Significance
Chronic heart failure is associated with decreased cardiac myosin light chain kinase (MLCK; cMLCK) expression and myosin regulatory light chain (RLC) phosphorylation, similar to heart failure associated with mutations in numerous sarcomeric proteins. Although ablation of cMLCK expression reduces RLC phosphorylation sufficiently to cause heart failure, the residual phosphorylation indicates that another kinase also phosphorylates RLC. We find that MLCK4 is also expressed abundantly in cardiac muscle, and structural analyses indicate that it is a Ca2+/calmodulin (CaM)-independent kinase, in contrast to Ca2+/CaM-stimulated cMLCK. Biochemical kinetic analyses confirmed these structural predictions. These studies define distinct regulation of cMLCK and MLCK4 activities to affect RLC phosphorylation, and lay the foundation for RLC phosphorylation as a therapeutic target for heart failure.
Keywords: cMLCK, MLCK4, kinase, calcium/calmodulin, crystallography
Abstract
The well-known, muscle-specific smooth muscle myosin light chain kinase (MLCK) (smMLCK) and skeletal muscle MLCK (skMLCK) are dedicated protein kinases regulated by an autoregulatory segment C terminus of the catalytic core that blocks myosin regulatory light chain (RLC) binding and phosphorylation in the absence of Ca2+/calmodulin (CaM). Although it is known that a more recently discovered cardiac MLCK (cMLCK) is necessary for normal RLC phosphorylation in vivo and physiological cardiac performance, information on cMLCK biochemical properties are limited. We find that a fourth uncharacterized MLCK, MLCK4, is also expressed in cardiac muscle with high catalytic domain sequence similarity with other MLCKs but lacking an autoinhibitory segment. Its crystal structure shows the catalytic domain in its active conformation with a short C-terminal “pseudoregulatory helix” that cannot inhibit catalysis as a result of missing linker regions. MLCK4 has only Ca2+/CaM-independent activity with comparable Vmax and Km values for different RLCs. In contrast, the Vmax value of cMLCK is orders of magnitude lower than those of the other three MLCK family members, whereas its Km (RLC and ATP) and KCaM values are similar. In contrast to smMLCK and skMLCK, which lack activity in the absence of Ca2+/CaM, cMLCK has constitutive activity that is stimulated by Ca2+/CaM. Potential contributions of autoregulatory segment to cMLCK activity were analyzed with chimeras of skMLCK and cMLCK. The constitutive, low activity of cMLCK appears to be intrinsic to its catalytic core structure rather than an autoinhibitory segment. Thus, RLC phosphorylation in cardiac muscle may be regulated by two different protein kinases with distinct biochemical regulatory properties.
Unlike most smooth and skeletal muscles, the muscle of a continuously beating heart never relaxes for an extended period as a result of cyclic increases and decreases in [Ca2+] with activation/deactivation of myofilament contractile proteins. The heart is intricately linked to signaling modules, which regulate cardiac performance to meet the circulatory demands of the body. Within the contractile apparatus of the cardiac myocyte, fine tuning of cardiac contractile performance is partly achieved by posttranslational modifications of myofilament proteins. The main driver of muscle contractions is the ATPase activity of myosin molecules, which are hexamers comprised of two each of heavy chain, essential light chain, and regulatory light chain (RLC) subunits. RLCs of myosins are phosphorylated by myosin light chain kinases (MLCKs) to activate or modulate the myosin ATPase activity (1).
The MLCK family is comprised of four distinct kinases, MLCK1, -2, -3, and -4, which are encoded by distinct genes (1). Based on their muscle type-specific expression and activities, MLCK1 is known as smooth muscle MLCK (smMLCK), MLCK2 as skeletal muscle MLCK (skMLCK), and MLCK3 as cardiac muscle MLCK (cMLCK). MLCK4 remains uncharacterized, and tissue distribution of MLCK4 protein has not been reported. Without available crystal structures of any of the MLCKs, the mechanism of autoregulation has been defined by structural studies of other Ca2+/calmodulin (CaM)-dependent kinases combined with biochemical investigations of different MLCKs (2, 3). The smMLCK and skMLCK are Ca2+/CaM-dependent protein kinases (CaMKs) with autoregulatory segments C-terminal of the catalytic core. In the absence of Ca2+/CaM, the autoinhibitory sequence N-terminal to the CaM binding sequence binds to the surface of the kinase C domain toward the catalytic cleft, blocking RLC binding. Ca2+/CaM binding to the CaM-binding sequence of the autoregulatory segment displaces the autoregulatory segment, exposing the catalytic cleft for RLC binding (4, 5). Thus, the catalytic activity of these two MLCKs are completely dependent on Ca2+/CaM (1). As cMLCK also has an autoregulatory segment, and RLC is dephosphorylated in nonbeating hearts, presumably because of reduced [Ca2+]i (6, 7), expectations are that it is Ca2+/CaM-dependent like smMLCK and skMLCK. However, conflicting reports on the Ca2+/CaM-dependency of cMLCK activity necessitate further investigation (8, 9).
The extent of RLC phosphorylation in muscle cells is determined by balanced activities of MLCK and myosin light chain phosphatase(s) (1). When heart extracts are processed by using procedures that minimize loss of RLC phosphorylation and maximize solubilization of myofilament proteins, cardiac RLC (cRLC) shows an average phosphorylation of 0.40 ± 1 mol phosphate/mol RLC in a variety of animals (6, 7, 10–20). Selective ablation of the gene for cMLCK, but not skMLCK, decreases cRLC phosphorylation from 0.45 to 0.10 mol of phosphate/mol of RLC, showing kinase-specific effects in the heart (6, 11, 12, 21). The decreased cardiac performance and dilation of the adult mouse heart associated with the attenuation of RLC phosphorylation in the cMLCK-KO animals is similar to the results obtained with knock-in mutant mice containing a nonphosphorylatable cRLC (18).
RLC phosphorylation is reduced in human heart failure (22–25) and in heart failure animal models (8, 26, 27). Overexpression of MLCK in the heart increases the extent of RLC phosphorylation, which is associated with improved cardiac performance and attenuation of hypertrophic responses to stress (15, 27). In addition, overexpression of an RLC phosphomimetic mutation S15D in the background of a hypertrophic cardiomyopathy-associated mutation D166V was recently shown to prevent the development of a disease phenotype associated with the overexpression of D166V mutant RLC (28). Although animal studies have solidified the requirement of RLC S15 phosphorylation for normal cardiac performance (29), and have confirmed that cMLCK is the primary protein kinase responsible for phosphorylation of RLC in ventricular muscle, information on the regulation of RLC phosphorylation and the enzymatic properties of cMLCK is conflicting (8, 9). In addition, residual RLC phosphorylation (20–25% of normal RLC phosphorylation) in cMLCK-KO animals indicates that another MLCK could phosphorylate RLC.
Here we report that MLCK4 protein is expressed in the mouse heart at levels higher than in skeletal and smooth muscles. We also report the resolved structure of MLCK4, to our knowledge the first MLCK family member to be crystallized, and biochemically establish MLCK4 as a kinase for cRLC. Distinct from other MLCK family members, MLCK4 has a “pseudoregulatory helix” (PRH) in place of a full autoregulatory segment. From the crystal structure of MLCK4 and sequence comparisons with other MLCK family members, we hypothesized that MLCK4 would have Ca2+/CaM-independent kinase activity and cMLCK would be Ca2+/CaM-dependent. In vitro kinase assays with purified MLCK4 showed that it is a constitutive kinase as predicted by the crystal structure, but purified recombinant cMLCK and endogenous cMLCK immunoprecipitated from mouse hearts showed that cMLCK has Ca2+/CaM-independent and -dependent activities, contrary to predictions. MLCK chimeras were generated from cMLCK and skMLCK sequences, and activities compared with WT kinases to determine the contribution of the specific autoregulatory segment sequences to Ca2+/CaM-dependency. We report that cMLCK has high affinity for cRLC, ATP, and CaM, comparable to other MLCKs, and that the Vmax value of cMLCK is orders of magnitude lower than those of other MLCK family members.
Results
MLCK4 Is Highly Expressed in Cardiac Myocytes.
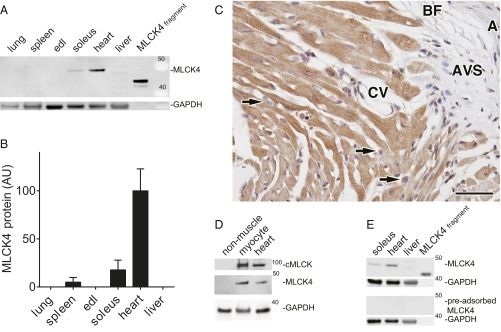
Tissue extracts from WT mice were prepared, and MLCK4 protein and GAPDH in lung, spleen, extensor digitorum longus muscle, soleus muscle, heart apex, and liver were compared. There was one band in the region of expected mass (44 kDa; Fig. 1A), and the relative MLCK4 expression normalized to GAPDH was greatest in the heart (Fig. 1B). Compared with the purified MLCK4 protein standard, MLCK4 was expressed at 0.12 µM based on previously published assumptions (6). Expression of MLCK4 in cardiac myocytes was confirmed by immunohistochemistry (Fig. 1C) of mouse heart, and immunoblotting isolated adult mouse cardiac myocytes and cardiac nonmuscle cells (Fig. 1D). Relative to the GAPDH-normalized MLCK4 content in heart tissue extracts, 98 ± 7% of the MLCK4 is expressed in cardiac myocytes. Preadsorption of the antibody mixture with purified MLCK4 removed MLCK4 band from all samples (Fig. 1E), confirming specificity of the antibody.
Fig. 1.
Tissue expression of MLCK4. (A) Representative immunoblot of MLCK4 and GAPDH expression in supernatant fractions of mouse tissue homogenates. Purified MLCK4 (0.02 pmol) was used as control. (B) Comparison of MLCK4 protein expression in mouse tissues (mean ± SE; n = 3). (C) Immunohistochemistry for MYLK4 in adult mouse myocardium. Micrograph of basal junction of right ventricular wall and ventricular septum shows MYLK4 protein expression conserved to cardiomyocytes (brown DAB Chromogen). In the micrograph, staining is absent in fibroblasts (arrowheads), wall of aorta (marked as “A”), endothelium and basal lamina of cardiac veins (CV), aortic valve spongiosa (AVS), and basal brown fat (BF). Nuclei are counterstained with Harris hematoxylin (blue). (Scale bar: 40 µm.) (D) Representative immunoblot of cMLCK, MLCK4, and GAPDH protein expression in homogenates of mouse heart, isolated cardiac myocytes, and cardiac nonmuscle cells. (E) Immunoadsorption test for MLCK4 antibody. MLCK4 antibody (Upper) and a solution of the same antibody after preadsorption with purified MLCK4 protein (Lower) were used to blot MLCK4 protein in tissues and the purified MLCK4 fragment. Tissues are indicated. Apparent molecular weights are noted.
MLCK4 Assumes an Active Conformation in the Absence of Ca2+/CaM.
To obtain structural insight into the regulation of MLCKs, we crystallized MLCK4 and determined its structure at 2.7-Å resolution. By using several truncation constructs, an N-terminal truncation starting at residue K40 yielded stable protein and readily formed crystals. The structure was refined, maintaining favorable bond geometry to an R/Rfree of 19.2%/24.1% (Table 1). The final model included residues D80–Q373; thus. 40 residues at the N terminus as well as 11 residues at the C terminus were not visible in electron density maps and have been assumed to be disordered. In addition, no density was visible for the tip of the phosphate binding loop (P-loop) G113–Q119, and these residues have therefore not been modeled. The protein was crystallized in complex with a nonspecific ATP mimetic inhibitor [4-(2 amino-4-methyl-1,3-thiazol-5-yl)-n-(3-dioxaziridin-3-yl phenyl)pyrimidine-2-amine], which was used to stabilize the protein during crystallization.
Table 1.
Data collection and refinement statistics for the crystallization of MLCK4
| Data collection | MLCK4 |
| Space group | C 1 2 1 |
| Cell dimensions a, b, c (Å) | 148.44, 70.65, 96.57 |
| α, β, γ (°) | 90.0, 118.96, 90.0 |
| Resolution (Å) | 2.67 (2.67–2.81) |
| Unique observations | 23,801 (1,729) |
| Completeness (%) | 99.78 (99.8) |
| Redundancy | 3.7 (3.7) |
| I/σI | 8.4 |
| Refinement | |
| Rwork/Rfree (%) | 19.0/24.1 |
| No. of atoms (protein/other/solvent) | 4,568/79/252 |
| Mean B-factors (Å2) | 20.7 |
| rmsd bond (Å) | 0.015 |
| rmsd angle (o) | 1.526 |
| PDB ID code | 2X4F |
Values in parentheses are last shell.
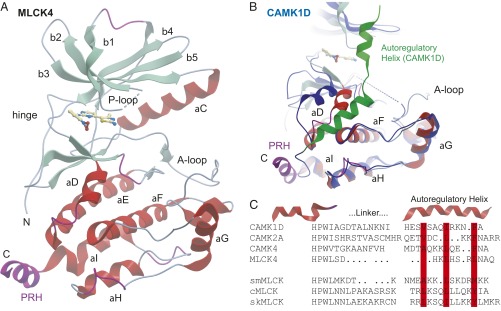
As expected, the structure of MLCK4 revealed the typical bilobal architecture of protein kinase catalytic domains. The N terminus assumed an extended conformation containing also an extra-short β-sheet segment. Sheet β1 was highly twisted, including a short turn. The activation segment was fully ordered. The inhibitor bound to the ATP binding site formed a hydrogen bond to the hinge main chain (residue V183). A structural overview showing the main structural elements is shown in Fig. 2A.
Fig. 2.
Structure of MLCK4. (A) Ribbon diagram of the catalytic domain. The main structural elements are labeled. The cocrystallized inhibitor is shown in ball-and-stick representation. The C-terminal PRH is highlighted in magenta. (B) Superimposition of the MLCK4 structure with autoinhibited CaMK1D (PDB ID code ID 2JC6) colored in blue. (C) Structure-based sequence alignment of the linker and the regulatory helices of CaMK1D, CaMK2A, CaMK4, and MLCK4. The alignment of smMLCK, cMLCK, and skMLCK was based on sequence only. Conserved hydrophobic residues that anchor the regulatory helix are highlighted in red.
The C terminus formed a short helix typical for autoinhibited CaMKs. Interestingly, in contrast to autoinhibited CaMKs, this helix was oriented away from the kinase domain. Superimposition with the autoinhibited structure of CaMK1D (Fig. 2B) showed that the extended linker region that allows the regulatory helix to interact with a deep groove formed by αD and αF is lacking in MLCK4, whereas at least two of the three hydrophobic residues that anchor the amphipathic regulatory helix to the kinase core (L366, L370) are still present in MLCK4. Thus, a short C-terminal helix is also conserved in MLCK4. This helix, however, cannot inhibit the catalytic domain because of missing linker regions. We therefore named this structural element PRH. In comparison with the autoinhibited CaMK1D structure, helix αD shifted inward, closing the binding groove of the regulatory helix. This rearrangement of αD has been observed in CaMKII as a result of CaM binding and is consistent with a constitutively active kinase (30).
Structure-based alignments revealed the main residue conservation shared among CaMKs (Fig. 2C). This analysis revealed a conserved helix insertion (“HPW”) present in all compared CaMKs and a diverse linker region. The autoregulatory helices present in MLCKs were predicted based on the structure-based alignment and conserved hydrophobic residues anchoring the autoregulatory helix. Also, the missing linker region and the first turn of the autoregulatory helix is evident, suggestive of Ca2+/CaM-independent activity.
The Kinase Activity of MLCK4 Is Distinct from skMLCK.
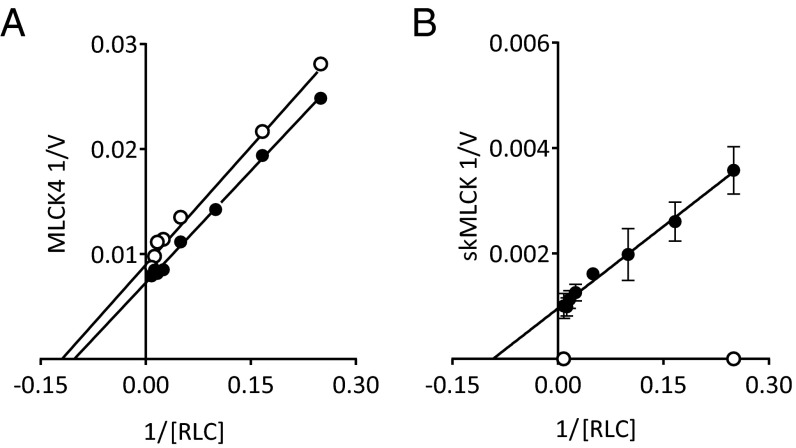
Kinase rates were measured to compare the Ca2+/CaM-dependent and -independent activities of MLCK4 to skMLCK. MLCK4 activity was not different in the presence and absence of Ca2+/CaM. The averaged Vmax value was 129 ± 6 mol phosphate/min/mol kinase, and the RLC Km value was 7.6 ± 1.1 µM (Fig. 3A). The skMLCK has catalytic activity only in the presence of Ca2+/CaM; the Vmax value was 1,132 ± 54 mol phosphate/min/mol kinase and the Km value was 11.6 ± 2 µM (Fig. 3B).
Fig. 3.
Comparison of Ca2+/CaM dependency of MLCK4 activity to skeletal MLCK. Representative Lineweaver–Burk plot of (A) MLCK4 and (B) skMLCK rates in the presence of EGTA (open circle) and Ca2+/CaM (filled circle).
Immunoprecipitated cMLCK from Mouse Ventricles Has both Ca2+/CaM-Dependent and -Independent Activities.
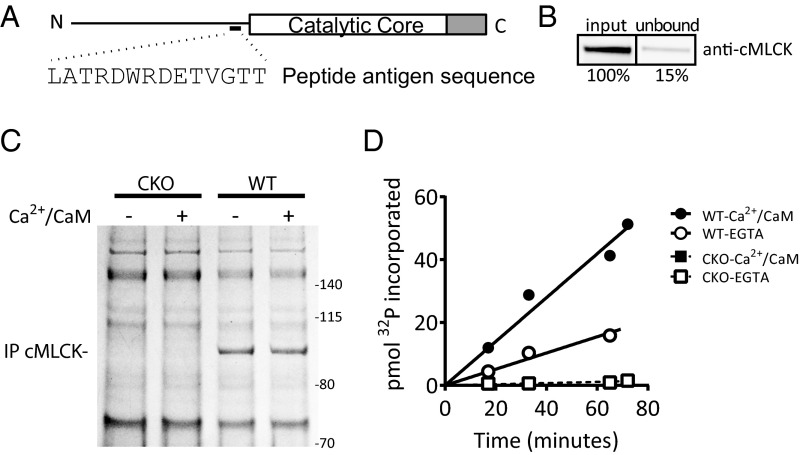
To resolve the discrepancy in the reported Ca2+/CaM dependency of cMLCK activity, a new custom antibody was generated against a peptide fragment N terminus of the catalytic core (Fig. 4A). Comparison of cMLCK in input and unbound fractions showed an immunoprecipitation efficiency 85% (Fig. 4B). Comparison of immunoprecipitated proteins from the hearts of cMLCK-KO and WT mice by Coomassie-stained gel analysis showed specific immunoprecipitation of cMLCK expressed in the heart (Fig. 4C). Coimmunoprecipitation of other proteins specifically with cMLCK was not observed. Immunoprecipitated cMLCK was assayed for activity by measurement of 32P incorporated over time, with purified cRLC (20 µM) in the presence and absence of Ca2+/CaM (Fig. 4D). The initial kinase rate in EGTA normalized to cMLCK contents in assay mixtures was 50 ± 8% of the rate in the presence of Ca2+/CaM.
Fig. 4.
Kinase activity of endogenous cMLCK immunoprecipitated from mouse hearts. (A) Illustration of cMLCK depicting the location of the peptide antigen sequence used to generate the custom antibody used in immunoprecipitations. C terminus of the catalytic core (gray bar) represents the autoregulatory segment. (B) Immunoblot of cMLCK detected from equivalent volumes of heart homogenate before (input) and after (unbound) immunoprecipitation. Quantified amount of cMLCK in unbound fraction is shown as percentage of input. Representative gel bands are shown, taken from the same immunoblot image. (C) Coomassie-stained gel of immunoprecipitated proteins from cMLCK-KO (CKO) and WT hearts. Proteins used to assay for kinase activity in the presence (+) or absence (−) of Ca2+/CaM are shown. Apparent molecular weights are noted. (D) Representative time course of 32P incorporated into purified cRLC in vitro by immunoprecipitated proteins shown in C.
The Properties of GST-Tagged Truncated cMLCK Purified from Sf9 Insect Cell Expression System Are Similar to Full-Length cMLCK Found in Mouse Hearts.
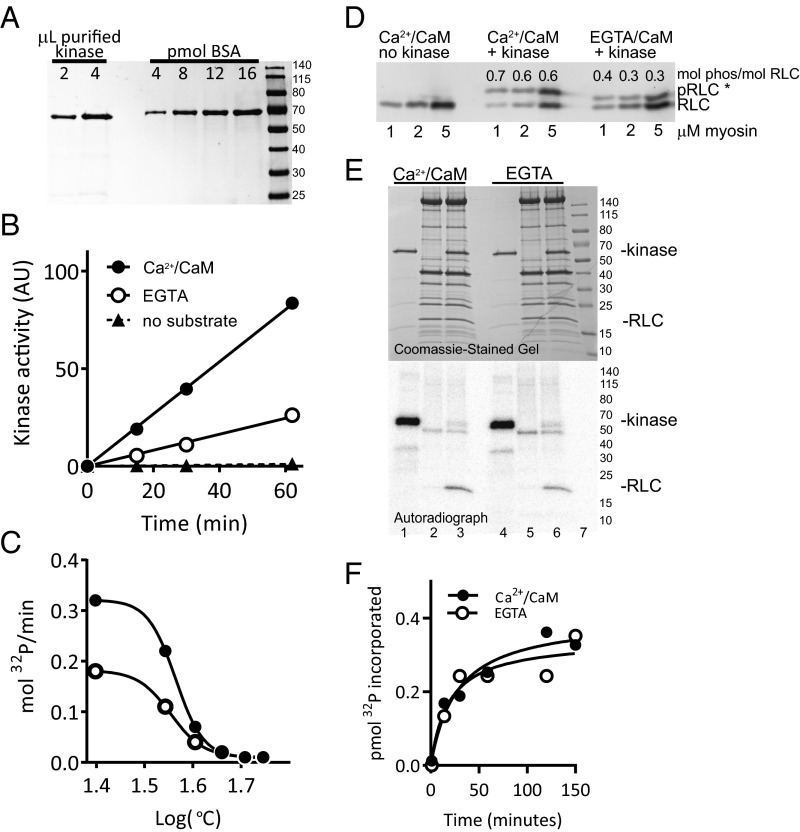
To facilitate rigorous measurements of the enzymatic properties of cMLCK in vitro, and to confirm Ca2+/CaM-independent activity measured with immunoprecipitated endogenous cMLCK, a truncation mutant was generated whereby the sequences N-terminal of the catalytic core were replaced with a GST-tag. The kinase was overexpressed by using the Sf9 insect cell system and affinity-purified in tandem with glutathione-agarose and CaM-Sepharose resins. cMLCK was purified to >90% purity (Fig. 5A) and assayed for activity in the presence and absence of Ca2+/CaM (Fig. 5B). Kinase that selectively bound to CaM-Sepharose had Ca2+/CaM-dependent and -independent activities, consistent with the activities of full-length cMLCK immunoprecipitated from mouse ventricles. Kinase activity measured after thermal denaturation showed equivalent sensitivity to increased temperature in the presence and absence of Ca2+/CaM (Fig. 5C). These results are not consistent with the possibility of two distinct populations of purified kinase accounting for the observed Ca2+/CaM-dependent and -independent activities. Validating the use of the truncated kinase to gain insight into the properties of cMLCK found in vivo, the purified kinase had Ca2+/CaM-dependent and -independent activities when whole myosin or cardiac myofibrils were used as substrate (Fig. 5 D and E). Phosphorylation of cMLCK was detected by autoradiograph from purified cMLCK incubated alone in assay conditions (Fig. 5E, lanes 1 and 4). Extent of maximum cMLCK phosphorylation was measured by two methods, autoradiography of proteins on the same film as a 32P standard curve and liquid scintillation spectrometry of incorporated 32P in an extended assay (5–150 min) of kinase phosphorylation (Fig. 5F). Autoradiography and calculation from a standard curve showed that 7.5% of total cMLCK protein in Ca2+/CaM and 6.6% in EGTA were phosphorylated in the absence of other specific substrates. Addition of BSA did not affect the extent of phosphorylation. Liquid scintillation spectrometry measurements showed that the maximum extent of phosphorylation was 2.6% of total kinase, and the calculated rate of autophosphorylation was 0.008 mol 32P incorporated/min/mol kinase. The low extents and rates of autophosphorylation, along with the absence of autophosphorylation in the presence of RLC, indicate that cMLCK is probably not significantly autophosphorylated in vivo.
Fig. 5.
Kinase activity of expressed cMLCK purified from Sf9 cells. (A) Coomassie-stained gel of purified GST-cMLCK (residues 447–795) with BSA loading curve. (B) Kinase activity of purified GST-cMLCK in EGTA or Ca2+/CaM. (C) Kinase activity of purified GST-cMLCK in EGTA (open circle) or Ca2+/CaM (filled circle) after thermal denaturation. (D) Immunoblot for total cRLC after separation of proteins by Phos-tag-PAGE. Cardiac myosin purified from mouse ventricles was phosphorylated by purified GST-cMLCK in vitro in EGTA or Ca2+/CaM. Numbers above bands are moles of phosphate incorporated per mole of RLC. (*Faster migration by phosphorylated RLC in EGTA reaction set is affected by EGTA in the manganese-Phos-tag acrylamide gel system.) (E) Phosphorylation of RLC in mouse cardiac myofibrils. Kinase assay mixtures of GST-cMLCK alone (lanes 1 and 4), myofibrils alone (lanes 2 and 5), or combined (lanes 3 and 6) were incubated in EGTA or Ca2+/CaM. Assay mixtures were separated by SDS/PAGE, then Coomassie-stained (Upper) and autoradiographed (Lower) to identify phosphorylated proteins. (F) Representative time course of 32P incorporated into GST-cMLCK by autophosphorylation in EGTA (open circles) or Ca2+/CaM (filled circles).
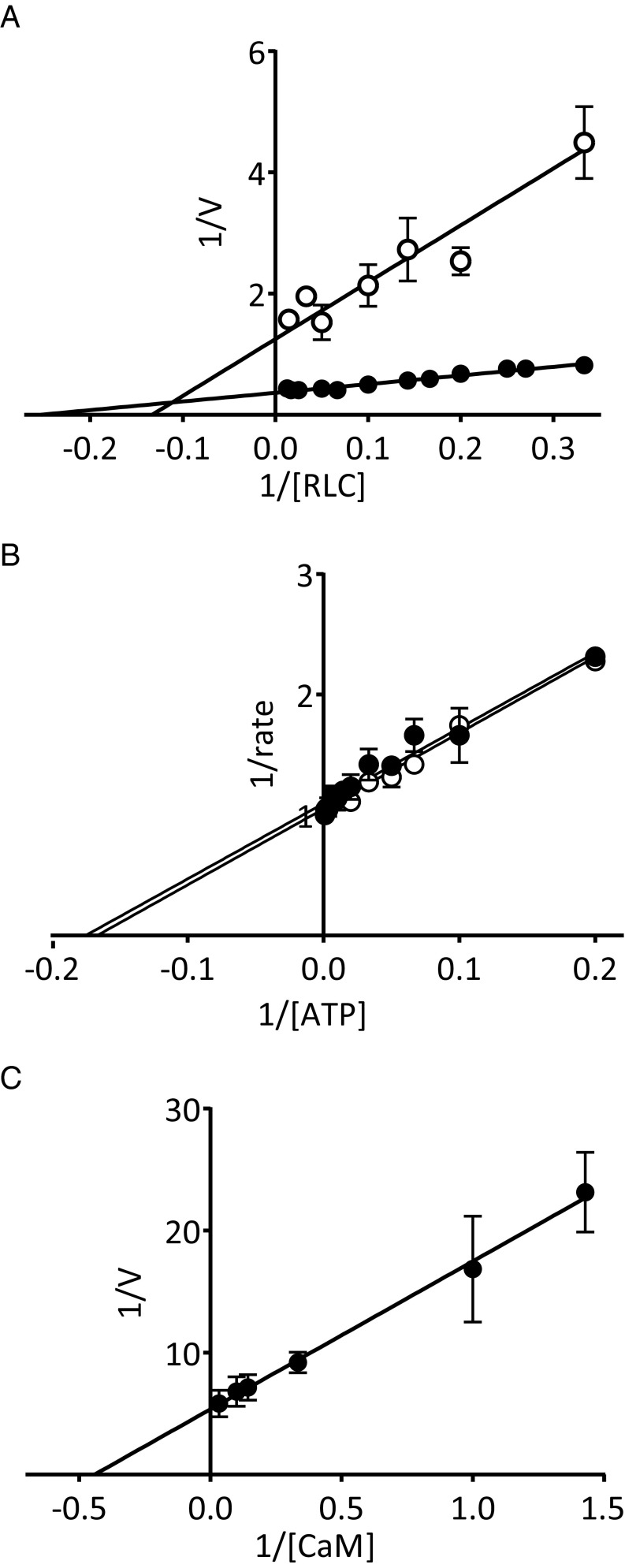
cMLCK Has a Low Vmax Value with RLC and ATP Km and KCaM Values Comparable to Other MLCKs.
In the presence of Ca2+/CaM and cRLC, cMLCK had a Vmax value of 2.6 ± 0.1 mol phosphate/min/mol kinase and a Km of 3.4 ± 0.4 µM. In EGTA, the Vmax value was 0.7 ± 0.1 mol phosphate/min/mol kinase and Km was 4.4 ± 0.9 µM (Fig. 6A). The ATP Km value in the presence and absence of Ca2+/CaM was not significantly different, at 6.3 ± 0.5 µM in Ca2+/CaM and 6.2 ± 0.1 µM in EGTA (Fig. 6B). KCaM was 2.3 ± 0.4 nM for cMLCK (Fig. 6C), comparable to values obtained for smooth and skeletal MLCK, which are absolutely dependent on Ca2+/CaM for activation (1).
Fig. 6.
Measurement of the enzymatic properties of cMLCK. Lineweaver–Burk plots of cMLCK rates in the presence of EGTA (open circle) and Ca2+/CaM (filled circle) with (A) RLC (in µM), (B) ATP (in µM), and (C) CaM (in nM).
Low Ca2+/CaM-Dependent and Independent Activities of cMLCK Are Not Caused by the Autoregulatory Sequence.
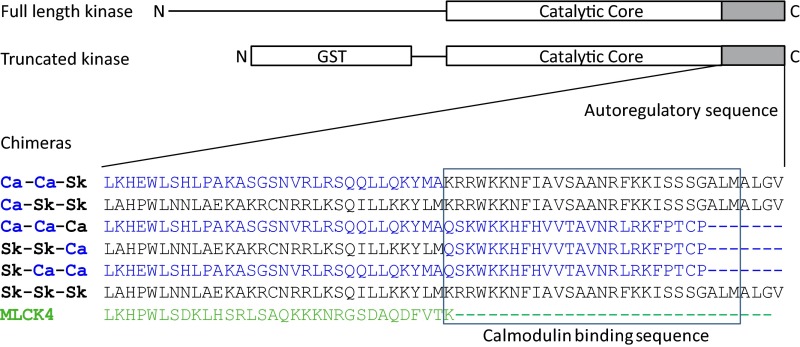
Chimeras of cMLCK and skMLCK were designed to determine contributions of distinct autoregulatory sequences to the unique properties of cMLCK (Fig. 7). The cMLCK chimeras were generated whereby the whole autoregulatory segment, or CaM-binding sequence only, were exchanged with that of skMLCK. In addition, skMLCK chimeras with cardiac autoregulatory or CaM binding sequences were generated to determine effects of the cardiac sequences on the Ca2+/CaM-dependency of skMLCK. Chimeras were compared with WT cMLCK and skMLCK.
Fig. 7.
Scheme of chimeric kinases designed to test contributions of the autoregulatory and CaM-binding sequences to the Ca2+/CaM-independent activity of cMLCK. Names of chimeras correspond to sequence origin for interchanged segments. Ca-Ca-Sk is truncated cMLCK with skMLCK CaM binding sequence; Ca-Sk-Sk is truncated cMLCK with skMLCK autoinhibitory and CaM binding sequence; Ca-Ca-Ca is WT cMLCK; Sk-Sk-Ca is truncated skMLCK with cMLCK CaM binding sequence; Sk-Ca-Ca is truncated skMLCK with cMLCK autoinhibitory and CaM binding sequence; and Sk-Sk-Sk is WT skMLCK.
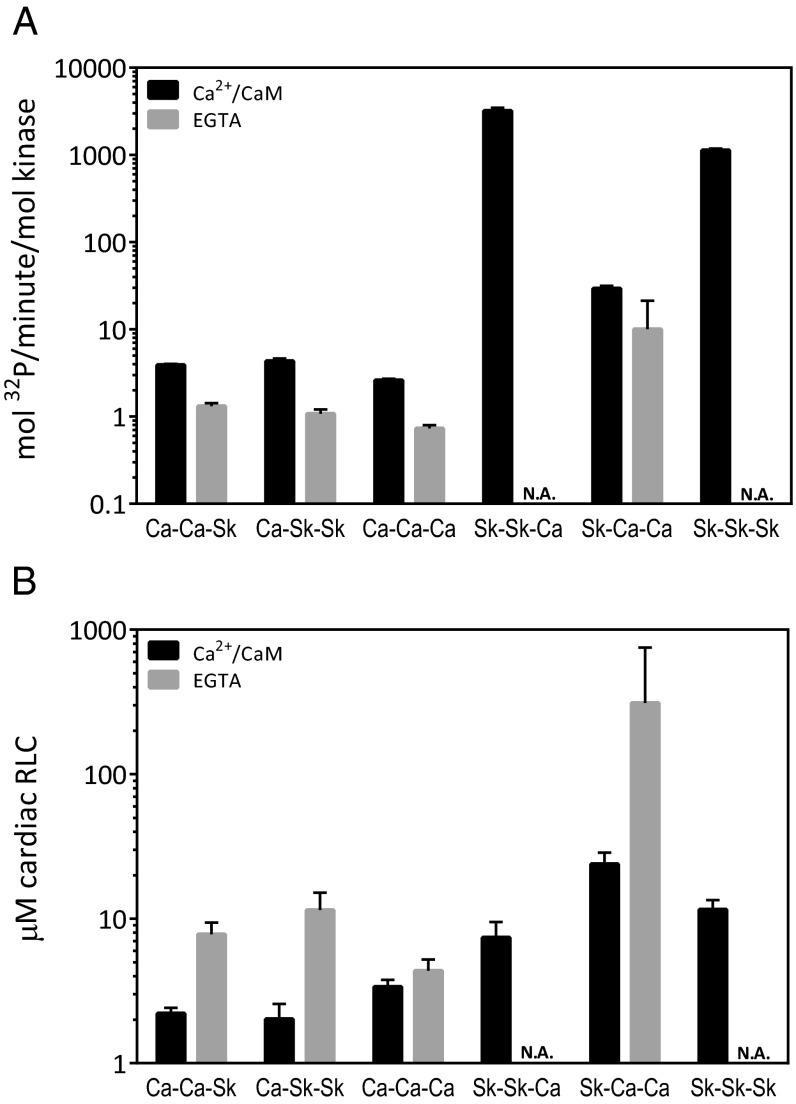
Comparisons of Vmax and Km values of cMLCK chimeras vs. WT controls showed that the low activity of cMLCK did not increase with skMLCK sequences (Fig. 8 and Table 1). In addition, the presence of skMLCK autoregulatory sequences did not affect the Ca2+/CaM-independent activity of cMLCK (Fig. 8). Comparisons of Vmax and Km values of skMLCK chimeras vs. WT controls showed no effect of cardiac CaM binding sequence on skMLCK activity, consistent with the comparable KCaM values for cMLCK and skMLCK. Exchange of the whole autoregulatory segment of skMLCK for cardiac sequence caused a 100-fold decrease in Vmax, from 3,204 ± 283 mol phosphate/min/mol kinase. When only the CaM binding sequence was from cMLCK, the Vmax value was 29 ± 2 mol phosphate/min/mol kinase. The Km increased from 7.4 ± 2 µM with cardiac CaM-binding sequence only to 24 ± 5 µM with the whole cardiac autoregulatory segment. Exchange of the skMLCK’s autoregulatory sequence for that of cMLCK sequence caused the appearance of Ca2+/CaM-independent activity (Fig. 8). Because of the high Km value of >200 µM in EGTA, the Vmax value for Ca2+/CaM-independent activity could not be accurately measured by using cRLC. However, the significant kinase activity in EGTA suggests that replacement of the skMLCK autoregulatory segment with cardiac sequences partially activates the kinase, mimicking the constitutive activity of cMLCK.
Fig. 8.
Comparison of chimera activities with cRLC. Comparison of (A) Vmax values and (B) cRLC Km values in the presence of EGTA (gray) or Ca2+/CaM (black). Bars are plotted on a logarithmic scale to accommodate values that vary by multiple orders of magnitude.
Discussion
Increased cRLC phosphorylation in vivo is associated with improved cardiac performance and resistance to maladaptive hypertrophy (15, 27). Moreover, RLC phosphorylation is decreased in several models of heart failure, underscoring the importance of cRLC phosphorylation for normal cardiac function. With the exceptions of two recent reports (31, 32), ventricular cRLC is reported to be phosphorylated in normal beating hearts, at approximately 0.40 mol phosphate/mol RLC when tissue extraction minimizes phosphatase activity with reasonable recovery of total myofilament proteins (6, 7, 11–13, 15–20, 24, 27, 33–39). We and others have previously reported that cMLCK is the primary kinase responsible for RLC phosphorylation (8, 9, 40), but, in the conventional KO animal hearts, RLC phosphorylation at S15 is not completely abolished (6). Thus, another kinase is present in the heart that phosphorylates RLC in the absence of cMLCK.
We know skMLCK is not expressed in the heart (21), and smMLCK is not a kinase for cRLC (9). After careful evaluation of commercially available antibodies toward MLCK4 (Fig. S1), we have found that MLCK4 is a soluble protein kinase significantly expressed in the heart, so we explored its biochemical properties. We solved a crystal structure of MLCK4, providing what is, to our knowledge, a first model for the MLCK family. Surprisingly, we found that a short regulatory helix is still present in MLCK4. As a result of deletions in the linker region to helix αI and the first turn of the regulatory helix, this structural element forms a PRH that does not inhibit catalysis. Additionally, there is no Ca2+/CaM binding sequence. Enzymatic assays confirmed that MLCK4 is indeed a constitutively active kinase with no Ca2+/CaM-dependency. The amount of MLCK4 expressed in cardiac myocytes is 20-fold less than cMLCK (6), but MLCK4 has a Vmax value that is 50-fold higher than cMLCK in vitro (Table 1). Thus, MLCK4 is potentially a significant contributor to RLC phosphorylation in intact cells, indicating that MLCK4 is a candidate kinase responsible for the residual 20–25% of the 0.4 mol phosphate/mol RLC in cMLCK-KO hearts. This hypothesis needs to be tested, and potential regulatory mechanisms for MLCK4 remain to be investigated. Potential roles of MLCK4 in noncardiac tissues also need exploration.
Fig. S1.
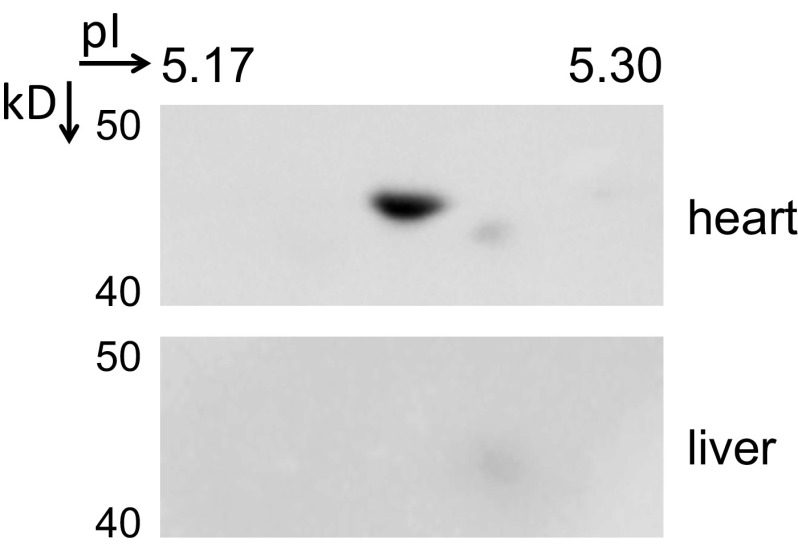
Immunoblot for MLCK4 in heart and liver tissue extracts after separation by 2D PAGE. A protein of similar size as MLCK4 that is detected in heart and liver extracts using an antibody from Proteintech does not focus to the expected isoelectric point in the liver extract. Molecular weights and isoelectric points are denoted.
RLC is dephosphorylated in nonbeating hearts and rephosphorylated with restoration of rhythmic contractions (6, 7, 41). Thus, cMLCK activity may be regulated by cytosolic [Ca2+], similar to the regulation of smMLCK and skMLCK activities (1). However, there are two conflicting reports on the Ca2+/CaM-dependency of cMLCK activity (8, 9). One study added no magnesium to form MgATP as a kinase substrate, and thus addition of EGTA probably inhibited cMLCK activity as a result of chelation of all divalent metal ions (9). The other study used RLC concentrations too low to accurately assess Vmax values (8). Whether cMLCK is Ca2+/CaM-dependent is important to resolve biochemically to identify and understand the regulatory mechanisms for RLC phosphorylation in vivo.
We took two approaches to investigate whether cMLCK activity is Ca2+/CaM-dependent. First, we expressed and purified the kinase domain of cMLCK, which is comprised of the catalytic core and autoregulatory segment. Control experiments confirmed activities measured were from the expressed kinase (Fig. S2). Kinase assays were performed with purified RLC, intact cardiac myosin and cardiac myofibrils. Second, we generated a custom antibody raised to an epitope N-terminal of the catalytic core which efficiently immunoprecipitates cMLCK directly from heart extract for measurement of cMLCK for kinase activity. The immunoprecipitation of cMLCK provides purification to control for any nonspecific protein kinase activity that may be comparable to the low cMLCK activity in total cell lysates. With both approaches, we measured low cMLCK activity in EGTA that was stimulated by Ca2+/CaM. The Km values for RLC and ATP were similar to those reported for skMLCK and smMLCK (3, 42–44). Additionally, the KCaM value of the Ca2+/CaM stimulated activity of cMLCKwas also similar to values reported for skMLCK and smMLCK (3). The primary difference in Ca2+/CaM-independent and Ca2+/dependent activities are in the Vmax values. We were careful to use RLC concentrations that were greater and less than the Km values to accurately determine these Vmax values (45).
Fig. S2.
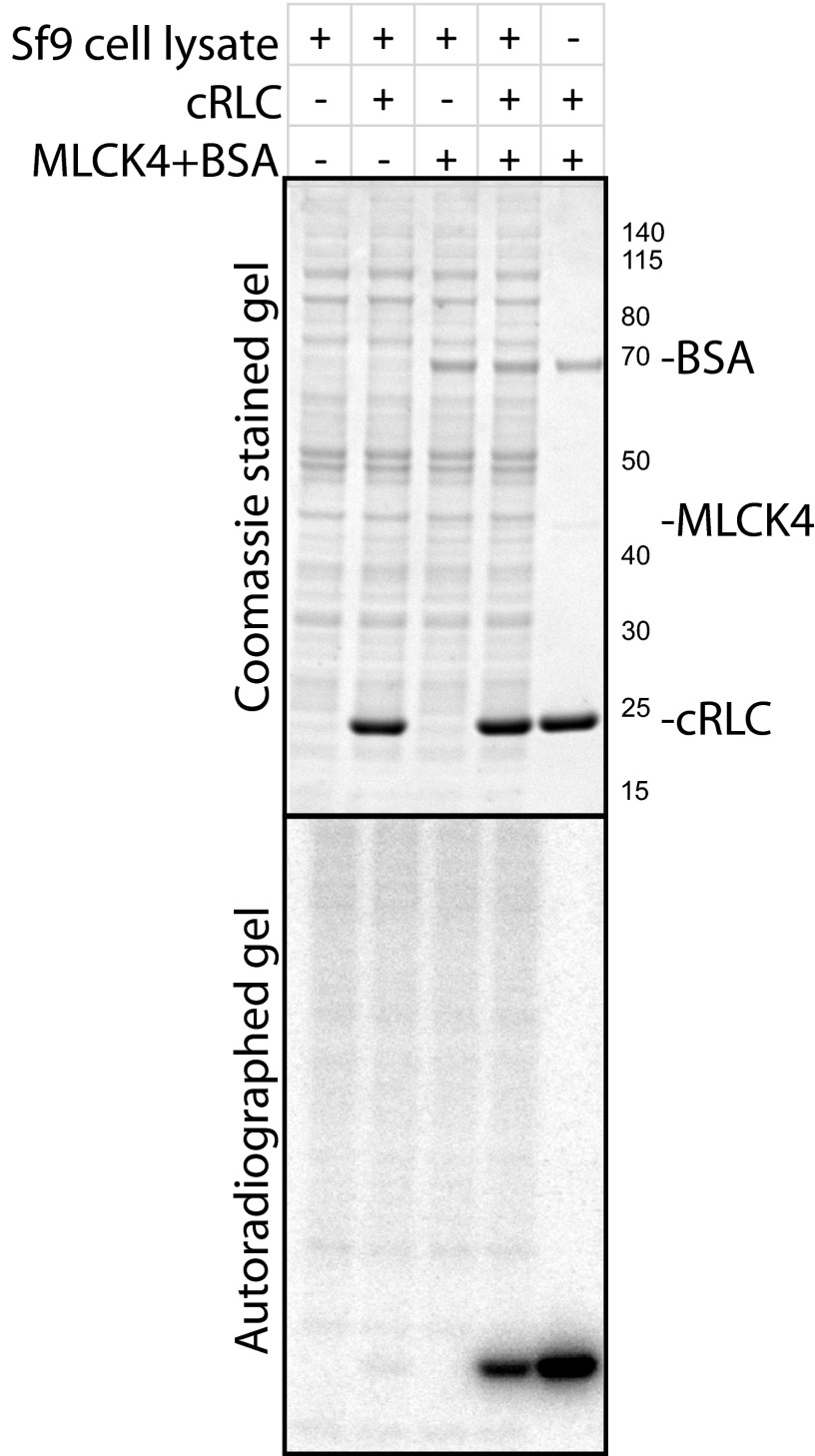
Specific phosphorylation of cRLC by MLCK4 purified from Sf9 cells. In vitro kinase assays performed with purified MLCK4 and cRLC in the presence and absence of Sf9 cell lysates showed that cRLC phosphorylation is specific to the presence of MLCK4.
Comparison of the Coomassie-stained proteins that were immunoprecipitated from WT and cMLCK-KO hearts did not reveal any coimmunoprecipitated proteins associated specifically with cMLCK. This result is consistent with our previous reports that cMLCK is soluble in heart homogenates, and that ectopically overexpressed GFP-tagged cMLCK diffuses out of detergent-treated live adult cardiac myocytes (6, 12). Collectively, these results suggest that cMLCK does not form a complex with another protein in the heart that might affect its activity.
cMLCK was reported to be autophosphorylated (8), and we have also observed autophosphorylation. skMLCK is known to be rapidly autophosphorylated by an intramolecular mechanism, but the phosphorylation does not affect its catalytic activity, and a potential function remains unrecognized (13). Measurements of the low extent and rate of cMLCK autophosphorylation in vitro, and lack of autophosphorylation in the presence of its physiological substrate RLC, indicates that it is not likely to be a significant physiological regulatory mechanism. This conclusion does not negate regulation of phosphorylation from other protein kinases.
The low kinase activity (i.e., Vmax value) and Ca2+/CaM-independent activity that is stimulated by Ca2+/CaM are two properties that distinguish cMLCK from smMLCK and skMLCK (Table 2). Unlike smooth and skeletal muscles with defined contraction and relaxation mechanisms, the heart beats continuously and never fully relaxes for any extended period. Based on the apparent high affinity of Ca2+/CaM for cMLCK, the rate of dissociation of CaM from cMLCK is predicted to be slow (on the order of sec−1) with diffusion-limited binding and activation (46). In a beating heart, cMLCK may thus be saturated with Ca2+/CaM, with maximal activity. However, limited RLC phosphorylation in beating hearts suggest that the low activity of cMLCK is in balance with slow dephosphorylation by myosin light chain phosphatase activity, consistent with a slow RLC phosphate turnover rate (14). In nonbeating hearts, cMLCK activity in the absence of Ca2+/CaM is not sufficient to exceed the phosphatase activity to maintain 40% phosphorylation. Thus, stimulation by Ca2+/CaM allows fine tuning of RLC phosphorylation by cMLCK.
Table 2.
Kinetic parameters of skMLCK, smMLCK, and cMLCKs
| MLCK | skRLC | smRLC | cRLC | ATP | |||||||
| Km | Vmax | Vmax/Km | Km | Vmax | Vmax/Km | Km | Vmax | Vmax/Km | Km | KCaM | |
| smMLCK | 100 | 600 | 6 | 9 | 2,055 | 228 | 63 | 1,800 | 50 | 50 | 1 |
| skMLCK | 10 | 3,300 | 330 | 15 | 4,817 | 321 | 9 | 3,563 | 396 | 340 | 1 |
| cMLCK | 14 | 0.5 | 0.04 | 5 | 0.8 | 0.17 | 3 | 2.6 | 0.76 | 6 | 2 |
| cMLCK (EGTA) | 54 | 0.3 | 0.01 | 21 | 0.4 | 0.02 | 4 | 0.7 | 0.08 | 6 | — |
| MLCK4 | 14 | 42 | 3 | 8 | 134 | 16.75 | 8 | 129 | 14.44 | — | — |
Average Km values for skeletal RLC (skRLC), smooth (smRLC), and cRLC (in µM) and ATP (in µM) and Vmax (in mol phosphate/min/mol kinase) and KCaM (in nM) values were obtained from double-reciprocal plots of at least three independent experiments for cMLCK and MLCK4. For cMLCK, Ca2+/CaM-dependent and independent (EGTA) values are shown. Assays were performed as described in Materials and Methods. Measured values are summarized in the table without SE values. Ca2+/CaM-dependent values for smMLCK and skMLCK were previously published (3, 42–44).
A shared property between autoregulated MLCKs is the presence of an autoinhibitory sequence N-terminal of the CaM binding sequence. We argued that, if cMLCK was constitutively partially autoinhibited, it would have low kinase rates and activity in the absence of Ca2+/CaM. This possibility was tested by comparing kinase rates of chimeric cMLCK and skMLCK, whereby the autoregulatory sequences were exchanged. The Ca2+/CaM-independent activity of cMLCK did not disappear when the autoregulatory sequence was changed to sequences of skMLCK, and the Vmax value did not increase, which suggests that the low kinase rate and constitutive activity is intrinsic to the catalytic core sequence. Interestingly, exchanging the autoregulatory sequence of skMLCK for cardiac sequences caused an increase in Ca2+/CaM-independent activity and a large decrease in Vmax for the skMLCK catalytic domain. The distinct effects of exchanging the autoregulatory sequences of cMLCK and skMLCK could be caused by differential destabilization of the catalytic core. In the absence of Ca2+/CaM, the autoinhibitory sequence of skMLCK may bind into the catalytic cleft, stabilizing the catalytic core in a closed conformation (5, 47). The cardiac autoregulatory sequence in the skMLCK chimera may not effectively inhibit the catalytic domain in the absence of Ca2+/CaM. For both kinases, detailed structural information is necessary to determine the intermolecular interaction between the catalytic core and autoregulatory segment.
In summary, the low constitutive activity of cMLCK stimulated by Ca2+/CaM is a distinct property that is not shared by other MLCK family members, and it appears to be intrinsic to the catalytic core sequence. This Ca2+/CaM-stimulated activity over Ca2+/CaM-independent cMLCK appears to maintain RLC phosphorylation at 0.40 mol phosphate/mol RLC, which is necessary for normal cardiac performance. Additionally, MLCK4 may also contribute to the extent of RLC phosphorylation in cardiac muscle.
Materials and Methods
Animals.
All procedures were performed in accordance with the institutional animal care and use guidelines at University of Texas Southwestern Medical Center, with all animal experimental procedures reviewed and approved by the institutional animal care and use committee. Animals were housed under standard conditions in the rodent facility.
Statistical Analyses.
Data are expressed as mean ± SE. Statistical evaluation was carried out in GraphPad Prism using ANOVA with Dunnett’s posttest for comparison with control. Significance was accepted at a value of P < 0.05.
Quantification of MLCK4 Protein.
Tissues from WT anesthetized mice were homogenized in 30× volume of homogenization buffer (50 mM Tris, pH 8.0, 50 mM NaF, 1% Nonidet P-40, 2 mM EGTA, 0.1% sodium deoxycholate, 0.1% Brij-35, 2× Halt Protease Inhibitor mixture, 10 µM E-64) and lysed on ice for 15 min, and the supernatant fraction was collected after centrifugation at 20,000 × g for 2 min. Adult cardiac myocytes and cardiac nonmuscle cells were isolated as previously described (6). Cells were lysed in the tissue homogenization buffer. Protein concentration was determined by Bradford assay, and 10 µg of total protein was boiled in 1× LDS Buffer (Invitrogen) with reducing reagent (Invitrogen) and separated by 4–12% Bolt gradient gel (Invitrogen). Separated proteins were immunoblotted for MLCK4 and GAPDH. Antibody to MLCK4 was from Abcam (Ab179395), and antibody to GAPDH was from Santa Cruz (sc25778). Antibody to cMLCK was previously described (6).
Tissue Harvest and Preparation.
Heart for immunohistochemistry was harvested from anesthetized mice and fixed via retrograde perfusion with 4% (wt/vol) paraformaldehyde, freshly prepared in PBS solution. Subsequent paraffin processing, embedding, and sectioning were performed by standard procedures (48, 49).
Immunohistochemistry.
Rabbit anti-sera used for MYLK4 immunolabeling of paraffin heart sections was obtained from Abcam (Ab179395). Following deparaffinization and heat antigen retrieval with 10 mM Tris/1 mM EDTA, 0.05% Tween-20 (pH 9.0), sections were blocked against endogenous peroxidase activity and secondary antibody host-serum affinity. Serial sections were then subjected to primary antibody (1:33 dilution of commercially supplied stock) or normal rabbit serum and incubated overnight at 4 °C. Subsequent biotin/streptavidin HRP detection of bound primary was conducted the following day according to previously described immunoperoxidase methods (50, 51).
Immunoprecipitation.
Ventricles from WT or cMLCK-KO anesthetized mice were rapidly frozen in liquid nitrogen and stored at −80 °C. Frozen ventricles were homogenized/thawed for 1 min by using a ground-glass homogenizer in 10× volume of homogenization buffer (50 mM Tris, pH 8.0, 50 mM NaF, 1% Nonidet P-40, 2 mM EGTA, 0.1% sodium deoxycholate, 0.1% Brij-35, 2× Halt Protease Inhibitor mixture, 10 µM E-64). Homogenates were lysed on ice for 15 min, and then supernatant fraction was collected after centrifugation at 20,000 × g for 2 min. Protein-A agarose (Thermo Fisher) prebound with a polyclonal antibody raised to a peptide N terminal to the catalytic core of mouse cMLCK, designed and produced by Genscript, was used to immunoprecipitate endogenous cMLCK from the supernatant fraction. Antibody-bound beads were incubated with the supernatant fraction for 2 h, rocked at 4 °C, then washed three times in PBS solution. Immunoprecipitated proteins were eluted by boiling in 1× LDS buffer (Invitrogen) with reducing reagent (Invitrogen) and separated by 4–12% Bolt gradient gel (Invitrogen). Separated proteins were visualized by staining with Coomassie (Sigma).
Immunoblot of Phosphorylated Myosin.
Myosin was purified from mouse ventricles by using low-salt precipitation steps at 4 °C, similar to the original protocol by Murakami and Uchida (52). Purified mouse cardiac myosin was phosphorylated in vitro with purified GST-cMLCK for 15 min at 30 °C. Reactions were terminated with addition of 10% trichloroacetic acid containing 10 mM DTT. Precipitated protein was washed free of acid with three 5-min washes in ethyl ether and resuspended by vigorous agitation in urea sample buffer (8 M urea, 20 mM Tris base, 23 mM glycine, 0.2 mM EDTA, 10 mM DTT) by using an orbital shaker (IKA Vibrax VXR) set at 1,400 rpm for 30 min at room temperature. Complete denaturation and solubilization was achieved by further addition of urea crystals and prolonged agitation. Solubilized proteins were subjected to 30 µM Phostag-10% SDS/PAGE after boiling in Laemmli buffer and transferred to PVDF (Immobilon-P; Millipore). Proteins were fixed onto the PVDF membrane with 0.4% glutaraldehyde/PBS solution for 15 min at room temperature. The membrane was then rinsed three times in PBS solution and immunoblotted with antibody to cRLC (Enzo).
Kinase Activity Assay.
Kinase activities were assayed in 10 mM Mops, pH 7.4, 5 mM MgCl2, 100 mM NaCl, 0.3 mM CaCl2 or 3 mM EGTA, 1.5 µM CaM, 1 mM DTT, and 0.2 mM [γ-32P]ATP (100–300 cpm/pmol) with purified proteins in 40 μL total volume. Reaction mixtures were preincubated for 5 min, and the kinase activity was measured at 30 °C by the addition of [γ-32P]ATP as described previously (53). For measurement of ATP Km values, 0.1 µM cMLCK was assayed with 15 µM RLC in 1–150 µM [γ-32P]ATP. For measurement of RLC Km and Vmax values, 1–50 nM kinase (empirically determined for each kinase used) was assayed with 0.1–120 µM RLC. Km and Vmax values were calculated by nonlinear fit to the Michaelis–Menten equation or linear fit to Lineweaver–Burk plots by using GraphPad Prism 6.0 software. Kinase concentrations were verified by silver stain of assay mixtures. For measurement of Ca2+/CaM required for half-maximal activation of cMLCK (KCaM values), assays were performed at 25 °C for 4–8 h with 0.5 nM cMLCK to measure the rate of cMLCK activity at a concentration below the measured KCaM value (46).
Expression and Purification of Kinases.
All kinases were expressed in Sf9 cells and affinity-purified. Cells were lysed for 20 min in 20 mM Mops, pH 7.4, 0.5 mM EGTA, 1% Nonidet P-40, 1 mM DTT, 1× Halt Protease Inhibitor mixture (Pierce), and 10 µM E-64. Lysates were centrifuged for 30 min at 20,000 × g (Beckman Coulter), and supernatant fractions were collected for purification procedures. GST-tagged cMLCK and chimeras were purified with glutathione-agarose (ThermoFisher) and CaM-Sepharose (GE Healthcare) by using procedures found in the instruction manual. skMLCK was a gift from Kathy Trybus, University of Vermont, Burlington, VT.
Crystallization of MLCK4.
MYLK4 (residues K40–K388) in frame with an N-terminal His tag and TEV protease site (MGHHHHHHSSGVDLGTENLY FQ^SM) was expressed in Sf9 cells at 27 °C. After 48 h infection by the virus, the cells were harvested by centrifugation, washed once with PBS solution, and resuspended in 3 three volumes of lysis buffer (50 mM Hepes, 300 mM NaCl, 5% glycerol, pH 7.5, 1 mM TCEP, 1:1,000 dilution of protease inhibitor mixture; Promega). The cell suspension was then lysed by sonication, and the lysate was cleared by centrifugation in a Beckman JA-17 rotor at 36,000 × g for 45 min. The supernatant fraction was mixed with 10 mL of 50% Ni-NTA slurry. The mixture was rotated for 1 h at 4 °C and then loaded on a column, which was washed with 150 mL of wash buffer (50 mM Hepes, 300 mM NaCl, 5% glycerol, pH 7.5) and wash buffer with 25 mM imidazole. The protein was eluted using 300 mM imidazole in wash buffer. The eluate of the nickel-affinity column was diluted in 50 mM Hepes, 5% glycerol to achieve a final NaCl concentration of 150 mM and loaded onto an anion-exchange HiTrap Q FF column. The protein was eluted by using a liner gradient ranging from 150 mM to 2 M NaCl in 50 mM Hepes, 5% glycerol, pH 7.5. The fractions containing recombinant MLCK4 were pooled, concentrated, and applied to a Superdex 75 GF column equilibrated in GF Buffer (50 mM Hepes, 300 mM NaCl, 5% glycerol, pH 7.5). The eluted protein was more than 95% pure as judged by SDS/PAGE. Liquid chromatography electrospray ionization MS (time-of-flight) revealed the expected mass of the protein (42,433 Da) as predicted from the expressed sequence after TEV cleavage. This purified kinase was used for kinase assays herein.
Crystals were grown at 4 °C in 300-nL sitting drops from a 2:1 ratio of protein (10 mg/mL) to reservoir solution containing 2 M ammonium sulfate and 2.5 wt/vol PEG400 in Hepes buffer (50 mM), pH 8.0. The ATP mimetic inhibitor [4-(2 amino-4-methyl-1,3-thiazol-5-yl)-n-(3-dioxaziridin-3-yl phenyl)pyrimidine-2-amine] was added to the concentrated protein from a 50-mM DMSO stock solution. For data collection, crystals were cryoprotected by using the well solution supplemented with 2 M Li2SO4 and flash-frozen in liquid nitrogen. Diffraction data were collected from a single crystal on Diamond beamline IO2 at a single wavelength of 0.9802 Å, and the structure was refined to 2.8 Å. Indexing and integration was carried out by using MOSFLM (54), and scaling was performed with SCALA (55). The structure was solved by molecular replacement (PHASER) using the structure of Protein Data Bank (PDB) ID code 1KOB as a starting model (56). COOT was used for model building, and refinement was carried out in REFMAC5 (57, 58). Thermal motions were analyzed using TLSMD, and defined domains were used in later cycles of refinement. The coordinates have been deposited in the PDB with the accession code 2X4F.
MLCK4 Is Not Expressed in Liver
Significant expression of a ∼44-kDa protein was detected in heart and liver extracts by immunoblot after separation by SDS/PAGE, with an MLCK4 antibody from Proteintech (no. 24309-1-AP). To confirm expression of MLCK4 in these tissues, heart and liver tissue extracts were separated by 2D gel electrophoresis and immunoblotted with the same MLCK4 antibody from Proteintech. Tissues extracts were prepared as described in Materials and Methods, then precipitated with the addition of 10% trichloroacetic acid to remove buffers and detergents that would interfere with separation of proteins in an immobilized pH gradient (IPG) strip. Precipitated proteins were washed three times for 5 min in ethyl ether, dried, and then resuspended in 2D sample buffer (ReadyPrep; Bio-Rad). Solubilized 2D samples were separated in a pH 4–7 linear 24-cm IPG strip (GE Healthcare) for 100 kVh, following procedures found in the instruction manual for the Protean IEF cell (Bio-Rad). Region corresponding to pH 4.78–5.70 on the linear IPG strip was separated by 10% SDS/PAGE, then transferred to nitrocellulose membrane for immunoblotting procedures. The ∼44-kDa protein in the liver extract did not migrate to the same isoelectric point as the protein in the heart extract when separated by 2D gel electrophoresis (Fig. S1), confirming that immunoblot signal in liver extract is from nonspecific proteins. An affinity-purified antibody from Abcam (no. 179395) is more selective and was used in all other immunoblots for MLCK4.
Sf9 Cell Lysates Do Not Phosphorylate cRLC in the Absence of MLCK4 Protein
Partially purified titin kinase (peptide substrate) (59) and a contaminating lipid kinase (lipid substrate) (60) have been shown to phosphorylate substrates in vitro when overexpressed in Sf9 cells. Nonspecific phosphorylation of cRLC was not expected because striated muscle RLCs are not promiscuous substrates as a result of primary and tertiary structural constraints (61–63), consistent with the single phosphorylation site in vivo for cardiac (53) and skeletal muscle (64) RLC. The peptide substrate used for the titin kinase (59) has the sequence for the smooth muscle RLC containing the phosphorylation site [KKRARAATS(P)NVF]. This peptide can be phosphorylated by numerous protein kinases, including CaM-dependent protein kinase II as a result of the positively charged residues N-terminal of the phosphorylatable serine.
To confirm that in vitro kinase activity measurements toward cRLC reported here are not artifacts resulting from nonspecific activities, control experiments were performed with Sf9 cell lysates (Fig. S2). Sf9 cells were collected by centrifugation and lysed in 10× volume (microliters lysis buffer per milligram cells) of lysis buffer [20 mM Mops, pH 7.4, 0.5 mM EGTA, 1% Nonidet P-40, 1 mM DTT, 1× Halt Protease Inhibitor mixture (Pierce), 10 µM E-64]. Supernatant fraction was used at 1:10 dilution in kinase assay, along with 100 nM MLCK4 that was diluted with BSA as carrier and 8 µM cRLC, as indicated in Fig. S2. Equivalent volume of Sf9 lysis buffer was added to the control sample, and assay conditions were the same as detailed in Materials and Methods. After incubation for 3 min, SDS sample buffer was added to reaction mixtures and boiled for SDS/PAGE, followed by protein staining and autoradiography. Radioactive signal from cRLC in Sf9 lysate-added mixture was not different from the background. The amount of radioactivity detected was less than 1.5% of the phosphorylation by MLCK4. Thus, there was no significant kinase activity toward cRLC in Sf9 cell lysates in the absence of MLCK4.
Acknowledgments
We thank John Shelton for help with immunohistochemistry images of MLCK4 expression in mouse hearts and Kathy Trybus for the generous gift of purified skMLCK. This work was supported by an American Heart Association Postdoctoral Fellowship (to A.N.C.), the Leducq Foundation (H.L.S.), NIH Grant HL080536 (to J.T.S.), the Moss Heart Fund (J.T.S.), and the Fouad A. and Val ImmBashour Distinguished Chair in Physiology (J.T.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2X4F).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1600633113/-/DCSupplemental.
References
- 1.Kamm KE, Stull JT. Signaling to myosin regulatory light chain in sarcomeres. J Biol Chem. 2011;286(12):9941–9947. doi: 10.1074/jbc.R110.198697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stull JT, Nunnally MH, Michnoff CH. The Enzymes, Control by Phosphorylation Part A. 3rd revised Ed. Academic; London: 1986. Calmodulin-dependent protein kinases; pp. 113–166. [Google Scholar]
- 3.Gallagher PJ, Herring BP, Trafny A, Sowadski J, Stull JT. A molecular mechanism for autoinhibition of myosin light chain kinases. J Biol Chem. 1993;268(35):26578–26582. [PMC free article] [PubMed] [Google Scholar]
- 4.Krueger JK, et al. Structures of calmodulin and a functional myosin light chain kinase in the activated complex: A neutron scattering study. Biochemistry. 1997;36(20):6017–6023. doi: 10.1021/bi9702703. [DOI] [PubMed] [Google Scholar]
- 5.Krueger JK, Zhi G, Stull JT, Trewhella J. Neutron-scattering studies reveal further details of the Ca2+/calmodulin-dependent activation mechanism of myosin light chain kinase. Biochemistry. 1998;37(40):13997–14004. doi: 10.1021/bi981311d. [DOI] [PubMed] [Google Scholar]
- 6.Chang AN, et al. Constitutive phosphorylation of cardiac myosin regulatory light chain in vivo. J Biol Chem. 2015;290(17):10703–10716. doi: 10.1074/jbc.M115.642165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.High CW, Stull JT. Phosphorylation of myosin in perfused rabbit and rat hearts. Am J Physiol. 1980;239(6):H756–H764. doi: 10.1152/ajpheart.1980.239.6.H756. [DOI] [PubMed] [Google Scholar]
- 8.Chan JY, et al. Identification of cardiac-specific myosin light chain kinase. Circ Res. 2008;102(5):571–580. doi: 10.1161/CIRCRESAHA.107.161687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seguchi O, et al. A cardiac myosin light chain kinase regulates sarcomere assembly in the vertebrate heart. J Clin Invest. 2007;117(10):2812–2824. doi: 10.1172/JCI30804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akiyama K, et al. Myocardial infarction and regulatory myosin light chain. J Mol Cell Cardiol. 1997;29(10):2641–2652. doi: 10.1006/jmcc.1997.0493. [DOI] [PubMed] [Google Scholar]
- 11.Chang AN, et al. The effects of neuregulin on cardiac Myosin light chain kinase gene-ablated hearts. PLoS One. 2013;8(6):e66720. doi: 10.1371/journal.pone.0066720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ding P, et al. Cardiac myosin light chain kinase is necessary for myosin regulatory light chain phosphorylation and cardiac performance in vivo. J Biol Chem. 2010;285(52):40819–40829. doi: 10.1074/jbc.M110.160499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fitzsimons DP, Bodell PW, Baldwin KM. Myocardial functional correlates of cardiac myosin light chain 2 phosphorylation. J Appl Physiol. 1990;68(6):2426–2433. doi: 10.1152/jappl.1990.68.6.2426. [DOI] [PubMed] [Google Scholar]
- 14.Herring BP, England PJ. The turnover of phosphate bound to myosin light chain-2 in perfused rat heart. Biochem J. 1986;240(1):205–214. doi: 10.1042/bj2400205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang J, Shelton JM, Richardson JA, Kamm KE, Stull JT. Myosin regulatory light chain phosphorylation attenuates cardiac hypertrophy. J Biol Chem. 2008;283(28):19748–19756. doi: 10.1074/jbc.M802605200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klaiman JM, Fenna AJ, Shiels HA, Macri J, Gillis TE. Cardiac remodeling in fish: Strategies to maintain heart function during temperature change. PLoS One. 2011;6(9):e24464. doi: 10.1371/journal.pone.0024464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morano I. Effects of different expression and posttranslational modifications of myosin light chains on contractility of skinned human cardiac fibers. Basic Res Cardiol. 1992;87(Suppl 1):129–141. doi: 10.1007/978-3-642-72474-9_11. [DOI] [PubMed] [Google Scholar]
- 18.Sheikh F, et al. Mouse and computational models link Mlc2v dephosphorylation to altered myosin kinetics in early cardiac disease. J Clin Invest. 2012;122(4):1209–1221. doi: 10.1172/JCI61134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taniguchi M, et al. New isoform of cardiac myosin light chain kinase and the role of cardiac myosin phosphorylation in α1-adrenoceptor mediated inotropic response. PLoS One. 2015;10(10):e0141130. doi: 10.1371/journal.pone.0141130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Toepfer C, et al. Myosin regulatory light chain (RLC) phosphorylation change as a modulator of cardiac muscle contraction in disease. J Biol Chem. 2013;288(19):13446–13454. doi: 10.1074/jbc.M113.455444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhi G, et al. Myosin light chain kinase and myosin phosphorylation effect frequency-dependent potentiation of skeletal muscle contraction. Proc Natl Acad Sci USA. 2005;102(48):17519–17524. doi: 10.1073/pnas.0506846102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morano I. Tuning the human heart molecular motors by myosin light chains. J Mol Med (Berl) 1999;77(7):544–555. doi: 10.1007/s001099900031. [DOI] [PubMed] [Google Scholar]
- 23.van der Velden J, et al. The effect of myosin light chain 2 dephosphorylation on Ca2+ -sensitivity of force is enhanced in failing human hearts. Cardiovasc Res. 2003;57(2):505–514. doi: 10.1016/s0008-6363(02)00662-4. [DOI] [PubMed] [Google Scholar]
- 24.van der Velden J, et al. Increased Ca2+-sensitivity of the contractile apparatus in end-stage human heart failure results from altered phosphorylation of contractile proteins. Cardiovasc Res. 2003;57(1):37–47. doi: 10.1016/s0008-6363(02)00606-5. [DOI] [PubMed] [Google Scholar]
- 25.Riise J, et al. Prostanoid-mediated inotropic responses are attenuated in failing human and rat ventricular myocardium. Eur J Pharmacol. 2012;686(1-3):66–73. doi: 10.1016/j.ejphar.2012.04.022. [DOI] [PubMed] [Google Scholar]
- 26.AitMou Y, et al. Beneficial effects of SR33805 in failing myocardium. Cardiovasc Res. 2011;91(3):412–419. doi: 10.1093/cvr/cvr096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warren SA, et al. Myosin light chain phosphorylation is critical for adaptation to cardiac stress. Circulation. 2012;126(22):2575–2588. doi: 10.1161/CIRCULATIONAHA.112.116202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan CC, et al. Constitutive phosphorylation of cardiac myosin regulatory light chain prevents development of hypertrophic cardiomyopathy in mice. Proc Natl Acad Sci USA. 2015;112(30):E4138–E4146. doi: 10.1073/pnas.1505819112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheikh F, Lyon RC, Chen J. Functions of myosin light chain-2 (MYL2) in cardiac muscle and disease. Gene. 2015;569(1):14–20. doi: 10.1016/j.gene.2015.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rellos P, et al. Structure of the CaMKIIdelta/calmodulin complex reveals the molecular mechanism of CaMKII kinase activation. PLoS Biol. 2010;8(7):e1000426. doi: 10.1371/journal.pbio.1000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kampourakis T, Irving M. Phosphorylation of myosin regulatory light chain controls myosin head conformation in cardiac muscle. J Mol Cell Cardiol. 2015;85:199–206. doi: 10.1016/j.yjmcc.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng Y, et al. Top-down proteomics reveals concerted reductions in myofilament and Z-disc protein phosphorylation after acute myocardial infarction. Mol Cell Proteomics. 2014;13(10):2752–2764. doi: 10.1074/mcp.M114.040675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fewell JG, et al. A treadmill exercise regimen for identifying cardiovascular phenotypes in transgenic mice. Am J Physiol. 1997;273(3 pt 2):H1595–H1605. doi: 10.1152/ajpheart.1997.273.3.H1595. [DOI] [PubMed] [Google Scholar]
- 34.Fitzsimons DP, Bodell PW, Baldwin KM. Phosphorylation of rodent cardiac myosin light chain 2: Effects of exercise. J Appl Physiol. 1989;67(6):2447–2453. doi: 10.1152/jappl.1989.67.6.2447. [DOI] [PubMed] [Google Scholar]
- 35.Holroyde MJ, Small DA, Howe E, Solaro RJ. Isolation of cardiac myofibrils and myosin light chains with in vivo levels of light chain phosphorylation. Biochim Biophys Acta. 1979;587(4):628–637. doi: 10.1016/0304-4165(79)90014-x. [DOI] [PubMed] [Google Scholar]
- 36.Mizutani H, et al. Overexpression of myosin phosphatase reduces Ca(2+) sensitivity of contraction and impairs cardiac function. Circ J. 2010;74(1):120–128. doi: 10.1253/circj.cj-09-0462. [DOI] [PubMed] [Google Scholar]
- 37.Morano I, Adler K, Agostini B, Hasselbach W. Expression of myosin heavy and light chains and phosphorylation of the phosphorylatable myosin light chain in the heart ventricle of the European hamster during hibernation and in summer. J Muscle Res Cell Motil. 1992;13(1):64–70. doi: 10.1007/BF01738429. [DOI] [PubMed] [Google Scholar]
- 38.Morano I, Osterman A, Arner A. Rate of active tension development from rigor in skinned atrial and ventricular cardiac fibres from swine following photolytic release of ATP from caged ATP. Acta Physiol Scand. 1995;154(3):343–353. doi: 10.1111/j.1748-1716.1995.tb09918.x. [DOI] [PubMed] [Google Scholar]
- 39.Wei B, Wei H, Jin JP. Dysferlin deficiency blunts β-adrenergic-dependent lusitropic function of mouse heart. J Physiol. 2015;593(23):5127–5144. doi: 10.1113/JP271225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding HL, Ryder JW, Stull JT, Kamm KE. Signaling processes for initiating smooth muscle contraction upon neural stimulation. J Biol Chem. 2009;284(23):15541–15548. doi: 10.1074/jbc.M900888200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silver PJ, Buja LM, Stull JT. Frequency-dependent myosin light chain phosphorylation in isolated myocardium. J Mol Cell Cardiol. 1986;18(1):31–37. doi: 10.1016/s0022-2828(86)80980-4. [DOI] [PubMed] [Google Scholar]
- 42.Nunnally MH, Rybicki SB, Stull JT. Characterization of chicken skeletal muscle myosin light chain kinase. Evidence for muscle-specific isozymes. J Biol Chem. 1985;260(2):1020–1026. [PubMed] [Google Scholar]
- 43.Padre RC, Stull JT. Functional assembly of fragments from bisected smooth muscle myosin light chain kinase. J Biol Chem. 2000;275(35):26665–26673. doi: 10.1074/jbc.M001769200. [DOI] [PubMed] [Google Scholar]
- 44.Stull JT, Nunnally MH, Moore RL, Blumenthal DK. Myosin light chain kinases and myosin phosphorylation in skeletal muscle. Adv Enzyme Regul. 1985;23:123–140. doi: 10.1016/0065-2571(85)90043-3. [DOI] [PubMed] [Google Scholar]
- 45.Segel IH. Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems. Wiley; New York: 1993. [Google Scholar]
- 46.Blumenthal DK, Stull JT. Activation of skeletal muscle myosin light chain kinase by calcium(2+) and calmodulin. Biochemistry. 1980;19(24):5608–5614. doi: 10.1021/bi00565a023. [DOI] [PubMed] [Google Scholar]
- 47.Krueger JK, et al. Activation of myosin light chain kinase requires translocation of bound calmodulin. J Biol Chem. 2001;276(7):4535–4538. doi: 10.1074/jbc.C000857200. [DOI] [PubMed] [Google Scholar]
- 48.Sheehan DC, Hrapchak BB. Theory and Practice of Histotechnology. Mosby; St. Louis: 1980. [Google Scholar]
- 49.Woods AE, Ellis RC. Laboratory Histopathology: A Complete Reference. Churchill Livingstone; London: 1994. [Google Scholar]
- 50.Borvak J, et al. Functional expression of the MHC class I-related receptor, FcRn, in endothelial cells of mice. Int Immunol. 1998;10(9):1289–1298. doi: 10.1093/intimm/10.9.1289. [DOI] [PubMed] [Google Scholar]
- 51.Cianga P, Medesan C, Richardson JA, Ghetie V, Ward ES. Identification and function of neonatal Fc receptor in mammary gland of lactating mice. Eur J Immunol. 1999;29(8):2515–2523. doi: 10.1002/(SICI)1521-4141(199908)29:08<2515::AID-IMMU2515>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 52.Murakami U, Uchida K. Contents of myofibrillar proteins in cardiac, skeletal, and smooth muscles. J Biochem. 1985;98(1):187–197. doi: 10.1093/oxfordjournals.jbchem.a135257. [DOI] [PubMed] [Google Scholar]
- 53.Chang AN, Chen G, Gerard RD, Kamm KE, Stull JT. Cardiac myosin is a substrate for zipper-interacting protein kinase (ZIPK) J Biol Chem. 2010;285(8):5122–5126. doi: 10.1074/jbc.C109.076489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leslie AGW, Powell HR. 2007. Processing diffraction data with mosflm. Evolving Methods for Macromolecular Crystallography, NATO Science Series, eds Read RJ, Sussman JL (Springer, Dordrecht, The Netherlands), Vol 245, pp 41–51.
- 55.Evans P. SCALA-Scale Together Multiple Observations of Reflections. 3.3.0 Ed MRC Laboratory of Molecular Biology; Cambridge, UK: 2007. [Google Scholar]
- 56.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61(Pt 4):458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 57.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(pt 12 pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 58.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53(pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 59.Bogomolovas J, et al. Titin kinase is an inactive pseudokinase scaffold that supports MuRF1 recruitment to the sarcomeric M-line. Open Biol. 2014;4(5):140041. doi: 10.1098/rsob.140041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shymanets A, Ahmadian MR, Nürnberg B. Gbetagamma-copurified lipid kinase impurity from Sf9 cells. Protein Pept Lett. 2009;16(9):1053–1056. doi: 10.2174/092986609789055340. [DOI] [PubMed] [Google Scholar]
- 61.Herring BP, Gallagher PJ, Stull JT. Substrate specificity of myosin light chain kinases. J Biol Chem. 1992;267(36):25945–25950. [PubMed] [Google Scholar]
- 62.Zhi G, Abdullah SM, Stull JT. Regulatory segments of Ca2+/calmodulin-dependent protein kinases. J Biol Chem. 1998;273(15):8951–8957. doi: 10.1074/jbc.273.15.8951. [DOI] [PubMed] [Google Scholar]
- 63.Zhi G, Herring BP, Stull JT. Structural requirements for phosphorylation of myosin regulatory light chain from smooth muscle. J Biol Chem. 1994;269(40):24723–24727. [PubMed] [Google Scholar]
- 64.Ryder JW, Lau KS, Kamm KE, Stull JT. Enhanced skeletal muscle contraction with myosin light chain phosphorylation by a calmodulin-sensing kinase. J Biol Chem. 2007;282(28):20447–20454. doi: 10.1074/jbc.M702927200. [DOI] [PubMed] [Google Scholar]