Significance
β1 integrins form the largest and most diverse integrin subfamily, with 12 distinct αβ heterodimers. Together, the seven other integrin β-subunits form an equal number of 12 heterodimers. Despite speculation that different integrin subfamilies may differ in activation mechanism or conformation, we show that α5β1, similar to β2 and β3 integrins, can adopt the bent-closed, extended-closed, and extended-open overall global conformational states. We characterize nine function-perturbing antibodies for the subunit and domain they bind and the conformation they stabilize. Only the extended-open α5β1 conformation mediates adhesion to fibronectin. Our results enable many cell biological studies on the 12 β1 integrins to be interpreted in terms of the integrin conformational states on cell surfaces that are stabilized.
Abstract
Whether β1 integrin ectodomains visit conformational states similarly to β2 and β3 integrins has not been characterized. Furthermore, despite a wealth of activating and inhibitory antibodies to β1 integrins, the conformational states that these antibodies stabilize, and the relation of these conformations to function, remain incompletely characterized. Using negative-stain electron microscopy, we show that the integrin α5β1 ectodomain adopts extended-closed and extended-open conformations as well as a bent conformation. Antibodies SNAKA51, 8E3, N29, and 9EG7 bind to different domains in the α5 or β1 legs, activate, and stabilize extended ectodomain conformations. Antibodies 12G10 and HUTS-4 bind to the β1 βI domain and hybrid domains, respectively, activate, and stabilize the open headpiece conformation. Antibody TS2/16 binds a similar epitope as 12G10, activates, and appears to stabilize an open βI domain conformation without requiring extension or hybrid domain swing-out. mAb13 and SG/19 bind to the βI domain and βI–hybrid domain interface, respectively, inhibit, and stabilize the closed conformation of the headpiece. The effects of the antibodies on cell adhesion to fibronectin substrates suggest that the extended-open conformation of α5β1 is adhesive and that the extended-closed and bent-closed conformations are nonadhesive. The functional effects and binding sites of antibodies and fibronectin were consistent with their ability in binding to α5β1 on cell surfaces to cross-enhance or inhibit one another by competitive or noncompetitive (allosteric) mechanisms.
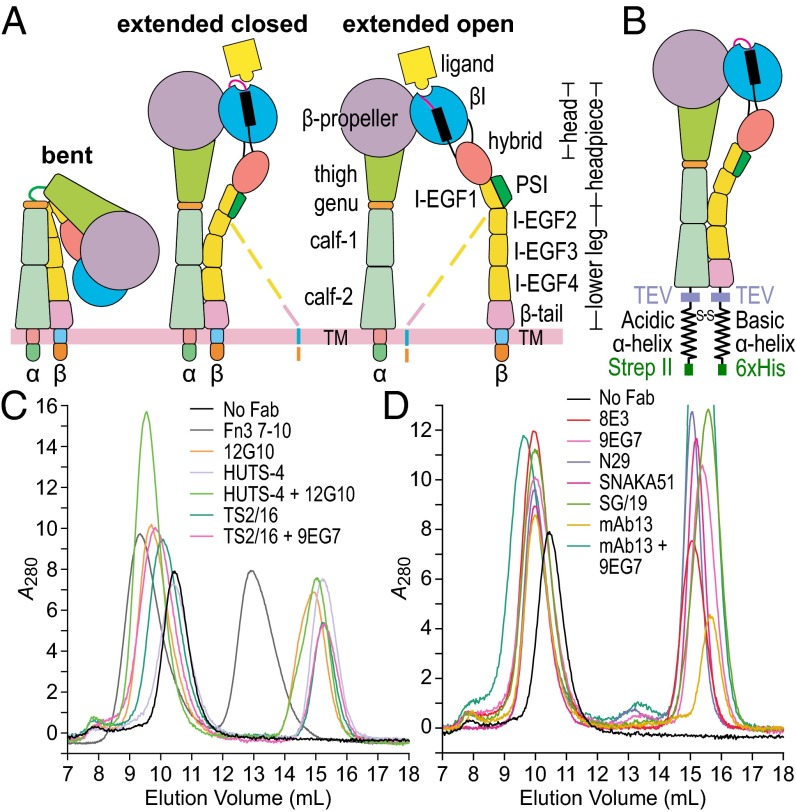
Integrins comprise a family of 24 adhesion receptors that transmit bidirectional signals across the cell membrane (1). Integrin α- and β-subunits are both type I transmembrane glycoproteins, with a large N-terminal extracellular domain, a single-span transmembrane domain, and usually a short C-terminal cytoplasmic domain. β2 and β3 integrins adopt three global conformational states: bent-closed, extended-closed, and extended-open (Fig. 1A) (2–10). With β2 and β3 integrins, high concentrations of ligands induce the extended-open conformation of intact integrin and ectodomain preparations—and the open conformation of headpiece preparations—showing that the open conformation has higher affinity than closed conformations (7, 10–12). Electron microscopy (EM) studies and cell-surface affinity measurements with αXβ2 and αLβ2 integrins and conformation-specific Fabs show that the bent-closed and extended-closed conformations have low affinity for ligand whereas the extended-open conformation has high affinity for ligand (8, 9, 13).
Fig. 1.
Integrin overview and complexes for EM. (A) Three integrin conformational states. Dashed lines represent alternative conformations of the flexible lower β-leg. (B) Clasped α5β1 ectodomain construct. (C and D) Gel filtration of α5β1 alone and in complex with Fabs or Fn3 7–10. Peak integrin fractions were subjected to EM. TM, transmembrane.
We know less about β1 integrin conformational states. The β1 subunit associates with 12 integrin α-subunits, including those that contain or lack αI domains, to form by far the largest and most diverse integrin subfamily. β1 integrins recognize ligands including collagens, laminins, ligands with Arg-Gly-Asp (RGD) motifs including fibronectin, and Ig superfamily members including cell-adhesion molecules (1). Cells localized in tissues basally adhere through β1 integrins to the extracellular matrix. In contrast, β2 and β3 integrins are commonly expressed on cells that circulate in the bloodstream in a state in which these integrins are nonadhesive. Only upon cellular activation in the bloodstream do β2 integrins on leukocytes or β3 integrins on platelets become active and bind their ligands (1). A common discussion point among integrin biologists is the concept that β1 integrins may be regulated differently from β2 and β3 integrins. One proposal is that β1 integrins may not have a bent conformation and may be constitutively extended. Small-angle X-ray scattering (SAXS) and EM studies on the α5β1 headpiece have shown that it can adopt a closed conformation with the hybrid domain swung in, and an open conformation with the hybrid domain swung out (11, 14, 15) (Fig. 1A). However, whether β1 integrins have both bent and extended conformations like β2 and β3 integrins and the specific conformational states of the β1 integrin ectodomain that correspond to active and inactive states remains elusive.
A large number of activating and inhibitory antibodies are mainstays in the β1 integrin field (14, 16–22). Some have been surmised to be specific for conformational states, particularly of α5β1, by indirect means such as their ability to alter ligand binding coupled with mutagenesis interpreted with respect to the integrin αVβ3 structure (20, 23, 24). However, the lack of evidence that α5β1 has a bent conformation and paucity of structural information on β1 integrin complexes with antibodies have limited our ability to define the structural basis of β1 integrin biology. Among such antibodies, only SG/19 has been structurally characterized, and its EM and crystal structures are limited to headpiece complexes. These have shown that whereas binding of fibronectin fragments and RGD peptides stabilizes the open conformation of the α5β1 headpiece, binding of this inhibitory Fab stabilizes the closed headpiece conformation (11, 14, 25).
Here we characterize the conformational states that are accessible to integrin α5β1 in the absence and presence of α5β1 Fabs, and the role of these states in α5β1-mediated adhesiveness to fibronectin substrates. By comparing nine function-perturbing Fabs for their binding sites on specific integrin domains and the conformational states they stabilize, we enable a large number of previous cell biological studies on the 12 β1 integrins to be interpreted in terms of the integrin conformational states on cell surfaces that were stabilized. We demonstrate that activating β1 mAb can perturb the conformational equilibrium either by stabilizing extension or headpiece opening, that α5β1 can adopt a bent conformation, and that the extended-open and not the bent-closed or extended-closed conformations of α5β1 mediates adhesion to fibronectin.
Results
EM, Integrin α5β1, and Fibronectin.
The α5β1 ectodomain was expressed in mammalian cells using a clasped protein construct (Fig. 1B) in which ACID and BASE coiled-coils provide the close association between the α- and β-subunit ectodomain C termini provided in intact cells by the associating transmembrane domains (4, 6–9, 26). Purified clasped α5β1 was incubated with or without Fab or a fibronectin fragment with Fn3 domains 7–10 (Fn3 7–10) and subjected to gel filtration (Fig. 1 C and D). Peak fractions were immediately adsorbed to electrostatically charged (glow-discharged) carbon grids, negatively stained (and fixed) with uranyl formate, and subjected to EM. Over 5,000 particles were picked from images (SI Appendix, Fig. S1) and subjected to alignment, classification, and averaging using two different algorithms (Methods). Representative classes with good structural detail (see below) and all class averages (SI Appendix, Figs. S2–S14) are shown.
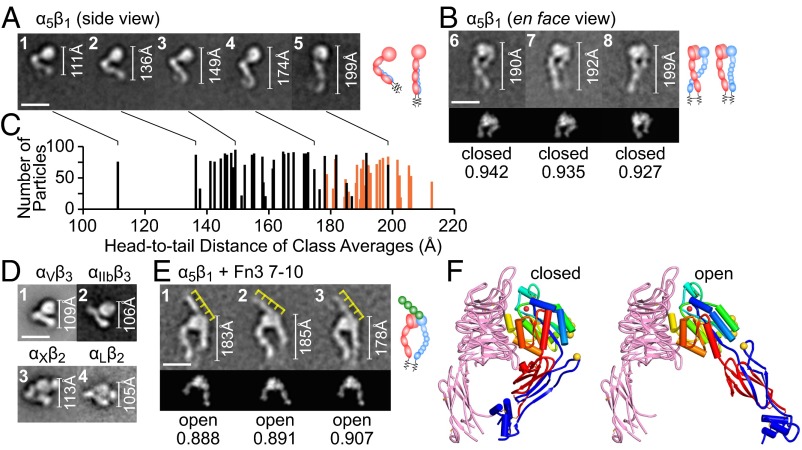
The α5β1 ectodomain adsorbed onto EM grids in side and en face orientations (Fig. 2 A and B, respectively). The α- and β-legs lie together in side views and cannot be distinguished. Side-view particles are V-shaped and comprise a globular end containing the headpiece and a lower leg, each ∼100 Å long (Fig. 2A). These sum to ∼200 Å, very similar to the 190–200 Å that en face, extended particles span (Fig. 2B). Side-view α5β1 particles were present in a range of head-to-tail distances from 110 to 200 Å (Fig. 2 A and C). Most had head-to-tail distances of 135–150 Å (Fig. 2A, panels 2 and 3 and Fig. 2C) and an appearance that closely resembled compact, bent particles of four other integrins (4, 7, 8) (Fig. 2D). Although other integrins including αIIbβ3 show a range of V angles in the bent conformation (4), the angle is on average greater in α5β1.
Fig. 2.
Integrin α5β1 alone and in complex with Fn3 7–10. (Scale bars, 10 nm.) (Right) Interpretative cartoons show α5, β1, and Fn3 7–10 in red, blue, and green, respectively. Headpiece portions of en face views were cross-correlated with 2D projections of closed and open headpiece crystal structures; the best-correlating projection and its correlation coefficient are shown below each class average. White rules show the total distance from head to tail; yellow rules interpret the four Fn3 domains in Fn3 7–10. (A and B) α5β1 class averages; the classes represented in A are marked in C by connecting lines. (C) All α5β1 class averages showing head-to-tail distance and particles per class, with side views in black and en face views in orange. (D) Class averages of αVβ3, αIIbβ3, αXβ2, and αLβ2 in their bent conformation (4, 7, 8). (E) α5β1–Fn3 7–10 complex. (F) Closed and open headpiece crystal structures (1, 25) rendered as described in Fig. 8.
En face α5β1 was clearly extended (Fig. 2B). Although some side-view particles were extended (Fig. 2A, panel 5), there was little overlap in head-to-tail distance between side-view and en face particles (Fig. 2C), suggesting that most side-view α5β1 particles are bent (Fig. 2A, panels 1–4). In en face α5β1 the two subunits can be clearly distinguished by the larger size in the head of the α-subunit β-propeller domain than the β-subunit βI domain. Thus, the α-subunit β-propeller domain and upper leg lie to the left in Fig. 2B (Right; schematics). The α- and β-legs tended to cross below the knees. The α-leg has larger domains than the β-leg and was often resolved to its last domain, calf-2, whereas the β-leg is flexible and was often “averaged out” or faint in class averages (Fig. 2B). These leg properties resemble those of other integrins (4, 7, 8). En face α5β1 predominantly exhibited the closed conformation of the headpiece; the headpiece moiety cross-correlated better with 2D projections from closed headpiece than open headpiece crystal structures (Fig. 2B, Lower and Methods).
α5β1 ectodomain complexes with Fn3 7–10 exclusively exhibited en face views and the extended open conformation (Fig. 2E); the headpiece region best-correlated with 2D projections from open headpiece integrin crystal structures (Fig. 2E, Lower). In the open conformation, the hybrid domain swings out. This appears to result in slightly but consistently lower head-to-tail distances (Fig. 2E) than for extended-closed particles (Fig. 2B), as the legs adopt a more curved conformation to meet at the clasp (even though the clasp, along with the lower β-leg, is not visible). Fn3 7–10 bound to an interface formed by the α5 β-propeller and β1 βI domain. Fn3 7–10 adopted an extended conformation and had a length of ∼145 Å, as seen in its crystal structure (27). Four individual Fn3 modules were moderately resolved as globules; these globules and their equal lengths allowed their positions to be deduced (yellow rules, Fig. 2E). Fn3 domains 9 and 10, known to be sufficient for binding (28), each bound the integrin. Fn3 domain 10 with its RGD largely laid over the βI domain, whereas domain 9 with its synergy site appeared to partially contact the β-propeller domain and partially extend beyond it (11, 15). Fn3 domains 7 and 8, although not essential for binding, were helpful for visualizing and orienting the ligand in EM.
Fabs That Stabilize Extension.
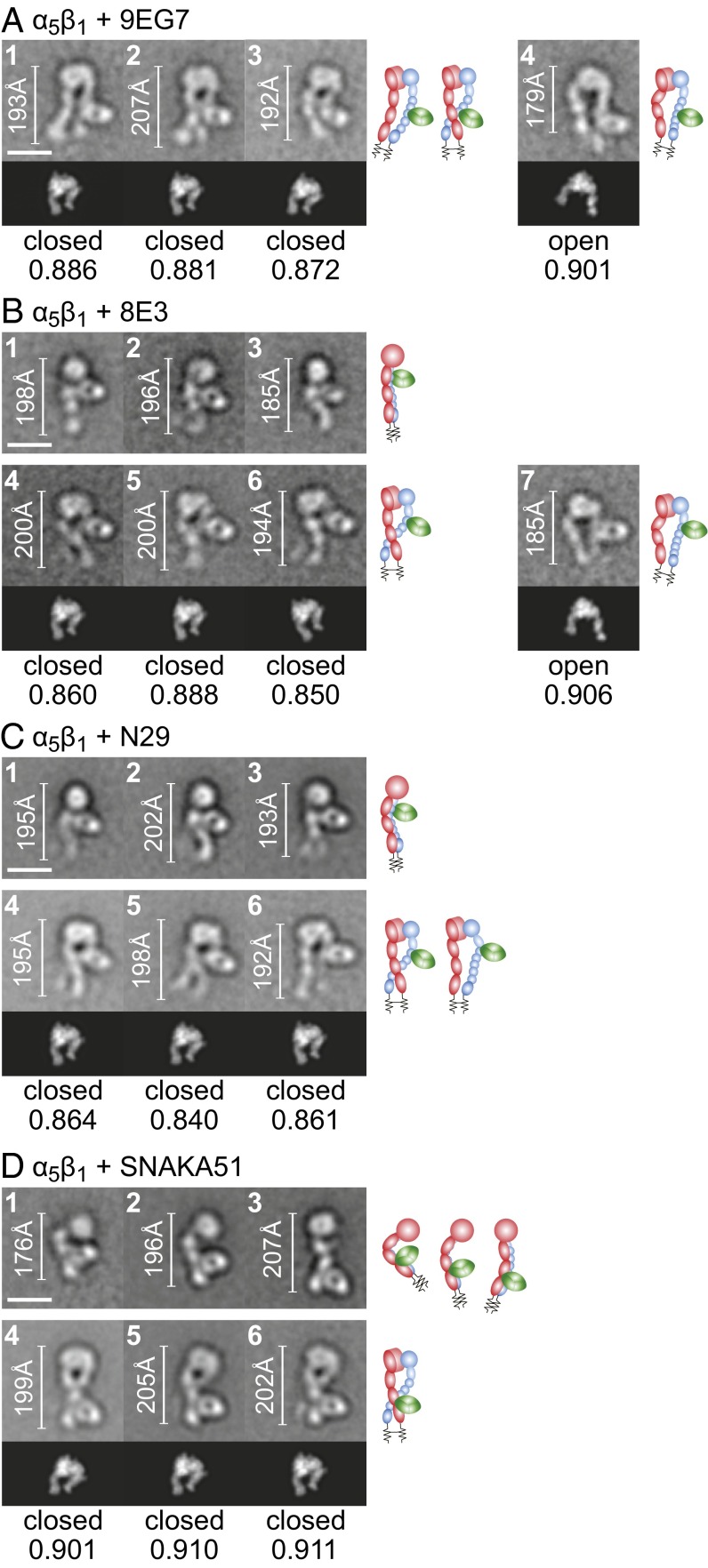
Four activating Fabs, 9EG7, 8E3, N29, and SNAKA51, all stabilized extension and yielded side or en face views of the α5β1 ectodomain with similar head-to-tail lengths of 180–200 Å (Fig. 3). The great majority of en face views had the closed conformation, as shown by better correlation of their headpiece moieties with the closed conformation (Fig. 3), whereas a small minority of class averages, at least for two of the Fabs, displayed the open headpiece (Fig. 3 A and B, Right). 8E3 and N29 Fabs bound to the lower portion of the upper β-leg (Fig. 3 B and C), confirming previous mapping to the plexin–semaphorin–integrin (PSI) domain (20). 9EG7 bound to the upper portion of the lower β-leg. Individual integrin-epidermal growth factor (I-EGF) domains were not resolved; therefore, 9EG7-binding position was measured as % of head-to-tail distance in EM. Comparison with distances in extended integrin ectodomain models (4) suggested that 9EG7 bound to I-EGF2, in agreement with abolition of the epitope by a D522E mutation in I-EGF2 (19, 29). SNAKA51 bound to the lower α5 leg at the interface between the calf-1 and calf-2 domains (Fig. 3D), in agreement with the sufficiency of a fragment containing both the calf-1 and calf-2 domains, but not either domain alone, for SNAKA51 binding (30).
Fig. 3.
Complexes of α5β1 with Fabs that stabilize extension. Representative class averages of complexes with Fabs 9EG7 (A), 8E3 (B), N29 (C), and SNAKA51 (D). Scales, rules, cross-correlations, and cartoons are described in the legend for Fig. 1.
Fabs That Stabilize the Open Headpiece.
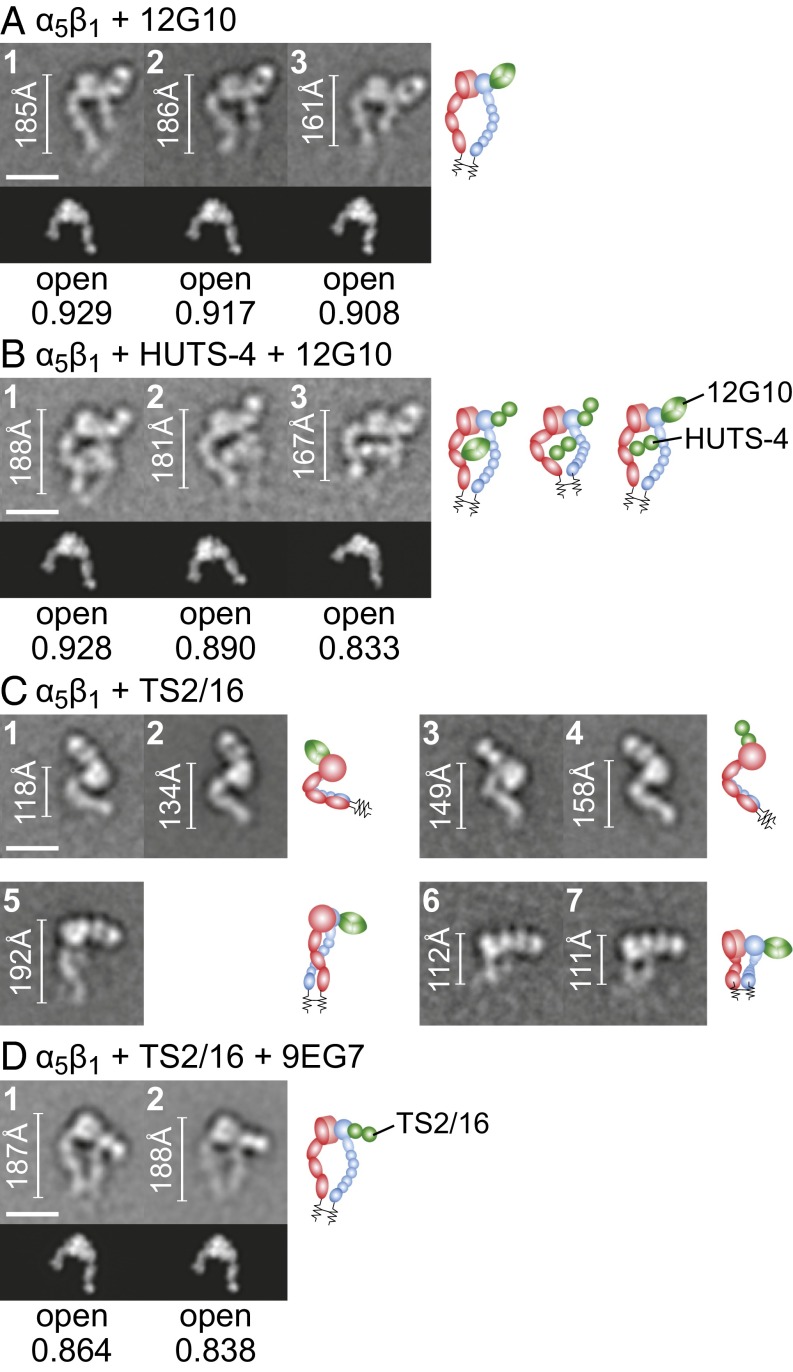
The activating antibodies 12G10 and HUTS-4 stabilized the open α5β1 headpiece, as shown by EM (Fig. 4 A and B). All class averages of the α5β1–12G10 complex showed the en face view of α5β1 in the extended-open conformation with 12G10 bound to the outer side of the βI domain (Fig. 4A), consistent with epitope mapping. HUTS-4 and α5β1 did not form a stable complex that can be purified by gel filtration; the elution peak coincided with that of α5β1 alone (Fig. 1C). However, it formed a stable ternary complex with α5β1 and 12G10, consistent with selection of HUTS-4 for recognition of activated integrin (16). Class averages of the ternary complex all showed the en face view of α5β1 in the extended-open conformation (Fig. 4B). HUTS-4 bound to the hybrid domain and laid between the α5 and β1 subunits (Fig. 4B), consistent with inclusion in the epitope of β1 residues 355–425 (16).
Fig. 4.
Complexes of α5β1 with Fabs that stabilize the open headpiece. Representative class averages of complexes with Fabs 12G10 (A), HUTS-4 + 12G10 (B), TS2/16 (C), and TS2/16 + 9EG7 (D). Scales, rules, cross-correlations, and cartoons are described in the legend for Fig. 1.
TS2/16 is an unusual activating antibody that may stabilize an open conformation of the βI domain while favoring but not requiring the hybrid domain to swing out or the integrin to extend. Most class averages of the α5β1–TS2/16 complex showed a side view where α5β1 exhibited both bent and extended conformations with TS2/16 bound to the head (Fig. 4C, panels 1–5) that resembled α5β1 class averages (Fig. 2 A and B). A few class averages showed a view with the head–Fab complex on the grid and the legs missing and perhaps farther from the grid and thus not well-stained (Fig. 4C, panels 6 and 7). En face views were absent, and thus none of the class averages with TS2/16 alone defined whether the headpiece was open or closed. Ternary complexes of α5β1 with TS2/16 and the extension-specific Fab 9EG7 yielded orientations on the grid with little overlap with TS2/16–α5β1 class averages (SI Appendix, Figs. S10 and S11) even though 9EG7 Fab was not visualized in most ternary class averages (Fig. 4D and SI Appendix, Fig. S11). In contrast to TS2/16–α5β1 complexes, en face views were predominant in TS2/16–9EG7–α5β1 complexes. The best-resolved and most-populated en face class averages clearly showed the extended-open conformation of α5β1 (Fig. 4D) and all class averages appeared to have the extended-open conformation, particularly when prepared with iterative stable alignment and clustering (SI Appendix, Fig. S11B); however, we could not rule out the presence of a minority of extended-closed class averages (SI Appendix, Fig. S11). The en face class averages showed that TS2/16 bound to the βI domain (Fig. 4D), consistent with its localization in side-view complexes (Fig. 4C) and epitope mapping (14, 31). Saturation of α5β1 with TS2/16, 12G10, or HUTS-4 Fabs induces indistinguishable high affinity for cyclic RGD peptide. These results and more described below suggest that TS2/16 induces the open conformation of the βI domain but binds in such a way that βI domain opening (conformational change in ligand-binding regions) favors but does not require hybrid domain swing-out, namely headpiece opening as visualized in EM (Fig. 4C).
Fabs That Stabilize the Closed Headpiece.
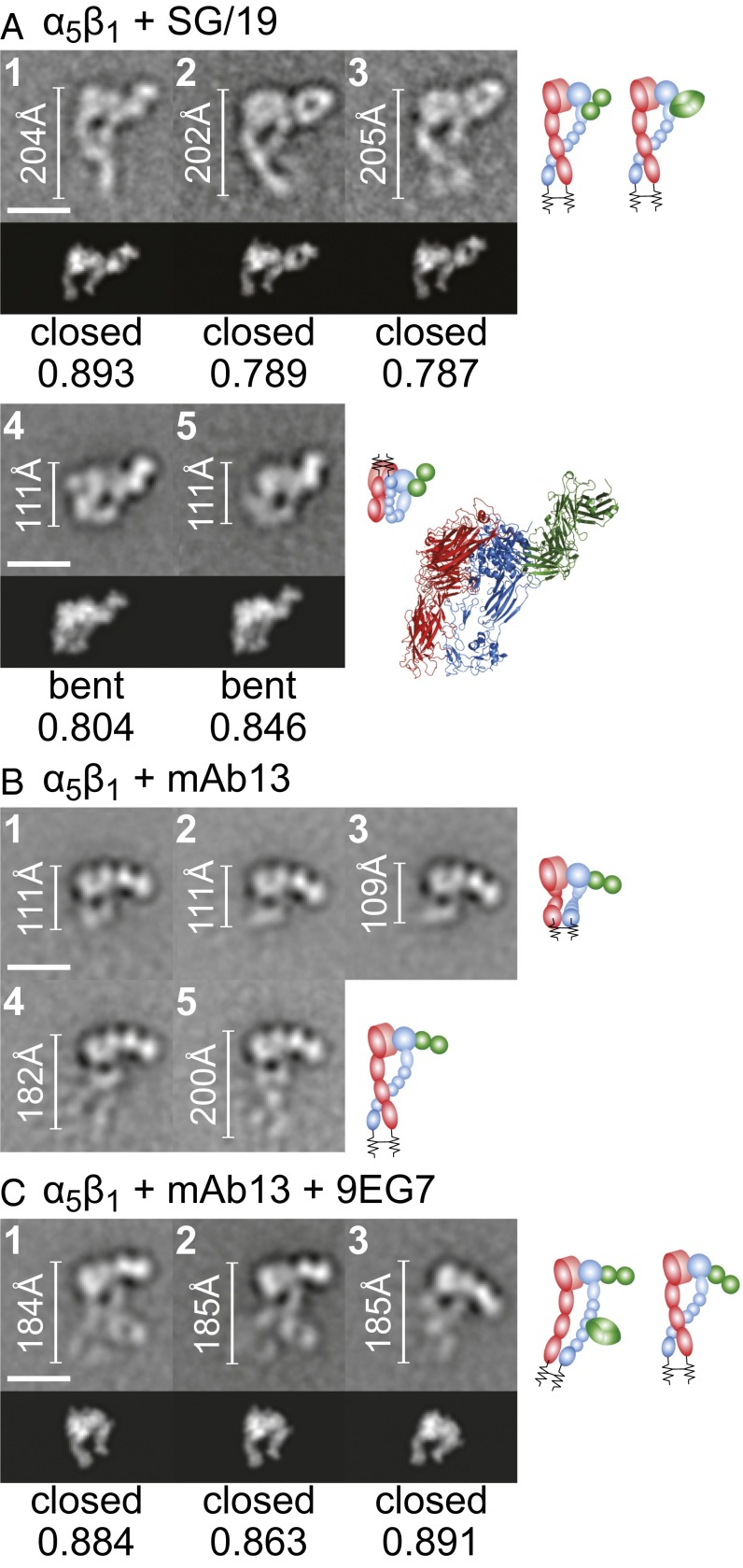
Complexes with inhibitory Fabs SG/19 and mAb13 showed α5β1 in the bent or extended-closed conformation (Fig. 5). The α5β1–SG/19 complex was mostly in the en face orientation, showing a fully extended ectodomain with closed headpiece (Fig. 5A, panels 1–3). SG/19 bound at the interface between the βI and hybrid domains, consistent with EM and crystal structures (14, 25). Some class averages also showed SG/19 Fab bound to the compact, bent conformation of α5β1 (Fig. 5A, panels 4 and 5). We made a model of an SG/19 Fab–α5β1 ectodomain complex by superimposing α5β1 headpiece–SG/19 Fab complex and αVβ3 ectodomain crystal structures (6, 25). The ectodomain–SG/19 Fab complex model cross-correlated well with class averages (Fig. 5A, panels 4 and 5, Lower and Right), providing support for a compact, bent α5β1 ectodomain conformation similar to that of αVβ3.
Fig. 5.
Complexes of α5β1 with Fabs that stabilize the closed headpiece. Representative class averages of SG/19 (A), mAb13 (B), and mAb13 + 9EG7 (C). Scales, rules, cross-correlations, and cartoons are described in the legend for Fig. 1, except the cross-correlated model or crystal structure included Fab SG/19 (A, classes 1–3) or a structure-based model of SG/19 Fab bound to the bent, compact ectodomain (A, classes 4 and 5).
mAb13 clearly bound to the head of α5β1, but class averages tended not to show the legs (Fig. 5B). Many class averages showed the head–Fab bound to the grid with the legs perhaps farther from the grid (Fig. 5B, panels 1–3), as shown for some TS2/16 complexes (Fig. 4C, panels 6–7). Some mAb13 complex class averages showed an en face view consistent with the closed headpiece (Fig. 5B, panels 4 and 5). Ternary complexes with 9EG7 yielded more en face views with the α5β1 ectodomain clearly in the extended-closed conformation and with mAb13 bound to the βI domain (Fig. 5C), consistent with epitope mapping (31).
Functional Effects and Compatibility Between Fabs.
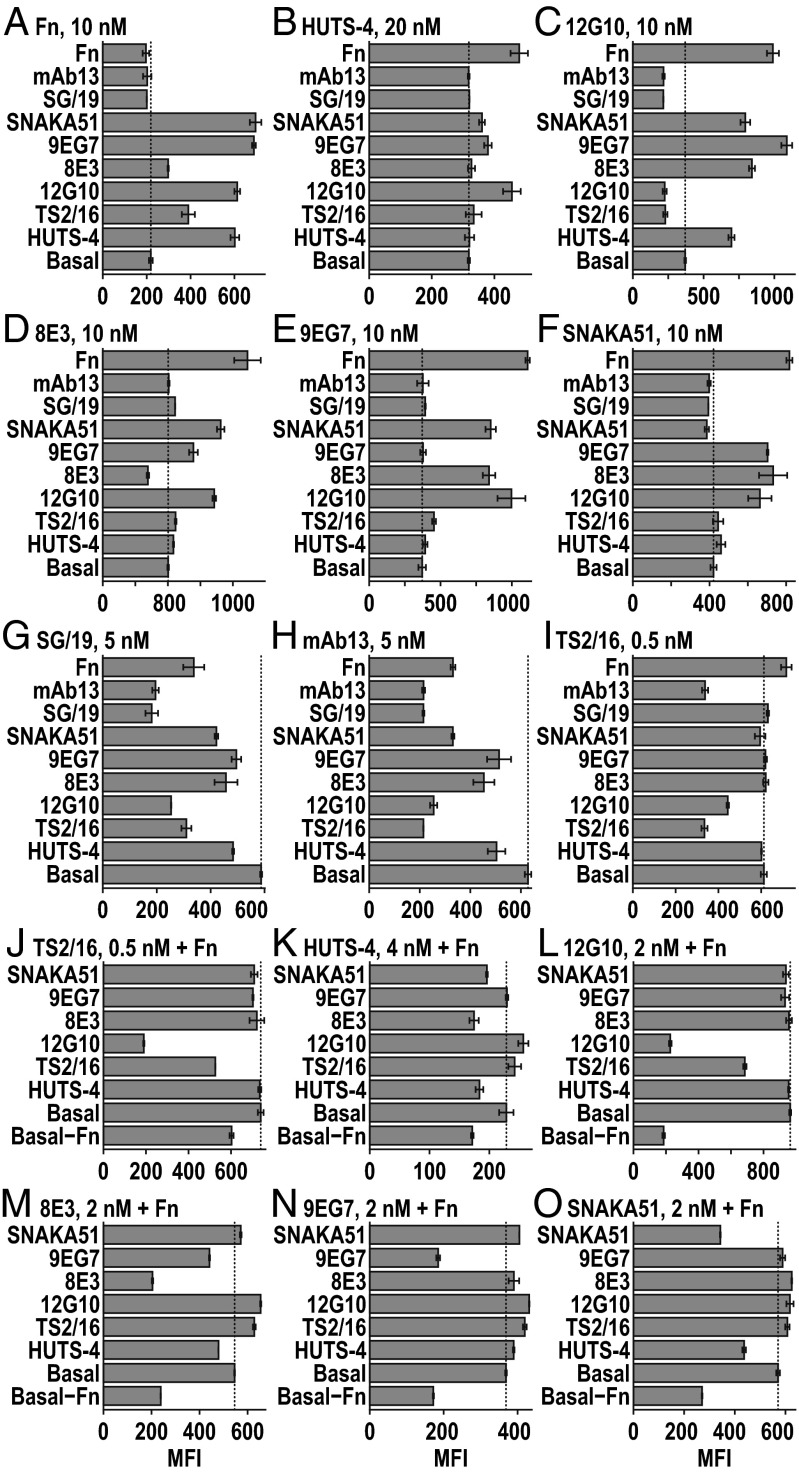
Fab cross-competition experiments corroborated conformational specificities of Fabs and examined competition among them for binding sites (Fig. 6). K562 cells, which abundantly express α5β1, were pretreated with high concentrations of one Fab or Fn3 9–10 as “effector” or “competitor” in the absence (Fig. 6 A–I) or presence of high concentrations of Fn3 9–10 as a second effector (Fig. 6 J–O), and then labeled with a lower concentration of Alexa Fluor 488-labeled second Fab or Fn3 9–10. Subsequent fluorescence flow cytometry was performed in the absence of washing to avoid removal of the effectors/competitors. Results are informed by findings that on K562 cells, α5β1 is >99% bent-closed; that once extended, the extended-open conformation is more stable than the extended-closed conformation; and that Fn3 9–10 binds much more strongly to the extended-open than bent- or extended-closed conformations.
Fig. 6.
Competition or synergy in binding of Fabs and fibronectin. K562 cells were incubated with high concentrations of Fabs or Fn3 9–10 (Fn) effectors for 90 min (labeled on left axis) without (A–I) or with (J–O) 15 µM Fn, and then with a second Alexa Fluor 488-labeled Fab or Fn reporter at the concentrations shown (Top) for 30 min and subjected to immunofluorescence flow cytometry without any washing. Effector concentrations are (A–I) 3 μM for TS2/16 and 12G10; 6 μM for 8E3, 9EG7, SNAKA51, SG19, and mAb13; 15 μM for HUTS-4 and Fn; and (J–O) 20 nM for TS2/16 and 12G10; 40 nM for 8E3, 9EG7, and SNAKA51; and 100 nM for HUTS-4. Error bars show standard deviation.
Self-competition results were consistent with the predominance of the bent conformation of α5β1 on K562 cells. The two Fabs specific for the closed conformation, SG/19 and mAb13, each showed robust self-competition in the absence of Fn3 9–10 (Fig. 6 G and H). Among extension-specific and open-specific Fabs, HUTS-4, 12G10, 8E3, 9EG7, and SNAKA51 showed binding augmented by Fn3 9–10 and good self-competition in the presence of Fn3 9–10 (Fig. 6 K–O) but less or no self-competition in the absence of Fn3 9–10 (Fig. 6 B–F), consistent with the importance of Fn3 9–10 augmentation. In agreement with its ability to stabilize βI domain opening without requiring extension or hybrid domain swing-out, TS2/16 showed good self-competition in the absence of Fn3 9–10 (Fig. 6I) and bound better basally to α5β1 than other activating Fabs, as shown by its ability to give good fluorescent labeling at a lower concentration (0.5 nM) than other activating Fabs (5–20 nM) (Fig. 6 A–I).
Among effectors that stabilize the open headpiece, synergy was seen among all three, Fn3 9–10, HUTS-4, and 12G10 (Fig. 6 A–C). This is consistent with binding to three nonoverlapping sites on the α5β1 headpiece in EM (Figs. 2E and 4 A and B). Moreover, the open headpiece-specific effector Fn3 9–10 also enhanced TS2/16 binding, consistent with TS2/16 specificity for the open conformation of the βI domain (Fig. 6I). Among effectors that induce extension, synergy was seen among 8E3, 9EG7, and SNAKA51, and also with Fn3 9–10 (Fig. 6 A and D–F). Synergy was also seen between effectors that stabilized headpiece opening and extension. Open headpiece-specific Fab 12G10 stabilized binding of extension-specific Fabs 8E3, 9EG7, and SNAKA51 (Fig. 6 D–F). In contrast, the weaker, opening-reporting (16) Fab HUTS-4 more modestly stabilized binding of SNAKA51 and 9EG7 and lacked effect on 8E3 (Fig. 6 D–F). Conversely, extension-specific Fabs 8E3, 9EG7, and SNAKA51 all enhanced binding of the open headpiece-specific Fab 12G10 (Fig. 6C) and of Fn3 9–10 (Fig. 6A).
Fabs that destabilized conformations recognized by other mAbs also showed competition. Thus, extension-specific Fabs SNAKA51, 9EG7, and 8E3 and open headpiece-specific Fab HUTS-4 all inhibited mAb13 and SG/19 binding (Fig. 6 G and H).
Competition was also evident among Fabs that bind to overlapping epitopes; many of the open headpiece-specific and closed headpiece-specific Fabs compete by binding to overlapping epitopes on the βI domain in addition to binding to alternative conformations. The activating Fabs TS2/16 and 12G10 and inhibitory Fab mAb13 were raised in mice or rats against human β1 integrins. These and many other β1 integrin function-modulating antibodies are specific for human residues at positions 207, 208, 211, and 218 in the βI domain; indeed, introduction of these four human residues into the chicken β1 integrin subunit is sufficient for epitope reconstitution (14, 31, 32). Thus, cross-blocking of activating TS2/16 and 12G10 and closed headpiece-specific mAb13 Fabs (Fig. 6 C, H–J, and L) is explicable by binding to this epitope. Fab SG/19 binds to an epitope that includes residues 151, 154, and 155 at the C-terminal end of the α1-helix, which are immediately adjacent to residues 207–211 at the beginning of the α2-helix in the βI domain. The close spatial proximity of the SG/19 and mAb13 epitopes is consistent with their cross-blocking (Fig. 6 G and H) (14), and similar proximity of the SG/19 epitope to the TS2/16 and 12G10 epitopes likely explains cross-blocking among these Fabs (Fig. 6 C, G, and I). When TS2/16 prebinding stabilized the open βI domain conformation it competed with SG/19 binding (Fig. 6G), whereas when SG/19 prebinding stabilized the closed conformation it failed to compete with TS2/16 binding (Fig. 6I). This lack of reciprocity might mean that the epitopes recognized by these two Fabs overlap in the open βI domain but not the closed βI domain conformation.
In the presence of Fn3 9–10, open headpiece-specific Fab HUTS-4 and extension-specific Fab 8E3 cross-competed with one another (Fig. 6 K and M). These Fabs bind to the hybrid and PSI domains, respectively, which are adjacent in the upper β-leg, and simultaneous Fab binding might be forbidden by steric overlap. Fab HUTS-4 also competed with SNAKA51 in the presence of Fn3 9–10 (Fig. 6 K and O). Although these Fabs bind to different domains in different integrin subunits, when bound to β1, HUTS-4 Fab extends toward α5 (Fig. 4B) and might sterically overlap with SNAKA51 Fab in the extended-open conformation.
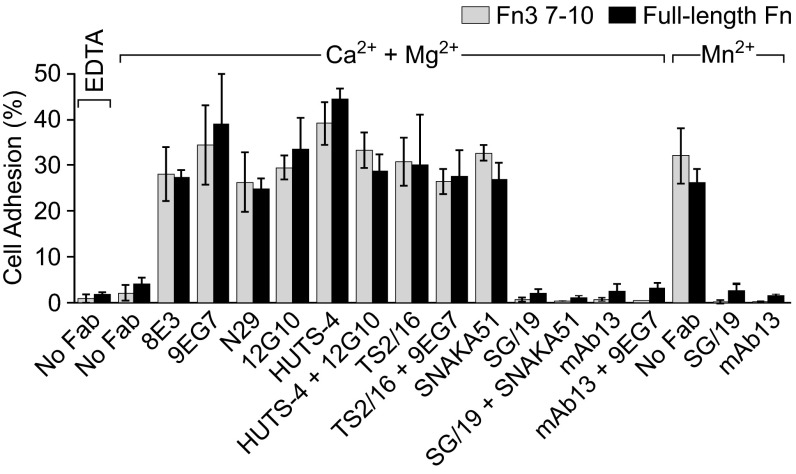
The functional relevance of the three conformational states of α5β1 was characterized by measuring the effect of Fabs on adhesion of K562 cells to substrates precoated with Fn3 7–10 or plasma fibronectin. Basal binding to the two substrates by K562 cells was low in Mg2+/Ca2+ and much greater in Mn2+ (Fig. 7). Extension-specific Fabs 8E3, 9EG7, N29, and SNAKA51; open headpiece-specific Fabs 12G10 and HUTS-4; and open βI domain-specific Fab TS2/16 all enhanced adhesion strongly. Closed headpiece-specific Fabs SG/19 and mAb13 completely suppressed activation by Mn2+ (Fig. 7).
Fig. 7.
Effects of Fabs on adhesion of K562 cells to Fn3 7–10 (gray) and full-length Fn (black) substrates in the indicated divalent cations. Error bars show standard deviation.
To address the mechanism of activation by extension-specific Fabs, we used combinations of extension-specific and closed headpiece-specific Fabs that showed no interference with one another in cross-competition assays (Fig. 6 and Table 1), SNAKA51 plus SG/19 and 9EG7 plus mAb13 (Fig. 7). In each case, the closed headpiece-specific Fab completely suppressed the activating effect of the extension-specific Fab, demonstrating that α5β1 ectodomain extension alone is not sufficient for adhesion and that the open headpiece conformation is required. In contrast, combinations of extension-specific and open headpiece-specific Fabs were activating. Taken together, these results demonstrate that the bent-closed and extended-closed states of integrin α5β1 are nonadhesive, whereas the extended-open conformation is highly adhesive. Furthermore, the low basal adhesiveness of K562 cells suggests the presence of little extended-open α5β1 on the cell surface.
Table 1.
Antibody specificity and interaction with other antibodies and fibronectin
| Number* | Name | Species | Domain | Function | Conformational specificity | ⊘† | +‡ | −§ |
| 1 | 8E3 | Mouse | PSI | Activating | Extended | 5 | 3, 4, 6, 10 | |
| 2 | N29 | Mouse | PSI | Activating | Extended | |||
| 3 | 9EG7 | Rat | I-EGF2 | Activating | Extended | 1, 4, 5, 6, 10 | ||
| 4 | SNAKA51 | Mouse | Calf-1/Calf-2 | Activating | Extended | 5 | 1, 3, 6, 10 | |
| 5 | HUTS-4 | Mouse | hybrid | Activating | Open headpiece | 1, 4 | 3, 6, 10 | |
| 6 | 12G10 | Mouse | βI | Activating | Open headpiece | 7, 8, 9 | 1, 3, 4, 5, 10 | 9 |
| 7 | TS2/16 | Mouse | βI | Activating | Open βI | 6, 8 | 10 | |
| 8 | mAb13 | Rat | βI | Inhibitory | Closed headpiece | 6, 7, 9 | 1, 3, 4, 5, 10 | |
| 9 | SG/19 | Mouse | βI/hybrid | Inhibitory | Closed headpiece | 6, 8 | 1, 3, 4, 5, 6, 7, 10 | |
| 10 | Fibronectin | βI/β-Propeller | Ligand | Open headpiece | 1, 3, 4, 5, 6 |
Antibody number used in the three right-hand columns.
Numbers of antibodies that inhibit binding, presumably by competing for binding.
Numbers of antibodies that enhance binding, presumably by affecting conformational equilibria.
Numbers of antibodies that inhibit binding, presumably by affecting conformational equilibria.
Discussion
Our major finding is that integrin α5β1 has three conformational states including a bent-closed state. Furthermore, we demonstrate that the bent-closed and extended-closed states are nonadhesive and the extended-open state is adhesive on K562 cells. Basally, K562 cells are nonadhesive. These results correlate with our EM observations that α5β1 ectodomain preparations predominantly populate the bent-closed conformation and that complex formation with Fn3 9–10 induces the extended-open conformation. By defining which of the three conformational states of α5β1 that nine different function-perturbing antibodies stabilize (Table 1), we enable extensive structure–function correlations. Previously, many of these antibodies have been inferred to be conformation-specific by their effects on ligand binding, mapping to regions known to shift conformation in other integrins, effects of conformation-constraining disulfide bonds, and reference to conformations defined in other integrins (16, 20, 29). Such methods have been used to show that α5β1 can be stabilized in a bent conformation with a disulfide bond introduced between the calf-2 and β-tail domains and is extended and active in focal contacts but bent in other locations on the cell surface; however, functional differences between extended-closed and extended-open conformations were not resolved (29). Although previous studies have been helpful, the paucity of information on conformational change in β1 integrins compared with β2 and β3 integrins has been emphasized even by laboratories that have made leading contributions to characterizing β1 integrin antibodies (22, 29).
The β1 integrin subfamily has many more distinct αβ heterodimers (12) than the β2 (4) or β3 (2) integrin subfamilies. Work with β1 integrins has shown heterogeneity in their ability to be activated and has suggested subunit-dependent differences among integrins (19). Furthermore, it has been proposed that there are only two types of integrin activation-dependent antibodies, (i) ligand-mimetic antibodies and (ii) cation and ligand-induced binding-site antibodies (33). Our findings show that the latter consist of distinct groups, those (i) that stabilize the extended conformation and (ii) that stabilize the open conformation—of the βI domain only or of both the βI and hybrid domains. Despite speculation that different subfamilies of integrins may differ in activation mechanism or conformation, the three overall global conformational states of α5β1 seen here with EM—bent-closed, extended-closed, and extended-open—are similar to those of αVβ3, αIIbβ3, αXβ2, and αLβ2 (4, 7–9). Characterization of function-perturbing β3 integrin antibodies in EM is limited (34), and the extended-open conformation has been induced by ligand binding (7, 10, 12) rather than correlated with function using antibodies as done here for α5β1. The extended-open conformation has been shown to be the only adhesive and high-affinity conformation of β2 integrins using sets of Fabs analogous to those used here (9, 13); however, the correlation is less direct than demonstrated here because β2 integrins bind ligand through an αI domain inserted into their α-subunits. αI domain conformation cannot be visualized in EM, and conformational change must be transmitted to the αI domain from the βI domain in the headpiece.
Despite global similarities among the three overall conformational states possessed by β1, β2, and β3 integrins, there are notable differences among these integrins in their behavior in negative-stain EM and among the antibodies that define β1, β2, and β3 integrin conformational states. The strength of negative-stain EM is that particles become oriented; certain orientations selectively adsorb to the negatively charged carbon grid (perhaps those that present larger areas for adsorption), and this limited set of orientations makes it easier to distinguish alternative conformations from alternative orientations (in projection averages) than in vitreous ice with cryo-EM, where orientation is usually random (35). However, in contrast to αVβ3, αIIbβ3, and αXβ2, which when extended largely adsorbed in en face views, α5β1 largely adsorbed in side views [similar to αLβ2 (8)], in which the α- and β-subunits and their conformations were difficult to distinguish. Perhaps related to this difference in adsorption, or perhaps because the compact form of the bent conformation is less stable in α5β1 than in these other integrins, it exhibited a wide range of bending angles, ranging from a compact conformation resembling the compact, bent conformations of αVβ3, αIIbβ3, αXβ2, and αLβ2 (4, 7, 8) to fully extended. A large number of EM grids and Fab complexes were examined for α5β1 and each was subjected to two independent methods of particle classification and averaging that gave similar results (SI Appendix, Figs. S2–S14). The conclusion that α5β1 exists in a bent conformation is also supported by class averages of SG/19 Fab–ectodomain complexes that show a compact, bent conformation similar to that of the αVβ3 ectodomain modeled bound to SG/19 Fab.
How do the Fabs studied here stabilize particular conformational states? Models of the bent conformation of α5β1 made using bent conformations of αVβ3 and αIIbβ3 suggest that extension-specific Fabs bind to epitopes that are buried in headpiece–tailpiece or α-leg–β-leg interfaces in the bent conformation. Integrins are likely to equilibrate rapidly among their conformational states; extension-specific Fabs would bind when the integrin was extended, and then trap (stabilize) it in that state. 9EG7 recognizes a single species-specific difference in the β1 I-EGF2 domain, Asp-522 in human and mouse, which is Glu in rat (29). The equivalent residue in the bent conformation of αIIbβ3 is surrounded and protected from Fab by the α-subunit thigh and calf-1 domains and β-subunit PSI and I-EGF1 domains (Fig. 1A) (4). These domains move away from I-EGF2 during extension and thus permit 9EG7 binding. Extension-specific KIM127 Fab to β2 integrins similarly binds to I-EGF2 (8). 8E3 and N29 Fabs bind to the PSI domain. The bend at the β-knee between the upper and lower β-legs places the PSI domain between I-EGF1 in the upper leg and I-EGF2 and I-EGF3 in the lower leg. Leg extension removes the proximity of I-EGF2 and I-EGF3 to PSI, and thus permits binding of antibodies specific for the extended conformation to PSI (Fig. 1A). SNAKA51 binds to the interface between the calf-1 and calf-2 domains in the lower α-leg, near both the lower β-leg and the headpiece in the bent conformation. Thus, SNAKA51 is predicted to bind to a site that is exposed in the extended but not bent conformations.
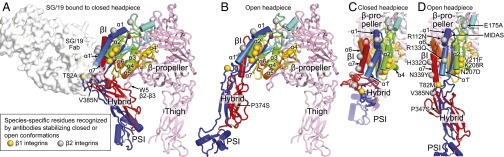
Fabs that stabilize the closed headpiece bind to regions of the closed headpiece that differ in conformation from the open headpiece. SG/19 recognizes a single species-specific residue, Thr-82, in a hybrid domain loop that only becomes ordered when bound to SG/19 (Fig. 8A) (25, 36). SG/19 simultaneously recognizes human–mouse invariant residues at the end of the α1-helix in the βI domain, and thus recognizes an orientation between the hybrid and βI domains present only in the closed conformation. Similarly, 7E4 binds to hybrid domain residue V385 in this interface and stabilizes the closed conformation of this interface in β2 integrins (Fig. 8A) (9).
Fig. 8.
Species-specific residues recognized by antibodies specific for open or closed integrin conformations. Cartoon ribbon diagrams of the indicated domains are shown in en face (A and B) or side (C and D) views with α-helices as cylinders (PyMOL; Schrödinger). The closed conformation uses α5β1 (25), and A includes SG/19 Fab as a white, transparent surface. The open conformation uses α5 with the open β3 subunit (1). The order of α-helices in the βI domain sequence is emphasized by coloring the βI domain sequence by rainbow from blue (N) to red (C); PSI is blue and the N- and C-terminal portions of the hybrid domain are blue and red, respectively. Species-specific β1 and β2 integrin residues are shown as Cβ atom spheres (key) labeled with human and rodent amino acids before and after the residue number, respectively; the positions shown are the equivalents in β1 or β3 structures. The MIDAS metal ion is shown as a red sphere.
The β1 subunit is unusually conserved between mouse and human among integrins, and many function-perturbing antibodies recognize species-specific differences at βI domain α2-helix residues 207, 208, 211, and 218 (14, 31, 32); however, the α2-helix moves little in integrin allostery (Fig. 8). Instead, the α2-helix neighbors the α1-helix, which in turn neighbors the α7-helix. Movements of the α1- and α7-helices transmit conformational change in the βI domain between its ligand-binding site and interface with the hybrid domain (Fig. 8) (12). α1-Helix side chains form important coordinations to the metal ion dependent adhesion site (MIDAS) and adjacent to MIDAS (ADMIDAS) metal ions that alter in the high-affinity state, and pistoning of the βI domain α7-helix C terminus toward the hybrid domain causes pivoting at the N-terminal connection of the βI domain to the hybrid domain. In other words, headpiece opening comes about because the βI domain is inserted into the hybrid domain, and pistoning at one of the two connections between these domains causes pivoting at the other.
At the adjacent α7-, α1-, and α2-helices in open and closed β3 structures, backbone Cα atoms differ 4.7–9 Å at the α7-helix, 4–6.5 Å at the α1-helix, and only 0.7–2.7 Å at the equivalents of residues 207, 208, 211, and 218 in the α2-helix (12) (Fig. 8). Thus, the closed headpiece-specific Fab mAb13 and the open headpiece-specific Fabs 12G10 and TS2/16 likely become conformation-specific by recognizing not only species-specific residues in the α2-helix but also conformation-specific locations of residues in the adjacent α1-helix. Among these three Fabs, the en face views with 12G10 suggest it binds more toward the α1-helix because it projects from the side of the headpiece similarly to SG/19, which binds the α1-helix, whereas the lack of clear views of the main plane of the headpiece with mAb13 and TS2/16 suggests they interfere with the binding of the main plane of the headpiece to the grid by projecting more orthogonally and binding more toward the α3-helix (see species-specific residue locations in Fig. 8).
HUTS-4 binds to the inner, α-subunit proximal face of the hybrid domain and stabilizes its open conformation. βI domain insertion splits the hybrid domain into N- and C-terminal portions (colored blue and red, respectively, in Fig. 8). The C-terminal sequence forms the inner, α-subunit proximal half of the hybrid domain, consistent with mapping of the species specificity of HUTS-4 to this segment (16). In the closed conformation, this face of the hybrid domain contacts a long β-ribbon in the β-propeller W5 β2-β3 loop (Fig. 8A), explaining why HUTS-4 is specific for the open conformation. MEM148 to β2 integrins recognizes the P374S substitution in the C-terminal segment of the hybrid domain (Fig. 8B) to similarly stabilize the open conformation (9).
In the high-affinity conformational state of α5β1, the ligand-binding site at the β-propeller and βI domain interface is normally linked through α7-helix motion to headpiece opening, visualized in EM by hybrid domain swing-out, as shown here with Fn3 7–10 and Fabs 12G10 and HUTS-4 bound to the ectodomain. TS2/16 appears not to require hybrid domain swing-out, but nonetheless appears to favor hybrid domain swing-out. In EM, TS2/16–α5β1 complexes resembled those of α5β1 alone. Most ternary complexes of TS2/16 and α5β1 with the extension-stabilizing Fab 9EG7 showed en face views with the open headpiece, although we could not rule out the presence of a minority of closed headpieces (SI Appendix, Fig. S11), in contrast to 9EG7 complexes with α5β1 alone, in which the closed headpiece predominated. Immunofluorescence flow cytometry showed cooperation between Fn and TS2/16 binding; however, cooperation with HUTS-4 or extension-stabilizing Fabs was not seen, again suggesting that TS2/16 does not require hybrid domain swing-out. Linkage of Fn3 7–10 and 12G10 but not TS2/16 binding to headpiece opening also correlates with regulation by divalent cations of their binding (20). Saturation binding of TS2/16, 12G10, and HUTS-4 Fabs induces nearly identical intrinsic, high affinities for cyclic RGD peptide, suggesting they stabilize identical conformations of the ligand-binding site in the βI domain.
These findings suggest that TS2/16 might enable a “cocked” conformation of the βI domain in which the α1-helix moves, enabling high-affinity ligand binding, despite the absence of movement or alteration of α7-helix movement from the pathway it normally takes in headpiece opening. A cocked conformation was seen in a crystal structure of αXβ2 in which the “internal ligand” of the αI domain bound to the βI domain MIDAS and relayed allostery between the αI and βI domains, despite the absence of α7-helix motion and presence of a bent-closed ectodomain conformation (37), as seen here in EM with bound TS2/16. In each case, the cocked conformation may represent a metastable intermediate, namely, in energy landscape parlance, a local energy minimum lying between two global, lower-energy minima representing the open and closed headpiece conformations. In this scenario, TS2/16 would be able to bind both the cocked and open conformations but not the closed conformation. A cocked conformation, with the βI domain open around the ligand-binding site but the hybrid domain not swung out, may be on the pathway between the closed and open conformations. However, this cocked conformation is not seen under physiologic ligand-binding conditions, because all class averages with Fn3 7–10 had extended-open conformations (SI Appendix, Fig. S3).
The results with TS2/16 force us to distinguish between βI domain opening and headpiece opening. We cannot visualize βI domain opening by negative-stain EM. Usually βI domain opening is linked to headpiece movement by C-terminal pistoning of the βI domain α7-helix, which forces the hybrid domain to pivot about its other connection to the βI domain and to swing out, which is readily apparent in EM as the “open headpiece.” When α5β1 is extended, TS2/16 complexes exhibit the open headpiece conformation. Thus, TS2/16 stabilizes the open conformation of the βI domain and is distinct from Fabs that bind to the βI domain and stabilize the closed conformation of the βI domain such as mAb13, which show the extended-closed conformation in en face views.
Evolution has resulted in great variation in the types of conformation-specific, species-specific antibodies that are available for different integrin β-subunits. In integrin β1 the α1- and α7-helices have essentially invariant sequences in mammals and vertebrates, respectively, whereas in integrin β2 the α1- and α7-helices each have two mouse–human differences that elicit conformation-specific antibodies (Fig. 8) (38, 39). Antibodies to these epitopes not only directly bind the elements that communicate allostery between the βI domain and headpiece but also project from the side of the headpiece (9), making recognition of headpiece conformation in EM easier with β2 than β1 integrins. The integrin β3 subunit also has little variation in α1- and α7-helix sequence in mammals, except near the MIDAS, where antibodies would likely block the ligand-binding site and therefore not be allosteric. There are currently no known open headpiece-specific or closed headpiece-specific allosteric antibodies to β3 integrins; β3 integrin ligand-induced binding-site antibodies appear to be extension-specific (34).
In summary, we have shown that integrin α5β1 possesses at least three distinct global conformational states. We have defined the states stabilized by nine function-perturbing antibodies widely used in the β1 integrin field. Basally under conditions in which the bent-closed conformation likely predominates, α5β1 on K562 cells is not adhesive for fibronectin. With Fabs to stabilize extension and the closed or open states of the βI domain and the headpiece, we demonstrated that the extended-open conformation and not the extended-closed conformation of α5β1 on K562 cells is adhesive for fibronectin. Among the remaining important questions is how β1 integrins with different α-subunits differ in the set points for their conformational equilibria on cell surfaces (19).
Methods
Integrin α5β1 Glycoprotein.
DNA constructs were exactly as described previously, except for replacement of αX and β2 with α5 and β1, respectively (8). The α5 ectodomain with signal peptide and residues F1–Y954 with a C-terminal tobacco etch virus (TEV) protease site, acidic coiled-coil with a cysteine for disulfide-bond formation, and Strep II tag was inserted into the XbaI and AgeI sites of pcDNA3.1/Hygro(−). The β1 ectodomain cDNA with signal peptide and mature residues Q1–D708 with a C-terminal TEV site, basic coiled-coil with a cysteine, and hexahistidine tag was subcloned into BamHI and XbaI sites of pEF1-puro. Stable HEK293S GnTI−/− transfectants were selected. Purification was as for the α5β1 headpiece (36), except for omission of TEV cleavage. Sources of SG/19 and TS2/16 (14) and 12G10, 8E3, SNAKA51, HUTS-4, and 9EG7 (29) were as described. N29 (40) and mAb13 (41) hybridomas were kind gifts of J. Wilkins (University of Manitoba, Winnipeg, MB) and K.M. Yamada (NIH, Bethesda, MD), respectively. IgGs were purified by protein G affinity, and Fab fragments were prepared by papain digestion.
Fibronectin Fragments.
Human Fn3 7–10 (mature residues 1,142–1,509), Fn3 9–10 (mature residues 1,326–1,509 with an N-terminal His6 tag), or the Fn3 9–10 Ser-1417 to Cys mutant were expressed and purified from Escherichia coli as described (26), except His tag affinity purification was used for Fn3 9–10. Fn3 7–10 was used in EM because domains 7 and 8 helped localize the α5β1-binding site in Fn3 domains 9 and 10. Fn3 9–10 was obtained in higher yield and could easily be concentrated highly, and was therefore used in Fab cross-competition assays. The Fn3 9–10 S1417C mutant was specifically labeled at C1417 with Alexa Fluor 488 C5 maleimide (Thermo Fisher Scientific) for use as a reporter in Fab cross-competition assays; Fn3 7–10 has a native unpaired Cys residue (C1201) in Fn3 domain 7.
EM.
α5β1 (100 μg/mL) alone or with five molar equivalents of Fab (100 μg/mL) or Fn3 7–10 (80 μg/mL) in 20 mM Tris buffered saline (pH 7.4) supplemented with 1 mM CaCl2, 1 mM MgCl2 (α5β1 and Fab complexes) or with 1 mM MnCl2 (Fn3 7–10 complex) was held for 20 min at 21 °C and then subjected to Superdex 200 gel filtration in the same buffer. Peak fractions were applied to glow-discharged grids, stained with 0.75% (wt/vol) uranyl formate (8), and imaged with an FEI Tecnai T12 microscope and Gatan 4K×4K CCD camera at 67,000× magnification (1.68 Å pixel size) and -1.5 μm defocus. Well-separated particles of reasonable sizes for integrins were interactively picked using EMAN2 (42) as illustrated in SI Appendix, Fig. S1. Class averages were calculated by (i) multireference alignment followed by K-means clustering using SPIDER (9, 43, 44) and also (ii) iterative stable alignment and clustering using SPARX (45). Cross-correlations were with 2D projections generated at 4° intervals from 20-Å-filtered α5β1 closed crystal structures, models of the open headpiece made by substituting the open β3 subunit from Protein Data Bank (PDB) ID code 2VDR for the β1 subunit in PDB ID code 3VI3, or by superposition of SG/19-bound α5β1 in PDB ID code 3VI3 onto the αVβ3 ectodomain in PDB ID code 4G1E.
Cell Adhesion.
Microtiter plate wells adsorbed with Fn3 7–10 or plasma fibronectin (10 μg/mL in 50 μL) at 4 °C overnight were blocked with 3% (wt/vol) BSA in PBS. K562 cells (kept in PBS with 0.5% BSA) were fluorescently labeled with 2′,7′-bis-(2-carboxyethyl)-5-(and-6)-carboxyfluorescein, acetoxymethyl ester (BCECF AM) at 37 °C for 30 min and washed with 5 mM EDTA. Cells (2 × 106 per mL, 50 μL) in PBS with 1% BSA and 25 μg/mL Fabs in either 1 mM CaCl2/1 mM MgCl2, 1 mM MnCl2, or 10 mM EDTA were incubated at 37 °C for 1 h and washed thrice. Adherence was measured as the ratio of fluorescence intensity post and before wash.
Fab Cross-Competition.
K562 cells (2 × 106 per mL) in phenol red-free L15 medium with 1% BSA were incubated with high, specified concentrations of unlabeled Fabs with or without Fn3 9–10 at 21 °C for 90 min, mixed with lower, specified concentrations of Alexa Fluor 488-labeled Fabs or Fn3 9–10 for 30 min, and then without washing subjected to immunofluorescence flow cytometry. Mean fluorescence intensity (MFI) was measured.
Supplementary Material
Acknowledgments
We thank Li-Zhi Mi for help with EM data collection and processing. This work was supported by NIH Grant HL-108248.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1605074113/-/DCSupplemental.
References
- 1.Luo B-H, Carman CV, Springer TA. Structural basis of integrin regulation and signaling. Annu Rev Immunol. 2007;25:619–647. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xiong J-P, et al. Crystal structure of the extracellular segment of integrin αVbeta3. Science. 2001;294(5541):339–345. doi: 10.1126/science.1064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xiong JP, et al. Crystal structure of the extracellular segment of integrin αVbeta3 in complex with an Arg-Gly-Asp ligand. Science. 2002;296(5565):151–155. doi: 10.1126/science.1069040. [DOI] [PubMed] [Google Scholar]
- 4.Zhu J, et al. Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces. Mol Cell. 2008;32(6):849–861. doi: 10.1016/j.molcel.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie C, et al. Structure of an integrin with an alphaI domain, complement receptor type 4. EMBO J. 2010;29(3):666–679. doi: 10.1038/emboj.2009.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong X, et al. α(V)β(3) integrin crystal structures and their functional implications. Biochemistry. 2012;51(44):8814–8828. doi: 10.1021/bi300734n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takagi J, Petre BM, Walz T, Springer TA. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell. 2002;110(5):599–611. doi: 10.1016/s0092-8674(02)00935-2. [DOI] [PubMed] [Google Scholar]
- 8.Nishida N, et al. Activation of leukocyte β2 integrins by conversion from bent to extended conformations. Immunity. 2006;25(4):583–594. doi: 10.1016/j.immuni.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 9.Chen X, et al. Requirement of open headpiece conformation for activation of leukocyte integrin alphaXbeta2. Proc Natl Acad Sci USA. 2010;107(33):14727–14732. doi: 10.1073/pnas.1008663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eng ET, Smagghe BJ, Walz T, Springer TA. Intact alphaIIbbeta3 integrin is extended after activation as measured by solution X-ray scattering and electron microscopy. J Biol Chem. 2011;286(40):35218–35226. doi: 10.1074/jbc.M111.275107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takagi J, Strokovich K, Springer TA, Walz T. Structure of integrin α5β1 in complex with fibronectin. EMBO J. 2003;22(18):4607–4615. doi: 10.1093/emboj/cdg445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu J, Zhu J, Springer TA. Complete integrin headpiece opening in eight steps. J Cell Biol. 2013;201(7):1053–1068. doi: 10.1083/jcb.201212037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schürpf T, et al. The RGD finger of Del-1 is a unique structural feature critical for integrin binding. FASEB J. 2012;26(8):3412–3420. doi: 10.1096/fj.11-202036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo B-H, Strokovich K, Walz T, Springer TA, Takagi J. Allosteric β1 integrin antibodies that stabilize the low affinity state by preventing the swing-out of the hybrid domain. J Biol Chem. 2004;279(26):27466–27471. doi: 10.1074/jbc.M404354200. [DOI] [PubMed] [Google Scholar]
- 15.Mould AP, et al. Structure of an integrin-ligand complex deduced from solution X-ray scattering and site-directed mutagenesis. J Biol Chem. 2003;278(41):39993–39999. doi: 10.1074/jbc.M304627200. [DOI] [PubMed] [Google Scholar]
- 16.Luque A, et al. Activated conformations of very late activation integrins detected by a group of antibodies (HUTS) specific for a novel regulatory region (355–425) of the common β1 chain. J Biol Chem. 1996;271(19):11067–11075. doi: 10.1074/jbc.271.19.11067. [DOI] [PubMed] [Google Scholar]
- 17.Mould AP, Akiyama SK, Humphries MJ. The inhibitory anti-beta1 integrin monoclonal antibody 13 recognizes an epitope that is attenuated by ligand occupancy. Evidence for allosteric inhibition of integrin function. J Biol Chem. 1996;271(34):20365–20374. doi: 10.1074/jbc.271.34.20365. [DOI] [PubMed] [Google Scholar]
- 18.Lenter M, et al. A monoclonal antibody against an activation epitope on mouse integrin chain β1 blocks adhesion of lymphocytes to the endothelial integrin α6β1. Proc Natl Acad Sci USA. 1993;90(19):9051–9055. doi: 10.1073/pnas.90.19.9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bazzoni G, Ma L, Blue M-L, Hemler ME. Divalent cations and ligands induce conformational changes that are highly divergent among β1 integrins. J Biol Chem. 1998;273(12):6670–6678. doi: 10.1074/jbc.273.12.6670. [DOI] [PubMed] [Google Scholar]
- 20.Mould AP, et al. Evidence that monoclonal antibodies directed against the integrin β subunit plexin/semaphorin/integrin domain stimulate function by inducing receptor extension. J Biol Chem. 2005;280(6):4238–4246. doi: 10.1074/jbc.M412240200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mould AP, Garratt AN, Askari JA, Akiyama SK, Humphries MJ. Identification of a novel anti-integrin monoclonal antibody that recognises a ligand-induced binding site epitope on the β1 subunit. FEBS Lett. 1995;363(1–2):118–122. doi: 10.1016/0014-5793(95)00301-o. [DOI] [PubMed] [Google Scholar]
- 22.Byron A, et al. Anti-integrin monoclonal antibodies. J Cell Sci. 2009;122(Pt 22):4009–4011. doi: 10.1242/jcs.056770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mould AP, et al. Integrin activation involves a conformational change in the alpha1 helix of the beta subunit A-domain. J Biol Chem. 2002;277(22):19800–19805. doi: 10.1074/jbc.M201571200. [DOI] [PubMed] [Google Scholar]
- 24.Mould AP, et al. Conformational changes in the integrin βA domain provide a mechanism for signal transduction via hybrid domain movement. J Biol Chem. 2003;278(19):17028–17035. doi: 10.1074/jbc.M213139200. [DOI] [PubMed] [Google Scholar]
- 25.Nagae M, et al. Crystal structure of α5β1 integrin ectodomain: Atomic details of the fibronectin receptor. J Cell Biol. 2012;197(1):131–140. doi: 10.1083/jcb.201111077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takagi J, Erickson HP, Springer TA. C-terminal opening mimics ‘inside-out’ activation of integrin α5β1. Nat Struct Biol. 2001;8(5):412–416. doi: 10.1038/87569. [DOI] [PubMed] [Google Scholar]
- 27.Leahy DJ, Aukhil I, Erickson HP. 2.0 Å crystal structure of a four-domain segment of human fibronectin encompassing the RGD loop and synergy region. Cell. 1996;84(1):155–164. doi: 10.1016/s0092-8674(00)81002-8. [DOI] [PubMed] [Google Scholar]
- 28.Schwarzbauer JE, DeSimone DW. Fibronectins, their fibrillogenesis, and in vivo functions. Cold Spring Harb Perspect Biol. 2011;3(7):1–19. doi: 10.1101/cshperspect.a005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Askari JA, et al. Focal adhesions are sites of integrin extension. J Cell Biol. 2010;188(6):891–903. doi: 10.1083/jcb.200907174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clark K, et al. A specific alpha5beta1-integrin conformation promotes directional integrin translocation and fibronectin matrix formation. J Cell Sci. 2005;118(Pt 2):291–300. doi: 10.1242/jcs.01623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takada Y, Puzon W. Identification of a regulatory region of integrin β1 subunit using activating and inhibiting antibodies. J Biol Chem. 1993;268(23):17597–17601. [PubMed] [Google Scholar]
- 32.Mould AP, Garratt AN, Puzon-McLaughlin W, Takada Y, Humphries MJ. Regulation of integrin function: Evidence that bivalent-cation-induced conformational changes lead to the unmasking of ligand-binding sites within integrin alpha5beta1. Biochem J. 1998;331(Pt 3):821–828. doi: 10.1042/bj3310821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bazzoni G, Hemler ME. Are changes in integrin affinity and conformation overemphasized? Trends Biochem Sci. 1998;23(1):30–34. doi: 10.1016/s0968-0004(97)01141-9. [DOI] [PubMed] [Google Scholar]
- 34.Lin FY, Zhu J, Eng ET, Hudson NE, Springer TA. β-Subunit binding is sufficient for ligands to open the integrin αIIbβ3 headpiece. J Biol Chem. 2016;291(9):4537–4546. doi: 10.1074/jbc.M115.705624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ohi M, Li Y, Cheng Y, Walz T. Negative staining and image classification—Powerful tools in modern electron microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xia W, Springer TA. Metal ion and ligand binding of integrin α5β1. Proc Natl Acad Sci USA. 2014;111(50):17863–17868. doi: 10.1073/pnas.1420645111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sen M, Yuki K, Springer TA. An internal ligand-bound, metastable state of a leukocyte integrin, αXβ2. J Cell Biol. 2013;203(4):629–642. doi: 10.1083/jcb.201308083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang C, Zang Q, Takagi J, Springer TA. Structural and functional studies with antibodies to the integrin β2 subunit. A model for the I-like domain. J Biol Chem. 2000;275(28):21514–21524. doi: 10.1074/jbc.M002286200. [DOI] [PubMed] [Google Scholar]
- 39.Lu C, Shimaoka M, Zang Q, Takagi J, Springer TA. Locking in alternate conformations of the integrin alphaLbeta2 I domain with disulfide bonds reveals functional relationships among integrin domains. Proc Natl Acad Sci USA. 2001;98(5):2393–2398. doi: 10.1073/pnas.041618598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ni H, Li A, Simonsen N, Wilkins JA. Integrin activation by dithiothreitol or Mn2+ induces a ligand-occupied conformation and exposure of a novel NH2-terminal regulatory site on the beta1 integrin chain. J Biol Chem. 1998;273(14):7981–7987. doi: 10.1074/jbc.273.14.7981. [DOI] [PubMed] [Google Scholar]
- 41.Akiyama SK, Yamada SS, Chen WT, Yamada KM. Analysis of fibronectin receptor function with monoclonal antibodies: Roles in cell adhesion, migration, matrix assembly, and cytoskeletal organization. J Cell Biol. 1989;109(2):863–875. doi: 10.1083/jcb.109.2.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rees I, Langley E, Chiu W, Ludtke SJ. EMEN2: An object oriented database and electronic lab notebook. Microsc Microanal. 2013;19(1):1–10. doi: 10.1017/S1431927612014043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frank J, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116(1):190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 44.Mi LZ, et al. Simultaneous visualization of the extracellular and cytoplasmic domains of the epidermal growth factor receptor. Nat Struct Mol Biol. 2011;18(9):984–989. doi: 10.1038/nsmb.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Z, Fang J, Chittuluru J, Asturias FJ, Penczek PA. Iterative stable alignment and clustering of 2D transmission electron microscope images. Structure. 2012;20(2):237–247. doi: 10.1016/j.str.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.