Significance
All heme-dependent functions require the mobilization of labile heme (LH), of which there is little understanding of its nature and dynamics. To probe LH pools, we developed genetically encoded fluorescent heme sensors and deployed them in the unicellular eukaryote Saccharomyces cerevisiae. We find that LH is relatively abundant in the cytosol, but exceedingly low in the mitochondria and nucleus. Further, we find that LH can be mobilized by signaling molecules like nitric oxide. We also find that the glycolytic enzyme glyceraldehyde phosphate dehydrogenase constitutes a major heme buffer and is responsible for regulating the activity of a heme-dependent transcription factor. Altogether, our work will have profound implications for understanding the mechanisms of heme utilization.
Keywords: heme sensors, heme trafficking, heme dynamics, nitric oxide, glyceraldehyde phosphate dehydrogenase
Abstract
Heme is an essential cofactor and signaling molecule. Heme acquisition by proteins and heme signaling are ultimately reliant on the ability to mobilize labile heme (LH). However, the properties of LH pools, including concentration, oxidation state, distribution, speciation, and dynamics, are poorly understood. Herein, we elucidate the nature and dynamics of LH using genetically encoded ratiometric fluorescent heme sensors in the unicellular eukaryote Saccharomyces cerevisiae. We find that the subcellular distribution of LH is heterogeneous; the cytosol maintains LH at ∼20–40 nM, whereas the mitochondria and nucleus maintain it at concentrations below 2.5 nM. Further, we find that the signaling molecule nitric oxide can initiate the rapid mobilization of heme in the cytosol and nucleus from certain thiol-containing factors. We also find that the glycolytic enzyme glyceraldehyde phosphate dehydrogenase constitutes a major cellular heme buffer, and is responsible for maintaining the activity of the heme-dependent nuclear transcription factor heme activator protein (Hap1p). Altogether, we demonstrate that the heme sensors can be used to reveal fundamental aspects of heme trafficking and dynamics and can be used across multiple organisms, including Escherichia coli, yeast, and human cell lines.
Heme (iron protoporphyrin IX) is an essential protein cofactor and signaling molecule (1–11). The canonical view of heme is that it is a static cofactor buried in the active sites of hemoproteins. This view point is irreconcilable with the fact that all heme-dependent processes, from heme acquisition by proteins to heme signaling, require the dynamic mobilization of heme. However, heme mobilization has never been monitored, and the mechanisms that mediate it are poorly understood. The hydrophobicity and cytotoxicity of heme necessitate that its concentration is tightly regulated and buffered to low levels, creating an apparent paradox when trying to conceptualize the movement of heme to client hemoproteins or for heme-based signal transduction (5, 12).
The total heme quota is the sum of the exchange inert and labile heme (LH) pools. Inert heme, which is unavailable for new heme-dependent processes, is more abundant and represents the large fraction of heme that is associated with high-affinity hemoproteins like cytochromes and globins. The LH pool, which is available for hemoproteins and heme signaling, is far less abundant and buffered by unknown factors (12). The properties of LH pools, including concentration, speciation, oxidation state, distribution, and dynamics, are paramount for understanding how cells assimilate this essential nutrient, but are poorly understood. The current lack of understanding of LH is, in large part, due to the dearth of tools available to probe it.
Herein, we report genetically encoded ratiometric fluorescent heme sensors and deploy them in the unicellular eukaryote Saccharomyces cerevisiae (Baker’s yeast) to elucidate the nature and dynamics of LH. We find that LH is buffered at a concentration of 20–40 nM in the cytosol and less than 2.5 nM in the nucleus and mitochondria. Further, we find that the signaling molecule nitric oxide (NO) can initiate the rapid mobilization of heme in the cytosol and nucleus from certain thiol-containing factors. By integrating our heme sensors with genetic screens, we also find that the glycolytic enzyme glyceraldehyde phosphate dehydrogenase (GAPDH) is responsible for buffering intracellular heme and regulating the activity of the nuclear heme-dependent transcription factor heme activator protein (Hap1p). Altogether, these results reveal fundamental aspects of heme trafficking and dynamics, providing fresh insight into the cellular management of this essential nutrient.
Results
Design and Characterization of Heme Sensors.
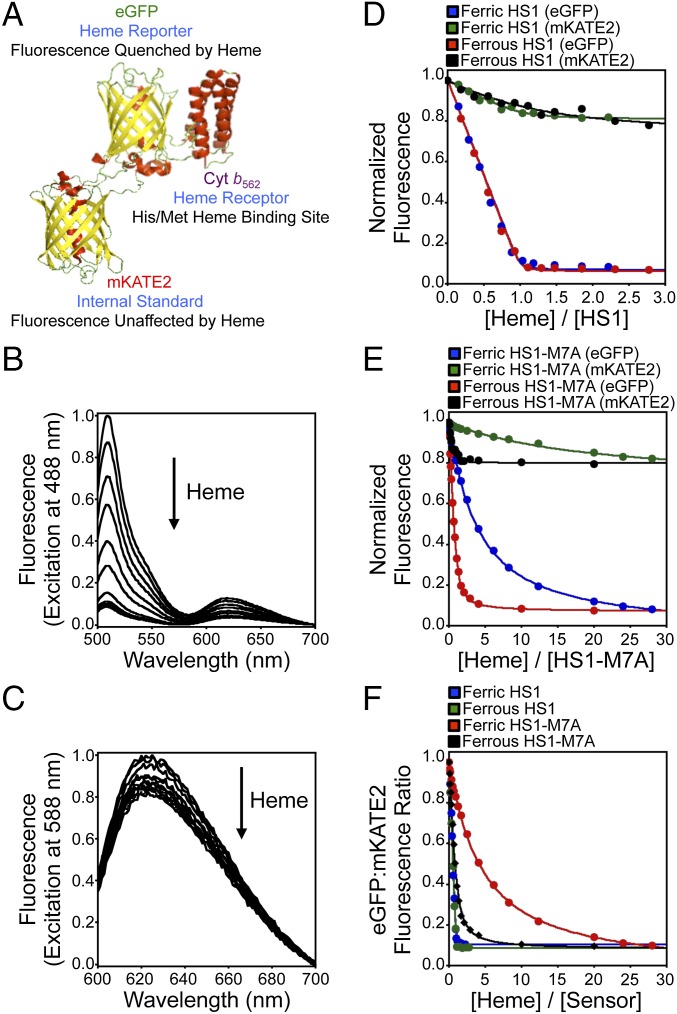
The first-generation heme sensor, HS1, consists of a heme-binding domain, the His/Met coordinating 4-alpha-helical bundle hemoprotein cytochrome b562 (Cyt b562) (13), fused to a pair of fluorescent proteins, EGFP and Katushka 2 (mKATE2), that are expected to exhibit heme-sensitive and -insensitive fluorescence, respectively (Fig. 1A). Holo-Cyt b562 is a fluorescence resonance energy transfer (FRET) acceptor for EGFP (14, 15) [excitation (ex.) = 488 nm, emission (em.) = 510 nm] but not mKATE2 (16) (ex. = 588 nm, em. = 620 nm) (SI Appendix, Fig. S1). Thus, HS1 was designed as an excitation-emission ratiometric probe (17) in which the ratio of heme-sensitive EGFP fluorescence to heme-insensitive mKATE2 fluorescence provides a readout of cellular heme independent of sensor concentration.
Fig. 1.
Design and heme-dependent fluorescence properties of the heme sensors. (A) Molecular model and design principles of the heme sensor, HS1. The model is derived from the X-ray structures of mKATE [Protein Data Bank (PDB) ID code 3BXB] and CG6 (PDB ID code 3U8P). Ferric heme-dependent changes in the normalized fluorescence emission spectra of HS1 at pH 8.0 upon excitation of EGFP (B, ex. = 488 nm) and mKATE2 (C, ex. = 588 nm) are illustrated. Normalized changes in EGFP (ex. = 488 nm, em. = 510 nm) and mKATE2 (ex. = 588 nm, em. = 620 nm) fluorescence upon titration of heme into 0.5 μM HS1 (D) and HS1-M7A (E) at pH 8.0 are illustrated. (F) Change in EGFP/mKATE2 fluorescence ratios for HS1 and HS1-M7A upon titration of heme. All titration data are fit to 1:1 heme/protein binding models.
HS1 was adapted from a previously reported EGFP-Cyt b562 integral fusion protein, CG6, that exhibits >99% efficient FRET between EGFP and heme (18). mKATE2 was appended to the N terminus of CG6 with a GlySer linker. Titration of ferric and ferrous heme into an aqueous buffered solution [20 mM NaPi, 100 mM NaCl (pH 8.0)] of 0.5 μM purified HS1 resulted in visible absorbance spectra indicative of heme coordination (SI Appendix, Fig. S2), and quenched EGFP fluorescence (∼10-fold decrease in signal), with minimal perturbation to mKATE2 fluorescence (∼20% decrease in signal) (Fig. 1 B–D). Both oxidized and reduced heme quench the fluorescence of EGFP and mKATE2 to similar degrees and the change in fluorescence as a function of [heme] evinces a 1:1 heme/HS1 stoichiometry (Fig. 1 D and F).
HS1-ferric heme dissociation constants, KdIII, were determined by direct titration of hemin chloride into HS1 over a broad pH range (pH 5–8), and found to be 3 nM between pH 6.0 and 7.5 (SI Appendix, Fig. S3 A–C). At pH 8.0, the 10 nM KdIII value for HS1 is identical to the KdIII values previously reported for Cyt b562 (13) and CG6 (18). HS1-ferrous heme dissociation constants, KdII values, were too tight to measure by direct titration and are less than 1 nM.
Because previous estimates for the concentration of the LH pool are highly varied between nanomolar and micromolar values (12), we generated heme sensors with altered heme binding affinities. One variant, HS1-M7A, which was generated by replacing the heme axial Met7 ligand of Cyt b562 with Ala (SI Appendix, Fig. S2), exhibits fluorescence properties similar to the fluorescence properties of HS1 and binds heme in a 1:1 stoichiometry (Fig. 1 E and F). HS1-M7A binds ferrous heme with a KdII value of 25 nM between pH 6.0 and 7.5 (SI Appendix, Fig. S3 D–F). The HS1-M7A KdIII values are very weak, determined to be 0.5–2.0 μM over a pH range of 5.0–8.0 (SI Appendix, Fig. S3F). The KdII value of HS1-M7A is similar in magnitude to previous estimates of the “regulatory” heme pool, 10–100 nM (12, 19), as well as the affinities of proteins that may respond to this pool, including the heme-dependent transcription factor Rev-erbβ (20) and constitutive heme oxygenase-2 (HO-2) (21) (SI Appendix, Fig. S3G).
Both HS1 and HS1-M7A are selective for ferrous heme over other metals, protoporphyrin IX, and the heme degradation products bilirubin and biliverdin (SI Appendix, Fig. S4 A and B). Further, apo- and ferrous heme-bound HS1 and HS1-M7A exhibit pH-independent EGFP/mKATE2 fluorescence ratios between pH 6.5 and 9.0, but are markedly pH-dependent below pH 6.5 (SI Appendix, Fig. S4 C and D). The fluorescence ratio of HS1 and HS1-M7A is independent of protein concentration between 10 nM and 1 μM, suggesting that the sensors do not aggregate over these concentrations (SI Appendix, Fig. S4E). Ferrous heme binding to HS1 and HS1-M7A is reversible because competition with excess apo-Cyt b562 restores the fluorescence ratio to apo-sensor (SI Appendix, Fig. S5).
Cellular Heme Imaging: Cytosol.
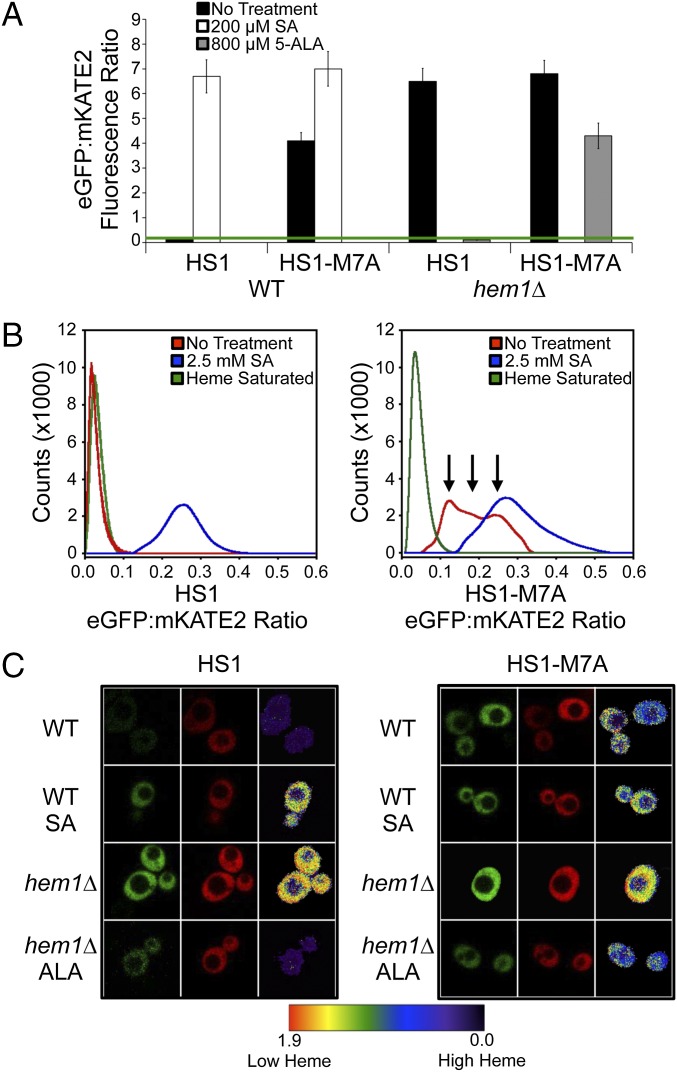
Using fluorimetry (Fig. 2A), flow cytometry (Fig. 2B), and fluorescence microscopy (Fig. 2C), we find that HS1 and HS1-M7A can be used to sense endogenous LH in a quantitative manner. Fluorimetry (Fig. 2A) and fluorescence microscopy (Fig. 2C) of WT yeast cells expressing cytosolic HS1 and HS1-M7A indicate an EGFP/mKATE2 ratio that is markedly reduced in comparison to heme-deficient hem1∆ cells, which lack 5-aminolevulinic acid (5-ALA) synthase, the first enzyme in the heme biosynthetic pathway (22). The difference in HS1 and HS1-M7A fluorescence ratios in WT cells reflects their differential heme binding affinities (vida infra).
Fig. 2.
Heme-dependent fluorescence ratios of HS1 and HS1-M7A in WT and hem1∆ yeast cells as measured by fluorimetry (A), flow cytometry (B), and confocal microscopy (C). Where indicated, cells were treated with SA or 5-ALA. Cells for microscopy experiments were treated with 0.2 mM SA or 1.5 mM 5-ALA. “Heme saturated” is the ratio recorded upon digitonin permeabilization of cells and incubation with excess heme as described in the main text for in situ calibration of the cytosolic sensors. Fluorimetry data represent the mean ± SD of triplicate cultures. The green bar indicates the eGFP/mKATE2 fluorescence ratio when the heme sensors are saturated with heme. The flow cytometry and microscopy data are representative of three independent mid-log-phase cultures grown in synthetic complete media lacking leucine and supplemented with ergosterol and tween-80 (SCE-LEU).
The HS1 and HS1-M7A EGFP/mKATE2 fluorescence ratios in hem1∆ cells decrease when heme synthesis is initiated due to supplementation with 5-ALA (Fig. 2 A and C) in a dose-dependent manner (SI Appendix, Fig. S6 C and D), which is consistent with increased heme binding. Excess 5-ALA does not reduce HS1-M7A fluorescence ratios to the same extent as HS1. This effect is because excess 5-ALA (1.5 mM) results in intracellular heme levels similar to WT cells (SI Appendix, Fig. S6E); strict control of heme biosynthesis does not allow for endogenous heme levels that are sufficient to saturate the low-affinity heme sensor HS1-M7A. Conversely, the HS1 and HS1-M7A EGFP/mKATE2 fluorescence ratios in WT cells increase upon supplementation with the heme biosynthesis inhibitor succinylacetone (SA) (Fig. 2 A–C) in a dose-dependent manner (SI Appendix, Fig. S6 A and B), which is consistent with decreased heme binding (2). A variant of HS1 with His102 and Met7 heme-coordinating ligands mutated to Ala, HS1-M7A,H102A, and an mKATE2-EGFP fusion protein lacking the Cyt b562 domain do not exhibit heme-dependent changes in fluorescence ratio, indicating that heme iron coordination to HS1 is required for cellular heme sensing (SI Appendix, Fig. S7).
The heme sensor does not itself perturb heme metabolism and is a reliable reporter for endogenous LH. Titration of HS1-M7A expression using weak (prADH1), medium (prTEF1), and strong (prGPD) promoters does not result in a change in the observed concentration of the bioavailable heme pool, and prGPD-HS1-M7A does not affect cell growth or heme-regulated functions like total heme, catalase activity, or respiration (SI Appendix, Fig. S8 A–F).
For quantitative heme monitoring, an in situ method to calibrate the sensor was developed (19). The concentration of [heme] accessible to the sensor is governed by the following expression (23):
| [1] |
where Kd is the heme-sensor dissociation constant, Rexpt is the EGFP/mKATE2 fluorescence ratio under any given condition, Rmin is the EGFP/mKATE2 fluorescence ratio when 0% of the sensor is bound to heme, Rmax is the EGFP/mKATE2 fluorescence ratio when 100% of the sensor is bound to heme, FminmKATE2 is the mKATE2 emission intensity when 0% of the sensor is bound to heme, and FmaxmKATE2 is the mKATE2 emission intensity when 100% of the sensor is bound to heme. Determination of Rmax and FmaxmKATE2 involves recording EGFP and mKATE2 fluorescence after digitonin permeabilization of cells and incubation with 50 μM heme. Determination of Rmin and FminmKATE2 involves recording EGFP and mKATE2 fluorescence after cells are treated with SA or from hem1∆ cells cultured in parallel. Because the ferrous heme binding affinities are tighter than ferric affinities for both HS1 and HS1-M7A, Rmax and FmaxmKATE2 are determined in the presence of the reducing agent ascorbate (1 mM) to drive heme saturation of the sensors.
Using fluorimetry, in situ calibration of HS1 in WT cells indicates that it is >95% saturated (Fig. 2A and SI Appendix, Fig. S9), which is not ideal for imaging. On the other hand, HS1-M7A is typically 20–50% bound in exponential-phase WT cells and well poised to monitor changes in heme availability (Fig. 2A and SI Appendix, Fig. S9). Estimation of the concentration of LH using Eq. 1 depends on assumptions made about its oxidation state. In the two limiting cases of LH being 100% reduced or oxidized, cytosolic LH can be estimated to be buffered between ∼30 nM and ∼1 μM, respectively, after considering the data depicted in Fig. 2A and the KdII or KdIII value of 25 nM or 1 μM at pH 7.0 (SI Appendix, Fig. S3), which is the pH of the yeast cytosol (24). Given the relatively reducing cellular environment, EmCytosol ∼ −320 mV vs. normal hydrogen electrode (NHE) (25), which is governed by the ratio of oxidized-to-reduced glutathione, and the reduction potential of aqueous heme, estimated to be between −50 mV and −220 mV vs. NHE (26, 27), we propose that LH is biased toward the reduced state and assume herein that LH is 100% reduced. However, the actual fraction of LH that is reduced is dependent on its speciation and the degree to which it equilibrates with the glutathione redox buffer, both of which are unknown. SI Appendix, Fig. S10G outlines estimates of [LH] for a sensor that is between 20% and 50% bound because the assumption in the oxidation state deviates from being 100% reduced. Given the weak ferric heme affinities of HS1-M7A, if [LH] is <100 nM, HS1-M7A cannot sense oxidized LH (SI Appendix, Fig. S10 A–F).
Flow cytometry of log-phase WT cells expressing HS1-M7A reveals that there are three populations of cells with distinct concentrations of cytosolic LH; 40 nM, 20 nM, and <1 nM heme (Fig. 2B). These LH concentrations correspond to 1,200, 600, and <30 molecules of heme, respectively, assuming a cytosolic volume (Vcyt) of 50 fL (28). By comparison, total cellular heme was determined to be ∼1 μM (SI Appendix, Fig. S8D). Although this trimodal distribution is highly reproducible, the relative abundance of cells with these three distinct heme levels is quite variable between experiments (SI Appendix, Fig. S11). This observation suggests that heme availability may be dynamically regulated. In contrast, HS1 exhibits only one population due to complete heme saturation as a result of its high heme binding affinity (Fig. 2B and SI Appendix, Fig. S12).
Cellular Heme Imaging: Nucleus and Mitochondria.
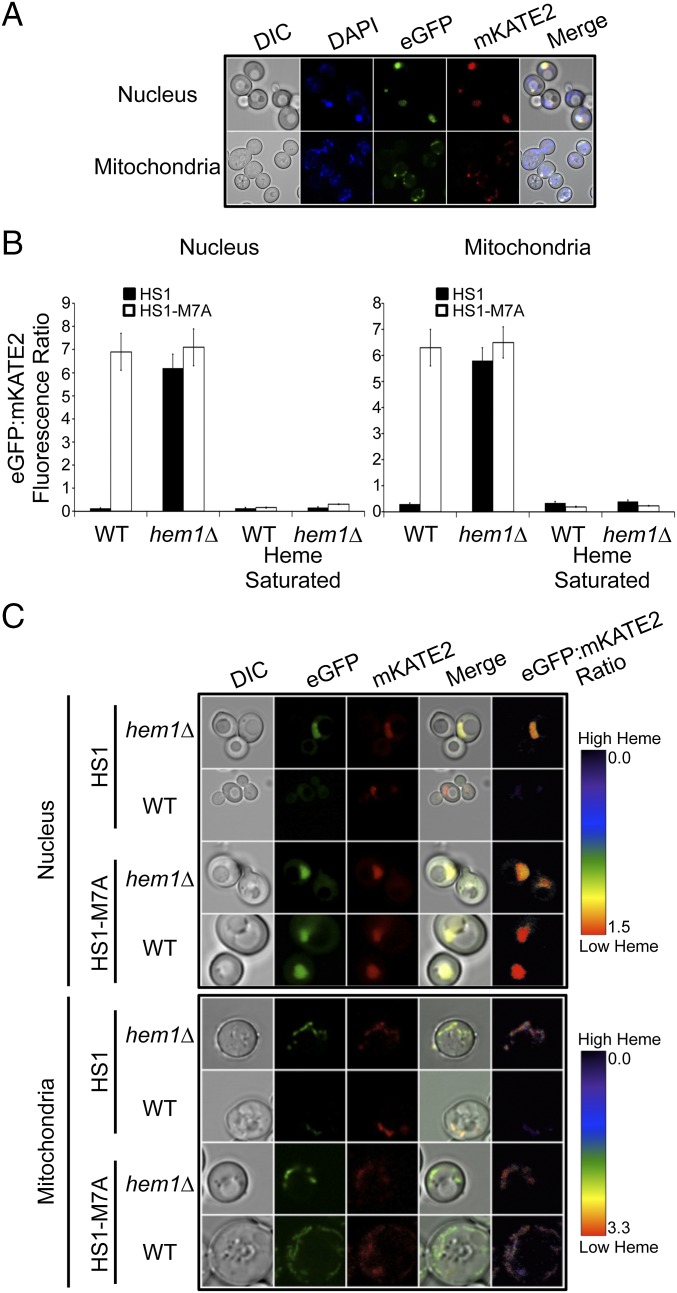
To probe the subcellular distribution of heme, we targeted HS1 and HS1-M7A to the mitochondria and nucleus by appending N-terminal COX4 mitochondrial matrix or C-terminal SV40 nuclear localization sequences, respectively (25, 29) (Fig. 3A). The localization tags do not affect heme binding to the sensors (SI Appendix, Fig. S13).
Fig. 3.
Nuclear and mitochondrial heme monitoring with HS1 and HS1-M7A. (A) To label DNA, hem1∆ cells expressing nuclear or mitochondrial targeted HS1-M7A were stained with DAPI. Live cells were imaged as described in Materials and Methods at a magnification of 63×. “Merge” is the merged images of DAPI, EGFP, and mKATE2. (B) Heme-dependent EGFP/mKATE2 fluorescence ratios of nuclear and mitochondrial targeted HS1 and HS1-M7A in WT and hem1∆ yeast cells as measured by fluorimetry. “Heme-saturated” is the ratio recorded upon Triton X-100 permeabilization of yeast spheroplasts and incubation with excess heme as described in the main text for in situ calibration for the mitochondrial and nuclear sensors. (C) Confocal microscopy of WT and hem1∆ cells expressing nuclear and mitochondrial targeted HS1 and HS1-M7A. Fluorimetry data represent the mean ± SD of triplicate mid-log-phase cultures grown in SCE-LEU. Microscopy images are representative of at least two independent cultures. DIC, differential interference contrast.
Fluorimetry (Fig. 3B) and fluorescence microscopy (Fig. 3C) indicate that nuclear and mitochondrial HS1 exhibit EGFP/mKATE2 fluorescence ratios in WT cells that are distinctly lower than in hem1∆ cells, indicative of heme binding. In contrast, nuclear and mitochondrial HS1-M7A EGFP/mKATE2 fluorescence ratios are unaltered between hem1∆ and WT cells (Fig. 3 B and C). To determine the fractional saturation of our sensors in these compartments, we developed an in situ sensor calibration method analogous to the one described for the cytosol. However, because digitonin cannot permeabilize nuclear or mitochondrial membranes (19), we first had to enzymatically digest the yeast cell wall with zymolyase and then incubate the resulting spheroplasts with 0.1% Triton X-100, 50 μM heme, and 1 mM ascorbate to heme-saturate the sensors. As shown in Fig. 3B, this in situ calibration method resulted in limiting Rmax values for HS1 and HS1-M7A that are identical within experimental error.
In total, the data indicate that HS1 is ∼100% heme-saturated in the mitochondria and nucleus, whereas HS1-M7A is ∼0% heme-saturated in these organelles. Given the HS1-M7A KdII of 25 nM, we estimate that heme is buffered at an upper limit of 2.5 nM in the mitochondria and nucleus. This concentration corresponds to fewer than six molecules in the nucleus [Vnuc ∼ 5 fL (30)] and ∼0.6 molecules in the mitochondria [Vmito ∼ 0.5 fL (28, 31)].
NO Mobilizes Cytosolic and Nuclear Heme.
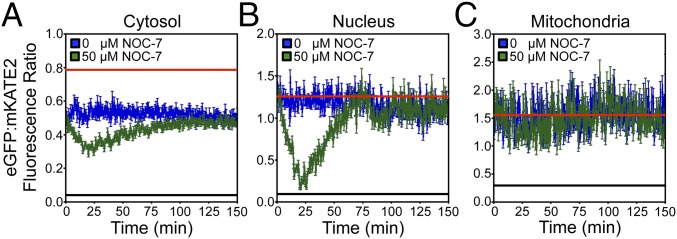
Previous work demonstrated that NO could regulate heme transfer between certain protein pairs (32). Could NO also mobilize LH pools? Using cytosolic, nuclear, and mitochondrial targeted HS1-M7A, we found that NO derived from the small molecule NOC-7 (33) rapidly increases cytosolic (Fig. 4A) and nuclear (Fig. 4B) LH in WT cells, but not in the mitochondria (Fig. 4C) or in heme-deficient hem1∆ cells (SI Appendix, Fig. S14A). In the cytosol, LH increases from 17 nM to 40 nM within 20 min of adding 50 μM NOC-7 (t1/2 = 10 min at 22 °C), which corresponds to an increase from 500 to 1,200 molecules of heme, assuming Vcyt ∼ 50 fL (28). This rapid increase is followed by the reestablishment of the initial steady-state heme levels over the course of ∼80 min. In the nucleus, there is an even more pronounced change in the HS1-M7A EGFP/mKATE2 ratio upon NOC-7 treatment. NO results in an increase in nuclear LH from <2.5 nM to a maximum of 218 nM, corresponding to a change from fewer than six to ∼650 heme molecules assuming Vnuc ∼ 5 fL (30). NO-mediated heme mobilization is dose-dependent (SI Appendix, Fig. S14B), and the observed changes in fluorescence ratios are not due to NO interactions within HS1-M7A because incubation of purified heme-bound HS1-M7A with NOC-7 does not alter sensor fluorescence (SI Appendix, Fig. S15).
Fig. 4.
NO-dependent mobilization of LH. WT cells expressing cytosolic (A), nuclear (B), or mitochondrial (C) HS1-M7A were incubated with 50 μM NOC-7, and eGFP/mKATE2 fluorescence ratios were monitored by fluorimetry. The data represent the mean ± SD of triplicate mid-log-phase cultures grown in SCE-LEU. The red and black lines indicate the fluorescence ratios of HS1-M7A when it is 0% and 100% saturated with heme as derived from hem1∆ cells expressing HS1-M7A or WT cells expressing HS1, respectively. This approach for determining Rmax is validated by the fact that the eGFP/mKATE2 fluorescence ratio of HS1-M7A that is saturated with heme from the in situ calibration methods is identical within experimental error to HS1 (Fig. 3B and SI Appendix, Fig. S9).
Given that cellular thiols are targets of NO, we sought to determine the effect of a thiol-specific alkylating agent [e.g., iodoacetamide (IAM)] on heme mobilization. Although IAM does not initiate the mobilization of heme, it does prevent the reestablishment of the steady-state LH pool after NO treatment (SI Appendix, Fig. S14C). This observation suggests that IAM blocks the rebinding of heme from sites of mobilization, presumably certain thiol-containing factors.
To differentiate the effects of NO and general oxidative stress on heme mobilization, we probed the role of various oxidative insults. Diamide, a thiol-specific oxidant; paraquat, a superoxide-generating agent; and H2O2 had no impact on LH pools, suggesting that heme mobilization is specific to NO (SI Appendix, Fig. S14D).
GAPDH Regulates Cytosolic Heme Availability and the Activity of the Nuclear Heme-Dependent Transcription Factor Hap1p.
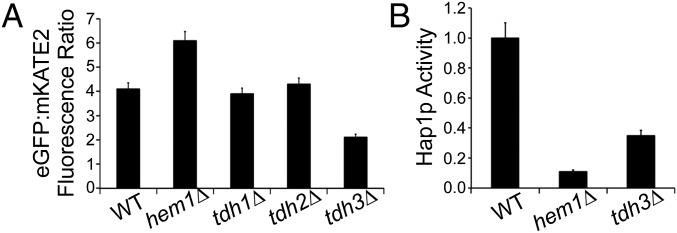
To identify genes that regulate LH, we transformed cytosolic HS1-M7A into the yeast knockout collection and screened for mutants that exhibited altered sensor fluorescence ratios. After partial screening, we identified TDH3, which encodes an isoform of the glycolytic enzyme GAPDH, to be a determinant of LH. The tdh3∆ cells exhibit an approximately fourfold greater concentration of labile cytosolic heme (13 nM in WT cells and 57 nM in tdh3∆ cells) (Fig. 5A) without affecting total heme levels (SI Appendix, Fig. S16B). Deletion of TDH3 homologs TDH1 and TDH2 does not affect LH pools (Fig. 5A), likely because these isoforms contribute to a small fraction of total GAPDH (SI Appendix, Fig. S16A). Episomal expression of TDH3 in tdh3∆ cells restores LH to WT levels (SI Appendix, Fig. S16 D and E).
Fig. 5.
Tdh3p regulates intracellular heme availability and Hap1p activity. (A) Fluorimetric determination of EGFP/mKATE2 fluorescence ratios of the indicated yeast strains expressing HS1-M7A. Rmin and Rmax were determined to be 6.1 (from the hem1∆ strain) and 0.2 (from in situ calibration with digitonin), respectively. (B) Hap1p activity in the indicated strains as measured by a transcriptional reporter that used EGFP driven by the CYC1 promoter, a Hap1p target gene. All data represent the mean ± SD of triplicate mid-log-phase yeast extract-peptone-dextrose media supplemented with ergosterol and tween-80 (YPDE) cultures.
To determine the role of Tdh3p on other heme-dependent functions, we measured the activity of cytosolic heme catalase, Ctt1p, and the activity of the heme-dependent transcription factor, Hap1p. The tdh3∆ cells have normal Ctt1p activity as determined by a native in-gel colorimetric assay for catalase (34) (SI Appendix, Fig. S16C). On the other hand, tdh3∆ cells have a defect in Hap1p activity as measured using a transcriptional reporter that employs EGFP driven by the CYC1 promoter, a Hap1p target gene (9) (Fig. 5B). In total, our data indicate GAPDH is involved in buffering cytosolic heme and regulates the activity of Hap1p, presumably by acting as a source for heme.
Validation of Sensors in Escherichia coli and HEK293 Cells.
To broaden the utility of HS1, we tested its applicability in other cell lines, including E. coli and human embryonic kidney HEK293 cells (SI Appendix, Fig. S17). In HEK293 cells, flow cytometry and fluorescence microscopy demonstrate that HS1 and HS1-M7A exhibit EGFP/mKATE2 fluorescence ratios that are distinctly lower when cells are cultured in heme-replete media relative to heme-deficient (HD) media containing SA (HD + SA) (SI Appendix, Fig. S17 A–E). Similarly, heme sensor fluorescence ratios in E. coli exhibit heme-dependent changes. A heme-deficient ∆hemA strain lacking glutamyl-tRNA reductase, which catalyzes the first step in porphyrin synthesis, is a 5-ALA auxotroph. The ∆hemA strain showed a higher EGFP/mKATE2 fluorescence ratio after growth shift to media lacking 5-ALA (SI Appendix, Fig. S17F). In total, HS1 can be used to probe heme physiology across multiple model organisms and cell lines.
Discussion
The mobilization of LH underlies all heme-dependent processes, including heme acquisition by client proteins and heme-based signal transduction. However, the properties of the LH pool are poorly understood. To address this fundamental gap in heme cell biology, we developed genetically encoded ratiometric heme sensors and deployed them in Baker’s yeast to probe key characteristics of the LH pool, including its concentration, subcellular distribution, buffering factors, and dynamics.
The cytosol has more LH (∼20–40 nM) than the nucleus or mitochondria (<2.5 nM). Further, the cell-to-cell heterogeneity in cytosolic heme suggests its availability may be dynamically regulated (Fig. 2B). Given the subcellular distribution of heme, we propose that once heme is biosynthesized in the mitochondrial matrix, a fraction of heme transits to the cytosol and acts as a reservoir for signaling and/or a heme source for proteins.
The mitochondria, which have a very high demand for heme and are the site of heme biosynthesis, have exceptionally low quantities of LH, less than 2.5 nM or fewer than one molecule. By comparison, total ferrous heme in the yeast mitochondria has been estimated to be ∼30 μM, or ∼9,000 molecules (28). Taken together, this low amount of mitochondrial LH suggests that mitochondrial heme is tightly regulated and trafficked in a manner that limits its availability. This observation is consistent with the identification of mitochondrial heme metabolism complexes that traffic heme via transient protein–protein interactions, thereby circumventing the LH pool (35).
The nucleus also limits LH to less than 2.5 nM or fewer than six molecules. This low amount of nuclear LH is not surprising, given the cytotoxicity of heme and proximity to genetic material (5, 12). However, this observation raises the question of how nuclear heme-regulated transcription factors acquire heme, given that the heme-regulatory motifs (HRMs) of many heme-dependent transcription factors, including Hap1p in yeast, exhibit micromolar affinities for heme (11). Factors like Hap1p may acquire heme from transient increases in LH due to active signaling processes. Indeed, this concept is supported by our demonstration that signaling molecules like NO can result in a >10-fold increase in nuclear LH (Fig. 4B). An alternative possibility is that the heme affinities of many HRM-containing proteins are mischaracterized. For instance, a reevaluation of the heme dissociation constants of the HRM-containing transcription factor Rev-erbβ found that heme binds 100-fold tighter than previously estimated, with KdIII and KdII of ∼20 nM vs. 2–6 μM (20).
Our observation that subcellular [LH] is heterogeneous stands in striking contrast to a recent report by He and coworkers (19) that used a heme chaperone-based FRET sensor (CISDY-9) for heme in human cell lines. In that work, cytosolic, mitochondrial, and nuclear heme were all reported to be ∼25 nM. The deviation between our two results needs to be clarified but could reflect organismal variation, difficulty in calibrating CISDY-9 in organelles, or uncertainties in LH oxidation state.
We demonstrate, for the first time to our knowledge, that LH is dynamic and can be mobilized by signaling molecules like NO (Fig. 4 and SI Appendix, Fig. S14). Although previous studies have indicated that H2O2 and NO can regulate heme transfer between specific protein pairs (32, 36), our results suggest that NO can mobilize cell-wide LH pools to regulate heme-dependent processes in multiple compartments. The mechanism of NO-mediated heme mobilization is unknown, but likely involves S-nitrosation of certain hemoproteins and heme dissociation (32, 37).
NO-mediated heme mobilization provides insights into NO physiology, including NO-dependent inflammatory responses. Both heme and NO regulate inflammation, but through distinct mechanisms (8, 38, 39). Our work indicates that NO can promote inflammation via heme mobilization. Indeed, previous observations in endothelial cells found that NO elevates cellular iron due to enhanced activity of the heme-degrading enzyme, HO (40). This observation was proposed to occur as a result of NO-induced heme release from hemoproteins. Our current studies are the first, to our knowledge, to provide direct support for this model.
Our findings that GAPDH and NO regulate heme availability and mobilization are highly reminiscent of and supported by previous studies demonstrating that GAPDH and NO cooperate to control heme insertion into nitric oxide synthase (32). As such, it is tempting to speculate that Tdh3p is the source of NO-mobilized heme. However, NO-dependent heme mobilization is unaffected in tdh3∆ cells (SI Appendix, Fig. S18).
Altogether, the HS1s can be applied across prokaryotes and eukaryotes to probe heme trafficking, signaling, and dynamics in various physiological contexts.
Materials and Methods
All cell lines, culture conditions, reagents, and methods are described in SI Appendix, SI Materials and Methods. Yeast strains used in this study were in the BY4741 genetic background. The ∆hemA E. coli strains were in the MG1655 background. HEK293 cells were obtained from A. Oyelere (Georgia Institute of Technology). The genes for mKATE2 and CG6 used to construct HS1 were purchased from GENESCRIPT and codon-optimized for expression in E. coli and S. cerevisiae or humans. Standard published methods for protein expression and purification (18), heme binding studies (20, 27), fluorimetry, UV/visible spectroscopy, microscopy, flow cytometry, immunoblotting (41), catalase activity (34), oxygen consumption (41), total heme analysis (42), and cell growth are outlined in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Profs. C. Fahrni, B. Gibney, and A. Medlock for critical reading of the manuscript, A. Shaw and Dr. D. Bourassa for assistance with microscopy, and J. Ashworth for help with preliminary studies. This work was supported by NIH Grant ES025661 (to A.R.R. and I.H.), National Science Foundation CAREER Award MCB 1552791 (to A.R.R.), and funds from the Georgia Institute of Technology (to A.R.R.).
Footnotes
Conflict of interest statement: I.H. is the founder and President of Rakta Therapeutics, Inc., a company involved in the development of heme transporter-related diagnostics. He declares no other competing financial interests.
This article is a PNAS Direct Submission.
See Commentary on page 7296.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1523802113/-/DCSupplemental.
References
- 1.Severance S, Hamza I. Trafficking of heme and porphyrins in metazoa. Chem Rev. 2009;109(10):4596–4616. doi: 10.1021/cr9001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shen J, et al. Iron metabolism regulates p53 signaling through direct heme-p53 interaction and modulation of p53 localization, stability, and function. Cell Reports. 2014;7(1):180–193. doi: 10.1016/j.celrep.2014.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raghuram S, et al. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol. 2007;14(12):1207–1213. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghosh S, et al. Extracellular hemin crisis triggers acute chest syndrome in sickle mice. J Clin Invest. 2013;123(11):4809–4820. doi: 10.1172/JCI64578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiabrando D, Vinchi F, Fiorito V, Mercurio S, Tolosano E. Heme in pathophysiology: A matter of scavenging, metabolism and trafficking across cell membranes. Front Pharmacol. 2014;5:61. doi: 10.3389/fphar.2014.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keel SB, et al. A heme export protein is required for red blood cell differentiation and iron homeostasis. Science. 2008;319(5864):825–828. doi: 10.1126/science.1151133. [DOI] [PubMed] [Google Scholar]
- 7.Haldar M, et al. Heme-mediated SPI-C induction promotes monocyte differentiation into iron-recycling macrophages. Cell. 2014;156(6):1223–1234. doi: 10.1016/j.cell.2014.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dutra FF, Bozza MT. Heme on innate immunity and inflammation. Front Pharmacol. 2014;5:115. doi: 10.3389/fphar.2014.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mense SM, Zhang L. Heme: A versatile signaling molecule controlling the activities of diverse regulators ranging from transcription factors to MAP kinases. Cell Res. 2006;16(8):681–692. doi: 10.1038/sj.cr.7310086. [DOI] [PubMed] [Google Scholar]
- 10.Hou S, Reynolds MF, Horrigan FT, Heinemann SH, Hoshi T. Reversible binding of heme to proteins in cellular signal transduction. Acc Chem Res. 2006;39(12):918–924. doi: 10.1021/ar040020w. [DOI] [PubMed] [Google Scholar]
- 11.Zhang L, Guarente L. Heme binds to a short sequence that serves a regulatory function in diverse proteins. EMBO J. 1995;14(2):313–320. doi: 10.1002/j.1460-2075.1995.tb07005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sassa S. Why heme needs to be degraded to iron, biliverdin IXalpha, and carbon monoxide? Antioxid Redox Signal. 2004;6(5):819–824. doi: 10.1089/ars.2004.6.819. [DOI] [PubMed] [Google Scholar]
- 13.Robinson CR, Liu Y, Thomson JA, Sturtevant JM, Sligar SG. Energetics of heme binding to native and denatured states of cytochrome b562. Biochemistry. 1997;36(51):16141–16146. doi: 10.1021/bi971470h. [DOI] [PubMed] [Google Scholar]
- 14.Takeda S, Kamiya N, Nagamune T. A novel protein-based heme sensor consisting of green fluorescent protein and apocytochrome b(562) Anal Biochem. 2003;317(1):116–119. doi: 10.1016/s0003-2697(03)00096-4. [DOI] [PubMed] [Google Scholar]
- 15.Takeda S, Kamiya N, Arai R, Nagamune T. Design of an artificial light-harvesting unit by protein engineering: cytochrome b(562)-green fluorescent Protein chimera. Biochem Biophys Res Commun. 2001;289(1):299–304. doi: 10.1006/bbrc.2001.5966. [DOI] [PubMed] [Google Scholar]
- 16.Shcherbo D, et al. Far-red fluorescent tags for protein imaging in living tissues. Biochem J. 2009;418(3):567–574. doi: 10.1042/BJ20081949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bregestovski P, Arosio D. Green fluorescent protein-based chloride ion sensors for in vivo imaging. In: Jung G, editor. Fluorescent Proteins II: Application of Fluorescent Protein Technology. Vol 12. Springer; Berlin: 2012. pp. 99–125. [Google Scholar]
- 18.Arpino JA, et al. Structural basis for efficient chromophore communication and energy transfer in a constructed didomain protein scaffold. J Am Chem Soc. 2012;134(33):13632–13640. doi: 10.1021/ja301987h. [DOI] [PubMed] [Google Scholar]
- 19.Song Y, et al. A Genetically Encoded FRET Sensor for Intracellular Heme. ACS Chem Biol. 2015;10(7):1610–1615. doi: 10.1021/cb5009734. [DOI] [PubMed] [Google Scholar]
- 20.Gupta N, Ragsdale SW. Thiol-disulfide redox dependence of heme binding and heme ligand switching in nuclear hormone receptor rev-erbbeta. J Biol Chem. 2011;286(6):4392–4403. doi: 10.1074/jbc.M110.193466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fleischhacker AS, et al. The C-terminal heme regulatory motifs of heme oxygenase-2 are redox-regulated heme binding sites. Biochemistry. 2015;54(17):2709–2718. doi: 10.1021/acs.biochem.5b00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ness F, et al. Sterol uptake in Saccharomyces cerevisiae heme auxotrophic mutants is affected by ergosterol and oleate but not by palmitoleate or by sterol esterification. J Bacteriol. 1998;180(7):1913–1919. doi: 10.1128/jb.180.7.1913-1919.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grynkiewicz G, Poenie M, Tsien RY. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J Biol Chem. 1985;260(6):3440–3450. [PubMed] [Google Scholar]
- 24.Orij R, Postmus J, Ter Beek A, Brul S, Smits GJ. In vivo measurement of cytosolic and mitochondrial pH using a pH-sensitive GFP derivative in Saccharomyces cerevisiae reveals a relation between intracellular pH and growth. Microbiology. 2009;155(Pt 1):268–278. doi: 10.1099/mic.0.022038-0. [DOI] [PubMed] [Google Scholar]
- 25.Hu J, Dong L, Outten CE. The redox environment in the mitochondrial intermembrane space is maintained separately from the cytosol and matrix. J Biol Chem. 2008;283(43):29126–29134. doi: 10.1074/jbc.M803028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conant JB, Tongberg CO. The oxidation-reduction potentials of hemin and related substances: I. The potentials of various hemins and hematins in the absence and presence of pyridine. J Biol Chem. 1930;86:733–741. [Google Scholar]
- 27.Reddi AR, Reedy CJ, Mui S, Gibney BR. Thermodynamic investigation into the mechanisms of proton-coupled electron transfer events in heme protein maquettes. Biochemistry. 2007;46(1):291–305. doi: 10.1021/bi061607g. [DOI] [PubMed] [Google Scholar]
- 28.Garber Morales J, et al. Biophysical characterization of iron in mitochondria isolated from respiring and fermenting yeast. Biochemistry. 2010;49(26):5436–5444. doi: 10.1021/bi100558z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsang CKL, Liu Y, Thomas J, Zhang Y, Zheng XF. Superoxide dismutase 1 acts as a nuclear transcription factor to regulate oxidative stress resistance. Nat Commun. 2014;5:3446. doi: 10.1038/ncomms4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jorgensen P, et al. The size of the nucleus increases as yeast cells grow. Mol Biol Cell. 2007;18(9):3523–3532. doi: 10.1091/mbc.E06-10-0973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rafelski SM, et al. Mitochondrial network size scaling in budding yeast. Science. 2012;338(6108):822–824. doi: 10.1126/science.1225720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chakravarti R, Aulak KS, Fox PL, Stuehr DJ. GAPDH regulates cellular heme insertion into inducible nitric oxide synthase. Proc Natl Acad Sci USA. 2010;107(42):18004–18009. doi: 10.1073/pnas.1008133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maragos CM, Wang JM, Hrabie JA, Oppenheim JJ, Keefer LK. Nitric oxide/nucleophile complexes inhibit the in vitro proliferation of A375 melanoma cells via nitric oxide release. Cancer Res. 1993;53(3):564–568. [PubMed] [Google Scholar]
- 34.Baureder M, Hederstedt L. Genes important for catalase activity in Enterococcus faecalis. PLoS One. 2012;7(5):e36725. doi: 10.1371/journal.pone.0036725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Medlock AE, et al. Identification of the Mitochondrial Heme Metabolism Complex. PLoS One. 2015;10(8):e0135896. doi: 10.1371/journal.pone.0135896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kathiresan M, Martins D, English AM. Respiration triggers heme transfer from cytochrome c peroxidase to catalase in yeast mitochondria. Proc Natl Acad Sci USA. 2014;111(49):17468–17473. doi: 10.1073/pnas.1409692111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weichsel A, et al. Heme-assisted S-nitrosation of a proximal thiolate in a nitric oxide transport protein. Proc Natl Acad Sci USA. 2005;102(3):594–599. doi: 10.1073/pnas.0406549102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bucci M, et al. Endothelial nitric oxide synthase activation is critical for vascular leakage during acute inflammation in vivo. Proc Natl Acad Sci USA. 2005;102(3):904–908. doi: 10.1073/pnas.0408906102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Predonzani A, Calì B, Agnellini AH, Molon B. Spotlights on immunological effects of reactive nitrogen species: When inflammation says nitric oxide. World J Exp Med. 2015;5(2):64–76. doi: 10.5493/wjem.v5.i2.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yee EL, Pitt BR, Billiar TR, Kim YM. Effect of nitric oxide on heme metabolism in pulmonary artery endothelial cells. Am J Physiol. 1996;271(4 Pt 1):L512–L518. doi: 10.1152/ajplung.1996.271.4.L512. [DOI] [PubMed] [Google Scholar]
- 41.Reddi AR, Culotta VC. SOD1 integrates signals from oxygen and glucose to repress respiration. Cell. 2013;152(1-2):224–235. doi: 10.1016/j.cell.2012.11.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woods JS, Simmonds PL. 2001. HPLC methods for analysis of porphyrins in biological media. Curr Protoc Toxicol Chapter 8:Unit 8.9.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.