Significance
Elucidating the genetic and biological substrates of social behavior serves to advance the way basic human nature is understood and improves the way genetic and biological markers can be used to prevent, diagnose, and treat people with impairments in social cognition and behavior. This study shows that epigenetic modification of the structural gene for oxytocin (OXT) is an important factor associated with individual differences in social processing, including self-report, behavior, and brain function and structure in humans.
Keywords: epigenetics, oxytocin, OXT, sociability, social cognition
Abstract
Across many mammalian species there exist genetic and biological systems that facilitate the tendency to be social. Oxytocin is a neuropeptide involved in social-approach behaviors in humans and others mammals. Although there exists a large, mounting body of evidence showing that oxytocin signaling genes are associated with human sociability, very little is currently known regarding the way the structural gene for oxytocin (OXT) confers individual differences in human sociability. In this study, we undertook a comprehensive approach to investigate the association between epigenetic modification of OXT via DNA methylation, and overt measures of social processing, including self-report, behavior, and brain function and structure. Genetic data were collected via saliva samples and analyzed to target and quantify DNA methylation across the promoter region of OXT. We observed a consistent pattern of results across sociability measures. People that exhibit lower OXT DNA methylation (presumably linked to higher OXT expression) display more secure attachment styles, improved ability to recognize emotional facial expressions, greater superior temporal sulcus activity during two social-cognitive functional MRI tasks, and larger fusiform gyrus gray matter volume than people that exhibit higher OXT DNA methylation. These findings provide empirical evidence that epigenetic modification of OXT is linked to several overt measures of sociability in humans and serve to advance progress in translational social neuroscience research toward a better understanding of the evolutionary and genetic basis of normal and abnormal human sociability.
Sociability is a central feature of the human species. Through one perspective, the complex array of human social-cognition and behavior serves to differentiate humans from other animals. However, there exist several core elemental components of the human sociobiological system that are present across many animal species, which may have remained relatively conserved throughout recent evolutionary history (1). Elucidating the genetic and biological substrates of social behavior serves to advance the way basic human nature is understood and improves the way genetic and biological markers can be used to prevent, diagnose, and treat people with impairments in social cognition and behavior.
Oxytocin is a neurohypophysial peptide synthesized in the hypothalamus in the brain, linked to a wide range of social behaviors in humans and other mammals. Administration of oxytocin in humans is associated with changes in social-approach behaviors, such as trust (2) and personal proximity (3). This evidence has motivated the search for genes within the oxytocin system that confer individual differences in social behavior and cognition. A burgeoning body of evidence highlights the role of several key genes within the oxytocin signaling pathway linked to sociability, including the oxytocin receptor gene (OXTR), CD38, and the structural gene for oxytocin (OXT) (4). Although mounting data strongly support the role of OXTR in the phenotypic expression of sociability in humans (5, 6) and animals (7), the roles of other oxytocin pathway genes has received relatively little attention. As such, the role that OXT plays in the expression of sociability in humans is currently unknown.
As with all neurotransmitter and neurohypophysial systems, there exist a host of genes that influence the performance of specific nodes within the oxytocin system (4). One gene that influences the oxytocin signaling pathway is OXT. OXT is located on chromosome 20p13 and codes for a precursor protein that is synthetized to produce oxytocin and neurophysin I (8, 9). OXT knockout mice (Oxt −/−) show aberrations in social behavior (10, 11). In humans, single nucleotide polymorphisms of OXT are associated with maternal bonding (12, 13) and psychiatric conditions, including autism (14, 15) and schizophrenia (16, 17). Although OXT is an important gene within the oxytocin system, there currently exists a scant amount of empirical evidence linking OXT with human sociability. Furthermore, it is currently unknown how epigenetic modification of OXT may influence the social brain and ultimately may confer individual differences in sociability in humans.
The function, and ultimately the end products, of genes are influenced by many endogenous and exogenous factors. DNA methylation is one epigenetic factor that affects the expression and function of genes. DNA methylation occurs when a methyl group forms a covalent attachment with the 5′ carbon of cytosine in the context of a cytosine phosphodiester guanine (CpG) dinucleotide, commonly called a CpG site. DNA methylation regulates gene expression by influencing the recruitment and binding of regulatory protein to DNA. Typically, an increase in DNA methylation is associated with a decrease in expression of that gene (18). Recent breakthroughs in cross-disciplinary research approaches show that epigenetic modification of OXTR, via DNA methylation, is associated with human social behavior and brain activity during social-cognitive processing (6, 19, 20). However, unlike genetic studies, epigenetic modifications are tissue-specific, prompting most studies of brain function to rely on DNA extracted from the proxy tissues. A recent study demonstrated that methylation of DNA extracted from saliva is more similar to the methylation patterns observed in brain tissues than those observed in blood (21). Thus, saliva may be a good proxy tissue for methylation studies of brain-based traits, such as sociability.
Here, we report on the results of a comprehensive investigation of the association between epigenetic modification of OXT and sociability in humans. Genetic, behavioral, and functional and structural neuroimaging data were collected from 121 healthy participants. Genetic data were collected via saliva samples and analyzed to target and quantify DNA methylation across the promoter region of OXT (Fig. 1 and Fig. S1). Based on prior evidence associating oxytocin with social bonding and attachment style (22), we measured individual differences in anxious and avoidant attachment style via self-report. Based on evidence associating oxytocin with emotional face processing (23), we measured individual differences in emotion recognition using a dynamic emotional face-recognition task. For functional neuroimaging (fMRI), we collected fMRI data while participants completed two social-cognitive tasks designed to engage neural activity within the mentalizing, empathy/theory of the mind network (emotional perspective-taking and emotion attribution tasks) (24). Finally, we collected structural MRI data and performed a whole-brain, voxel-based analysis designed to test for the association between DNA methylation of OXT and individual differences in regional gray matter volume. The overarching hypothesis for this body of research was that reduced DNA methylation of OXT (presumably yielding higher OXT expression) is associated with a greater amount of overt measures of human sociability.
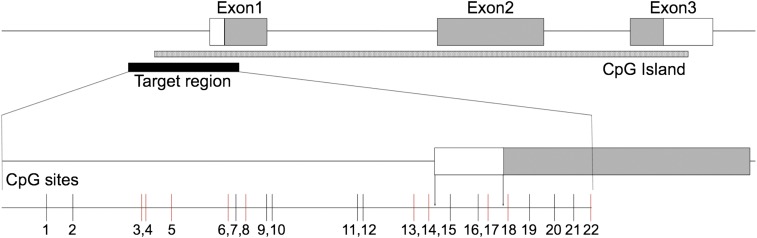
Fig. 1.
Gene structure of OXT (chr20:3,052,266–3,053,162; hg19). The location of the nine CpG sites in this study indicated in red were analyzed, whereas those indicated in black were not uniquely discriminated in the spectra or had low call rates. Assayed CpG sites correspond to the following genomic positions: chr20:3052043/3052058 (CpG1,2), chr20:3052098/3052100 (CpG3,4), chr20:3052115 (CpG5), chr20:3052147 (CpG6), chr20:3052151 (CpG7), chr20:3052157 (CpG8), chr20:3052169/3052172 (CpG9,10), chr20:3052221 (CpG11), chr20:3052224 (CpG12), chr20:3052253 (CpG13), chr20:3052262 (CpG14), chr20:3052274 (CpG15), chr20:3052290 (CpG16), chr20:3052296 (CpG17), chr20:3052307 (CpG18), chr20:3052319/3052334/3052345 (CpG19,20,21), chr20:3052355 (CpG22).
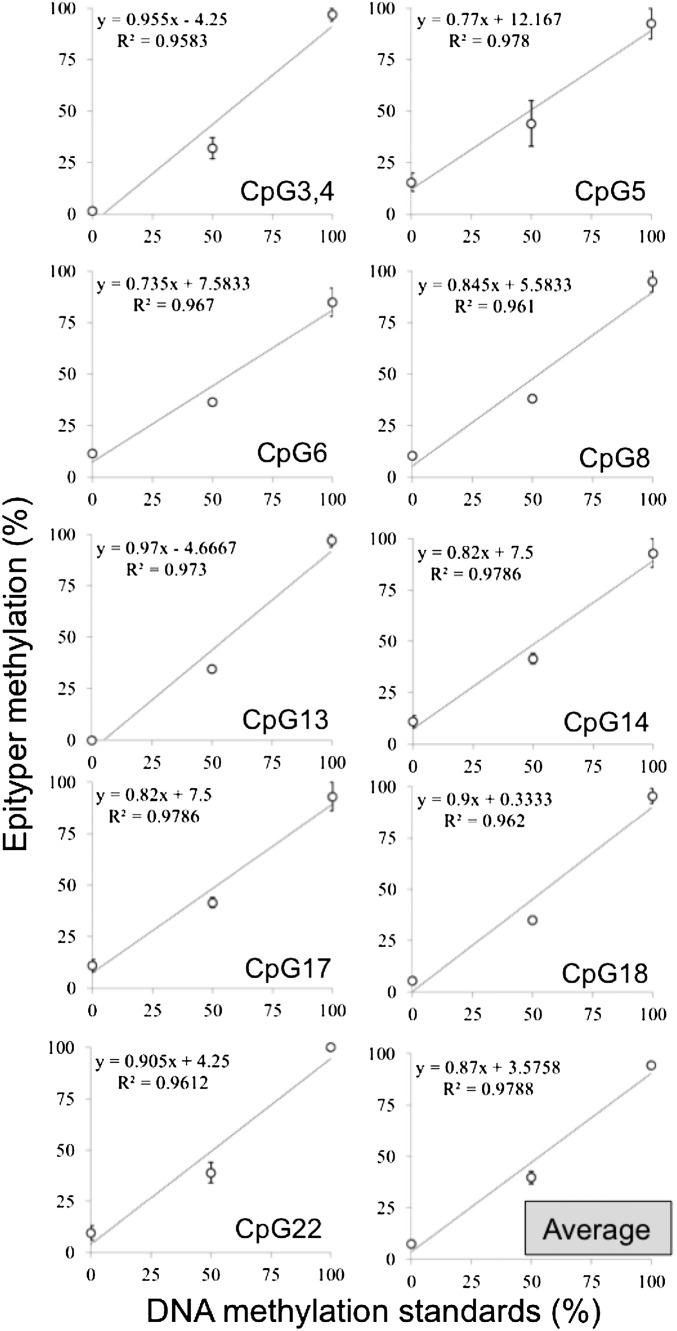
Fig. S1.
OXT assay validation. Linear association between unmethylated (0%), 50%, and 100% methylated positive controls for each CpG site in the Epityper assay (CpG 3,4-22) and average methylation across the target region (Right Bottom). The error bars are indicative of SE.
Results
Genetic, behavioral, and neuroimaging data were collected from a sample of healthy participants within the surrounding community in Athens, Georgia (Table S1). Statistical analyses were performed to test for associations between OXT DNA methylation values and self-reported attachment style, recognition of dynamic emotional facial expressions, brain activity during two social-cognitive tasks (Fig. S2), and regional gray matter volume.
Table S1.
Subgroup-specific demographic data
| Behavioral measure | n | Mean age, y (SD) | M/F | Racial distribution (n) |
| Attachment style | 129 | 21.37 (3.49) | 57/72 | White (62) |
| Black (26) | ||||
| Asian (19) | ||||
| Hispanic (5) | ||||
| Pacific Island (2) | ||||
| Mixed (12) | ||||
| Other (3) | ||||
| Emotion recognition | 128 | 21.38 (3.50) | 57/71 | White (61) |
| Black (26) | ||||
| Asian (19) | ||||
| Hispanic (5) | ||||
| Pacific Island (2) | ||||
| Mixed (12) | ||||
| Other (3) | ||||
| fMRI: Emotional perspective | 121 | 21.27 (3.48) | 52/69 | White (59) |
| Taking and MRI (VBM) | Black (24) | |||
| Asian (18) | ||||
| Hispanic (4) | ||||
| Pacific Island (2) | ||||
| Mixed (12) | ||||
| Other (2) | ||||
| fMRI: Emotion attribution | 120 | 21.28 (3.48) | 52/68 | White (59) |
| Black (23) | ||||
| Asian (18) | ||||
| Hispanic (4) | ||||
| Pacific Island (2) | ||||
| Mixed (12) | ||||
| Other (2) |
Fig. S2.
Social-cognitive fMRI tasks. During the emotional perspective-taking task (A), participants made two types of decisions. For emotional perspective-taking, participants were instructed to take into account the social interaction within a scene presented on the top of the screen, and to decide which of two emotional facial expressions best matches the face that is blanked out. During the shape-match condition (baseline control condition), participants were instructed to match the shape imbedded within the social scene with one of two shapes presented on the bottom of the screen. During the emotional attribution task (B), participants were informed that photographs were taken of people while they were reacting to images of social scenes. During the emotional attribution condition, participants were instructed to decide which of the two social scenes they believed the person was reacting to (view). During the gender-match (baseline control condition) condition, participants were instructed to select the social scene that contained the largest proportion of the gender of the target face on the top of the screen.
Attachment Style.
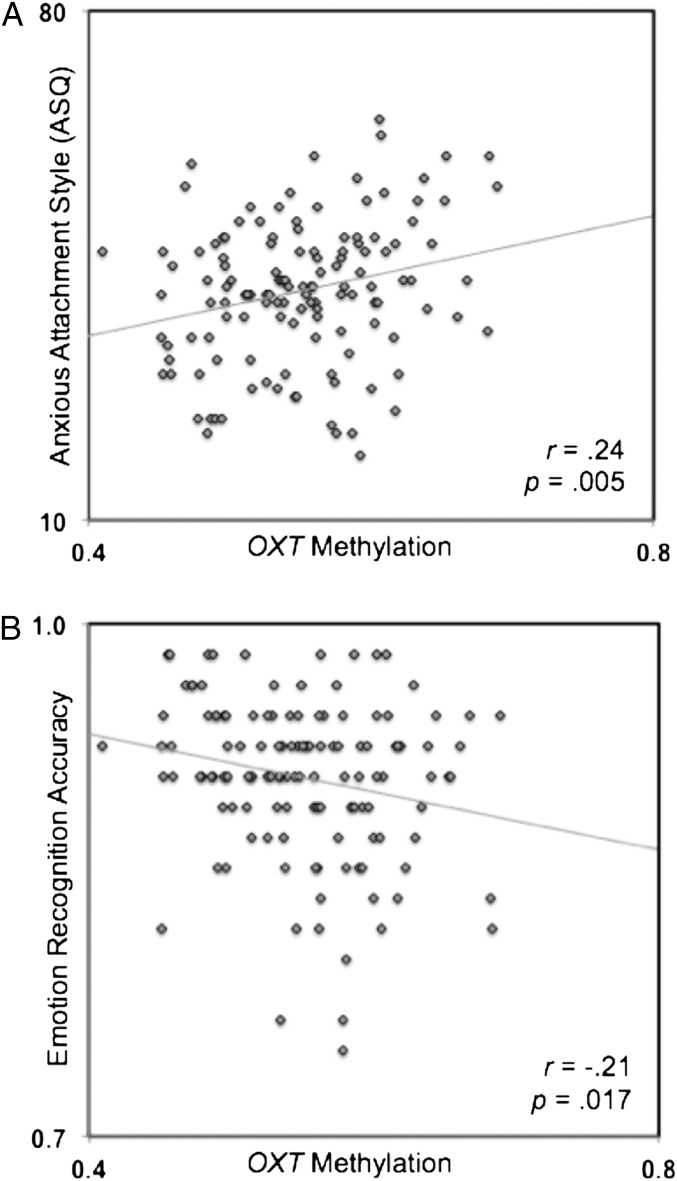
Regression analysis revealed a significant association between DNA methylation of OXT and self-reported anxious attachment style [R2 = 0.06, t(1, 128) = 2.83, P = 0.005, two-tailed] (Fig. 2A). Greater OXT DNA methylation was associated with higher anxious attachment-style scores (r = 0.24). People exhibiting greater DNA methylation of OXT report higher levels of anxious attachment style (i.e., insecure attachment). The association between DNA methylation of OXT and anxious attachment style remained statistically significant when participant’s sex and age were entered as covariates (P = 0.006). OXT methylation and avoidant attachment style were not significantly associated [r = 0.03, t(1, 128) = 0.38, P = 0.71].
Fig. 2.
Scatterplot displaying the association between OXT DNA methylation and ASQ (A) and emotion-recognition accuracy (B).
Emotion Recognition.
For the dynamic facial emotion-recognition task, we observed that greater OXT DNA methylation was associated with lower emotion recognition accuracy [r = −0.21, t(1, 127) = 2.41, P = 0.017, two-tailed] (Fig. 2B). People exhibiting greater DNA methylation of OXT tended to be less accurate when categorizing emotional facial expressions. The association between OXT DNA methylation and emotion recognition accuracy remained statistically significant when participant’s sex and age were entered as covariates (P = 0.014). Because we observed a significant effect for accuracy across all emotion categories, we also tested for associations between OXT DNA methylation and accuracy within each emotion expression category (happy, angry, sad, and fearful). We observed that greater OXT DNA methylation was associated with lower emotion recognition accuracy for angry (r = −0.21, P = 0.019) and sad (r = −0.20, P = 0.027) facial expressions, but not for happy (P = 0.91) or fearful (P = 0.95) facial expressions. The association between OXT DNA methylation and reaction time during the task was not statistically significant (P = 0.36).
Functional Neuroimaging.
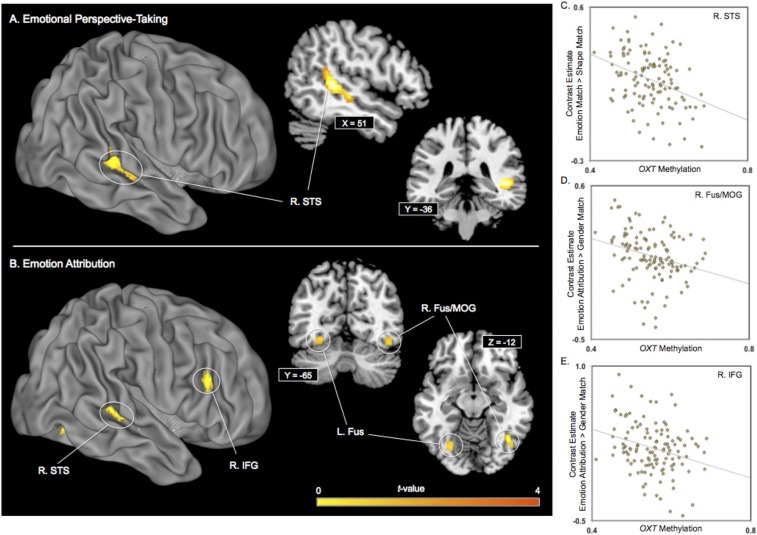
We tested for an association between OXT DNA methylation and brain activity during two social-cognitive tasks (Fig. S2). The emotional perspective-taking task engages the ability to think about views and feelings of other people within a social scene. For emotional perspective-taking, we contrasted blood-oxygen level-dependent (BOLD) response while participants performed the emotional perspective-taking condition to BOLD response collected during the shape-matching condition. Whole-brain regression analysis revealed that greater OXT DNA methylation values were associated with reduced right superior temporal sulcus (STS) activity during emotional perspective-taking [k = 407; peak voxel: Montreal Neurological Institute (MNI): 48, −38, 4; t = 4.59, P < 0.001] (Fig. 3A). People exhibiting greater DNA methylation of OXT show reduced STS activity while emotional perspective-taking. The association between OXT DNA methylation and right STS activity remained statistically significant when the participant’s sex and age were entered as covariates (P < 0.001). Analyses testing for sex differences and a Sex × Methylation interaction effect on right STS activity were not significant. Next, we tested for an association between greater OXT DNA methylation values and larger contrast values between emotional perspective-taking and shape matching. No clusters of increased brain activity were found to be associated with greater OXT DNA methylation (i.e., positive correlation). OXT DNA methylation was not associated with accuracy (P = 0.32) or reaction time (P = 0.22) during the emotion perspective-taking task.
Fig. 3.
OXT DNA methylation and brain activity during social-cognitive processing [(A) emotional perspective-taking, (B) emotion attribution]. Contrast estimates from three observed clusters [(C) emotional perspective-taking, (D and E) emotion attribution) are extracted (y axis) and plotted against OXT DNA methylation values (x axis). For the right superior temporal sulcus (STS), data were extracted from 407 voxels (peak voxel: MNI: 48, −38, 4), for the right fusiform gyrus (Fus) and middle occiptal gyrus (MOG), data were extracted from 106 voxels (peak voxel: MNI: 40, −60, −10) and for the right inferior frontal gyrus (IFG), data were extracted from 92 voxels (peak voxel: MNI: 58, 26, 24). L, left; R, right.
The emotion attribution task engages the ability to think about the reason why another person is emotionally reacting. For emotion attribution, we contrasted BOLD response collected while participants performed the emotion attribution condition to BOLD response collected during the gender-matching condition. Whole-brain regression analysis revealed that greater OXT DNA methylation values were associated with reduced right STS (k = 60; MNI: 42, −60, 2; t = 3.03, P = 0.002), right fusiform gyrus/middle occipital gyrus (Fus/MOG) (k = 106; MNI: 40, −60, −10; t = 3.26, P = 0.001), right inferior frontal gyrus (IFG) (k = 92; MNI: 58, 26, 24; t = 3.23, P = 0.001) and left fusiform (Fus) activity (k = 101; MNI: −26, −72, −8; t = 3.04, P = 0.001) (Fig. 3B). The association between OXT DNA methylation and right STS, Fus/MOG, IFG, and left Fus activity remained statistically significant when participant’s sex and age were entered as covariates (P < 0.005). Analyses testing for sex differences and a Sex × Methylation interaction effect on right STS, Fus/MOG, IFG, and left Fus activity were not significant. Next, we tested for an association between greater OXT DNA methylation values and larger contrast values between emotion attribution and gender matching. No clusters of increased brain activity were found to be associated with greater OXT DNA methylation during the emotion attribution task (i.e., positive correlation). OXT DNA methylation was not associated with accuracy (P = 0.28) or reaction time (P = 0.86) during the emotion attribution task.
Structural Neuroimaging.
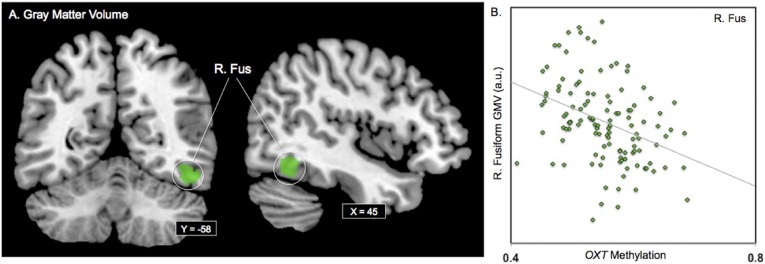
We performed a whole-brain analysis with OXT DNA methylation values entered as the predictor variable and gray matter volume as criterion variable, while total brain volume, sex, and age were entered as covariates using the VBM8 toolbox (www.fil.ion.ucl.ac.uk/spm/software/spm8/) in SPM8. The results of this analysis demonstrated that greater OXT DNA methylation values were associated with reduced gray matter volume within the right fusiform gyrus (Fus) (k = 354; MNI: 48, −57, −14; t = 4.44, P < 0.0005) (Fig. 4) (i.e., negative correlation). Analyses testing for sex differences and a Sex × Methylation interaction effect on right Fus gray matter volume were not significant. Next, we tested for an association between greater OXT DNA methylation values and increased gray matter volume. Greater OXT methylation values were not associated with increased gray matter volume in any brain regions (i.e., positive correlation). OXT DNA methylation values were not associated with total brain (P = 0.23), gray matter (P = 0.42), or white matter (P = 0.45) volume.
Fig. 4.
OXT DNA methylation and gray matter volume. (A) Areas of significant regional gray matter volume associations are overlaid on a coronal and sagittal slice of a standardized template brain. (B) Scatterplot showing the association between OXT DNA methylation values and regional gray matter volume. OXT DNA methylation values are plotted on the x axis and fusiform gyrus (Fus) gray matter volume estimates in arbitrary unites (a.u.) are plotted on the y axis. Data were extracted from 354 voxels (peak voxel: MNI: 48, −57, −14).
Discussion
Across several different metrics, including self-report, behavior, and the function and structure of the brain, we observed that epigenetic modification of OXT, via DNA methylation, is associated with individual differences in human sociability. We found that greater OXT DNA methylation, presumably linked to lower OXT expression, is associated with less-secure self-reported attachment (greater anxious attachment) and a reduced ability to recognize emotional facial expressions. Within the brain, we observed that greater OXT DNA methylation is associated with reduced neural activity within brain regions important for social-cognitive functioning, including the STS, fusiform gyrus, and IFG. Finally, we observed that greater OXT DNA methylation is associated with reduced gray matter volume within the right fusiform gyrus, a brain region important for face processing and social-cognition. Combined, these findings provide empirical, cross-modal evidence that OXT is a gene linked to human sociability and that epigenetic modification of OXT confers individual differences in human sociability.
These findings provide strong support that oxytocin pathway genes regulate the expression of social behavioral phenotypes in humans and other mammals. OXT is one gene among several that influences the oxytocin signaling pathway (4). OXT codes for a precursor protein synthetized to produce oxytocin and neurophysin I (8, 9). Neurophysin I is the carrier protein for oxytocin. The findings from this study are in accordance with animal studies demonstrating aberrations in social behavior in OXT knockout mice (11, 25). For example, Ferguson et al. (10) showed that OXT knockout mice fail to develop social memory and Winslow et al. (26) showed that OXT knockout mice exhibit reduced infant vocalizations and abnormal aggressive and fearful behavior. In humans, there is sparse evidence that single nucleotide polymorphisms of OXT are associated with social behavior, such as social bonding between mother and child and breastfeeding (12, 13). Love et al. (27) demonstrated a link between OXT (rs4813625) and anxious attachment style, anxiety, emotional well-being, and dopamine function. There also exists some evidence that OXT is associated with conditions characterized by aberrations in social-cognition, such as autism (14, 15). The current findings strengthen the link between OXT and human social behavior and suggest that the way OXT is methylated, and ultimately expressed, affects the function and structure of specific brain regions.
Unfortunately, within this study we did not measure OXT expression directly. However, there is evidence that OXT DNA methylation is associated with OXT expression in bovine cells. Bovine cells that express OXT have low levels of promoter methylation, whereas cells that do not express OXT have high methylation levels in this region (28). It is currently unclear how exogenous and endogenous factors may impact DNA methylation of the OXT gene. DNA methylation is modified in response to early childhood experience (29), neuronal activity (30), and social stress (31). It will be important for future epigenetic studies on OXT to include variables representative of individual life experience and psychological stress.
In this study, we characterized epigenetic modification of OXT via DNA methylation values derived from saliva samples. There exist several factors associated with this approach that warrant consideration. Despite its association with sociability, methylation patterns observed in saliva may not reflect DNA methylation patterns in the relevant brain regions, as we did not measure methylation in brain loci specifically. Some studies, however, indicate that DNA derived from saliva may be a good proxy for methylation within brain tissue (21, 32) and may reflect epigenetic programming that is not tissue-specific. In this study, salivary DNA may represent a noninvasive tissue for biomarker testing. Finally, methylation of the majority of CpG sites evaluated in this study were correlated, a common phenomenon in CpG islands. For this reason, average methylation across the region was analyzed, and a subset of these CpG sites may be just as informative as the region evaluated with this assay.
We observed that greater DNA methylation of OXT, presumably linked to lower OXT expression, is associated with higher anxious attachment-style scores. This finding is consistent with extant data showing that oxytocin administration facilitates social-approach behavior. Following oxytocin administration, people tend to trust one another more (2), stand in closer proximity to one another (3), and display a more secure attachment style (22). We measured both anxious and avoidant attachment styles, and observed that OXT methylation values were associated with the anxious form, but not with the avoidant form. An anxious attachment style reflects the tendency to display fear or anxiety about the value one has within relationships, and in extreme scenarios, to be overly dependent on relationship partners. The current finding indicates that OXT may influence social bonding during development and affects the way people form and maintain intimate social relationships with others.
We found that greater DNA methylation of OXT is associated with lower accuracy in determining the emotional expression from dynamic faces. This observation is in accordance with evidence associating oxytocin with social perception (33) and with emotional face recognition (23). Oxytocin may affect emotion recognition by increasing attention toward specific relevant cues, such as the eye region of faces (34). Because we observed an association between OXT methylation and overall accuracy during the task, we also tested for emotion category-specific associations and found that methylation is associated with accuracy during the angry and sad conditions, but not during the happy or fearful conditions. This emotion-specific pattern was unexpected, requires replication, and should be considered with caution. One factor, however, that may have influenced the likelihood of detecting an effect within the happy condition was the consistently high accuracy (happy accuracy: mean= 99%, SD = 2.6; all others: mean < 91%, SD > 9.0) observed during this condition. Taken together, these findings support the role of oxytocin signaling genes in the perception, processing, and recognition of emotional facial expressions.
Across two social-cognitive fMRI tasks, we observed that OXT DNA methylation is associated with STS activity. People exhibiting lower OXT methylation displayed greater right STS activity during an emotional perspective-taking task and an emotion attribution task. Both fMRI tasks are designed to elicit activation within brain regions encompassing the mentalizing/theory of mind networks. The emotional perspective-taking task requires a person to estimate the attitudes, beliefs, and behaviors of other people who are in different emotional situations. The emotion attribution task requires a person to estimate the cause of another person’s emotional reaction. Both the emotional perspective-taking and the emotion attribution task involve thinking about the mental states of other people (i.e., mentalizing). The STS is an important brain region within the mentalizing network (24) and is functionally and structurally aberrant in conditions characterized by deficits in social-cognition, such as autism (35).
An inspection of each of the STS clusters observed in this study indicates that each cluster is located relatively posterior within the sulcus. In reviewing characteristics of tasks that elicit STS activity, Hein el al. (36) found that the anterior portion of the STS is mainly involved in speech perception, whereas the posterior portion is mainly involved in cognitive tasks, such as theory of mind and face processing. The current finding is consistent with a prior neuroimaging study demonstrating that oxytocin administration enhances STS activity in individuals with autism (37), and with a recent finding that OXTR methylation is associated with STS activity (6). It is important to note however, that Puglia et al. (6) reported a positive association between OXTR methylation and STS activity, and we observed a negative association between OXT methylation and STS activity. These findings indicate that methylation of specific genes within the oxytocin signaling pathway affects the function of the STS differently. Combined, these data show that the STS plays an important role in the relationship between oxytocin signaling genes and the way humans process social-cognitive tasks.
For the emotion attribution fMRI task, we also found that OXT methylation is associated with IFG and fusiform gyrus activity. The IFG functions to regulate response tendencies during cognitive, emotional, and social decision-making (38). The emotion attribution task requires participants to decide which social-emotional scene they believed caused a person’s (depicted in a photograph) emotional reaction. Average reaction time during the emotion attribution condition (mean = 2324.13 ms, SD = 322.39) was considerably longer than during the gender-match condition (mean = 1804.32, SD = 281.32), indicating that additional neurocognitive resources were required. Taken together, these findings may indicate that individuals displaying greater IFG activity (associated with reduced OXT DNA methylation) may be allocating more neurocognitive resources when thinking about the emotional states of others, compared with those displaying less IFG activity.
The results of the structural MRI analysis demonstrated an association between OXT DNA methylation and gray matter volume of the fusiform gyrus. It is currently unknown how oxytocin pathway genes influence brain development and ultimately the structure of specific brain tissues. There exists limited evidence that other oxytocin genes, such as OXTR, correspond to individual differences in brain structure. Furman et al. (39) and Inoue et al. (40) showed a link between OXTR and amygdala volume, and Tost et al. (41) showed a link between OXTR and hypothalamus volume. The fusiform gyrus contains the fusiform face area and plays an import role in initial face representation and facial identity recognition (42). Prior research in various patient groups shows that greater fusiform gyrus gray matter volume is associated with an improved ability to recognize facial expressions (43, 44). OT administration is associated with increased fusiform gyrus activity (45) and OXTR methylation is associated with fusiform gyrus reactivity to emotional facial expressions (6). Combined, these findings suggest that fusiform gyrus structure and function may influence the relationship between OXT DNA methylation and the processing—and ultimately the recognition—of faces.
There exist several important limitations of this study that warrant consideration. First and foremost, the sample size for this study was modest (n ranged from 120–129). To our knowledge, this is the first empirical study on OXT methylation in humans. However, based on effect sizes reported in studies investigating other oxytocin genes (19, 20, 46, 47), namely OXTR, and behavior or brain associations, we estimated that a sample size of 66 or larger would be adequate to test each of our hypotheses. This effect size estimate, however, does not account for many other factors, such as publication bias; therefore, our results should currently be interpreted with caution until they can be replicated in an independent cohort. Second, there exist other oxytocin signaling genes that may influence human sociability in similar or different ways, such as CD38, that were not considered in this study. Third, there may not be a linear relationship between DNA methylation in this region and oxytocin protein levels, and further studies are warranted. Fourth, our sample was relatively homogeneous, and was comprised of people within a relatively narrow age range (18–35 y). Thus, it will be important to investigate how age and various life experiences may impact DNA methylation throughout the lifespan. Finally, our findings do not provide any direct empirical information regarding the production, function, or accessibility of oxytocin within the central nervous system.
In conclusion, this study provides comprehensive evidence that epigenetic modification of OXT is linked to several overt measures of sociability in humans. These findings serve to advance progress in translational social neuroscience research toward understanding the evolutionary and genetic basis of normal and abnormal human sociability.
Materials and Methods
Participants.
We recruited 129, fluent English-speaking (72 females, 57 males; mean age = 21.37 y, SD = 3.49 y) adults from the University of Georgia and surrounding community to participate in genetic and behavioral testing and neuroimaging. All participants were screened for neurological and psychiatric conditions (via self-report) and MRI contraindications. All participants provided written informed consent as detailed in the Declaration of Helsinki, and the University of Georgia Institutional Review Board approved all procedures within this study. From this total sample, 129 completed genetic testing, 129 completed the Attachment Style Questionnaire (ASQ), 128 completed the emotion recognition task, 121 completed the emotional perspective-taking fMRI task, 120 completed the emotion attribution fMRI task, and 121 completed structural neuroimaging. A complete listing of subgroup-specific demographic data are provided in Table S1, along with a description of factors that affected participant retention, data collection, and quality.
Saliva Collection and DNA Extraction.
Saliva samples were collected using Oragene Discover OGR-500 kits (DNA Genotek). DNA was extracted using prepIT•L2P reagent (DNA Genotek) and was quantified with PicoGreen (Quant-iT PicoGreen dsDNA Assay Kit, Thermo Fisher Scientific).
DNA Methylation of OXT.
One microgram of DNA was treated with bisulfite using the EpiTect Bisulfite Kit (Qiagen). DNA methylation of CpG sites (Fig. 1) in the promoter region of the OXT gene (chr20: 3,052,266–3,053,162; hg19 build) were analyzed using EpiTYPER (MassARRAY system; Agena Biosciences) according to the manufacturer’s instructions. Forward (aggaagagagTTTTTTTGTTTTATTTTAGTGGTTTAGG) and reverse (cagtaatacgactcactatagggagaaggctTCTTACCTCCCAAAAAACAATTCTA) primers corresponding to chr20:3,052,009–3,052,392 were designed using EpiDesigner (Agena Bioscience), and the spectrum characteristics were validated with RSeqMeth (48). Cycling conditions were: denaturation (94 °C for 15 min) then 50 cycles of amplification (94 °C for 30 s, 58 °C for 60 s, and 72 °C for 30 s) and a final extension step of 72 °C for 10 min. Samples were electrophoresed using 2% (wt/vol) agarose gel to confirm amplification. The CpG sites were unambiguously interrogated, and their genomic locations are detailed in Fig. 1. The mass spectra methylation ratios were generated using EpiTYPER v1.2 (Agena Biosciences).
Finally, we confirmed the reliability of the OXT methylation assay by using Epitect unmethylated (0%) and methylated (100%) DNA samples (Qiagen) as positive controls (Fig. S1). For each participant, average OXT DNA methylation values were calculated by averaging across a total of nine CpG cites.
Behavioral Measures.
Attachment style.
Each participant completed the ASQ (49). The ASQ consists of 40 items and is designed to measure adult attachment style in normative and clinical populations (50). The ASQ yields scores to characterize both anxious and avoidant attachment styles. The anxious attachment scale characterizes the tendency to exhibit an excessive need for reassurance, fear of rejection, and a desire to merge with relationship partners. The avoidant attachment scale characterizes the tendency to avoid intimacy and to be distrusting of others. Together, high scores of anxious and avoidance attachment style reflect an insecure attachment, whereas low scores reflect a secure attachment. For the current sample, reliability analysis for anxious and avoidant attachment scales yielded Cronbach’s α values of 0.72 and 0.80, respectively. We performed two regression analyses, with OXT methylation values entered as the predictor variable and attachment style (anxious and avoidant) entered as the criterion variable. We adopted a significance threshold of P = 0.025 to protect against type 1 errors.
Emotion recognition task.
Participants completed a dynamic facial emotion recognition task (23). Participants were presented with 10-s length video clips of neutral facial expressions gradually morphing to happy, angry, sad, or fearful facial expressions. Face stimuli were obtained from the NimStim set for Facial Expressions (51) and morphing was conducted using FantaMorph software (FantaMorph v5: www.fantamorph.com/index.html). To mimic the rapidity and the average course of natural facial change, each frame was presented for 100 ms, resulting in a frame ratio of 10 fps (52). Participants were presented with 56 dynamic stimuli, 14 for each emotion category. Each emotion category contained an equal number of male and female faces.
Participants were instructed to indicate, by pressing the spacebar, when they were confident what emotional expression the neutral face was morphing into. Then participants indicated via button press, which of the four emotion categories they believed the neutral face was morphing into. The outcome variables used for this task included emotion recognition accuracy and the speed required to reach “confidence” (reaction time), overall and for each emotion category. Further details of data processing and statistical analyses for the emotion recognition task are provided as SI Materials and Methods.
Neuroimaging Procedures.
fMRI tasks.
Participants underwent fMRI while completing a series of two social-cognitive tasks (emotional perspective-taking and emotion attribution) designed to evoke neural activity within the mentalizing, theory of mind/empathy brain network that includes the temporal partial junction, medial prefrontal cortex, and STS (24) (Fig. S2). For the emotional perspective-taking task, participants were presented a social scene with one person’s face “blanked out,” above two emotional expression face-response options (53). Participants were instructed to take into account the social interaction within the social scene and to decide which one of two emotional facial expressions is most appropriate for the person whose face is blanked out within the social scene. As a control condition, each participant was instructed to match the shape embedded within the social scene with one of two shapes presented on the bottom of the screen. There is no difference in the visual presentation of stimuli between the emotional perspective-taking and shape-matching conditions; however, participants were instructed to base their decision on different stimulus characteristics within each condition. The critical comparison used in this study is: emotional perspective taking > shape matching.
The emotion attribution task was designed to elicit the mental process of deciding the cause of another person’s emotional reaction (54). For the emotional attribution task, participates were informed that photos of people (target) were taken while they were presented with an image of a social-emotional scene. Participants were instructed to decide which one of two social-emotional scenes they believed “caused” the person’s (target) emotional reaction. As a control condition, each participant was instructed to match the gender of the person in the photograph (target) with the social-emotional scene that contained the highest proportion of that gender. There is no difference in the visual presentation of stimuli between the emotional attribution and gender-matching conditions; however, participants were instructed to base their decision on different stimulus characteristics within each condition. The critical comparison used in this study is: emotion attribution > gender matching.
fMRI data analysis.
fMRI data collection and preprocessing details are provided in SI Materials and Methods. Analyses of fMRI data during the emotional perspective-taking task were initiated by comparing the BOLD signal acquired during the emotion perspective-taking condition to the shape-matching condition. Each block, within each condition, was modeled based on a convolution to the hemodynamic response function and represented the BOLD signal acquired throughout each block (boxcar), including fixation-cues. Data within each model were high-pass–filtered using default settings (128 s). On a group level, t-contrast maps were entered into a random-effects model (emotion perspective taking > shape match).
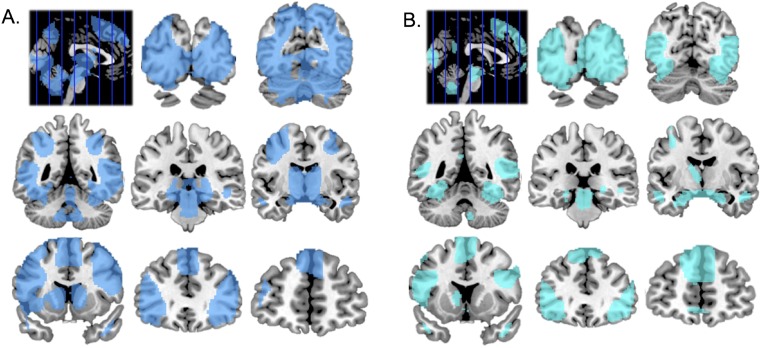
We used a two-tier statistical approach to identify brain loci that: (i) displayed a significant increase of activity during the emotional perspective-taking condition compared to the shape-matching condition, and (ii) exhibited activity significantly associated with OXT methylation. First, we identified brain regions exhibiting greater BOLD response during the emotional perspective-taking condition compared to the shape-match condition by performing a whole brain analysis contrasting emotion perspective-taking > shape match, using a family wise error (FWE), P < 0.05 (corrected) threshold (Fig. S3). All subsequent analyses were restricted to brain regions that surpassed the whole-brain–corrected FWE analysis. Next, we performed a regression analysis with OXT methylation values entered as the predictor variable and contrast estimates (emotion perspective-taking > shape match) as the criterion variable using a threshold of P < 0.005, voxel extent of k = 60, which is sufficient to preserve the balance between sensitivity and false-positive rates (55, 56). Confirmatory multiple regression analyses were performed while sex and age were entered as covariates using the same statistical procedures.
Fig. S3.
Neural activity elicited during each social-cognitive fMRI task, detected by a whole-brain, FWE, P < 0.05 (corrected) analysis, contrasting the emotional perspective-taking condition to the shape-matching condition (A), and the emotion attribution condition to the gender-matching condition (B). Brain loci are overlaid on a series of coronal slices (MNI: y = −88, −68, −48, −28, −8, 12, 32, 52) on a standardized template.
Analyses of fMRI data during the emotional attribution task were identical to the approach used for the emotional perspective-taking fMRI data in all procedures except that the contrast estimates used as the criterion variable were derived from emotion attribution > gender match (instead of emotion perspective-taking > shape match).
VBM analysis.
Structural MRI data collection and preprocessing details are provided in SI Materials and Methods. We used a multiple-regression analysis, using data processed through the VBM8 toolbox (www.fil.ion.ucl.ac.uk/spm/software/spm8/) within SPM8 software, to investigate the association between OXT methylation values and regional gray matter volume. For all analyses, age, sex, and total brain volume were entered as covariates. To protect against false-positive (type 1) errors, we used a combined nonparametric and parametric statistical approach. First, we used a nonparametric approach to create an inclusive mask for all areas that survived a whole brain [false-discovery rate (FDR)-corrected, P < 0.05] analysis, using threshold free-cluster enhancement (TFCE) (57). The TFCE analysis was implemented with the TFCE toolbox in SPM8. For TFCE, we calculated cluster-level statistics based on a total of 5,000 permutations. The TFCE analysis was performed on data smoothed using a 2-mm full-width at half-maximum kernel. Next, we used a parametric approach (restricted to areas within the nonparametric, TFCE, FDR-corrected mask), using data smoothed using a 10-mm full-width at half-maximum kernel, and statistical threshold consisting of a combined spatial extent and height threshold of 200 voxels and P < 0.0005 for each cluster, based on prior VBM analyses using similar parameters and simulations (58–60). For whole-brain analysis, only clusters that survived following both the nonparametric and parametric statistical procedures are reported (61).
SI Materials and Methods
Demographics and Subject Data Retention.
For the emotion recognition behavioral task, one participant failed to complete the task properly and was excluded from the analysis. For the emotional perspective-taking fMRI task and structural MRI data (VBM analysis), neuroimaging data were not collected for five participants and three participants exhibited structural brain anomalies and where therefore removed from analysis. For the emotion attribution fMRI task, neuroimaging data were not collected for five participants, three participants exhibited brain structural anomalies, and one participant exhibited excessive movement during the collection of data and were therefore removed from analysis.
This is part of a larger project to gather data on genes, cognition, sociability, and brain structure and function funded by the Office for the Vice President of Research at the University of Georgia. A subset of participants reported on in this article also completed an educational background form, birth-order questionnaire, mood questionnaire, and a control of attitudes fMRI task. Specific a priori hypothesis regarding associations among these variables and the OXT gene were not developed and thus statistical analyses designed to test for links among these variables were not carried out.
Emotion Recognition Task: Data Processing and Analysis.
Behavioral data were inspected to ensure participants followed instructions. Accuracy was calculated by dividing the total number of correct responses by the total number of stimuli. Average reaction time was calculated based on correct response trials only, overall and for each emotional expression condition (happy, angry, sad, and fearful). Regression analyses were performed using SPSS (v22: www.ibm.com/analytics/us/en/technology/spss/) software. A significance threshold of P = 0.025 was used for each of analysis (accuracy and reaction time) to protect against type I errors.
MRI Data Collection.
Whole-brain imaging data were acquired on a GE-Signa 3T scanner (General Electric) at the University of Georgia Bio-Imaging Research Center (birc.uga.edu/). Functional images (213 for emotional perspective-taking and 195 for emotion attribution) were acquired using a gradient echo T2*-weighted echoplanar imaging (EPI) scan and were obtained using a flip angle of 90°, repetition time (TR) = 2.0 s, echo time (TE) = 25 ms, 40 slices, and a field-of-view (FOV) = 220 mm × 64 matrix.
Structural MRI Data Collection.
High-resolution whole-brain imaging data using a T1-weighted spoiled grass gradient recalled sequence were acquired on a GE-Signa 3T scanner and the following parameters were used: repetition time, 24 ms; echo time, 4.5 ms; flip angle, 20°; matrix size, 256 × 256; FOV, 25.6 cm; slice thickness, 1.0 mm; 164 contiguous slices.
fMRI Data Preprocessing.
fMRI data were preprocessed and statistically analyzed using SPM8 software (Wellcome Department of Imaging Neuroscience, London, United Kingdom) and implemented through Matlab R2013a (www.mathworks.com/?requestedDomain=www.mathworks.com). The images were temporally realigned to the middle slice, spatially realigned to the first in the time series. The images were then coregistered and spatially normalized into standard stereotactic space (MNI template) and spatially smoothed with an 8-mm full-width half-maximum isotropic Gaussian filter.
MRI Data Preprocessing.
Structural MRI data were initially visually inspected for artifacts or structural abnormalities. VBM analyses were performed using SPM8 (www.fil.ion.ucl.ac.uk/spm/software/spm8/) as implemented through the VBM8 toolbox. First, the origin of each participant’s structural image was manually set to the anterior commissure. Next, each image was segmented into gray matter, white matter, and cerebrospinal fluid, and then transformed to MNI stereotactic space using affine and nonlinear spatial normalization. The segmented images were then iteratively registered by the Diffeomorphic Anatomical Registration Through Exponentiated Lie Algebra toolbox (62). This process created a template for the group of individuals. The resulting template image was transformed to MNI stereotactic space using affine and nonlinear spatial normalization with intensity modulation by the Jacobian determinant of the deformation flow field computed for each image. The resulting images were transformed to MNI space and smoothed with an isotropic Gaussian kernel.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.T.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1602809113/-/DCSupplemental.
References
- 1.McCall C, Singer T. The animal and human neuroendocrinology of social cognition, motivation and behavior. Nat Neurosci. 2012;15(5):681–688. doi: 10.1038/nn.3084. [DOI] [PubMed] [Google Scholar]
- 2.Kosfeld M, Heinrichs M, Zak PJ, Fischbacher U, Fehr E. Oxytocin increases trust in humans. Nature. 2005;435(7042):673–676. doi: 10.1038/nature03701. [DOI] [PubMed] [Google Scholar]
- 3.Scheele D, et al. Oxytocin modulates social distance between males and females. J Neurosci. 2012;32(46):16074–16079. doi: 10.1523/JNEUROSCI.2755-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feldman R, Monakhov M, Pratt M, Ebstein RP. Oxytocin pathway genes: Evolutionary ancient system impacting on human affiliation, sociality, and psychopathology. Biol Psychiatry. 2016;79(3):174–184. doi: 10.1016/j.biopsych.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 5.Skuse DH, et al. Common polymorphism in the oxytocin receptor gene (OXTR) is associated with human social recognition skills. Proc Natl Acad Sci USA. 2014;111(5):1987–1992. doi: 10.1073/pnas.1302985111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Puglia MH, Lillard TS, Morris JP, Connelly JJ. Epigenetic modification of the oxytocin receptor gene influences the perception of anger and fear in the human brain. Proc Natl Acad Sci USA. 2015;112(11):3308–3313. doi: 10.1073/pnas.1422096112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.King LB, Walum H, Inoue K, Eyrich NW, Young LJ. Variation in the oxytocin receptor gene predicts brain region specific expression and social attachment. Biol Psychiatry. 2015 doi: 10.1016/j.biopsych.2015.12.008. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rao VV, Löffler C, Battey J, Hansmann I. The human gene for oxytocin-neurophysin I (OXT) is physically mapped to chromosome 20p13 by in situ hybridization. Cytogenet Cell Genet. 1992;61(4):271–273. doi: 10.1159/000133420. [DOI] [PubMed] [Google Scholar]
- 9.Xu Y, et al. Variation of the oxytocin/neurophysin I (OXT) gene in four human populations. J Hum Genet. 2008;53(7):637–643. doi: 10.1007/s10038-008-0292-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferguson JN, et al. Social amnesia in mice lacking the oxytocin gene. Nat Genet. 2000;25(3):284–288. doi: 10.1038/77040. [DOI] [PubMed] [Google Scholar]
- 11.Higashida H, et al. Oxytocin signal and social behaviour: Comparison among adult and infant oxytocin, oxytocin receptor and CD38 gene knockout mice. J Neuroendocrinol. 2010;22(5):373–379. doi: 10.1111/j.1365-2826.2010.01976.x. [DOI] [PubMed] [Google Scholar]
- 12.Jonas W, et al. MAVAN Research Team Genetic variation in oxytocin rs2740210 and early adversity associated with postpartum depression and breastfeeding duration. Genes Brain Behav. 2013;12(7):681–694. doi: 10.1111/gbb.12069. [DOI] [PubMed] [Google Scholar]
- 13.Mileva-Seitz V, et al. Interaction between oxytocin genotypes and early experience predicts quality of mothering and postpartum mood. PLoS One. 2013;8(4):e61443. doi: 10.1371/journal.pone.0061443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hovey D, et al. Associations between oxytocin-related genes and autistic-like traits. Soc Neurosci. 2014;9(4):378–386. doi: 10.1080/17470919.2014.897995. [DOI] [PubMed] [Google Scholar]
- 15.Yrigollen CM, et al. Genes controlling affiliative behavior as candidate genes for autism. Biol Psychiatry. 2008;63(10):911–916. doi: 10.1016/j.biopsych.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Souza RP, de Luca V, Meltzer HY, Lieberman JA, Kennedy JL. Schizophrenia severity and clozapine treatment outcome association with oxytocinergic genes. Int J Neuropsychopharmacol. 2010;13(6):793–798. doi: 10.1017/S1461145710000167. [DOI] [PubMed] [Google Scholar]
- 17.Teltsh O, et al. Oxytocin and vasopressin genes are significantly associated with schizophrenia in a large Arab-Israeli pedigree. Int J Neuropsychopharmacol. 2012;15(3):309–319. doi: 10.1017/S1461145711001374. [DOI] [PubMed] [Google Scholar]
- 18.Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330(6004):612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jack A, Connelly JJ, Morris JP. DNA methylation of the oxytocin receptor gene predicts neural response to ambiguous social stimuli. Front Hum Neurosci. 2012;6:280. doi: 10.3389/fnhum.2012.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ziegler C, et al. Oxytocin receptor gene methylation: Converging multilevel evidence for a role in social anxiety. Neuropsychopharmacology. 2015;40(6):1528–1538. doi: 10.1038/npp.2015.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith AK, et al. DNA extracted from saliva for methylation studies of psychiatric traits: Evidence tissue specificity and relatedness to brain. Am J Med Genet B Neuropsychiatr Genet. 2015;168B(1):36–44. doi: 10.1002/ajmg.b.32278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buchheim A, et al. Oxytocin enhances the experience of attachment security. Psychoneuroendocrinology. 2009;34(9):1417–1422. doi: 10.1016/j.psyneuen.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lischke A, et al. Intranasal oxytocin enhances emotion recognition from dynamic facial expressions and leaves eye-gaze unaffected. Psychoneuroendocrinology. 2012;37(4):475–481. doi: 10.1016/j.psyneuen.2011.07.015. [DOI] [PubMed] [Google Scholar]
- 24.Frith CD, Frith U. The neural basis of mentalizing. Neuron. 2006;50(4):531–534. doi: 10.1016/j.neuron.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Winslow JT, Insel TR. The social deficits of the oxytocin knockout mouse. Neuropeptides. 2002;36(2-3):221–229. doi: 10.1054/npep.2002.0909. [DOI] [PubMed] [Google Scholar]
- 26.Winslow JT, et al. Infant vocalization, adult aggression, and fear behavior of an oxytocin null mutant mouse. Horm Behav. 2000;37(2):145–155. doi: 10.1006/hbeh.1999.1566. [DOI] [PubMed] [Google Scholar]
- 27.Love TM, et al. Oxytocin gene polymorphisms influence human dopaminergic function in a sex-dependent manner. Biol Psychiatry. 2012;72(3):198–206. doi: 10.1016/j.biopsych.2012.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kascheike B, Ivell R, Walther N. Alterations in the chromatin structure of the distal promoter region of the bovine oxytocin gene correlate with ovarian expression. DNA Cell Biol. 1997;16(10):1237–1248. doi: 10.1089/dna.1997.16.1237. [DOI] [PubMed] [Google Scholar]
- 29.Essex MJ, et al. Epigenetic vestiges of early developmental adversity: Childhood stress exposure and DNA methylation in adolescence. Child Dev. 2013;84(1):58–75. doi: 10.1111/j.1467-8624.2011.01641.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo JU, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci. 2011;14(10):1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Unternaehrer E, et al. Dynamic changes in DNA methylation of stress-associated genes (OXTR, BDNF) after acute psychosocial stress. Transl Psychiatry. 2012;2(8):e150. doi: 10.1038/tp.2012.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith AK, et al. Methylation quantitative trait loci (meQTLs) are consistently detected across ancestry, developmental stage, and tissue type. BMC Genomics. 2014;15:145. doi: 10.1186/1471-2164-15-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Theodoridou A, Rowe AC, Penton-Voak IS, Rogers PJ. Oxytocin and social perception: Oxytocin increases perceived facial trustworthiness and attractiveness. Horm Behav. 2009;56(1):128–132. doi: 10.1016/j.yhbeh.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 34.Guastella AJ, et al. Intranasal oxytocin improves emotion recognition for youth with autism spectrum disorders. Biol Psychiatry. 2010;67(7):692–694. doi: 10.1016/j.biopsych.2009.09.020. [DOI] [PubMed] [Google Scholar]
- 35.Zilbovicius M, et al. Autism, the superior temporal sulcus and social perception. Trends Neurosci. 2006;29(7):359–366. doi: 10.1016/j.tins.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 36.Hein G, Knight RT. Superior temporal sulcus—It’s my area: Or is it? J Cogn Neurosci. 2008;20(12):2125–2136. doi: 10.1162/jocn.2008.20148. [DOI] [PubMed] [Google Scholar]
- 37.Gordon I, et al. Oxytocin enhances brain function in children with autism. Proc Natl Acad Sci USA. 2013;110(52):20953–20958. doi: 10.1073/pnas.1312857110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aron AR, Robbins TW, Poldrack RA. Inhibition and the right inferior frontal cortex: One decade on. Trends Cogn Sci. 2014;18(4):177–185. doi: 10.1016/j.tics.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 39.Furman DJ, Chen MC, Gotlib IH. Variant in oxytocin receptor gene is associated with amygdala volume. Psychoneuroendocrinology. 2011;36(6):891–897. doi: 10.1016/j.psyneuen.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inoue H, et al. Association between the oxytocin receptor gene and amygdalar volume in healthy adults. Biol Psychiatry. 2010;68(11):1066–1072. doi: 10.1016/j.biopsych.2010.07.019. [DOI] [PubMed] [Google Scholar]
- 41.Tost H, et al. A common allele in the oxytocin receptor gene (OXTR) impacts prosocial temperament and human hypothalamic-limbic structure and function. Proc Natl Acad Sci USA. 2010;107(31):13936–13941. doi: 10.1073/pnas.1003296107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanwisher N, McDermott J, Chun MM. The fusiform face area: A module in human extrastriate cortex specialized for face perception. J Neurosci. 1997;17(11):4302–4311. doi: 10.1523/JNEUROSCI.17-11-04302.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baggio HC, et al. Structural correlates of facial emotion recognition deficits in Parkinson’s disease patients. Neuropsychologia. 2012;50(8):2121–2128. doi: 10.1016/j.neuropsychologia.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 44.Onitsuka T, et al. Fusiform gyrus volume reduction and facial recognition in chronic schizophrenia. Arch Gen Psychiatry. 2003;60(4):349–355. doi: 10.1001/archpsyc.60.4.349. [DOI] [PubMed] [Google Scholar]
- 45.Domes G, et al. Effects of intranasal oxytocin on emotional face processing in women. Psychoneuroendocrinology. 2010;35(1):83–93. doi: 10.1016/j.psyneuen.2009.06.016. [DOI] [PubMed] [Google Scholar]
- 46.Rubin LH, et al. Sex and diagnosis-specific associations between DNA methylation of the oxytocin receptor gene with emotion processing and temporal-limbic and prefrontal brain volumes in psychotic disorders. Biol Psychiatry Cogn Neurosci Neuroimaging. 2015;1(2):141–151. doi: 10.1016/j.bpsc.2015.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim Y-R, Kim JH, Kim MJ, Treasure J. Differential methylation of the oxytocin receptor gene in patients with anorexia nervosa: A pilot study. PLoS One. 2014;9(2):e88673. doi: 10.1371/journal.pone.0088673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coolen MW, Statham AL, Gardiner-Garden M, Clark SJ. Genomic profiling of CpG methylation and allelic specificity using quantitative high-throughput mass spectrometry: Critical evaluation and improvements. Nucleic Acids Res. 2007;35(18):e119. doi: 10.1093/nar/gkm662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feeney JA, Noller P, Hanrahan M. Assessing adult attachment. In: Sperling MB, Berman WH, editors. Attachement in Adults: Clinical and Developmental Perspectives. Guilford; New York: 1994. [Google Scholar]
- 50.Karantzas GC, Feeney JA, Wilkinson R. Is less more? Confirmatory factor analysis of the Attachment Style Questionnaires. J Soc Pers Relat. 2010;27(6):749–780. [Google Scholar]
- 51.Tottenham N, et al. The NimStim set of facial expressions: Judgments from untrained research participants. Psychiatry Res. 2009;168(3):242–249. doi: 10.1016/j.psychres.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Niedenthal PM, et al. When did her smile drop? Facial mimicry and the influences of emotional state on the detection of change in emotional expression. Cogn Emotion. 2001;15(6):853–864. [Google Scholar]
- 53.Haas BW, et al. I know how you feel: The warm-altruistic personality profile and the empathic brain. PLoS One. 2015;10(3):e0120639. doi: 10.1371/journal.pone.0120639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haas BW, Anderson IW, Filkowski MM. Interpersonal reactivity and the attribution of emotional reactions. Emotion. 2015;15(3):390–398. doi: 10.1037/emo0000053. [DOI] [PubMed] [Google Scholar]
- 55.Lieberman MD, Cunningham WA. Type I and type II error concerns in fMRI research: Re-balancing the scale. Soc Cogn Affect Neurosci. 2009;4(4):423–428. doi: 10.1093/scan/nsp052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Woo C-W, Krishnan A, Wager TD. Cluster-extent based thresholding in fMRI analyses: Pitfalls and recommendations. Neuroimage. 2014;91:412–419. doi: 10.1016/j.neuroimage.2013.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smith SM, Nichols TE. Threshold-free cluster enhancement: Addressing problems of smoothing, threshold dependence and localisation in cluster inference. Neuroimage. 2009;44(1):83–98. doi: 10.1016/j.neuroimage.2008.03.061. [DOI] [PubMed] [Google Scholar]
- 58.Hayasaka S, Phan KL, Liberzon I, Worsley KJ, Nichols TE. Nonstationary cluster-size inference with random field and permutation methods. Neuroimage. 2004;22(2):676–687. doi: 10.1016/j.neuroimage.2004.01.041. [DOI] [PubMed] [Google Scholar]
- 59.Valente AA, Jr, et al. Regional gray matter abnormalities in obsessive-compulsive disorder: A voxel-based morphometry study. Biol Psychiatry. 2005;58(6):479–487. doi: 10.1016/j.biopsych.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 60.Rüsch N, et al. A voxel-based morphometric MRI study in female patients with borderline personality disorder. Neuroimage. 2003;20(1):385–392. doi: 10.1016/s1053-8119(03)00297-0. [DOI] [PubMed] [Google Scholar]
- 61.Silver M, Montana G, Nichols TE. Alzheimer’s Disease Neuroimaging Initiative False positives in neuroimaging genetics using voxel-based morphometry data. Neuroimage. 2011;54(2):992–1000. doi: 10.1016/j.neuroimage.2010.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ashburner J, Friston KJ. Voxel-based morphometry—The methods. Neuroimage. 2000;11(6 Pt 1):805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]