Significance
An average human lung is composed of 14 million airway tips, conducting air to 300 million gas-exchange units. An organ of such complex architecture is nevertheless constructed with robust precision, the result of a largely stereotypical branching sequence. Although regulation at the transcript level is known to be critical, how control at the protein level may play a role remains poorly understood. The function of the ubiquitin proteasome system in the lung has primarily been studied in pathological settings in the adult. Here, we show that inactivation of Ring finger and WD domain 2 (RFWD2) led to a profound lung branching defect through misregulation of ETV transcription factors. These findings predict a protein-level regulatory network essential for the construction of a functional lung.
Keywords: lung branching, RFWD2, COP1, ETV transcription factors, E3 ubiquitin ligases
Abstract
The mammalian lung is an elaborate branching organ, and it forms following a highly stereotypical morphogenesis program. It is well established that precise control at the transcript level is a key genetic underpinning of lung branching. In comparison, little is known about how regulation at the protein level may play a role. Ring finger and WD domain 2 (RFWD2, also termed COP1) is an E3 ubiquitin ligase that modifies specific target proteins, priming their degradation via the ubiquitin proteasome system. RFWD2 is known to function in the adult in pathogenic processes such as tumorigenesis. Here, we show that prenatal inactivation of Rfwd2 gene in the lung epithelium led to a striking halt in branching morphogenesis shortly after secondary branch formation. This defect is accompanied by distalization of the lung epithelium while growth and cellular differentiation still occurred. In the mutant lung, two E26 transformation-specific (ETS) transcription factors essential for normal lung branching, ETS translocation variant 4 (ETV4) and ETV5, were up-regulated at the protein level, but not at the transcript level. Introduction of Etv loss-of-function alleles into the Rfwd2 mutant background attenuated the branching phenotype, suggesting that RFWD2 functions, at least in part, through degrading ETV proteins. Because a number of E3 ligases are known to target factors important for lung development, our findings provide a preview of protein-level regulatory network essential for lung branching morphogenesis.
The vital role of the mammalian lung as a gas-exchange organ is built on the basis of a remarkably elaborate branching sequence during lung development. In the mouse, lung development initiates at embryonic day (E) 9.5 with the formation of two primary lung buds (1, 2). Subsequently, these lung buds elongate and undergo multiple rounds of branching, following a largely stereotyped routine (3, 4). Extensive feedback regulations at the level of transcription have been shown to play central roles in branching (1, 2). In comparison, little is known on how control at the protein level is required for the process in vivo. One study showed that overexpression of SMAD specific ubiquitin protein ligase 1 (Smurf1) led to down-regulation of SMAD family members and a branching defect in lung explant culture (5).
The ubiquitin proteasome system is a fundamental molecular mechanism that controls protein homeostasis. Selective degradation of protein substrates by this system involves sequential function of three classes of enzymes: ubiquitin-activating enzyme E1, ubiquitin-conjugating enzyme E2, and ubiquitin-protein ligase E3 (6, 7). In a mammalian organism such as the mouse, although there are only a handful of E1 and E2 enzymes that act in the core pathway to mark all protein substrates, there are several hundred E3 ubiquitin ligases, each with a small set of substrates, thus providing specificity (8).
E3 ubiquitin ligase Ring finger and WD domain 2 (RFWD2, also known as COP1) was first identified as a regulator in the light signaling pathway in Arabidopsis thaliana (9). Subsequently, RFWD2 was shown to be conserved in mammals and to function as a tumor suppressor that regulates the degradation of various oncogenic factors, including JUN, p53, C/EBPα, MTA1, and the PEA3 family of ETS transcription factors (ETV1, ETV4, and ETV5) (10–16). We recently showed that two of the ETV factors, ETV4 and ETV5, play critical roles in the lung epithelium to control branching (17). In the present study, we tested the hypothesis that RFWD2, acting through ETVs, regulates lung branching morphogenesis.
Herein, we report that conditional inactivation of Rfwd2 gene in murine lung epithelium halted lung branching morphogenesis shortly after secondary branch formation. The mutant lungs showed increased ETV5 protein despite decreased Etv5 transcript. The lung branching defect can be partially reversed by inactivating Etv5 in the Rfwd2 mutant background. Our data demonstrate that protein-level regulation via the ubiquitin proteasome system is critical for lung branching morphogenesis, and that RFWD2 acts in this process, at least in part, through controlling ETV5.
Results
Rfwd2 Is Highly Expressed in the Lung Epithelium During Active Branching.
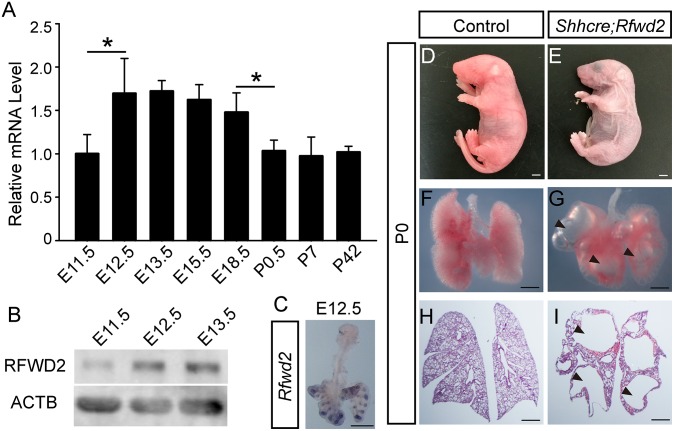
To address the requirement for Rfwd2 in lung development, we examined its spatial and temporal expression patterns in embryonic mouse lungs. By quantitative real-time PCR (qRT-PCR), we found that the Rfwd2 transcript level was significantly increased from E11.5 to E12.5, as the lung initiates secondary branching (Fig. 1A). This increase was verified at the protein level by Western blot analysis (Fig. 1B). Interestingly, Rfwd2 mRNA expression was reduced by ∼30% after birth compared with E18.5, indicating that the highest Rfwd2 expression is during fetal lung development. By whole-mount RNA in situ hybridization, we found that Rfwd2 expression was primarily detected in the lung epithelium at E12.5, when the lung is undergoing branching morphogenesis (Fig. 1C).
Fig. 1.
Rfwd2 is expressed in the developing lung epithelium, and inactivation leads to lung defects and lethality at birth. (A) qRT-PCR of Rfwd2 mRNA levels in wild-type lungs from E11.5 to postnatal day (P) 42 (adult). Data were normalized to Actb (also termed β-actin) and then to the expression level at E11.5. The quantification was carried out in n ≥ 3 samples for each stage. *P < 0.05. (B) Western blot of RFWD2 protein levels in wild-type lungs at E11.5, E12.5, and E13.5. ACTB was used as a loading control. (C) Representative whole-mount RNA in situ hybridization of Rfwd2 in E12.5 wild-type lung indicating expression primarily in the lung epithelium. Intact pups (D and E), whole lung (F and G), or H&E-stained sections (H and I) of controls and Shhcre;Rfwd2 mutants at birth are shown. Arrowheads indicate large air sacs. (Scale bars: 500 μm.)
Inactivation of Rfwd2 Resulted in Lung Branching Defects.
Rfwd2 global knockout mutants die by E11.5, before lung branching (12). To determine if Rfwd2 is required in the lung epithelium for proper lung branching, we conditionally inactivated Rfwd2 using sonic hedgehog cre (Shhcre), which is active in the early embryonic lung epithelium (18). We crossed a floxed allele of Rfwd2 (Rfwd2fl) to Shhcre, generating Shhcre/+;Rfwd2Δ/fl (hereafter Shhcre;Rfwd2) mutants (16). These mutants survived embryogenesis but exhibited respiratory distress shortly after birth, and all died with a cyanotic phenotype (Fig. 1 D and E). Mutant lungs were similar in overall size compared with controls (Fig. 1 F and G). However, there were large air-filled saccules in the Shhcre;Rfwd2 mutant lungs, leading to a drastic reduction of gas-exchange surface area, a likely reason for lethality at birth (Fig. 1 H and I). These results indicate that Rfwd2 is essential in the epithelium for the formation of a functional lung.
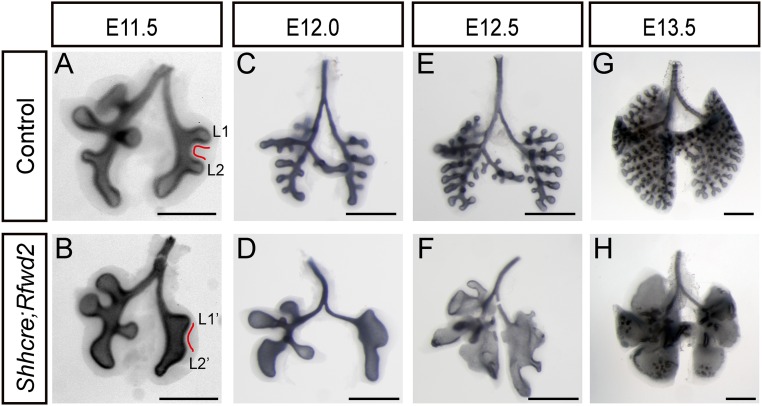
In tracing back the origin of lung defects, we found that Shhcre;Rfwd2 mutant lungs are morphologically distinct from controls as early as E11.5, at the start of secondary branching (Fig. 2 A and B). In the E11.5 control lung, the lateral secondary branches of the left lobe are perpendicular to the main bronchi and close to parallel with each other (Fig. 2A, L1 and L2). However, in the mutant, L1 and L2 have a wider base, and the angle between them becomes obtuse (Fig. 2B, L1′ and L2′). Several other tips were also dilated in the mutant lungs compared with controls. Between E11.5 and E13.5, as control lungs underwent stereotyped branching, mutant lungs stopped new branch generation but continued with lumen enlargement, such that the overall lung size is not different from controls (Fig. 2 C–H). Taken together, these results indicate that Rfwd2 is essential in the lung epithelium for proper branching morphogenesis.
Fig. 2.
Inactivation of Rfwd2 in the lung epithelium results in defects in lung branching morphogenesis. Representative control (A, C, E, and G) and Shhcre;Rfwd2 mutant (B, D, F, and H) whole lungs from E11.5 to E13.5 with the epithelium outlined by anti-CDH1 (also termed E-cadherin) immunohistochemical staining. (A and B) Red lines outline the epithelium between lateral secondary branch (L) 1 and L2. (Scale bars: 500 μm.)
Inactivation of Rfwd2 Altered Proximal–Distal Airway Patterning.
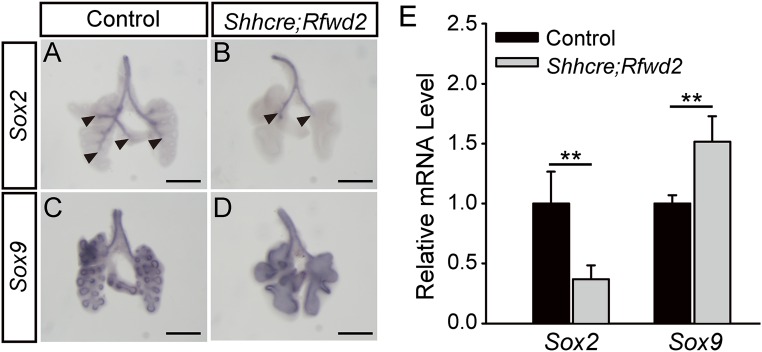
To investigate if Rfwd2 plays a role in lung patterning, we carried out expression analysis using the earliest markers: SRY-box 2 (Sox2) for proximal epithelium and SRY-box 9 (Sox9) for distal epithelium (19–22). In the E12.5 control lung, Sox2 was observed in the trachea, extrapulmonary bronchi, and proximal intrapulmonary epithelium (Fig. 3A). In contrast in the E12.5 Shhcre;Rfwd2 mutant lung, Sox2 was primarily restricted to the trachea and extrapulmonary bronchi (Fig. 3B). Conversely, although Sox9 was expressed in the distal epithelium in the control lung, it was expanded to most of the lung epithelium in the mutant (Fig. 3 C and D). These changes detected by RNA in situ hybridization were further supported by data from qRT-PCR showing decreased Sox2 and increased Sox9 mRNA levels in Shhcre;Rfwd2 mutants compared with controls (Fig. 3E).
Fig. 3.
Inactivation of Rfwd2 altered proximal–distal airway patterning. (A–D) Representative RNA in situ hybridization for Sox2 or Sox9 at E12.5 in Shhcre;Rfwd2 mutants and littermate controls. Arrowheads indicate the distal ends of the Sox2 domain. Sox9 expression in the trachea and extrapulmonary bronchi is in the mesenchymal precursors of the cartilage but not in the epithelial cells, as is its distal expression domain. (Scale bars: 500 μm.) (E) mRNA quantification by qRT-PCR of E12.5 lungs (Sox2: 1.00 ± 0.26 for controls, 0.37 ± 0.12 for Shhcre;Rfwd2 mutants, P = 0.004; Sox9: 1.00 ± 0.068 for controls, 1.51 ± 0.21 for Shhcre;Rfwd2 mutants, P = 0.005). Quantification was carried out in n ≥ 3 samples. **P < 0.01.
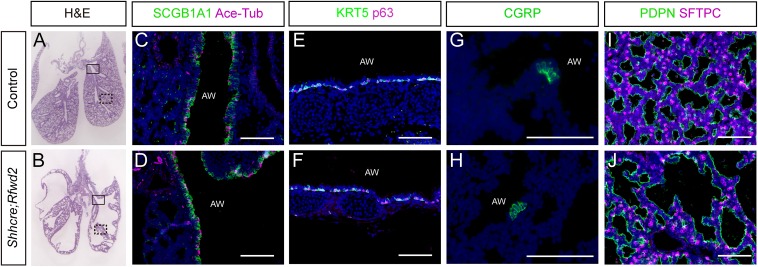
We next addressed whether loss of Rfwd2 affects epithelial cell differentiation at E18.5. All analyzed cell types, including airway cell types, such as club cells, ciliated cells, pulmonary neuroendocrine cells, and basal cells, as well as alveolar cell types, such as type I and type II cells, appear present in the Shhcre;Rfwd2 mutants (Fig. S1). These results indicate that despite overall distalization of the epithelium, loss of Rfwd2 does not prevent lung epithelial cell differentiation.
Fig. S1.
Cell differentiation occurs in Shhcre;Rfwd2 mutants. (A and B) H&E-stained lung sections from E18.5 lungs. Boxes delineate approximate areas stained for C and D (box with solid lines) or I and J (box with dashed lines). (C–J) Immunostaining of markers for club cells (SCGB1A1), ciliated cells (Ace-Tub), basal cells (KRT5, p63), and pulmonary neuroendocrine cells (CGRP) in the airways, and for type I (PDPN) and type II (SFTPC) cells in the alveolar regions at E18.5. AW, airway. (Scale bars: 100 μm.)
ETS Transcription Factors Are RFWD2 Substrates in Lung Epithelium.
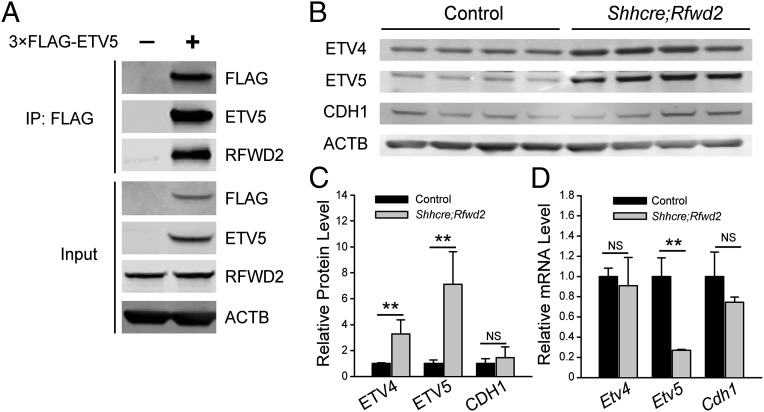
To determine the molecular mechanism underlying RFWD2 function in lung development, we addressed if its target substrates are affected in Shhcre;Rfwd2 mutants. ETS transcription factors, including ETV1, ETV4, and ETV5, are reported substrates of RFWD2 in adult prostate and pancreas (16, 23). Etv4 and Etv5, but not Etv1, are expressed in the developing lung epithelium, similar to Rfwd2, raising the possibility that they may be RFWD2 substrates in the lung (Fig. 1C) (24–26). To test this possibility, we first investigated if ETV and RFWD2 interact at the protein level. Mouse lung epithelial MLE-12 cells were transfected with expression vector encoding 3× FLAG-tagged ETV5 or an empty vector control. Immunoprecipitation of 3× FLAG-ETV5 protein with an anti-FLAG antibody led to coprecipitation of RFWD2 as detected on immunoblots with an anti-RFWD2 antibody, suggesting that ETV5 binds to RFWD2 in cultured lung epithelial cells (Fig. 4A).
Fig. 4.
Inactivation of Rfwd2 led to increased expression of ETV4 and ETV5 proteins. (A) ETV5 is immunoprecipitated in the same complex as RFWD2. MLE-12 cells were transfected with 3× FLAG-ETV5 (+) or 3× FLAG control (−) expression vector, immunoprecipitated with an anti-FLAG antibody. Either the immunoprecipitated products or inputs before immunoprecipitation (IP) were immunoblotted with the indicated antibodies. (B) Western blot of ETV4, ETV5, and CDH1 proteins in E12.5 lungs with the indicated genotypes. ACTB was used as a loading control. (C) Quantification of Western blot normalized to ACTB. The density of bands was measured using the Image Studio Lite program. **P < 0.01. NS, not significant. (D) Quantification of mRNA expression of Etv4, Etv5, and Cdh1 by qRT-PCR in E12.5 lungs. Quantification was carried out in n ≥ 3 samples. **P < 0.01.
To determine further if ETV proteins are RFWD2 substrates in vivo, we generated whole-lung extracts from mutant and control E12.5 lungs and examined protein levels of ETV4 and ETV5 by Western blot. As shown in Fig. 4B, ETV4 and ETV5 were more abundant in Shhcre;Rfwd2 lungs. ETV5 protein was increased about sixfold, whereas CDH1 (also termed E-cadherin), a general epithelial marker, remained unaltered (Fig. 4 B and C). Importantly, the increase in ETV4 and ETV5 protein was not due to up-regulation in transcript level, because Etv4 mRNA was unaltered, whereas the Etv5 mRNA level was actually reduced in the mutant to ∼30% of the control (Fig. 4D). These data indicate that RFWD2 is required for the degradation of ETV4 and ETV5 proteins in the branching lung.
Inactivation of Etv5 Partially Attenuates Shhcre;Rfwd2 Branching Phenotype.
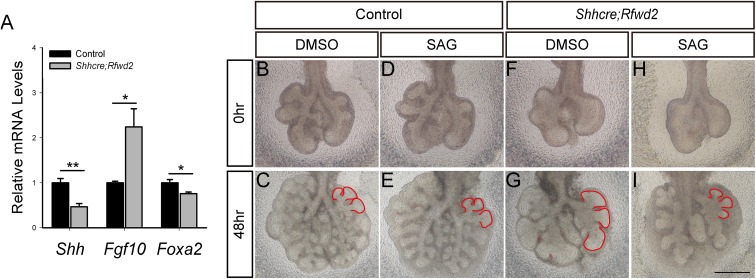
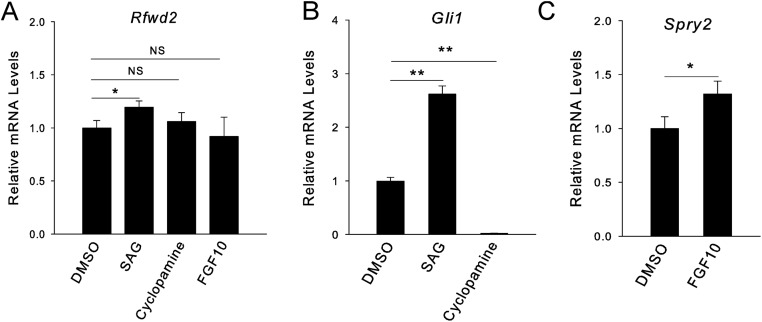
Previous studies showed that Etv genes are required in the lung epithelium for proper branching morphogenesis, raising the possibility that by being a substrate, they may mediate RFWD2 function in the lung (17, 25). Our previous study in the Etv mutant lungs showed that ETV can directly promote Shh expression in the distal epithelium, which, in turn, inhibits fibroblast growth factor 10 (Fgf10) expression in the surrounding mesenchyme (17). With the ETV increase in the Shhcre;Rfwd2 mutant, we expected an increase of Shh expression and a decrease of Fgf10 expression in this mutant. On the contrary, we found a decrease of Shh and an increase of Fgf10, suggesting that these genes may also be regulated by additional RFWD2 downstream transcription factors independent of ETV (Fig. S2A). Indeed, the known Shh activator genes Sox2 and forkhead box A2 (Foxa2) were both down-regulated in Shhcre;Rfwd2 mutants, suggesting that changes such genes may counteract the effect of ETV increase on Shh expression (27, 28) (Fig. 3E and Fig. S2A). Nevertheless, we found that treatment of Shhcre;Rfwd2 lungs with SHH signaling agonist (SAG) in culture attenuated the epithelial tip dilation phenotype, suggesting that the decrease of SHH in the Shhcre;Rfwd2 mutant contributes to the branching phenotype (Fig. S2 B–I). Conversely, wild-type lungs cultured in the presence of SAG show elevated Rfwd2 transcripts, suggesting that there is indirect feedback regulation between Rfwd2 and SHH signaling (Fig. S3).
Fig. S2.
SAG treatment of Shhcre;Rfwd2 mutant lungs led to attenuation of the tip dilation phenotype. (A) qRT-PCR analysis of genes Shh, Fgf10, and Foxa2 in Shhcre;Rfwd2 mutants compared with control at E13.5. n = 3 in each group. *P < 0.05; **P < 0.01. Representative images of E11.5 lungs cultured for 48 h in either DMSO (B, C, F, and G) or SAG (50 nM) (D, E, H, and I) are shown. Red lines outline the branch tips. (Scale bars: 500 μm.)
Fig. S3.
SHH-induced Rfwd2 expression in wild-type cultured lungs. A qRT-PCR analysis of the transcript level of Rfwd2 (A), Gli1 (B), and Spry2 (C) of E11.5 wild-type lungs cultured for 48 h in SAG (1 μM), SHH signaling inhibitor cyclopamine (500 nM), or FGF10 recombinant protein (500 ng/mL) was performed. Gli1 and Spry2 transcripts served as readouts of SHH or FGF activity, respectively. *P < 0.05; **P < 0.01. NS, not significant.
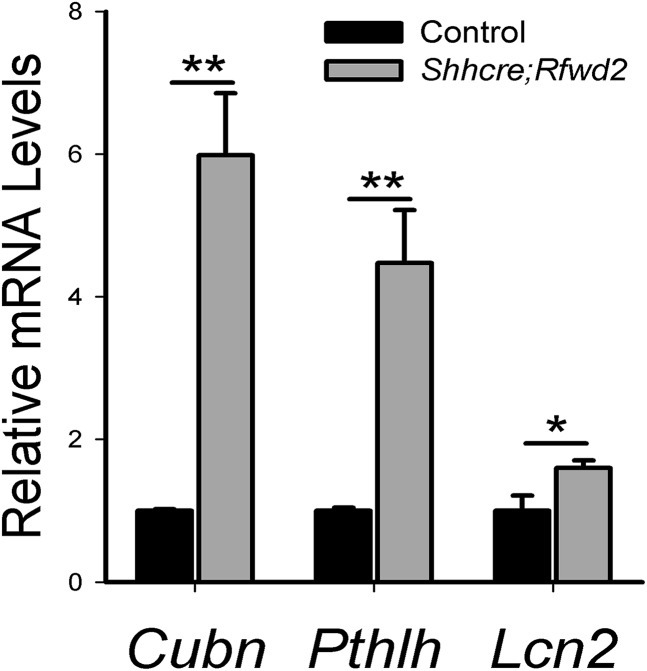
Because Shh does not show the expected change in the Rfwd2 mutant lung, we searched for other potential ETV targets that may act downstream of Rfwd2 function. We reasoned that these genes should show opposite expression changes in the Rfwd2 mutants versus the Etv mutants compared with their respective controls [Gene Expression Omnibus (GEO) accession nos. GSE80707 and GSE80734]. Comparison of the transcriptomes yielded 45 genes that are changed in opposite directions (Table S1). Among them, 35 genes contained the common ETV binding motif (5′-GGAA/T-3′) in their 5′ UTR regions (29) (Table S1). We also selected three of these genes, cubilin (Cubn), parathyroid hormone-like peptide (Pthlh), and lipocalin 2 (Lcn2), and confirmed their expression change by qRT-PCR (Fig. S4). All three genes have been shown to be expressed in the lung epithelium, and Pthlh has been implicated in lung development (30–33). This large cohort of genes with the expected opposite expression changes in the two mutants is consistent with the possibility that ETV may be a key mediator of RFWD2 function in lung branching.
Table S1.
Genes with opposite regulated directions in Shhcre;Rfwd2 and Shhcre;Etv4;5 mutants
| Gene symbol | FPKM of RNA-sequencing | Microarray read | Putative ETV binding sites | ||
| Control | Shhcre;Rfwd2 | Control | Shhcre;Etv4;5 | ||
| Cubn | 0.57 | 3.34 | 207.94 | 81.01 | 5′-TCAGGAAGGA-3′; 5′-CAAGGATTCC-3′ |
| Pthlh | 3.52 | 16.82 | 374.81 | 274.37 | 5′-GCAGGAATGC-3′ |
| Lcn2 | 0.57 | 1.90 | 151.17 | 77.71 | 5′-CCAGGATAAT-3′; 5′-GGAGGATGAC-3′ |
| Habp2 | 1.91 | 9.34 | 198.09 | 159.79 | 5′-GGAGGATTGC-3′ |
| Kcnt2 | 0.13 | 0.62 | 88.03 | 67.18 | 5′-TCAGGAAAGT-3′ |
| Slc39a8 | 5.48 | 18.40 | 354.59 | 272.48 | 5′-TCAGGAAGGG-3′; 5′-ACAGGAAAGT-3′; 5′-TGCGGATATC-3′ |
| Cfi | 11.24 | 29.22 | 190.02 | 116.16 | 5′-CCAGGAAGAA-3′; 5′-ACAGGAAGGG-3′; 5′-AAAGGAAAAC-3′ |
| Id2 | 121.83 | 270.55 | 5,673.41 | 4,608.24 | 5′-CGCGGAAGAA-3′ |
| Sftpa1 | 5.20 | 11.40 | 396.18 | 321.80 | 5′-AAAGGATGCC-3′ |
| Pla2g7 | 4.25 | 9.27 | 290.02 | 240.52 | 5′-TCAGGATTGG-3′; 5′-AGAGGATGTT-3′ |
| Rxfp1 | 0.26 | 0.53 | 70.03 | 56.10 | 5′-AGAGGAAAGA-3′; 5′-AAAGGAAGGG-3′; 5′-AAAGGATTTT-3′ |
| Tmem100 | 7.39 | 14.59 | 333.14 | 247.28 | 5′-TCCGGAATTC-3′; 5′-TAAGGAAGAG-3′ |
| Sh3bgrl2 | 6.66 | 12.79 | 975.50 | 588.13 | 5′-CCAGGAAAAA-3′; 5′-AGAGGATGCT-3′; 5′-CAAGGAAACT-3′ |
| Slc26a9 | 1.08 | 1.89 | 335.46 | 209.38 | 5′-AGAGGAATCT-3′ |
| Map2 | 3.73 | 6.08 | 552.56 | 451.94 | 5′-GAAGGAAGGA-3′; 5′-AAAGGATTCA-3′ |
| Rcsd1 | 15.45 | 24.77 | 288.01 | 219.79 | 5′-GGAGGATAAC-3′; 5′-TCAGGATTGC-3′ |
| Otud1 | 5.08 | 8.08 | 294.07 | 243.88 | 5′-TCCGGAAGTG-3′; 5′-GGAGGAAACG-3′; 5′-CAAGGAAGGC-3′ |
| Slc2a3 | 5.32 | 7.88 | 377.41 | 284.05 | 5′-GGAGGATGGC-3′; 5′-AACGGAAAGC-3′ |
| Il1r1 | 2.19 | 3.12 | 265.03 | 218.27 | 5′-GCAGGAATAA-3′; 5′-GCAGGATTAA-3′ |
| Bex4 | 85.78 | 116.82 | 191.34 | 119.43 | 5′-GGCGGAAAGA-3′; 5′-GAAGGAAAAA-3′; 5′-TCAGGAAGTT-3′ |
| Npr3 | 11.73 | 15.81 | 1,964.57 | 1,468.37 | 5′-AGAGGAAAGA-3′; 5′-GGAGGAAAAG-3′; 5′-AGAGGAATCT-3′ |
| Afap1l1 | 37.03 | 48.04 | 781.44 | 600.49 | 5′-TCAGGAAAGA-3′; 5′-GGAGGAATGC-3′ |
| Tgfbr3 | 5.69 | 7.25 | 704.28 | 451.94 | 5′-CCAGGAATCC-3′; 5′-AAAGGAATTT-3′ |
| Pde4b | 6.03 | 7.64 | 364.56 | 300.25 | 5′-TGAGGAAACC-3′; 5′-AGAGGATTCA-3′ |
| Plb1 | 2.02 | 0.43 | 218.27 | 286.03 | 5′-ACAGGAATAA-3′; 5′-TCAGGATATG-3′; 5′-CCAGGATTGC-3′; 5′-TGCGGAAAAC-3′; 5′-TGAGGAAATA-3′; 5′-GAAGGAAAGA-3′ |
| Gal | 6.29 | 1.88 | 206.50 | 304.44 | 5′-GAAGGAAAGC-3′ |
| Cldn10 | 9.51 | 3.61 | 643.59 | 803.41 | 5′-ACAGGAAATT-3′ |
| Entpd3 | 9.25 | 3.96 | 744.43 | 916.51 | 5′-ACAGGATGTG-3′; 5′-AAAGGATTGG-3′ |
| Ankrd33b | 0.80 | 0.39 | 99.04 | 122.79 | 5′-GGCGGATTAC-3′ |
| Ces1d | 13.50 | 6.64 | 377.41 | 471.14 | 5′-GCAGGAATAC-3′; 5′-GAAGGAAGAG-3′; 5′-CAAGGATTGT-3′; 5′-CCAGGAAGGC-3′ |
| Slc27a6 | 13.15 | 7.34 | 445.72 | 576.03 | 5′-ACAGGAAACC-3′; 5′-GGAGGAAGAT-3′ |
| Tinag | 7.49 | 4.89 | 709.18 | 910.17 | 5′-AGAGGATTCT-3′ |
| Gsta4 | 63.55 | 44.47 | 955.43 | 1,478.58 | 5′-ACAGGAATCG-3′ |
| Aldh1a7 | 52.35 | 36.73 | 1,389.16 | 1,833.01 | 5′-ACAGGAAATA-3′ |
| Fam89a | 12.36 | 9.09 | 274.37 | 333.14 | 5′-GACGGAAGAC-3′ |
| Ano1 | 11.04 | 7.85 | 803.41 | 1,038.29 | — |
| Aard | 8.63 | 11.29 | 164.28 | 129.79 | — |
| Sox9 | 11.44 | 14.21 | 1,833.01 | 1,478.58 | — |
| Scgb3a2 | 14.25 | 1.83 | 66.72 | 81.01 | — |
| Ascl1 | 1.87 | 1.08 | 526.39 | 694.58 | — |
| Igfbp6 | 4.28 | 2.51 | 97.01 | 120.26 | — |
| Nptx1 | 0.44 | 0.11 | 114.56 | 144.01 | — |
| Ndp | 2.71 | 4.07 | 245.57 | 162.02 | — |
| Gfra1 | 4.72 | 2.78 | 430.54 | 526.39 | — |
| Dram1 | 11.44 | 24.36 | 699.41 | 552.56 | — |
FPKM, fragments per kilobase of exon every million reads mapped.
Fig. S4.
Expression of some ETV-regulated genes shows opposite change in Shhcre;Rfwd2 mutant lungs. A qRT-PCR analysis of genes Cubn, Pthlh, and Lcn2 in Shhcre;Rfwd2 mutants compared with control at E13.5 is shown. n = 3 in each group. *P < 0.05; **P < 0.01.
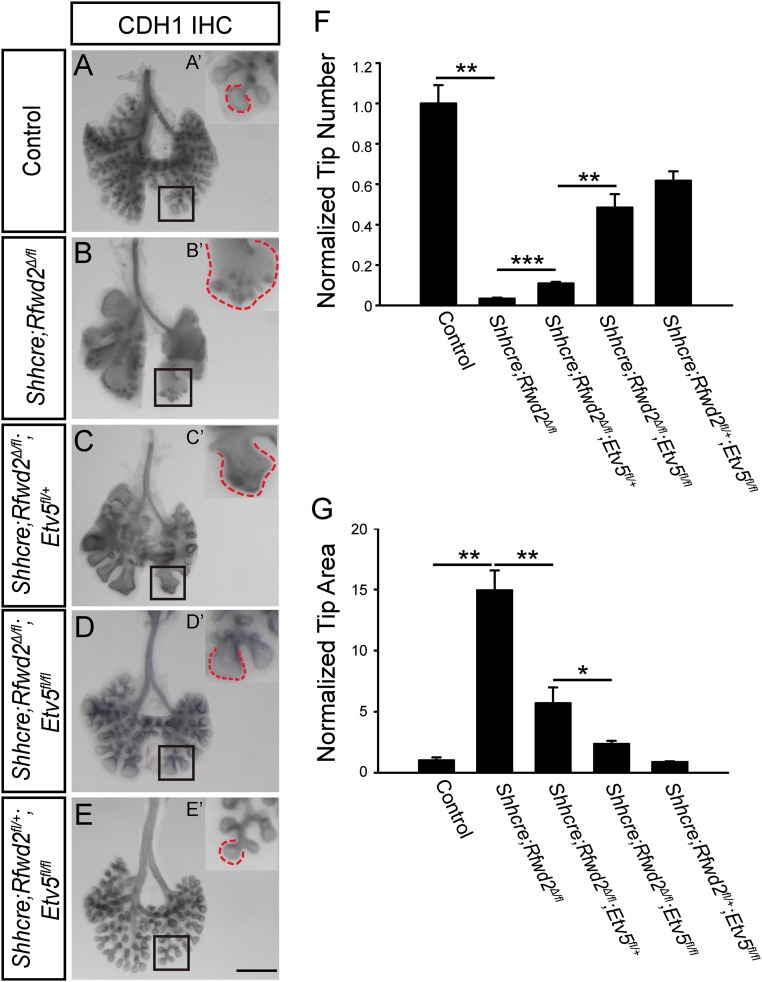
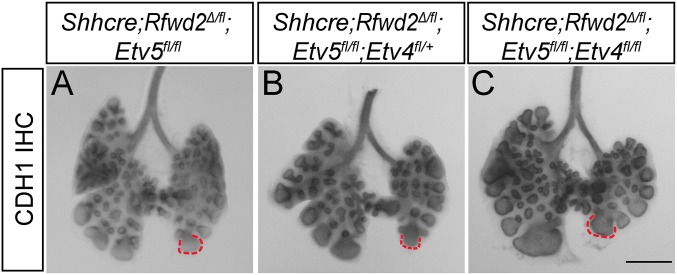
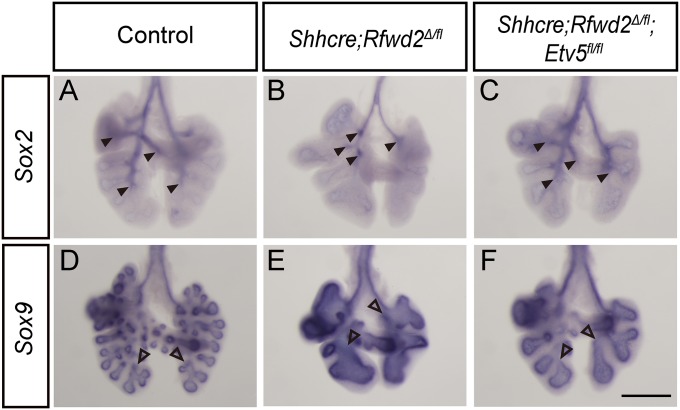
To test this possibility in vivo, we introduced Etv4 and Etv5 mutant alleles into the Rfwd2 mutant background (34, 35). Whole-mount CDH1 staining outlining the epithelium at E13.5 showed that introduction of Etv5 mutant alleles led to dosage-dependent increases in branch tip number and decreases in branch tip area, with two copies of Etv5 exerting stronger effects than one copy (Fig. 5). Further inactivation of Etv4 in addition to Etv5 did not further improve the branching phenotypes (Fig. S5). Inactivation of Etv5 also attenuated the proximal–distal airway patterning phenotype in Shhcre;Rfwd2 mutants (Fig. 6). These results demonstrate that the ETV5 increase observed in the Shhcre;Rfwd2 mutant significantly contributes to the branching and patterning defects.
Fig. 5.
Introduction of Etv5 mutation allele partially reversed the Shhcre;Rfwd2 branching phenotype. (A–E) Representative E13.5 lungs of indicated genotypes with the epithelium outlined by anti-CDH1 immunohistochemical staining. The boxed areas in A–E are shown at high magnification in A′–E′. Dashed lines indicated the baselines of branch tips. (Scale bar: 500 μm.) (F) Introducing the Etv5 mutant allele attenuated the branch tip number phenotype. For quantifying tip number, the epithelial tips of the left lobe were manually counted. Data were normalized to control samples. Quantification was carried out in n = 3 samples for each genotype (tip number: 1.00 ± 0.09 for controls, 0.03 ± 0.004 for Shhcre/+;Rfwd2Δ/fl, 0.11 ± 0.01 for Shhcre/+;Rfwd2Δ/fl;Etv5fl/+, 0.48 ± 0.07 for Shhcre/+;Rfwd2Δ/fl;Etv5fl/fl, 0.62 ± 0.05 for Shhcre/+;Rfwd2fl/+;Etv5fl/fl; P = 0.0014 for control versus Shhcre/+;Rfwd2Δ/fl, P = 0.0002 for Shhcre/+;Rfwd2Δ/fl versus Shhcre/+;Rfwd2Δ/fl;Etv5fl/+, P = 0.005 for Shhcre/+;Rfwd2Δ/fl;Etv5fl/+ versus Shhcre/+;Rfwd2Δ/fl;Etv5fl/fl). **P < 0.01; ***P < 0.001. (G) Introducing the Etv5 mutant allele also attenuated the branch tip area phenotype. For quantifying the lung tip areas, lungs were imaged following whole-mount CDH1 staining. ImageJ (NIH) was used to draw a free-form trace around each tip of left lobe such as the ones outlined, and the average area of the tips within the trace was measured. Data were normalized to control samples. Quantification was carried out in n = 3 samples for each genotype (tip area: 1.0 ± 0.26 for controls, 14.94 ± 1.63 for Shhcre/+;Rfwd2Δ/fl, 5.69 ± 1.31 for Shhcre/+;Rfwd2Δ/fl;Etv5fl/+, 2.36 ± 0.24 for Shhcre/+;Rfwd2Δ/fl;Etv5fl/fl, 0.88 ± 0.05 for Shhcre/+;Rfwd2fl/+;Etv5fl/fl; P = 0.002 for control versus Shhcre/+;Rfwd2Δ/fl, P = 0.0009 for Shhcre/+;Rfwd2Δ/fl versus Shhcre/+;Rfwd2Δ/fl;Etv5fl/+, P = 0.022 for Shhcre/+;Rfwd2Δ/fl;Etv5fl/+ versus Shhcre/+;Rfwd2Δ/fl;Etv5fl/fl). *P < 0.05; **P < 0.01.
Fig. S5.
Branching phenotype was not further attenuated by introduction of Etv4 mutant allele in the Shhcre;Rfwd2;Etv5 mutant background. (A–C) Representative E13.5 lungs of the indicated genotypes are shown, with the epithelium outlined by anti-CDH1 immunohistochemical staining. Red dashed lines outline equivalent branch tips. (Scale bar: 500 μm.)
Fig. 6.
Introduction of Etv5 mutant allele into the Shhcre;Rfwd2 mutant attenuated the proximal–distal patterning defect. RNA in situ hybridization for Sox2 or Sox9 at E12.5 in representative control (A and D), Shhcre;Rfwd2 mutant (B and E), and Shhcre;Rfwd2;Etv5 (C and F) mice. Filled arrowheads indicate the distal points of Sox2 expression domain. Open arrowheads indicate the proximal points of Sox9 expression domain. (Scale bar: 500 μm.)
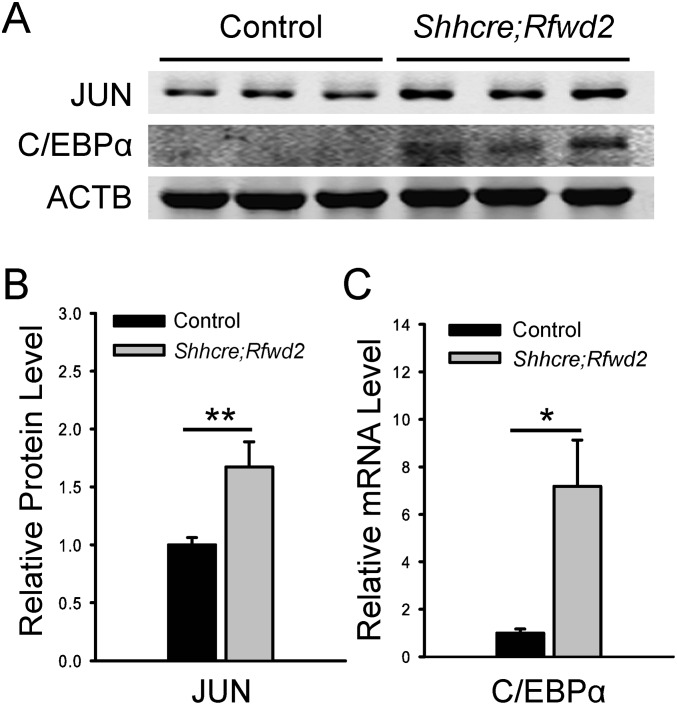
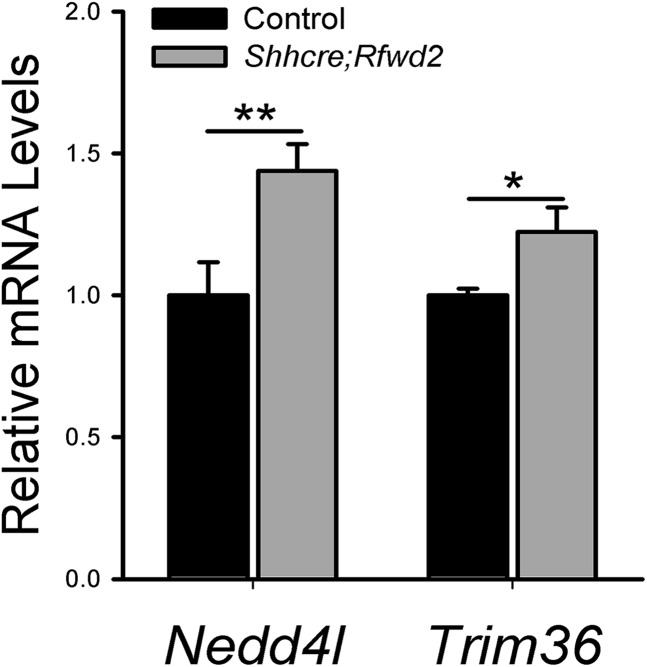
We note that inactivation of Etv5 does not fully rescue the branching defect, indicating that there must be additional genes affected in the Rfwd2 mutants that contribute to its phenotype. For example, in the Shhcre;Rfwd2 mutant lungs, the protein levels of two other known RFWD2 substrates, JUN and C/EBPAα, were increased (12, 14) (Fig. S6). The transcript levels of other E3 ligases, such as Nedd4l and Trim36, were increased (Fig. S7 and Table S2). These factors have been shown to control signaling pathways that function in lung development (36–39). Thus, their changes may have an impact on the Shhcre;Rfwd2 phenotype in addition to ETV.
Fig. S6.
Inactivation of Rfwd2 led to increased expression of JUN and C/EBPα proteins. (A) Western blot of JUN and C/EBPα proteins in E12.5 lungs with indicated genotypes. ACTB was used as a loading control. (B and C) Quantification of Western blot normalized to ACTB. The density of bands was measured using the Image Studio Lite program. *P < 0.05; **P < 0.01.
Fig. S7.
Expression of other E3 ligase genes are altered in Shhcre;Rfwd2 mutant lungs. qRT-PCR analysis of genes Nedd4l and Trim36 in Shhcre;Rfwd2 mutants compared with control at E13.5. n = 3 in each group. *P < 0.05; **P < 0.01.
Table S2.
E3 ligases regulated in Shhcre;Rfwd2
| Gene symbol | FPKM of control | FPKM of mutant | Log2 (mutant/control) | q_value |
| Pparg | 0.423601 | 1.30408 | 1.62225 | 0.00151629 |
| Asb4 | 13.1052 | 35.3307 | 1.43079 | 0.00151629 |
| Rnf144b | 0.745149 | 1.50675 | 1.01584 | 0.00151629 |
| Rnf152 | 0.81926 | 1.6023 | 0.967751 | 0.00151629 |
| Trim25 | 7.18052 | 11.0568 | 0.62278 | 0.00151629 |
| Trim36 | 1.08418 | 1.6664 | 0.620122 | 0.00151629 |
| Trim71 | 1.37725 | 2.00115 | 0.539038 | 0.00151629 |
| Sh3rf1 | 7.8295 | 11.0322 | 0.494734 | 0.00151629 |
| Nedd4l | 2.6594 | 3.58853 | 0.432292 | 0.00284004 |
| Aire | 0.587167 | 1.30104 | 1.14782 | 0.015378 |
| Ccnb1ip1 | 0.360814 | 0.809726 | 1.16618 | 0.0207246 |
| Sh3rf3 | 0.747062 | 0.229374 | −1.70352 | 0.00151629 |
| Neurl1b | 1.87735 | 1.36469 | −0.460128 | 0.0207246 |
| Fbxl22 | 3.58525 | 1.92798 | −0.894981 | 0.030527 |
| March9 | 4.31162 | 3.21138 | −0.425038 | 0.0358048 |
| Asb2 | 2.73453 | 1.14756 | −1.25272 | 0.0482745 |
q_value, the false-discovery rate adjusted P-value of the test statistic.
Discussion
Many of the studies of the ubiquitin proteasome system in the lung have been focused on the adult lung in pathological settings, where the E3 ligases are found to play essential roles in disease settings, such as acute respiratory distress syndrome and chronic obstructive pulmonary disease (40). Relatively little is known about how the ubiquitin proteasome system functions in lung development. In the present study, we show that genetic inactivation of E3 ligase gene Rfwd2 in the embryonic lung epithelium led to a halt in branching, respiratory failure, and death at birth. The mutant lung shows a clear increase of ETV proteins, and inactivation of Etv5 in Rfwd2 mutant background can partially rescue the branching defects. These findings demonstrate a critical requirement for the ubiquitin proteasome system in lung branching.
Before our study, RFWD2 has been shown to play important roles in pathogenesis in the adult. In humans, both elevated and, in rarer cases, reduced RFWD2 expression has been described in a wide spectrum of cancers in organs such as the breast, ovary, liver, pancreas, and lung (12, 41–44). In mice, two genetic studies provided strong evidence for a tumor suppressor role of RFWD2 (12, 16). Additionally, a recent study demonstrates that RFWD2 functions in the pancreas to facilitate insulin secretion from β cells and maintain normal glucose homeostasis (23). Furthermore, single-nucleotide polymorphism within Rfwd2 and decreased transcript level were associated with increased susceptibility to acrolein-induced acute lung injury in adult mice (45). Thus, in addition to its essential role in lung branching, RFWD2 may play important roles in adult lung diseases.
Our results support that ETV5 is a substrate that mediates RFWD2 function in lung branching. However, the partial rescue of the Rfwd2 phenotype by Etv5 suggests that there are additional substrates. We show here that the protein levels of JUN and C/EBPAα are both increased in the Shhcre;Rfwd2 mutant (Fig. S6). Genetic disruption of Jun in the alveolar epithelium promotes emphysema with enlarged air spaces (36). Deletion of Cebpa gene in respiratory epithelial cells delayed lung maturation and caused respiratory failure at birth (37, 46, 47). Further genetic experiments are needed to assess the specific contributions of these and additional RFWD2 substrates to the branching phenotype.
We expect that our findings are only revealing the tip of the iceberg for the role of the ubiquitin proteasome system in lung branching. It is well established that the stereotyped branching program is dependent on precise cross-talk among multiple signaling pathways, including SHH, FGF, WNT, BMP, and TGF-β (1, 2). A number of E3 ubiquitin ligases have been identified to regulate substrates within these signaling pathways, such as ITCH (targets protein patched homolog 1, SHH pathway), vHL [targets protein sprouty homolog 2 (SPRY2), FGF pathway], NEDD4 (targets fibroblast growth factor receptor 1, FGF pathway), RNF43 (targets transcription factor 4, canonical WNT pathway), NEDD4L (targets segment polarity protein dishevelled homolog DVL-2, canonical WNT pathway), TRIM36 (unknown, targets noncanonical WNT pathway), and SMURF1 and SMURF2 (target SMADs, TGF-β/BMP pathway) (5, 38, 39, 48–52). These factors may function together with RFWD2 to ensure order at the protein level during the construction of a functional lung.
Materials and Methods
Mice.
Embryos were harvested from time-mated mice, counting noon on the day when the vaginal plug was found as E0.5. The alleles used in this study have been described previously: Rfwd2fl (16), Etv5fl (34), Etv4fl (35), and Shhcre (53). For generating the conditional mutant, males were heterozygous for both cre and the conditional allele were mated to females homozygous for the conditional allele. Cre with heterozygous conditional allele littermate embryos were used as controls. All animal experimental procedures were approved by the University of Wisconsin Animal Care and Use Committee.
In Vitro Lung Culture.
Lungs were harvested at E11.5, placed on a Nuclepore Track-Etched membrane (8 μm; Whatman), and cultured in DMEM-F12 (Gibco) with penicillin and streptomycin (Sigma) at 37 °C in 5% CO2 for 48 h. To study the effect of signaling on Rfwd2 expression, a final concentration of SAG (1 μM), cyclopamine (SHH signaling inhibitor, 500 nM), or FGF10 recombinant protein (500 ng/mL) was added in the media. To study if an increase in SHH activity would have an impact on the Shhcre;Rfwd2 phenotype, SAG was added at a final concentration of 50 nM. DMSO was used as a vehicle control.
qRT-PCR.
Lungs from a minimum of thee animals per genotype were individually homogenized in TRIzol, and RNA was extracted using an RNeasy Plus Micro Kit (Qiagen). cDNA was prepared with a SuperScript-III First-Strand Synthesis System (Invitrogen) and PCR-quantified using SYBRgreen (Applied Biosystems) and in an ABI PRISM 7000 sequence detection system. Three technical and three biological replicates were performed per genotype. The mRNA levels were normalized to Actb mRNA levels and compared using a Student’s t test. Results are reported as mRNA quantity relative to control ± SEM and considered statistically significant if P < 0.05. The primers used are listed in SI Materials and Methods.
Western Blot Analysis.
Tissues were lysed in radioimmunoprecipitation assay buffer [50 mM Tris (pH 7.5), 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA] with protease inhibitor mixture (Roche). Soluble lysate was denatured in LDS-sample buffer (Invitrogen) containing 50 mM DTT and separated by 4–12% (wt/vol) NuPAGE Bis-Tris gel (Invitrogen). After transfer, the PVDF membrane was blocked in 5% nonfat milk and then incubated with a primary antibody overnight, followed by blotting with a secondary antibody. The membrane was then washed, and the proteins were visualized using a Licor Odyssey Infrared Imaging System. The density of the bands was quantified using the Image Studio Lite program. The antibodies used are listed in SI Materials and Methods.
Whole-Mount in Situ mRNA Hybridization.
Lungs were dissected in PBS, fixed in 4% paraformaldehyde overnight at 4 °C, and then dehydrated to 100% methanol. Whole-mount in situ hybridization was carried out using established protocols (35).
Sectional Immunofluorescence Staining.
Embryos were dissected in PBS solution, fixed in 4% paraformaldehyde overnight at 4 °C, processed for paraffin embedding, and sectioned at 5 μm. Immunofluorescence was carried out using standard protocols and citric acid antigen retrieval. The antibodies used are listed in SI Materials and Methods.
Whole-Mount Immunohistochemical Staining.
The staining was performed following a previously published protocol (54). The branching tip area quantification was carried out using a previously published method (17). The antibodies used are listed in SI Materials and Methods.
RNA-Sequencing.
Total RNA was extracted from E13.5 mouse lungs and used in the preparation of sequencing libraries with an Illumina Truseq RNA Library Preparation Kit. For each group, three biological replicates were obtained. Libraries were sequenced with an Illumina Hiseq 2500 Sequencer, and 100-bp single-end sequences were generated. RNA-sequencing reads were mapped to the mouse reference genome (GRCm38) using TopHat. Subsequently, expression quantification and differential expression analysis were performed using Cuffdiff. Expression values were output in the form of fragments per kilobase of exon every million reads mapped. The full-sequence dataset was deposited in the GEO database (accession no. GSE80707).
Immunoprecipitation.
MLE-12 cells were transfected with plasmids containing 3× FLAG-tagged, full-length mouse Etv5 or control 3× FLAG by Lipofectamine 2000 (Invitrogen) following the manufacturer’s protocol. For immunoprecipitation from cell lysates, we used immiscible filtration assisted by surface tension following a published protocol (55). Protein was eluted from the beads using SDS loading buffer, and Western blot analysis was performed.
SI Materials and Methods
Primary Antibodies.
Rabbit anti-RFWD2 (Santa Cruz Biotechnology), 1:50 dilution
Mouse anti-ACTB (Novus Biologicals), 1:5,000 dilution
Rabbit anti-CDH1 (Cell Signaling), 1:1,000 dilution for Western blot and 1:100 dilution for whole-mount immunohistochemistry
Rabbit anti-ETV5 (laboratory-made), 1:1,000 dilution
Rabbit anti-ETV4 (Abgent), 1:1,000 dilution
Rabbit anti-SCGB1A1 (Seven Hills Bioregents), 1:200 dilution
Rabbit anti-SFTPC (Millipore), 1:200 dilution
Syrian hamster anti-PDPN (Developmental Studies Hybridoma Bank), 1:200 dilution
Rabbit anti-CGRP (Sigma), 1:200 dilution
Rabbit anti-KRT5 (Covance), 1:200 dilution
Mouse anti-p63 4A4 (eBiosciences), 1:100 dilution
Mouse antiacetylated TUBULIN1A1 (Sigma), 1:100 dilution
Mouse anti-FLAG (Sigma), 1:1,000 dilution
Rabbit anti-JUN (GeneTex): 1:2,000 dilution
Rabbit anti-C/EBPα (Santa Cruz Biotechnology): 1:500 dilution.
Secondary Antibodies.
IRDye 800CW donkey anti-rabbit (LI-COR) for Western blot, 1:10,000 dilution
IRDye 680LT donkey anti-mouse (LI-COR) for Western blot, 1:15,000 dilution
Goat anti-rabbit–HRP (Sigma), 1:100 dilution
Goat anti-rabbit–FITC (Jackson ImmunoResearch), 1:200 dilution
Goat anti-rabbit–CY3 (Jackson ImmunoResearch), 1:200 dilution
Goat anti-mouse–CY3 (Jackson ImmunoResearch), 1:200 dilution
Goat anti-Syrian hamster–CY3 (Jackson ImmunoResearch), 1:200 dilution.
Quantitative Real-Time PCR Primers.
Actb-forward (F): 5′-CGGCCAGGTCATCACTATTGGCAAC-3′
Actb-reverse (R): 5′-GCCACAGGATTCCATACCCAAGAAG-3′
Rfwd2-F: 5′-GCGTGTGGAAGTGAGAACAAC-3′
Rfwd2-R: 5′-TCATCTTCTTTCCGATCTTTGTC-3′
Sox2-F: 5′-GCGGAGTGGAAACTTTTGTCC-3′
Sox2-R: 5′-CGGGAAGCGTGTACTTATCCTT-3′
Sox9-F: 5′-CGGAACAGACTCACATCTCTCC-3′
Sox9-R: 5′-GCTTGCACGTCGGTTTTGG-3′
Etv4-F: 5′-CGCACAGACTTCGCCTACG-3′
Etv4-R: 5′-CAGACATCATCTGGGAATGGTC-3′
Etv5-F: 5′-GGTATTTCTCCAGCAGCCATGAAGG-3′
Etv5-R: 5′-TCTCGGGTACCACGCAAGTATCATC-3′
Cdh1-F: 5′-GAGAACGGTGGTCAAAGAGC-3′
Cdh1-R: 5′-GCTGGCTCAAATCAAAGTCC-3′
Shh-F: 5′-GGCTGATGACTCAGAGGTGCAAAG-3′
Shh-R: 5′-GCTCGACCCTCATAGTGTAGAGAC-3′
Foxa2-F: 5′-AGAAGATGGCTTTCAGGCCC-3′
Foxa2-R: 5′-AGGTGAGACTGCTCCCTTGA-3′
Fgf10-F: 5′-GATTGAGAAGAACGGCAAGGTCAG-3′
Fgf10-R: 5′-TTGACGGCAACAACTCCGATTTCC-3′
Gli1-F: 5′-GCCTGGAGAACCTTAGGCTGGA-3′
Gli1-R: 5′-ACAGGTGCGCCAGCGTG-3′
Spry2-F: 5′-GATTCAAGGGAGAGGGGTTG-3′
Spry2-R: 5′-CTCCATCAGGTCTTGGCAGT-3′
Cubn-F: 5′-CACTTTAGGTTGTGGTGGAACA-3′
Cubn-R: 5′-TTGCTGTCAAAGCTAATCTCCC-3′
Pthlh-F: 5′-CATCAGCTACTGCATGACAAGG-3′
Pthlh-R: 5′-GGTGGTTTTTGGTGTTGGGAG-3′
Lcn2-F: 5′-GGGAAATATGCACAGGTATCCTC-3′
Lcn2-R: 5′-CATGGCGAACTGGTTGTAGTC-3′
Nedd4l-F: 5′-GAGTCAAGGGGTTTTTGAGGTT-3′
Nedd4l-R: 5′-TGGGAAGCTGAGTCGTTGGA-3′
Trim36-F: 5′-CGTTCAATGATGTGGCGTCAG-3′
Trim36-R: 5′-GGGTCAGTGAATTACGCTTCC-3′.
Acknowledgments
We thank members of the X.S. laboratory for constructive discussions and readings of the manuscript, Dr. Vishva M. Dixit (Genentech) for sharing the Rfwd2fl mice, and Dr. Scott Berry and Dr. David Beebe (University of Wisconsin–Madison) for guidance on immunoprecipitation experiments. This work was supported by American Heart Association Postdoctoral Fellowship 15POST25670070 (to Y.Z.) and by National Heart, Lung, and Blood Institute Grants RO1 HL113870 and HL097134 and Wisconsin Partnership Program Grant 2897 (to X.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE80707).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1603310113/-/DCSupplemental.
References
- 1.Cardoso WV, Lü J. Regulation of early lung morphogenesis: Questions, facts and controversies. Development. 2006;133(9):1611–1624. doi: 10.1242/dev.02310. [DOI] [PubMed] [Google Scholar]
- 2.Morrisey EE, Hogan BL. Preparing for the first breath: Genetic and cellular mechanisms in lung development. Dev Cell. 2010;18(1):8–23. doi: 10.1016/j.devcel.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Metzger RJ, Klein OD, Martin GR, Krasnow MA. The branching programme of mouse lung development. Nature. 2008;453(7196):745–750. doi: 10.1038/nature07005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Short K, Hodson M, Smyth I. Spatial mapping and quantification of developmental branching morphogenesis. Development. 2013;140(2):471–478. doi: 10.1242/dev.088500. [DOI] [PubMed] [Google Scholar]
- 5.Shi W, et al. Overexpression of Smurf1 negatively regulates mouse embryonic lung branching morphogenesis by specifically reducing Smad1 and Smad5 proteins. Am J Physiol Lung Cell Mol Physiol. 2004;286(2):L293–L300. doi: 10.1152/ajplung.00228.2003. [DOI] [PubMed] [Google Scholar]
- 6.Scheffner M, Nuber U, Huibregtse JM. Protein ubiquitination involving an E1-E2-E3 enzyme ubiquitin thioester cascade. Nature. 1995;373(6509):81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- 7.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol Rev. 2002;82(2):373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 8.Ardley HC, Robinson PA. E3 ubiquitin ligases. Essays Biochem. 2005;41:15–30. doi: 10.1042/EB0410015. [DOI] [PubMed] [Google Scholar]
- 9.McNellis TW, Torii KU, Deng XW. Expression of an N-terminal fragment of COP1 confers a dominant-negative effect on light-regulated seedling development in Arabidopsis. Plant Cell. 1996;8(9):1491–1503. doi: 10.1105/tpc.8.9.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yi C, Deng XW. COP1 - from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol. 2005;15(11):618–625. doi: 10.1016/j.tcb.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 11.Marine JC. Spotlight on the role of COP1 in tumorigenesis. Nat Rev Cancer. 2012;12(7):455–464. doi: 10.1038/nrc3271. [DOI] [PubMed] [Google Scholar]
- 12.Migliorini D, et al. Cop1 constitutively regulates c-Jun protein stability and functions as a tumor suppressor in mice. J Clin Invest. 2011;121(4):1329–1343. doi: 10.1172/JCI45784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dornan D, et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429(6987):86–92. doi: 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]
- 14.Yoshida A, Kato JY, Nakamae I, Yoneda-Kato N. COP1 targets C/EBPα for degradation and induces acute myeloid leukemia via Trib1. Blood. 2013;122(10):1750–1760. doi: 10.1182/blood-2012-12-476101. [DOI] [PubMed] [Google Scholar]
- 15.Li DQ, et al. E3 ubiquitin ligase COP1 regulates the stability and functions of MTA1. Proc Natl Acad Sci USA. 2009;106(41):17493–17498. doi: 10.1073/pnas.0908027106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vitari AC, et al. COP1 is a tumour suppressor that causes degradation of ETS transcription factors. Nature. 2011;474(7351):403–406. doi: 10.1038/nature10005. [DOI] [PubMed] [Google Scholar]
- 17.Herriges JC, et al. FGF-Regulated ETV Transcription Factors Control FGF-SHH Feedback Loop in Lung Branching. Dev Cell. 2015;35(3):322–332. doi: 10.1016/j.devcel.2015.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris KS, Zhang Z, McManus MT, Harfe BD, Sun X. Dicer function is essential for lung epithelium morphogenesis. Proc Natl Acad Sci USA. 2006;103(7):2208–2213. doi: 10.1073/pnas.0510839103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gontan C, et al. Sox2 is important for two crucial processes in lung development: Branching morphogenesis and epithelial cell differentiation. Dev Biol. 2008;317(1):296–309. doi: 10.1016/j.ydbio.2008.02.035. [DOI] [PubMed] [Google Scholar]
- 20.Rockich BE, et al. Sox9 plays multiple roles in the lung epithelium during branching morphogenesis. Proc Natl Acad Sci USA. 2013;110(47):E4456–E4464. doi: 10.1073/pnas.1311847110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang DR, et al. Lung epithelial branching program antagonizes alveolar differentiation. Proc Natl Acad Sci USA. 2013;110(45):18042–18051. doi: 10.1073/pnas.1311760110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abler LL, Mansour SL, Sun X. Conditional gene inactivation reveals roles for Fgf10 and Fgfr2 in establishing a normal pattern of epithelial branching in the mouse lung. Dev Dyn. 2009;238(8):1999–2013. doi: 10.1002/dvdy.22032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suriben R, et al. β-Cell Insulin Secretion Requires the Ubiquitin Ligase COP1. Cell. 2015;163(6):1457–1467. doi: 10.1016/j.cell.2015.10.076. [DOI] [PubMed] [Google Scholar]
- 24.Herriges JC, et al. Genome-scale study of transcription factor expression in the branching mouse lung. Dev Dyn. 2012;241(9):1432–1453. doi: 10.1002/dvdy.23823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y, Jiang H, Crawford HC, Hogan BL. Role for ETS domain transcription factors Pea3/Erm in mouse lung development. Dev Biol. 2003;261(1):10–24. doi: 10.1016/s0012-1606(03)00359-2. [DOI] [PubMed] [Google Scholar]
- 26.Lin S, Perl AK, Shannon JM. Erm/thyroid transcription factor 1 interactions modulate surfactant protein C transcription. J Biol Chem. 2006;281(24):16716–16726. doi: 10.1074/jbc.M602221200. [DOI] [PubMed] [Google Scholar]
- 27.Que J, Luo X, Schwartz RJ, Hogan BL. Multiple roles for Sox2 in the developing and adult mouse trachea. Development. 2009;136(11):1899–1907. doi: 10.1242/dev.034629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wan H, et al. Compensatory roles of Foxa1 and Foxa2 during lung morphogenesis. J Biol Chem. 2005;280(14):13809–13816. doi: 10.1074/jbc.M414122200. [DOI] [PubMed] [Google Scholar]
- 29.Wei GH, et al. Genome-wide analysis of ETS-family DNA-binding in vitro and in vivo. EMBO J. 2010;29(13):2147–2160. doi: 10.1038/emboj.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Assémat E, et al. Overlapping expression patterns of the multiligand endocytic receptors cubilin and megalin in the CNS, sensory organs and developing epithelia of the rodent embryo. Gene Expr Patterns. 2005;6(1):69–78. doi: 10.1016/j.modgep.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 31.Hastings RH. Parathyroid hormone-related protein and lung biology. Respir Physiol Neurobiol. 2004;142(2-3):95–113. doi: 10.1016/j.resp.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 32.Hastings RH, Duong H, Burton DW, Deftos LJ. Alveolar epithelial cells express and secrete parathyroid hormone-related protein. Am J Respir Cell Mol Biol. 1994;11(6):701–706. doi: 10.1165/ajrcmb.11.6.7946399. [DOI] [PubMed] [Google Scholar]
- 33.Saiga H, et al. Lipocalin 2-dependent inhibition of mycobacterial growth in alveolar epithelium. J Immunol. 2008;181(12):8521–8527. doi: 10.4049/jimmunol.181.12.8521. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Z, Verheyden JM, Hassell JA, Sun X. FGF-regulated Etv genes are essential for repressing Shh expression in mouse limb buds. Dev Cell. 2009;16(4):607–613. doi: 10.1016/j.devcel.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laing MA, et al. Male sexual dysfunction in mice bearing targeted mutant alleles of the PEA3 ets gene. Mol Cell Biol. 2000;20(24):9337–9345. doi: 10.1128/mcb.20.24.9337-9345.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reddy NM, et al. Targeted deletion of Jun/AP-1 in alveolar epithelial cells causes progressive emphysema and worsens cigarette smoke-induced lung inflammation. Am J Pathol. 2012;180(2):562–574. doi: 10.1016/j.ajpath.2011.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flodby P, Barlow C, Kylefjord H, Ahrlund-Richter L, Xanthopoulos KG. Increased hepatic cell proliferation and lung abnormalities in mice deficient in CCAAT/enhancer binding protein alpha. J Biol Chem. 1996;271(40):24753–24760. doi: 10.1074/jbc.271.40.24753. [DOI] [PubMed] [Google Scholar]
- 38.Ding Y, Zhang Y, Xu C, Tao QH, Chen YG. HECT domain-containing E3 ubiquitin ligase NEDD4L negatively regulates Wnt signaling by targeting dishevelled for proteasomal degradation. J Biol Chem. 2013;288(12):8289–8298. doi: 10.1074/jbc.M112.433185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cuykendall TN, Houston DW. Vegetally localized Xenopus trim36 regulates cortical rotation and dorsal axis formation. Development. 2009;136(18):3057–3065. doi: 10.1242/dev.036855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weathington NM, Sznajder JI, Mallampalli RK. The emerging role of the ubiquitin proteasome in pulmonary biology and disease. Am J Respir Crit Care Med. 2013;188(5):530–537. doi: 10.1164/rccm.201304-0754PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dornan D, et al. COP1, the negative regulator of p53, is overexpressed in breast and ovarian adenocarcinomas. Cancer Res. 2004;64(20):7226–7230. doi: 10.1158/0008-5472.CAN-04-2601. [DOI] [PubMed] [Google Scholar]
- 42.Lee YH, et al. Definition of ubiquitination modulator COP1 as a novel therapeutic target in human hepatocellular carcinoma. Cancer Res. 2010;70(21):8264–8269. doi: 10.1158/0008-5472.CAN-10-0749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Su CH, et al. 14-3-3sigma exerts tumor-suppressor activity mediated by regulation of COP1 stability. Cancer Res. 2011;71(3):884–894. doi: 10.1158/0008-5472.CAN-10-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ouyang M, et al. COP1, the negative regulator of ETV1, influences prognosis in triple-negative breast cancer. BMC Cancer. 2015;15:132. doi: 10.1186/s12885-015-1151-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leikauf GD, et al. Haplotype association mapping of acute lung injury in mice implicates activin a receptor, type 1. Am J Respir Crit Care Med. 2011;183(11):1499–1509. doi: 10.1164/rccm.201006-0912OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cassel TN, Nord M. C/EBP transcription factors in the lung epithelium. Am J Physiol Lung Cell Mol Physiol. 2003;285(4):L773–L781. doi: 10.1152/ajplung.00023.2003. [DOI] [PubMed] [Google Scholar]
- 47.Martis PC, et al. C/EBPalpha is required for lung maturation at birth. Development. 2006;133(6):1155–1164. doi: 10.1242/dev.02273. [DOI] [PubMed] [Google Scholar]
- 48.Chen XL, et al. Patched-1 proapoptotic activity is downregulated by modification of K1413 by the E3 ubiquitin-protein ligase Itchy homolog. Mol Cell Biol. 2014;34(20):3855–3866. doi: 10.1128/MCB.00960-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anderson K, et al. Regulation of cellular levels of Sprouty2 protein by prolyl hydroxylase domain and von Hippel-Lindau proteins. J Biol Chem. 2011;286(49):42027–42036. doi: 10.1074/jbc.M111.303222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Persaud A, et al. Nedd4-1 binds and ubiquitylates activated FGFR1 to control its endocytosis and function. EMBO J. 2011;30(16):3259–3273. doi: 10.1038/emboj.2011.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loregger A, et al. The E3 ligase RNF43 inhibits Wnt signaling downstream of mutated β-catenin by sequestering TCF4 to the nuclear membrane. Sci Signal. 2015;8(393):ra90. doi: 10.1126/scisignal.aac6757. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y, Chang C, Gehling DJ, Hemmati-Brivanlou A, Derynck R. Regulation of Smad degradation and activity by Smurf2, an E3 ubiquitin ligase. Proc Natl Acad Sci USA. 2001;98(3):974–979. doi: 10.1073/pnas.98.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harfe BD, et al. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118(4):517–528. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- 54.Domyan ET, et al. Roundabout receptors are critical for foregut separation from the body wall. Dev Cell. 2013;24(1):52–63. doi: 10.1016/j.devcel.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moussavi-Harami SF, et al. Characterization of molecules binding to the 70K N-terminal region of fibronectin by IFAST purification coupled with mass spectrometry. J Proteome Res. 2013;12(7):3393–3404. doi: 10.1021/pr400225p. [DOI] [PMC free article] [PubMed] [Google Scholar]