Inflammatory Phenotypes in Pip4k2c−/− Mice.
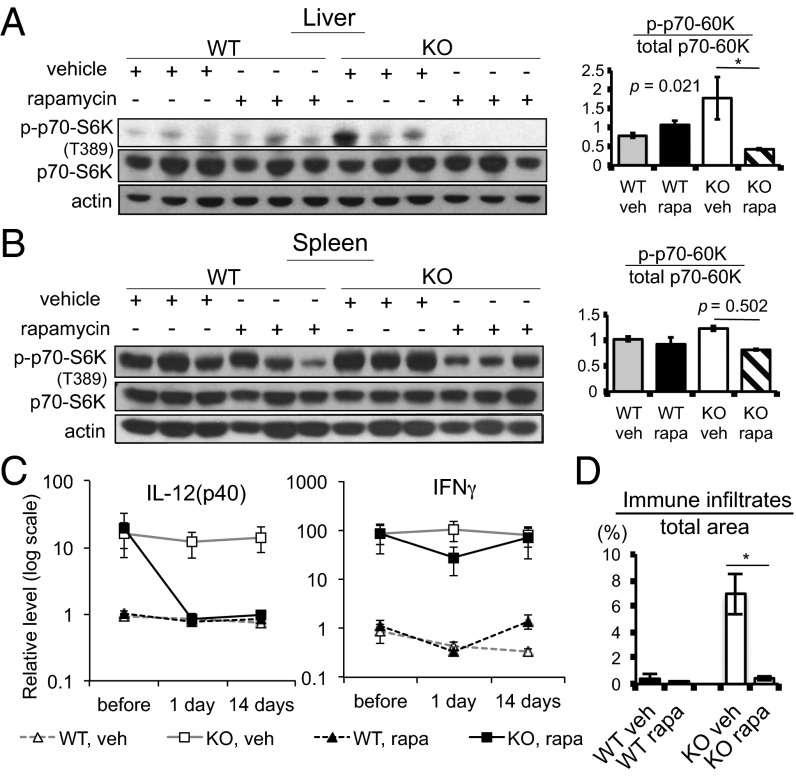
We next examined whether rapamycin, the allosteric mTORC1 inhibitor, could reduce the inflammatory phenotype of Pip4k2c−/− mice. Pip4k2c wild-type and knockout mice were intraperitoneally injected with either vehicle or rapamycin (3 mg⋅kg−1⋅d−1) once a day for 2 wk. Blood was withdrawn 2 wk before the treatment and 24 h after the first treatment. On the final day, blood, liver, and spleen were collected. Using protein lysates from the liver and spleen, we performed SDS/PAGE and Western blot. We found that Thr389 of p70-S6K was still hyperphosphorylated in liver and spleen tissues of Pip4k2c−/− mice treated with vehicle control for 2 wk. However, in the rapamycin-treated Pip4k2c−/− mice, p70-S6K phosphorylation on Thr389 was reduced to the levels seen in tissues from wild-type mice, indicating that rapamycin had suppressed mTORC1 signaling (Fig. 4 A and B).
Fig. 4.
(A and B) Rapamycin reduces the activation of mTORC1 signaling in Pip4k2c−/− mice. p70-S6K Thr389 phosphorylation, total p70-S6K, and actin were blotted for in liver (A) and spleen (B) from 12- to 14-mo-old wild-type and Pip4k2c−/− mice treated with vehicle or rapamycin. Daily i.p. injections were given for 2 wk (3 mg⋅kg−1⋅d−1, three mice per group). The bar graph shows the ratio of p70-S6K Thr389 phosphorylation over total p70-S6K. The results are presented as means ± SE of the means. *P < 0.05. (C) Changes in plasma cytokine levels after treatment with rapamycin. Plasma cytokines were detected using multiplex cytokine ELISA. Plasma was collected before the treatment (“before”), 24 h after the first treatment (“1 day”), and a day after the final treatment (“14 day”) from the following four groups: wild type with vehicle, wild type with rapamycin (3 mg⋅kg−1⋅d−1), knockout with vehicle, and knockout with rapamycin (3 mg⋅kg−1⋅d−1) [approximately four to eight mice (12–14 mo old) per group, daily i.p. injection for 2 wk]. Data were normalized to the mean plasma level of IL-12(p40) and IFNγ in wild-type mice before therapy, with 1 on the y axis indicating the mean of wild-type pretreatment (WT, vehicle, before): IL-12(p40): 46.1875 relative luminescence unit (RLU), IFNγ: 2.148 RLU. The results are presented as means ± SE of the means. Results for IL-2 and IL-12(p70) are shown in Fig. S1H. (D) Changes in immune cell infiltration after treatment with rapamycin. Mouse livers were collected after the final treatment (daily i.p. injections for 2 wk) from the following groups: wild type with vehicle, wild type with rapamycin (3 mg⋅kg−1⋅d−1), knockout with vehicle, and knockout with rapamycin (3 mg⋅kg−1⋅d−1), approximately four to eight mice (12–14 mo old) per group. The area of immune infiltrates of H&E-stained liver tissues was quantified using ImageJ software. Five images per mouse were examined. The y axis indicates the ratio of the area of immune infiltrates over total area of the tissue in each image. The results are presented as means ± SE of the means. *P < 0.05.
The levels of plasma cytokines were measured in the control and rapamycin-treated mice. In agreement with the results in Fig. 2A, before rapamycin treatment the Pip4k2c−/− mice exhibited very high plasma levels of IL-12(p40) and IFNγ compared with levels in plasma of wild-type mice. The plasma level of IL-12(p40) in Pip4k2c−/− mice was reduced to the level observed in wild-type mice at 24 h of rapamycin treatment and remained suppressed after 2 wk of treatment. Rapamycin had no significant effect on plasma IL-12(p40) in wild-type mice. IFNγ levels decreased somewhat in both wild-type and Pip4k2c−/− mice at 24 h after the first treatment with rapamycin. However, after 2 wk of treatment with rapamycin, IFNγ levels in both wild-type and Pip4k2c−/− mice returned to the basal levels (the level before any treatment). On the other hand, IL-2 and IL-12(p70) levels did not significantly change in the Pip4k2c−/− or wild-type mice in response to rapamycin treatment (Fig. S1H). More strikingly, we found that the area of immune infiltrates in the livers of rapamycin-treated Pip4k2c−/− mice was dramatically decreased after 2 wk of rapamycin treatment (Fig. 4D). These data collectively indicate that inhibition of mTORC1 by rapamycin partially reduced the inflammatory phenotypes in Pip4k2c−/− mice.
Discussion
Here we report the generation of Pip4k2c−/− mice and show that Pip4k2c−/− mice have hyperactivated immune systems. Pip4k2c−/− mice were viable with a normal lifespan and did not show any specific abnormality until they were older than 8 mo. But among the older mice at ages between 8 mo and 14 mo, Pip4k2c−/− mice displayed increased immune infiltrates in various tissues, including liver, intestine, kidney, and lungs. These infiltrating immune cells were mostly T cells and B cells. Moreover, we found that plasma of Pip4k2c−/− mice contained high levels of proinflammatory Th1-type cytokines such as IFNγ, IL-12, and IL-2. Importantly, the increase in Th cells and the decrease in Treg cells in Pip4k2c−/− mice reflect the inflammatory phenotype of these mice. Furthermore, the increase in CD44+ T-cell population (central memory T cells) in these mice also supports the hyperactivation of their immune system. Interestingly, mTORC1 downstream components p70-S6K and SREBP1 were activated in Pip4k2c−/− mouse tissues. Because mTORC1 signaling directs the immune system by regulating diverse immune cell types, a possibility that other immune cells in addition to T cells contribute to the inflammatory phenotype of these mice would be interesting to investigate further.
Moreover, after 2 wk of rapamycin treatment, the inflammatory phenotypes as well as the mTORC1 signaling that were enhanced in the Pip4k2c−/− mice decreased. These results suggested that increased activation of mTORC1 signaling in Pip4k2c−/− mice could be responsible for the chronic inflammation in Pip4k2c−/− mice. These results are in agreement with the correlation between the SNP (rs1678542) in the PIP4K2C locus and familial autoimmunity in humans and suggest that the SNP may suppress expression of PI5P4Kγ protein. Our current model is that loss of PI5P4Kγ results in activation of mTORC1 signaling and that this results in increased inflammation, partially through up-regulation of Th cells.
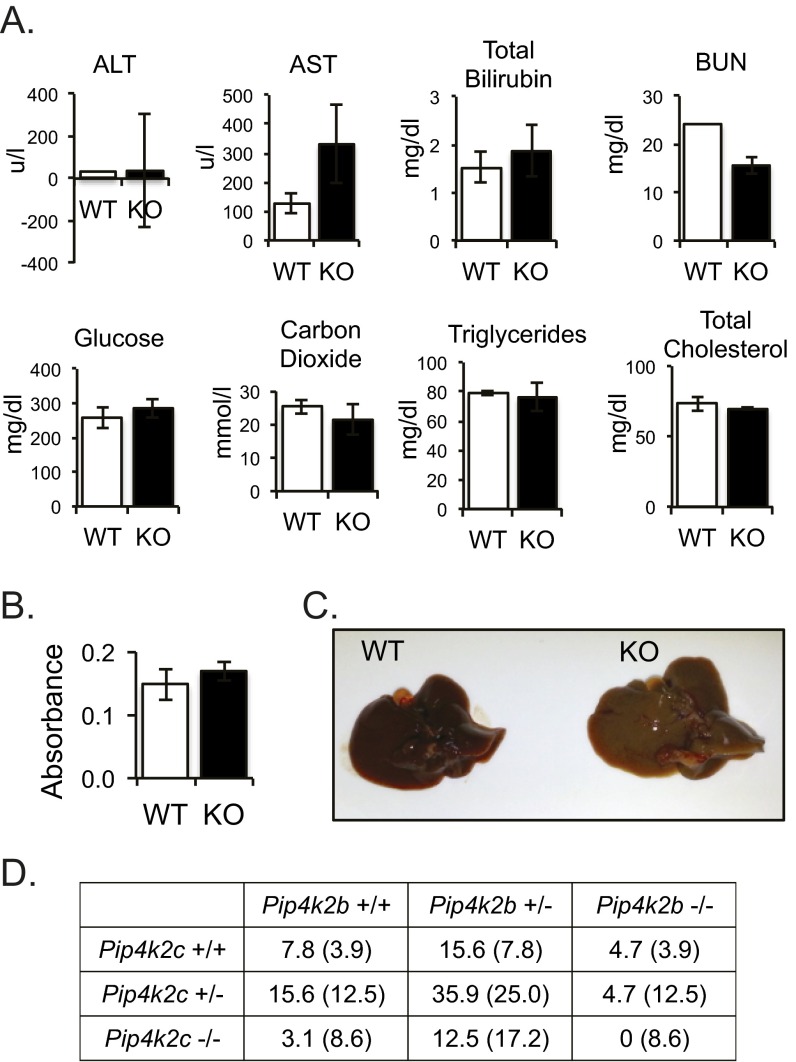
Whereas high rheumatoid factor levels are often associated with autoimmune diseases, they do not always correlate. We measured the levels of rheumatoid factor from Pip4k2c−/− mice and found that the levels of rheumatoid factor were not elevated in the case of Pip4k2c−/− mice (Fig. S3B). In addition, the sizes of organs of Pip4k2c−/− mice were not different from wild-type mice. However, we found that Pip4k2c−/− mice older than 12 mo occasionally exhibit pale livers (Fig. S3C). So we analyzed liver function parameter proteins and metabolites using plasma from Pip4k2c−/− mice (Fig. S3A). There was an increase in aspartate transaminase (AST) and a decrease in blood urea nitrogen (BUN), without a significant change in alanine transaminase (ALT) and other metabolites. These high AST and low BUN results suggest liver damage in Pip4k2c−/− mice, the mechanism of which would be intriguing to investigate further.
Fig. S3.
Additional phenotypes of Pip4k2c−/− mice. (A) Plasma levels of liver function parameter proteins and metabolites. Plasma of the mice were collected and analyzed for the level of the proteins and metabolites indicated (three 12-mo-old mice per genotype). The results are presented as means ± SE of the means. ALT, alanine transaminase; AST, aspartate transaminase; BUN, blood urea nitrogen. (B) Levels of rheumatoid factor in plasma. Rheumatoid factor was measured using mouse rheumatoid factor Ig ELISA kit (6200, Alpha Diagnostic International) according to the manufacturer’s protocol (three 12-mo-old mice per genotype). The results are presented as means ± SE of the means. (C) Gross observation of Pip4k2c−/− mouse livers. (Left) Wild type; (Right) Pip4k2c−/− (12 mo old). Pale color livers were often found in Pip4k2c−/− mice older than 12 mo. (D) Pip4k2b−/− Pip4k2c−/− mice are not viable at the developmental stage. Table showing the expected and observed genotype frequency (%) of 3-wk-old pups from breeding Pip4k2b+/− Pip4k2c+/− with Pip4k2b+/− Pip4k2c−/−, Pip4k2b+/− Pip4k2c+/−, or Pip4k2b−/− Pip4k2c+/− mice. Expected percent frequency is provided in parentheses. n = 64.
The mechanism by which the immune system is hyperactivated in Pip4k2c−/− mice has not yet been fully elucidated. Our studies suggest that mTORC1, which regulates the immune system in diverse aspects, including modulating T-cell differentiation and activation (12), is required to maintain the inflammatory phenotype in Pip4k2c−/− mice. The simplest explanation for the observed autoimmunity is that the protein encoded by Pip4k2c (PI5P4Kγ) plays a role in suppressing the function of the related and much more highly active enzymes PI5P4Kα and PI5P4Kβ when mTORC1 signaling is too high and that this provides feedback suppression of mTORC1 signaling to maintain homeostasis in the T-cell population. However, because these studies are based on germline deletion of Pip4k2c in all tissues, it is possible that non–T-cell autonomous events in other tissues contribute to the autoimmunity observed. This question is currently being addressed by generation of mice with T-cell–specific deletion of Pip4k2c.
The biochemical mechanisms by which PI5P4Kγ might provide feedback inhibition of mTORC1 signaling is not clear. The fact that PI5P4Kγ has very low activity compared with PI5P4Kα and PI5P4Kβ but forms heterodimeric complexes with these active enzymes suggests that its major role is to regulate the localization and/or activity of PI5P4Kα and PI5P4Kβ. As discussed in the introduction, it was reported by Mackey et al. (6) that PI5P4Kγ is phosphorylated by mTORC1 on S324 and S328. Interestingly, Mackey et al. (6) found that expression of a S324A/S328A double-mutant form of PI5P4Kγ in HeLa cells enhanced mTORC1 signaling, whereas expression of a phosphomimetic mutant (S324D/S328D) suppressed mTORC1 signaling. These results are consistent with a model in which PI5P4Kγ can facilitate mTORC1 signaling (perhaps by recruiting the more active enzymes, PI5P4Kα and/or PI5P4Kβ to lysosomes where mTORC1 is activated) and that phosphorylation of PI5P4Kγ at S324 and S328 by mTORC1 provides a negative feedback loop to shut off mTORC1 signaling (perhaps by preventing recruitment of PI5P4Kα and PI5P4Kβ to lysosomes). According to this model, deletion of PI5P4Kγ might impair basal mTORC1 signaling, although PI5P4Kα and/or PI5P4Kβ homodimers or heterodimers could be capable of localizing to lysosomes independent of PI5P4Kγ to maintain mTORC1 signaling in tissues from Pip4k2c−/− mice. In any event, in the absence of PI5P4Kγ the negative feedback control would be eliminated, thereby explaining increased mTORC1 activity in multiple tissues of the Pip4k2c−/− mice.
With respect to the abilities of PI5P4Kγ, PI5P4Kα, and PI5P4Kβ to heterodimerize with each other (17–20), the genetic interaction among PI5P4Kγ, PI5P4Kα, and PI5P4Kβ is particularly thought provoking. Interestingly, Pip4k2a−/− Pip4k2b−/− double KO mice (5) and Pip4k2b−/− Pip4k2c−/− double KO mice (Fig. S3D) are not viable, whereas Pip4k2a−/− Pip4k2c−/− mice are viable. Thus, mice that only have Pip4k2b are viable, but if this gene is deleted both Pip4k2a and Pip4k2c are critical for viability, indicating that these genes do not have redundant functions and must both be expressed to replicate the function of Pip4k2b. The respective roles for PI5P4Kγ, PI5P4Kα, and PI5P4Kβ are complex. Pip4k2b−/− mice exhibited enhanced insulin sensitivity, smaller body size, and decreased adiposity on a high-fat diet. In contrast, Pip4k2c−/− mice were not different from wild-type mice in these features (Fig. S1 A–E). In addition to the synthetic lethality for loss of Pip4k2a and Pip4k2b, and for loss of Pip4k2b and Pip4k2c, distinct phenotypes of each of the Pip4k2a−/−, Pip4k2b−/−, and Pip4k2c−/− mice indicate that each isoform has a unique role in vivo.
Of particular interest is the effect of the various knockouts on signaling through the PI3K–Akt–mTORC1 pathway. Deletion of Pip4k2b enhances Akt activation but, surprisingly, does not result in enhanced mTORC1 signaling (4, 5). Previous studies from many laboratories have shown that impaired mTORC1 activation results in smaller cells and smaller mice (10, 21). Consistent with the failure of Pip4k2b deletion to link Akt activation to mTORC1 activation, the Pip4k2b−/− mice are smaller than wild-type littermates. Although deletion of Pip4k2a results in no observable phenotypes, deletion of a single allele of Pip4k2a in the context of deletion of both alleles of Pip4k2b results in even smaller mice. These data indicate that Pip4k2a and PIP4k2b suppress PI3K-Akt activation but facilitate mTORC1 activation. This model is consistent with the observation that deletion of the single form of PIP4K2 in flies causes suppression of TORC1 activation and suppression of growth (7). To determine the epistasis among the multiple mammalian enzymes, phenotypes of the viable double knockout mice will need to be better examined.
Finally, the results that we present here support the association of a SNP in the PIP4K2C locus with autoimmunity, suggesting that PI5P4Kγ expression is probably low in autoimmune patients with the SNP near the PIP4K2C locus. Our observation that treating Pip4k2c−/− mice with rapamycin reduced the inflammatory phenotype by decreasing the activation of mTORC1 indicates that drugs that target mTORC1 signaling are likely to be effective for patients with familial autoimmunity that correlates with the SNP (rs1678542) near the PIP4K2C locus.
Methods
Generation of Pip4k2c−/− Mice.
Protocols approved by Beth Israel Deaconess Medical Center's Institutional Animal Care and Use Committee and Weill Cornell's Institutional Animal Care and Use Committee were used for the care and use of the mice in this research. Pip4k2c-targeted embryonic stem cell clones, BO1, BO2, and DO1, were obtained from the KOMP repository. These cells were grown in our laboratory and the conditional knockout allele of each clone was confirmed by genomic DNA PCR. The clones were then karyotyped and the normal clones BO1 and BO2 were selected and injected into blastocysts at the Beth Israel Deaconess Transgenic Facility. Three chimeric male mice were obtained and each was backcrossed with C57BL/6J mice. Cassette-bearing mice were mated to Rosa-eFLP1 mice to remove the lacZ reporter. The critical exons 3 and 4 were removed by mating the lacZ reporter-deleted mice to germline cytomegalovirus (CMV)-Cre deleter mice. Knockout mice were verified by PCR and Western blotting.
PCR Genotyping.
For genotyping, the four primers below were used to amplify regions of genomic DNA present in either wild-type samples or knockout samples:
pwtF: TGTCCCCAGGTCTTCAGGAACCT
pwtR: TGCCTTCAGTTTCGCTTGGGGG
pkoF: CACACCTCCCCCTGAACCTGAAAC
pkoR: AGCCGCTGGGGCCAGATGAT.
The primer pair pwtF/pwtR amplifies a fragment (∼0.5 kb) in wild type and the primer pair pkoF/pkoR amplifies a fragment (∼0.5 kb) in knockout.
Preparation of Mouse Tissues for Immunohistochemistry.
Tissues were removed from the killed mice and washed with PBS. The samples were fixed in 10% (vol/vol) buffered formalin for 24 h and paraffin embedded. H&E staining was performed at the Rodent Histopathology Core at the Dana-Farber/Harvard Cancer Center. Staining the immune infiltrates in liver tissue was performed at the Laboratory of Comparative Pathology at Memorial Sloan Kettering Cancer Center. The samples were microsectioned, deparaffinized, rehydrated, and heated with a pressure cooker to 125 °C for 30 s in citrate buffer for antigen retrieval and then incubated with peroxidase and protein blocking reagents, respectively, for 5 min. Sections were then incubated with anti-CD3, anti-B220, and anti-Mac2 antibodies, respectively.
Blood Collection from Mouse and Plasma Preparation.
Mouse tails were cut 1 mm from the tip with scissors and blood was collected into prechilled 1.5 mL EDTA-coated Eppendorf tubes (Microvette CB300, Sarstedt). The samples were centrifuged for 15 min at 825 × g at 4 °C. The supernatants were transferred to new Eppendorf tubes and passed through 0.22-μm filters (centrifuging for 1 min at 5,000 rpm at 4 °C).
Measurement of Plasma Cytokines.
Mouse plasma was prepared as described above. Plasma cytokine levels were measured using multiplex cytokine assay at Eve Technologies. To measure cytokines in our laboratory, the BD Bioscience ELISA kit for IL-12, IFNγ, and IL-2 (M1270, MIF00, and M2000, respectively) was used according to the manufacturer’s protocol.
T-Cell Proliferation.
Cells were grown in DMEM supplemented with 10% FCS, β-mercaptoethanol, l-glutamine, gentamicin sulfate, and penicillin/streptomycin. For the thymidine proliferation assay, 5 × 105 cells per milliliter purified naïve CD4+ T cells were cultured for 48 h in flat-bottom 96-well plates in the presence of various concentrations of anti-CD3 antibody (range, 0.1–10 μg/mL). Cells were pulsed with 1 μCi 3H thymidine for another 16 h of incubation. Mean thymidine incorporation in triplicate wells was measured using a β-counter (LS 5000; Beckman Coulter).
Rapamycin Treatment of the Mice.
For rapamycin treatment, stock solutions (50 mg/mL) were diluted into vehicle [5% (vol/vol) Tween-80, 5% (vol/vol) PEG 400 (polyethylene glycol, molecular weight 400)] in 1× PBS for 2 wk (3 mg⋅kg−1⋅d−1) treatments through i.p. injections. Mice were killed after 2 wk of treatment.
SI Methods
Western Blotting.
Tissues were collected from the killed mice and flash frozen in liquid nitrogen. Tissues were homogenized in prechilled Nonidet P-40 lysis buffer (50 mM Tris pH 7.8, 150 mM NaCl, 0.5% Nonidet P-40, Roche cOmplete EDTA-free protease inhibitor mixture tablet (1 tablet per 25 mL) added). Protein was quantified using Bradford assay (Bio-Rad), resolved with SDS-polyacrylamide gel electrophoresis, and transferred to nitrocellulose membranes. The membranes were probed overnight at 4 °C with the specified primary antibody. The image densitometries of the Western blots were quantified using ImageJ software. Antibodies used were as follows: PI5P4Kγ (HPA028658) (Sigma), p-p70-S6K (9206), p70-S6K (2708), PI5P4Kα (5527), (Cell Signaling Technology), and β-actin (ab6276) (Abcam). In addition, PI5P4Kγ antibody generated by Cell Signaling Technology (commercially not available) was used to detect mouse PI5P4Kγ.
Measurement of Cytokines Secreted from T Cells.
Secreted cytokines were measured by ELISA. All cytokine antibodies were purchased from Biolegend. Flow cytometric analysis was performed using FACSCalibur (Becton Dickinson).
Ig Isotyping.
Mouse plasma was prepared as described above. Ig isotyping was performed using BD Bioscience mouse Ig isotyping ELISA kit (550487) according to the manufacturer’s protocol.
Flow Cytometry.
For intracellular cytokine staining, cells were isolated and stimulated for 4 h at 37 °C in culture medium containing phorbol 12-myristate 13-acetate 50 ng/mL; Sigma), ionomycin (1 μg/mL; Sigma) and monensin (GolgiStop; 1 μg/mL; BD Biosciences). Surface markers were stained in PBS with 1% FCS for 20 min at room temperature, then subsequently fixed in Cytoperm/Cytofix (BD Biosciences), permeabilized with Perm/Wash Buffer (BD Biosciences), and stained with cytokine antibodies diluted in Perm/Wash. For Foxp3 stating, cells were fixed and permeabilized with the Foxp3 Staining Buffer Set, according to the manufacturer’s instructions (eBiosciences). The following antibodies were used in FACS and cell sorting: CD4 (H129.19, BD Pharmingen), CD8 (53-6.7, Biolegend), IL-17A (TC11-18H10.1, BD Pharmingen), IFNγ (XMG1.2, BD Pharmingen), CD44 (IM7, Biolegend), CD62L (MEL-14, Biolegend), and Foxp3 (FJK-16s, eBioscience). All flow cytometry data were acquired on a FACSCalibur (Becton Dickinson) and analyzed with FlowJo software (TreeStar).
Acknowledgments
We thank Gina DeNicola, Florian Kerrath, and other members of the L.C.C. laboratory for helpful discussions. L.C.C. is supported by NIH Grants R01 GM041890 and P01 CA120964. C.W. is supported by National Multiple Sclerosis Society Career Transition Award TA 3059-A-2 and R00 NIH Pathway to Independence Award 4R00AL110649-02.
Footnotes
Conflict of interest statement: L.C.C. owns equity in, receives compensation from, and serves on the board of directors and scientific advisory board of Agios Pharmaceuticals. Agios Pharmaceuticals is identifying metabolic pathways of cancer cells and developing drugs to inhibit such enzymes to disrupt tumor cell growth and survival. L.C.C. owns equity in, receives compensation from, and serves on the scientific advisory board of Petra Pharmaceuticals, a company that develops targeted therapies for cancer treatment. In addition, Petra Pharmaceuticals will be providing funds to support research in the L.C.C. laboratory, although the research described in this paper predated the existence of this collaborative agreement.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1600934113/-/DCSupplemental.
References
- 1.Clarke JH, Emson PC, Irvine RF. Localization of phosphatidylinositol phosphate kinase IIgamma in kidney to a membrane trafficking compartment within specialized cells of the nephron. Am J Physiol Renal Physiol. 2008;295(5):F1422–F1430. doi: 10.1152/ajprenal.90310.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ciruela A, Hinchliffe KA, Divecha N, Irvine RF. Nuclear targeting of the beta isoform of type II phosphatidylinositol phosphate kinase (phosphatidylinositol 5-phosphate 4-kinase) by its alpha-helix 7. Biochem J. 2000;346(Pt 3):587–591. [PMC free article] [PubMed] [Google Scholar]
- 3.Clarke JH, Emson PC, Irvine RF. Distribution and neuronal expression of phosphatidylinositol phosphate kinase IIgamma in the mouse brain. J Comp Neurol. 2009;517(3):296–312. doi: 10.1002/cne.22161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lamia KA, et al. Increased insulin sensitivity and reduced adiposity in phosphatidylinositol 5-phosphate 4-kinase beta-/- mice. Mol Cell Biol. 2004;24(11):5080–5087. doi: 10.1128/MCB.24.11.5080-5087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Emerling BM, et al. Depletion of a putatively druggable class of phosphatidylinositol kinases inhibits growth of p53-null tumors. Cell. 2013;155(4):844–857. doi: 10.1016/j.cell.2013.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mackey AM, Sarkes DA, Bettencourt I, Asara JM, Rameh LE. PIP4kγ is a substrate for mTORC1 that maintains basal mTORC1 signaling during starvation. Sci Signal. 2014;7(350):ra104. doi: 10.1126/scisignal.2005191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta A, et al. Phosphatidylinositol 5-phosphate 4-kinase (PIP4K) regulates TOR signaling and cell growth during Drosophila development. Proc Natl Acad Sci USA. 2013;110(15):5963–5968. doi: 10.1073/pnas.1219333110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raychaudhuri S, et al. Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat Genet. 2008;40(10):1216–1223. doi: 10.1038/ng.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fung EY, et al. Analysis of 17 autoimmune disease-associated variants in type 1 diabetes identifies 6q23/TNFAIP3 as a susceptibility locus. Genes Immun. 2009;10(2):188–191. doi: 10.1038/gene.2008.99. [DOI] [PubMed] [Google Scholar]
- 10.Laplante M, Sabatini DM. Regulation of mTORC1 and its impact on gene expression at a glance. J Cell Sci. 2013;126(Pt 8):1713–1719. doi: 10.1242/jcs.125773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saunders RN, Metcalfe MS, Nicholson ML. Rapamycin in transplantation: A review of the evidence. Kidney Int. 2001;59(1):3–16. doi: 10.1046/j.1523-1755.2001.00460.x. [DOI] [PubMed] [Google Scholar]
- 12.Delgoffe GM, et al. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat Immunol. 2011;12(4):295–303. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delgoffe GM, et al. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30(6):832–844. doi: 10.1016/j.immuni.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng Y, et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458(7236):351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mitsdoerffer M, et al. Proinflammatory T helper type 17 cells are effective B-cell helpers. Proc Natl Acad Sci USA. 2010;107(32):14292–14297. doi: 10.1073/pnas.1009234107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Düvel K, et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol Cell. 2010;39(2):171–183. doi: 10.1016/j.molcel.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bultsma Y, Keune WJ, Divecha N. PIP4Kbeta interacts with and modulates nuclear localization of the high-activity PtdIns5P-4-kinase isoform PIP4Kalpha. Biochem J. 2010;430(2):223–235. doi: 10.1042/BJ20100341. [DOI] [PubMed] [Google Scholar]
- 18.Rao VD, Misra S, Boronenkov IV, Anderson RA, Hurley JH. Structure of type IIbeta phosphatidylinositol phosphate kinase: A protein kinase fold flattened for interfacial phosphorylation. Cell. 1998;94(6):829–839. doi: 10.1016/s0092-8674(00)81741-9. [DOI] [PubMed] [Google Scholar]
- 19.Burden LM, et al. The flattened face of type II beta phosphatidylinositol phosphate kinase binds acidic phospholipid membranes. Biochemistry. 1999;38(46):15141–15149. doi: 10.1021/bi991571a. [DOI] [PubMed] [Google Scholar]
- 20.Wang M, et al. Genomic tagging reveals a random association of endogenous PtdIns5P 4-kinases IIalpha and IIbeta and a partial nuclear localization of the IIalpha isoform. Biochem J. 2010;430(2):215–221. doi: 10.1042/BJ20100340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shima H, et al. Disruption of the p70(s6k)/p85(s6k) gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J. 1998;17(22):6649–6659. doi: 10.1093/emboj/17.22.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]