Abstract
Sessile serrated colon adenoma/polyps (SSA/Ps) are found during routine screening colonoscopy and may account for 20–30% of colon cancers. However, differentiating SSA/Ps from hyperplastic polyps (HP) with little risk of cancer is challenging and complementary molecular markers are needed. Additionally, the molecular mechanisms of colon cancer development from SSA/Ps are poorly understood. RNA sequencing was performed on 21 SSA/Ps, 10 HPs, 10 adenomas, 21 uninvolved colon and 20 control colon specimens. Differential expression and leave-one-out cross validation methods were used to define a unique gene signature of SSA/Ps. Our SSA/P gene signature was evaluated in colon cancer RNA-Seq data from The Cancer Genome Atlas (TCGA) to identify a subtype of colon cancers that may develop from SSA/Ps. A total of 1422 differentially expressed genes were found in SSA/Ps relative to controls. Serrated polyposis syndrome (n=12) and sporadic SSA/Ps (n=9) exhibited almost complete (96%) gene overlap. A 51-gene panel in SSA/P showed similar expression in a subset of TCGA colon cancers with high microsatellite instability (MSI-H). A smaller seven-gene panel showed high sensitivity and specificity in identifying BRAF mutant, CpG island methylator phenotype high (CIMP-H) and MLH1 silenced colon cancers. We describe a unique gene signature in SSA/Ps that identifies a subset of colon cancers likely to develop through the serrated pathway. These gene panels may be utilized for improved differentiation of SSA/Ps from HPs and provide insights into novel molecular pathways altered in colon cancer arising from the serrated pathway.
Keywords: Sessile serrated adenoma/polyps, hyperplastic polyps, serrated polyposis syndrome, RNA-sequencing, gene signature, sporadic MSI-H colon cancer
Introduction
Colon cancer is the second leading cause of cancer-related deaths in United States and third most common cancer in men and women (1). Serrated colon polyps are found in 12–36% of patients undergoing routine screening colonoscopy (2–4). Serrated polyps are classified into three groups: Hyperplastic polyps (HPs), sessile serrated adenoma/polyps (SSA/Ps), and traditional serrated adenomas (TSAs) (5). Both SSA/Ps and relatively rare TSAs have malignant potential. Histologically, SSA/Ps often have basilar crypt dilation, which may present as an L-shaped or inverted T-shaped morphology. HPs lack these specific features (6). However, differentiating SSA/Ps from HPs by colonoscopy or histopathology remains difficult due to overlapping morphological and pathological features (7,8).
The serrated polyposis syndrome (SPS) is an extreme phenotype, with patients presenting with multiple SSA/Ps, and has a high risk of colon cancer (9–11). So far, no inherited gene mutation has been found in SPS. The risk of SSA/Ps progressing to colon cancer is not unique to SPS patients, and has also been described in patients with sporadic SSA/Ps (2,12).
The “serrated polyp pathway” has been described as an underlying mechanism in the development of colon cancer from SSA/Ps and may account for 20–30% of sporadic colon cancers (6,13–15). However, the molecular mechanisms or signaling pathways important in the progression of SSA/Ps to colon cancer are uncertain. DNA microsatellite instability, CpG island methylation and BRAF mutations are possible underlying molecular mechanisms in the development of SSA/Ps (14–17). At least a subset of proximal colorectal cancers have the CpG island methylator phenotype (CIMP) and high microsatellite instability (MSI-H), suggesting similar molecular backgrounds in serrated polyps and proximal cancer (18).
There is limited information on gene expression profiles differentiating SSA/Ps from traditional hyperplastic polyps. Two prior studies have described gene expression in SSA/Ps using microarray technologies (19, 20). We recently identified >1200 differentially expressed genes in SSA/Ps from patients with SPS using RNA sequencing (RNA-seq) and developed several immunohistochemical markers specific for SSA/Ps (21). However, comprehensive RNA-seq gene expression profiles have not been defined for sporadic SSA/Ps and HPs, and it is not known whether sporadic SSA/Ps differ from syndromic SSA/Ps that have a very high risk for progressing to colon cancer. The goals of our study were two-fold; first to identify a panel of differentially expressed genes that discriminate between SSA/P’s and HP’s and, second to characterize a subset of SSA/P genes that are also differentially expressed in colon cancers that likely develop through the serrated pathway. We compared gene expression in prospectively collected SSA/Ps from patients with SPS and sporadic SSA/Ps, HPs, tubular adenomas, and normal colon tissue to identify uniquely expressed genes in SSA/Ps. We report a 51-gene signature that differentiates SSA/Ps from HPs and shares a similar transcriptional profile with a subtype of colon cancers that may develop through the serrated pathway. Furthermore, our findings describe a novel seven-gene panel differentially expressed in SSA/Ps that has both high sensitivity and specificity for detection of BRAF mutant, CIMP-H and MLH1 silenced colon cancers.
Materials and Methods
Patients
Samples were obtained from patients visiting University of Utah Health Care and George Whalen Veterans Affairs Medical Center, Salt Lake City, Utah between age 45 and 75 for routine screening, surveillance or diagnostic colonoscopy. Patients with serrated polyposis syndrome were between 18 to 75 years of age. Subjects with family history of colon cancer, familial cancers including familial adenomatous polyposis and Lynch syndrome, history of inflammatory bowel disease and prior colonic resections were excluded. The samples were prospectively collected from 2008–2013 for RNA sequencing. All patients signed and agreed to informed consent as approved by the respective hospitals Institutional review boards (IRB). If polyps were found during colonoscopy, a biopsy of polyp tissue was collected in formalin for histopathological diagnosis. If additional polyp tissue remained, a small biopsy of polyp tissue was collected in RNAlater for RNA sequencing. If a polyp was too small to obtain a biopsy for both histology and RNA sequencing (RNA-Seq), a tissue sample for RNA-Seq was not collected for the study.
Twelve sessile serrated polyps were obtained from eight patients with serrated polyposis syndrome (ten right colon and two left colon) (21). SSA/Ps from these patients were previously analyzed for specific mRNA changes by qPCR but not analyzed by RNA sequencing. Uninvolved mucosa from right and left colon was also collected. Right colon was defined as colonic region from splenic flexure to cecum.
Sporadic sessile serrated polyps (n=9, six right colon, three left colon), hyperplastic polyps (n=10, two right colon, eight left colon) and adenomatous polyps (n=10, nine right colon, one left colon) were obtained along with uninvolved mucosa from patients undergoing routine colonoscopy. Normal colon tissue (n=20, ten right colon, ten left colon) was obtained from patients undergoing screening colonoscopy with no polyps found on exam. All samples were collected prospectively and placed in RNAlater (Invitrogen) immediately after tissue removal, stored at 4°C overnight and then at −80°C prior to performing RNA isolation. The demographics of sporadic SSA/Ps and hyperplastic polyps are presented in Supplementary Table 1A and 1B, respectively. The demographics of patients with adenoma and control colon tissues (analyzed using qPCR) have been described in our prior publication (21). Four retrospectively obtained frozen colon cancer samples (three right colon, one left colon) obtained from the University of Utah tissue bank were also sequenced.
Pathological classification
All biopsy specimens were reviewed by an expert GI pathologist. Serrated polyps were classified according to the recent recommendations of the Multi-Society Task Force on Colorectal Cancer for post-polypectomy surveillance and as described previously (21,22). Hyperplastic polyps were not subdivided into microvesicular hyperplastic polyps (MVHP) and goblet cell hyperplastic polyps (GCHP) since these classifications are not used clinically or discussed in the recent post-polypectomy colonoscopy surveillance guidelines (22). We decided to follow the classification which is most appropriate and practical in clinical practice with the aim to define clinically relevant and realistic gene signatures. Moreover, these two HP subtypes have not been shown to have different risks for development of colon cancer.
RNA isolation, RNA Sequencing and Differential Expression Analysis
Total RNA was isolated using TRIzol (Invitrogen) and quality of RNA assessed by an Agilent 2000 bioanalyzer as described previously (21,23,24). RNA sequencing was performed on 86 individual colon samples: 21 SSA/Ps (12 syndromic and 9 sporadic), 10 hyperplastic polyps, 10 adenomatous polyps, 21 uninvolved colon, 20 control colon, and 4 colon cancer samples. PCR amplified cDNA sequencing libraries were prepared using oligo dT-selected RNA according to the Illumina TruSeq library protocol. Single-end 50 bp sequence reads were performed on an Illumina HiSeq 2000 instrument and aligned to the GRCh37/Hg19 human reference genome using the Novoalign (Novocraft) application as described previously (21). Differentially expressed genes were determined using the USeq DefinedRegionsDifferentialSeq (DRDS) application and hierarchical clustering and principal component analysis of genes and samples performed using Cluster 3.0 as described previously (21). The RNA-Seq datasets described in this study have been deposited in the NCBI Gene Expression Omnibus (GEO) with accession number GSE76987.
Derivation of 51 SSA/P Gene signature and 7-Gene Panel
A 27-gene signature was developed to include genes with high fold change expression in SSA/Ps compared to HPs (see Supplementary Table 2). A separate 28-gene signature was obtained using a leave one out cross-validation method (see Selection and Cross Validation of a 28 Gene Signature section below). Combining the two gene signatures (27 and 28) resulted in a 55-gene signature unique for SSA/Ps. Four of these 55 genes were not found in colon cancer RNA-Seq datasets from the TCGA database resulting in a 51-gene signature to compare across all RNA-Seq datasets. We next looked at the correlation of increased expression of each of the 51 genes in BRAF mutant, CIMP-H and MLH1 silenced colon cancers (see Supplementary Tables 3 and 4). Seven of the 51 genes (ZIC5, SEMG1, TRNP1, MUC6, CRYBA2, FSCN1 and ZIC2) frequently overexpressed in MSI-H colon cancers were also frequently overexpressed in BRAF mutant, CIMP-H and/or MLH1 silenced colon cancers. We used this seven-gene panel for sensitivity and specificity calculations for identifying colon cancers that likely develop through the serrated pathway.
Selection and Cross Validation of a 28 Gene Signature
Sequencing data from 10 HP and 21 SSA/Ps samples were used to construct and cross validate a gene signature. Prior to analysis, genes differentially expressed between left and right colon (≥ 2-fold change, false discovery rate (FDR) < 0.01) were removed. An “unpaired” analysis was then performed on all 31 serrated polyp samples using DESeq2 negative binomial statistics with histology as the only predictor. The FDR threshold for the signature genes was set at 0.01. Twenty eight genes met these criteria and were used for cross validation. The average of log (count + 0.5) for the selected genes was used to form separate signatures for HP and SSP samples. A normalized Euclidean distance measure was constructed from the selected genes. Standard deviations < 0.05 were increased to 0.05 in the normalization so that genes with unrealistically low variability did not exert excess influence on the signature (25). The signature for each class is represented by the geometric average, or centroid, of the class. Samples are predicted to be in the class with the closest centroid. In order to evaluate the signature the entire process of selection of the genes to form the signature, construction of the centroid for each class, calculation of the Euclidean distance measure and classification was cross-validated. A principal component analysis was performed using Cluster 3.0 and a 3D plot constructed using the ‘rgl’ package in R.
Analysis of Signature Genes in Published Microarray Data of Serrated Polyps
No previously published RNA-Seq data of serrated polyps is available for comparison to our datasets. We evaluated the expression of each of our 51 signature genes in a previously published microarray dataset (GEO number GSE43841) (19). See Supplementary Methods.
Comparison to TCGA Colon Cancer RNA-Seq Datasets
Fifty-one SSA/P signature genes were used to interrogate 68 colon cancer RNA-Seq datasets from The Cancer Genome Atlas (TCGA, 36 specimens from Christiana Healthcare and 32 from Memorial Sloan Kettering) and four from the University of Utah (26). Raw sequencing data for each colon cancer dataset was downloaded from the TCGA database (27) and normalized by number of transcript reads per kilobase of gene length per million of total reads (RPKM). There was expression data for 18130 unique RefSeq genes in both the TCGA and University of Utah RNA-Seq datasets. One hundred and ninety-five TCGA colon cancer datasets were also evaluated for mRNA expression in the 51 signature genes using the cBioPortal for Cancer Genomics (28,29).
Mutual Exclusivity and Co-Occurrence Analysis
Mutual exclusivity and co-occurrence of genomic alterations in each of our 51 signature genes and incidence of BRAF mutations was evaluated using the cBioPortal for Cancer Genomics. This analysis uses a previously published statistical method, Mutual Exclusivity Modules (MEMo), to identify genes that may be involved in the same cancer pathway (30).
Sensitivity and Specificity of a Seven Gene Panel
The sensitivity and specificity of a seven-gene panel was evaluated in 182 TCGA colon cancer samples with gene expression, methylation and BRAF mutation data available. There were 31 MLH1 silenced, CIMP high and/or BRAF mutant samples out of 182 regarded as positive and the rest as negative. Cutoffs for each gene were set at 2 times the average expression of all samples. K-fold cross validation was used to get an estimate of sensitivity and specificity. In addition to individual expression, we also investigated panels of genes. For the panels, we considered the count of the number of genes above the 2-fold threshold as a predictor (see details in Supplementary Methods). PCR validation was performed on 4 of these genes FSCN1, ZIC5, SEMG1 and MUC6 (see Supplementary Methods).
Results
Differential Gene Expression Analysis
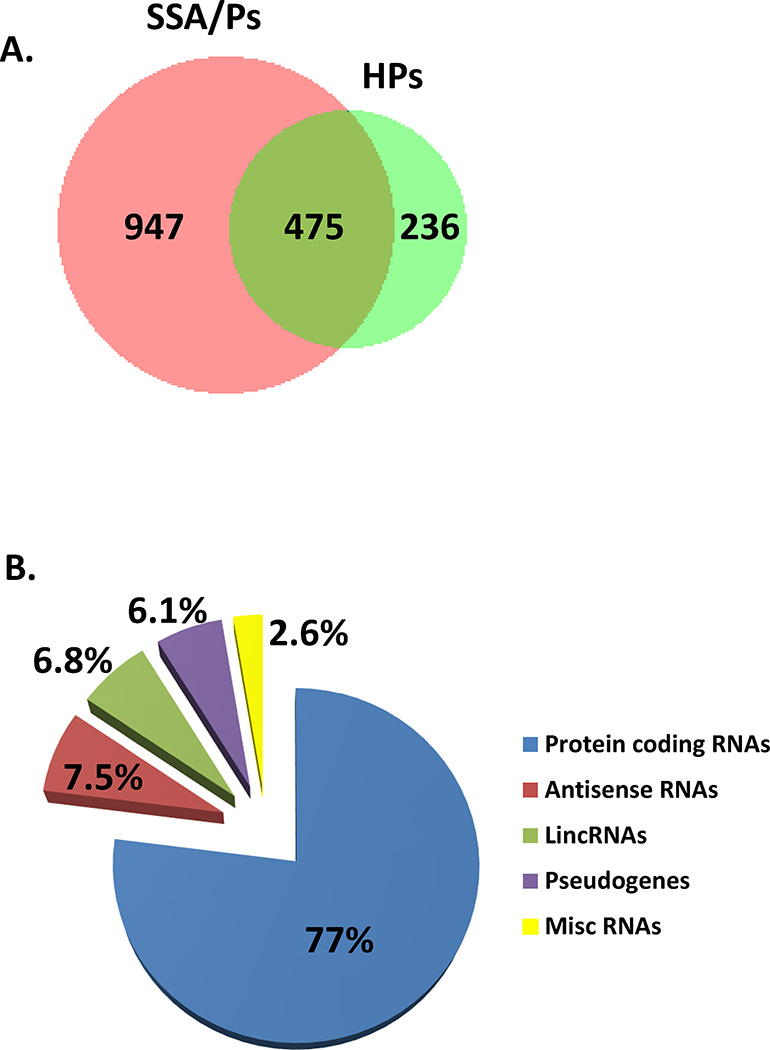
RNA sequencing was performed on 86 colon specimens with a mean sequence depth of 14.7 million mapped reads per sample. Comparing syndromic (n=12) and sporadic (n=9) SSA/P RNA-seq datasets to control right colon (n=10) we identified 1422 differentially expressed annotated genes (≥ 2-fold change, FDR < 0.05) by negative binomial statistical analysis (Figure 1, Panel A, Supplementary Table 2). Comparing hyperplastic polyps (HPs, n=10) to control left colon (n=10) we identified 711 differentially expressed genes using the same fold change and FDR cutoff. 475 genes were differentially expressed in both SSA/Ps and HPs. In the RNAs that were differentially expressed in SSA/Ps, 1095 (77%) were protein coding and 327 (23%) were non-coding (Figure 1, Panel B). A similar percentage of protein coding (80%) and non-coding (20%) RNAs was also significantly differentially expressed in HPs relative to control colon.
Figure 1.
Differentially expressed annotated protein coding and non-coding RNAs in SSA/Ps and traditional hyperplastic polyps (HPs) identified by RNA sequencing. Panel A – Differentially expressed genes with a ≥ 2-fold change and FDR < 0.05 in SSA/Ps (n=12 for syndromic and n=9 for sporadic) compared to control right colon (n=10) and HPs (n=10) compared to control left colon (n=10). Panel B – Relative abundance of protein coding and non-coding RNAs differentially expressed in SSA/Ps. Non-coding RNAs included antisense non-coding RNAs, long intergenic non-coding RNAs (lincRNAs), pseudogenes and other miscellaneous RNAs including immunoglobulin and intronic RNAs.
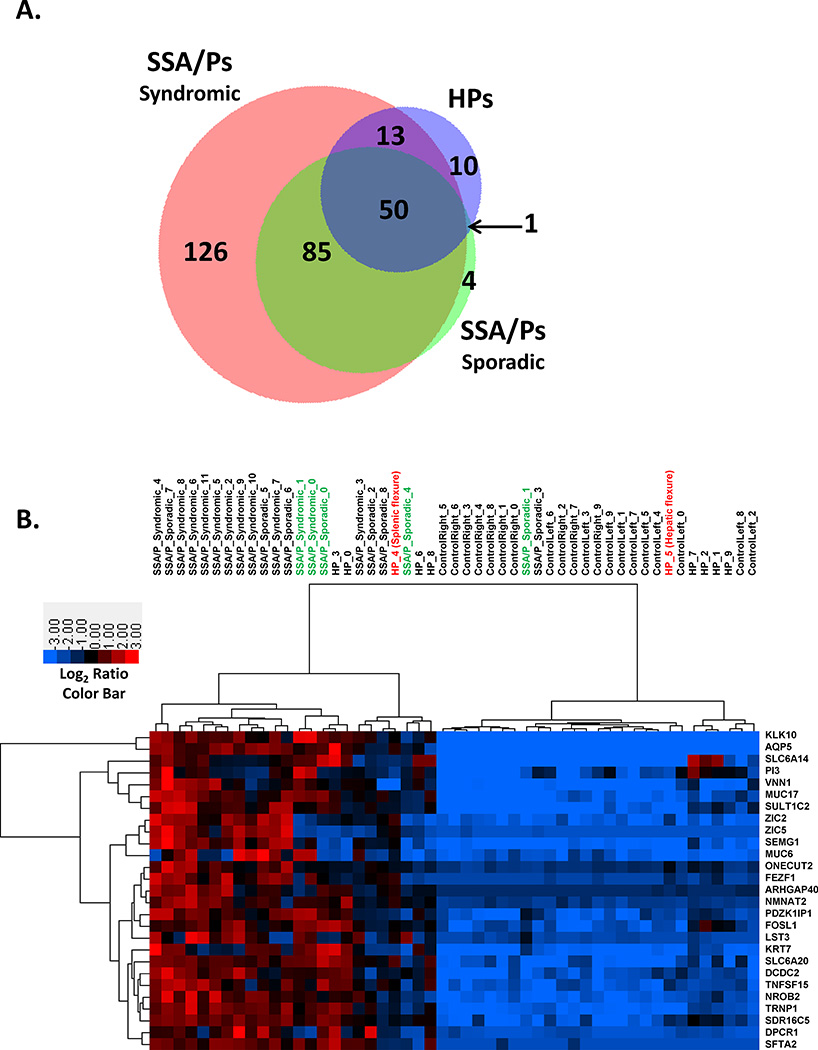
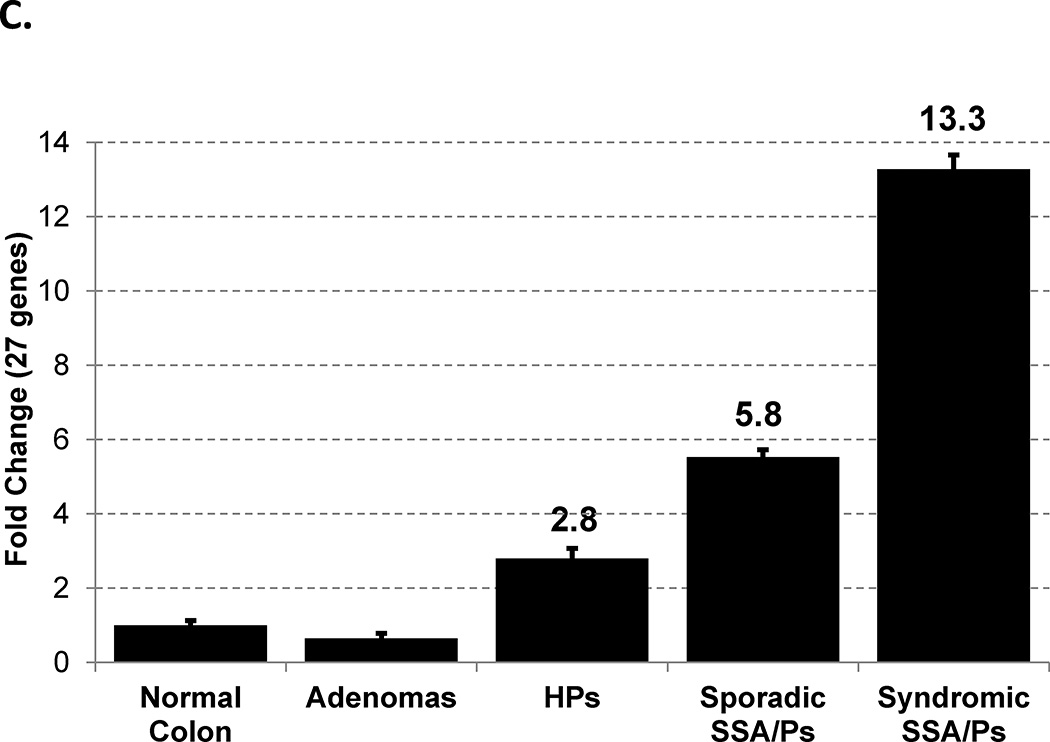
To determine if sporadic SSA/Ps had a gene expression profile similar to syndromic SSA/Ps, we compared differentially expressed genes with a ≥ 2- and 4-fold change in each group (Supplementary Figure 1 and Figure 2, Panel A, respectively). Greater than 89% (≥ 2 fold) and 96% (≥ 4 fold) of the differentially expressed genes observed in sporadic SSA/Ps were also differentially expressed in syndromic SSA/Ps. We are not aware of another gene expression comparison of sporadic and syndromic SSA/Ps and these results describe major molecular similarities in SSA/Ps from these two very different patient cohorts. 215 genes (77%) were uniquely differentially expressed ≥ 4-fold in SSA/Ps as compared to HPs (Figure 2, Panel A) while nearly 86% of the differentially expressed genes in HPs overlapped with SSA/Ps and only 10 genes (14%) were uniquely differentially expressed ≥ 4-fold in HPs. This suggests that the molecular phenotype in HPs (considered at little or no risk for progression to colon cancer) is surprisingly similar to that of SSA/Ps (considered high risk). One notable difference between SSA/Ps and HPs was the magnitude of fold-change in many differentially expressed genes. Hierarchical clustering of 27 protein-coding genes with average increased expression > 13 fold in SSA/Ps illustrates what was shared in gene expression changes among all but two of the SSA/Ps (Figure 2, Panel B, Supplementary Table 2). It should be noted that 2/10 (20%) HPs and 5/21 (24%) SSA/Ps were from right and left colon, respectively. Although our numbers of HPs from right colon and SSA/Ps from left colon are small we did not see appreciable differences in gene expression between left and right HPs or SSA/Ps. Increased expression of these 27 genes was not observed in adenomatous polyp RNA-seq datasets (Figure 2, Panel C).
Figure 2.
Differentially expressed genes in syndromic and sporadic SSA/Ps and HPs by RNA sequencing. Panel A – Genes with ≥ 4-fold change and FDR < 0.05 in syndromic SSA/Ps (n=12), sporadic SSA/Ps (n=9) and HPs (n=10). Syndromic and sporadic SSA/Ps were compared to control right colon and HPs were compared to control left colon. Panel B – Relative expression of 27 protein-coding genes in syndromic SSA/Ps, sporadic SSA/Ps, HPs and control left and right colon. Log2 ratios comparing each individual sample to the mean of all samples were used for hierarchical clustering. Two right-sided HPs are labeled in red and five left-sided SSA/Ps are labeled in green. Panel C – Mean fold change expression of the same 27 protein coding genes described in Panel B in normal colon, adenomas, HPs and sporadic and syndromic SSA/Ps.
We also compared gene expression in the uninvolved colon (n=10) of serrated polyposis syndrome (SPS) patients and patients with sporadic SSA/Ps with the control right colon (n=10) of patients undergoing screening colonoscopy with no polyps (Supplementary Figure 2). Surprisingly, 1922 genes were differentially expressed between the uninvolved colon of patients with SSA/Ps and control colon (≥ 2-fold change, FDR < 0.01). A significant overlap in the gene expression profile of uninvolved colon from patients with SPS and sporadic SSA/Ps was observed. However, the magnitude of fold change was small for most genes (< 3 fold) and the genes differentially expressed were not common to genes differentially expressed in SSA/Ps.
Selection and Cross Validation of a Gene Signature that Differentiates SSA/Ps from HPs
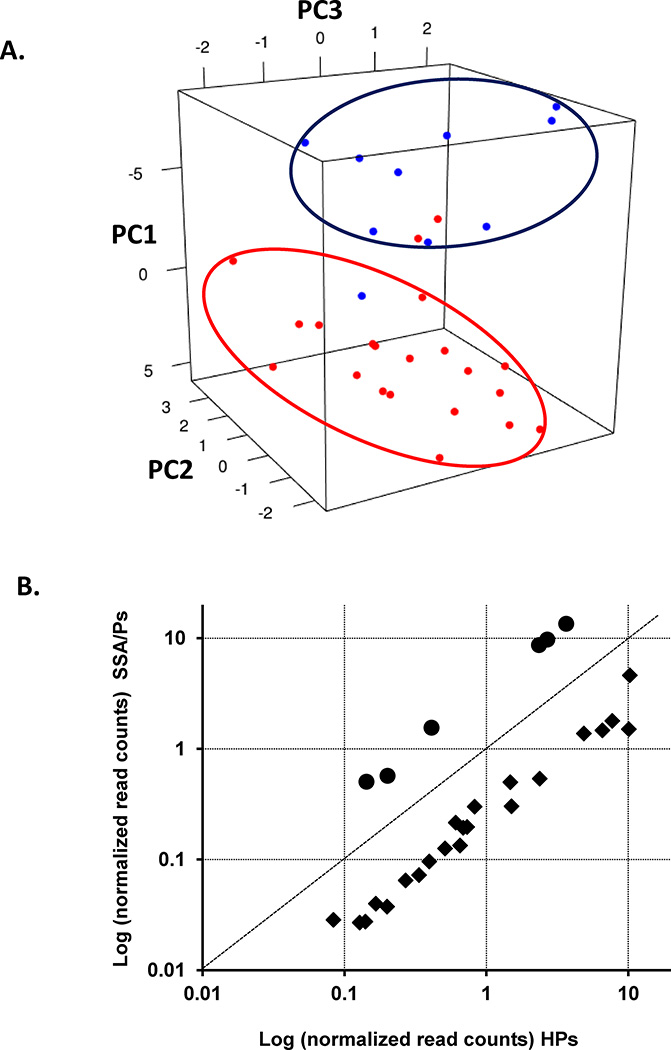
Count data from 31 serrated polyps (21 SSA/Ps and 10 HPs) were used in a leave-one-out cross-validation analysis. Twenty-eight genes with an FDR < 0.01 and ≥ 2-fold change (SSA/Ps vs HPs) defined the signature (Supplementary Table 2). 28 of 31 serrated polyps were classified correctly for a nominal error rate of 10%. After cross validating four times, the cross-validated error rate was 18%. Principal component analysis of the gene expression of each of the 28 genes in all 31 serrated polyps is shown in Figure 3, Panel A that demonstrates the misclassification of two SSA/Ps and one HP. The relative expression of each of the 28 genes in SSA/Ps and HPs is shown in Figure 3, Panel B. Six genes were overexpressed and twenty-two underexpressed in SSA/Ps relative to HPs.
Figure 3.
Evaluation of a 28-gene signature to distinguish SSA/Ps from HPs. The 28-gene panel was developed using a leave-one-out cross validation approach on 31 independent serrated polyps (21 SSA/Ps and 10 HPs) samples. Panel A – Principal component analysis of the 28 gene log2 ratios for each individual serrated polyp compared to the mean of all serrated polyps. Principal component 1 (PC1) accounted for 28% of the variation in the data and separated most SSA/Ps (red) from HPs (blue). Twenty-eight of thirty-one serrated polyps (~90%) clustered correctly similar to the nominal error rate found in the cross validation results. Panel B – Relative expression (log of normalized reads (RPKM) of the same 28 genes described in Panel A in SSA/Ps and HPs. Six genes (circles) were overexpressed in SSA/Ps compared to HPs (range 2.8 to 3.7 fold) and 22 genes (squares) were underexpressed in SSA/Ps compared to HPs (range −2.2 to −6.7).
Evaluation of Gene Signature in Published Microarray Data of Serrated Polyps
We compared the relative expression of each of our 51-gene signature in SSA/Ps, MVHPs and normal colon (left and right) from a previously published microarray study (19). Clear separation of SSA/Ps from MVHPs and control colon was observed by hierarchical clustering (Supplementary Figure 3). In fact, five out of six MVHPs, showed gene expression patterns more closely resembling control colon than SSA/Ps.
Identifying Colon Cancers with the SSA/P Gene Signature in The Cancer Genome Atlas
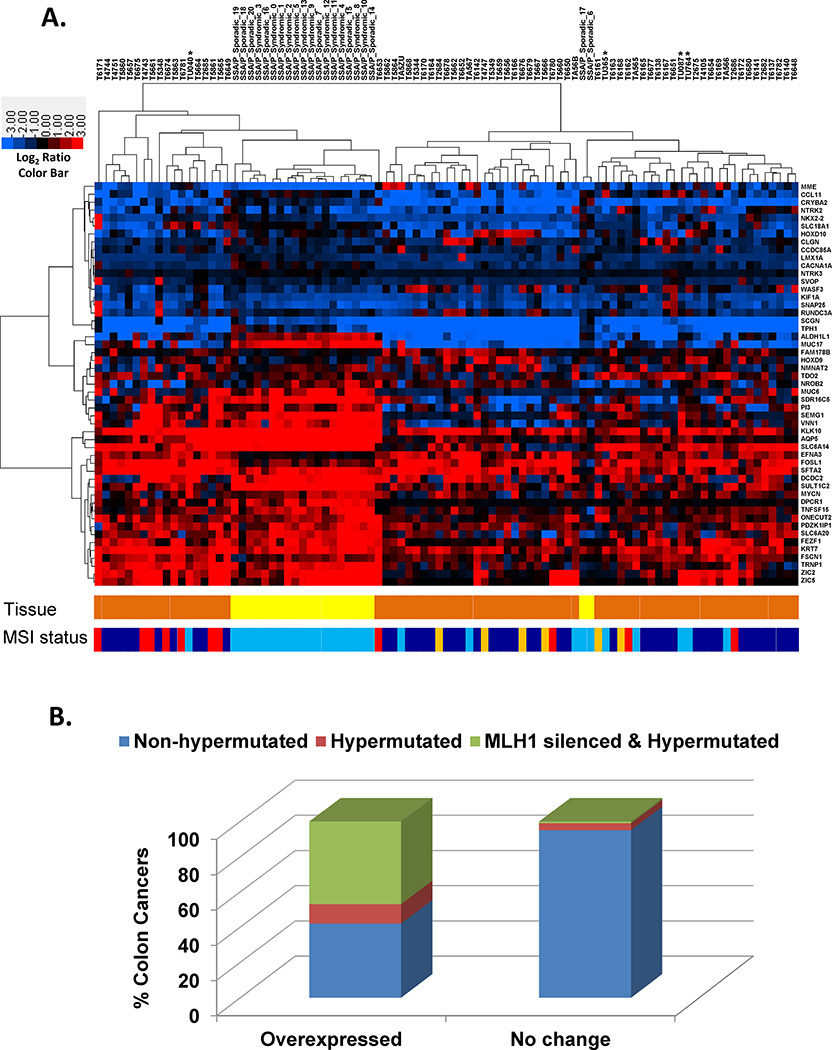
We compared our 51-gene SSA/P signature with sixty-eight colon cancer RNA-seq datasets available in the Cancer Genome Atlas and four colon cancers obtained from the University of Utah (Figure 4, Panel A, Supplementary Table 2). RNA-seq data from 4 of the 55 genes were not available in the TCGA datasets. We performed RNA sequencing on four colon cancers from the University of Utah to identify potential lab/batch effect differences in gene expression between our RNA-Seq datasets and the TCGA datasets. The 51 gene SSA/P signature showed similar expression patterns between syndromic and sporadic SSA/Ps and the MSI-H subset of colon cancers. No batch effects were observed between our colon cancer datasets and the TCGA datasets. Sixty-three out of 72 cancers had data on their MSI status with 11 cancers being MSI-H (MSI status unknown for 9 colon cancers). Eighteen colon cancers clustered with SSA/Ps and 8 of the 18 colon cancers (44%) were MSI-H. This is a significant finding since of the remaining 54 colon cancers that did not cluster with SSA/Ps only 3 were MSI-H (6%). This suggests that our SSA/P signature identifies MSI-H cancers.
Figure 4.
Evaluation of a 51 SSA/P gene signature in colon cancer RNA sequencing datasets from The Cancer Genome Atlas (TCGA). Panel A – Log2 ratios comparing individual colon cancers (n=72) and SSA/Ps (n=21) to the mean of 14 uninvolved and 10 control colon samples (n=24) were used for hierarchical clustering. “Tissue” color bar shows colon adenocarcinomas (orange) and SSA/Ps (yellow). “MSI status” color bar shows microsatellite stable (MSS) cancers (dark blue), MSI-H cancers (red) and MSI-L cancers (light orange). SSA/Ps and colon cancers not evaluated for MSI (light blue). Panel B – Percentage of TCGA colon cancers showing overexpression of FSCN1, ZIC2, ZIC5, CRYBA2, MUC6, TRNP1 and/or SEMG1 described in Table 1. 195 colon cancers with RNA expression and MLH1 methylation data in the TCGA database were evaluated using the cBioPortal for Cancer Genomics.
We also evaluated mRNA expression of each of our 51 SSA/P signature genes in 195 TCGA colon cancers using the cBioPortal for Cancer Genomics. Thirteen of the 51 signature genes had frequent increased mRNA expression in ≥ 10% of hypermutated colon cancers but not in non-hypermutated cancers (Table 1). Seven of these genes (FSCN1, ZIC2, ZIC5, CRYBA2, MUC6, TRNP1 and SEMG1) had increased mRNA expression in 13–30% of hypermutated and only 0–3% of non-hypermutated colon cancers with Fischer exact p-value < 0.01 (Table 1). Twenty-two of the thirty (73%) hypermutated colon cancers showed increased expression of at least one of the seven-gene panel. Seventeen of the twenty-two (77%) hypermutated colon cancers showing increased expression of at least one of the seven-gene panel also showed MLH1 silencing. (Figure 4, Panel B). Eleven of 51 genes showed frequent overexpression in CIMP-H and/or MLH1-silenced colon cancers including all seven that showed frequent increased expression in hypermutated cancers (Supplementary Table 3). We did not observe frequent increased expression of previous SSA/P markers (annexin A10 - ANXA10 and claudin 1 - CLDN1) in hypermutated, CIMP-H and/or MLH1 silenced colon cancers (Table 1 and Supplementary Table 3) (19, 31).
Table 1. Frequency of Increased mRNA Expression in SSA/P Signatures Genes in 30 Hypermutated and 165 Non-Hypermutated Colon Cancers from the Cancer Genome Atlas.
Frequency of increased mRNA expression in SSA/P signature genes in hypermutated and non-hypermutated colon cancers (CC) from the Cancer Genome Atlas (TCGA).
| Gene Symbol |
Gene Description | Hypermutated CC Incidence (%) |
Non- Hypermutated CC Incidence (%) |
Fisher Exact P-value |
|---|---|---|---|---|
| FSCN1 | Fascin actin-binding protein 1 | 9 (30) | 3 (2) | <0.001 |
| ZIC5 | Zic family member 5 | 7 (23) | 5 (3) | <0.001 |
| CRYBA2 | Crystallin, beta A2 | 5 (17) | 0 (0) | <0.001 |
| SEMG1 | Semenogelin | 4 (13) | 0 (0) | <0.001 |
| ZIC2 | Zic family member 2 | 6 (20) | 4 (2) | 0.001 |
| TRNP1 | TMF1-regulated nuclear protein 1 | 6 (20) | 5 (3) | 0.002 |
| MUC6 | Mucin 6 | 4 (13) | 1 (1) | 0.002 |
| FOSL1 | FOS-like antigen 1 | 3 (10) | 1 (1) | 0.012 |
| ALDH1L1 | Aldehyde dehydrogenase 1 family member L1 | 5 (17) | 7 (4) | 0.022 |
| KLK10 | Kallikrein related peptidase 10 | 3 (10) | 4 (2) | 0.075 |
| SLC18A1 | Solute carrier family 18, member 1 | 3 (10) | 4 (2) | 0.075 |
| VNN1 | Vanin 1 | 3 (10) | 4 (2) | 0.075 |
| MUC17 | Mucin 17 | 3 (10) | 13 (8) | 0.717 |
| ANXA10 | Annexin A10 | 1 (3) | 2 (1) | 0.396 |
| CLDN1 | Claudin 1 | 1 (3) | 11 (7) | 0.696 |
Incidence of increased mRNA in 195 colon cancers (30 hypermutated and 165 non-hypermutated) was obtained using TCGA data available in the cBioPortal for Cancer Genomics, Memorial Sloan-Kettering Cancer Center. Table lists 13/51 signature genes that show frequent (≥ 10%) increased mRNA expression in hypermutated colon cancers. Incidence of increased mRNA expression are also shown for two previously developed SSA/P gene markers, annexin A10 (ANXA10) and claudin 1 (CLDN1). Changes in mRNA expression were obtained by comparing normalized read counts (RPKM) for each gene across colon cancers diploid for each gene. Statistical significant differences between incidence of increased mRNA expression between hypermutated and non-hypermutated were determined using a Fisher Exact test. Nine genes showed statistically significant increased incidence of mRNA overexpression in hypermutated colon cancers.
Mutual Exclusivity and Co-occurrence Analysis
Using the cBioPortal we evaluated concurrent genomic alterations (RNA expression and somatic mutation) in each of our 51-gene panel and two genes from previous microarray studies (ANXA10 and CLDN1) with alterations in BRAF (19, 31). Thirteen of 51 genes showed statistically significant associations with BRAF mutation both by Fisher exact test and log odds ratio (Supplementary Table 4). Six of these genes (FSCN1, ZIC5, CRYBA2, MUC6, TRNP1 and SEMG1) were common to genes frequently overexpressed in hypermutated, CIMP-H and MLH1 silenced colon cancers. ZIC2 and CLDN1 did not show significant associations with BRAF mutation and ANXA10 showed a positive association by logs odds ratio but not the Fisher exact test.
Sensitivity and Specificity of a Seven Gene Panel
Using a seven-gene panel (FSCN1, ZIC2, ZIC5, CRYBA2, MUC6, TRNP1 and SEMG1) we determined the sensitivity and specificity of each gene in identifying 31 BRAF mutant, CIMP-H and/or MLH1 silenced colon cancers out of 182 total colon cancers from the TCGA database (Table 2A). The specificity of each gene in identifying this subset of cancers was very high, between 85 and 99%. SSA/P RNA markers ANXA10 and CLDN1 showed similar specificity to our seven gene panel. In contrast, the sensitivity of each gene in identifying BRAF mutant, CIMP-H and/or MLH1 silenced colon cancers was more variable between genes (26–68%) with ZIC5 showing the highest sensitivity at 68%. The two previously identified RNA markers for SSA/Ps were lower with 19% and 6% sensitivity for ANXA10 and CLDN1, respectively. Using a seven-gene panel our sensitivity increased to 94% if at least one of the seven genes showed a two-fold increase in expression (Table 2B). Using ANXA10 or CLDN1 with our seven-gene panel the sensitivity was 97% and 94%, and the specificity was 72% and 63%, respectively (Supplementary Table 5). qPCR validation was performed on 4 genes (FSCN1, ZIC5, SEMG1 and MUC6) and showed high expression in SSA/P’s compared to HPs, uninvolved or control colon consistent with our RNA-seq data (Supplementary Figure 4).
Table 2. Cross-Validated Sensitivity and Specificity of SSA/P Seven Gene Panel.
Sensitivity and specificity of a seven gene panel in identifying BRAF mutant, CIMP-H and/or MLH1 silenced colon cancers from the Cancer Genome Atlas (TCGA).
| A. Individual Genes | ||
|---|---|---|
| Gene | Sensitivity | Specificity |
| ZIC5 | 0.677 | 0.887 |
| ZIC2 | 0.548 | 0.854 |
| FSCN1 | 0.516 | 0.947 |
| SEMG1 | 0.484 | 0.960 |
| TRNP1 | 0.484 | 0.947 |
| CRYBA2 | 0.419 | 0.960 |
| MUC6 | 0.258 | 0.987 |
| ANXA10 | 0.194 | 0.974 |
| CLDN1 | 0.065 | 0.881 |
| B. Seven Gene Panel | ||
|---|---|---|
| Minimum # Genes Positive |
Sensitivity | Specificity |
| 1 | 0.935 | 0.722 |
| 2 | 0.839 | 0.874 |
| 3 | 0.613 | 0.960 |
| 4 | 0.419 | 0.987 |
| 5 | 0.290 | 1.000 |
| 6 | 0.194 | 1.000 |
| 7 | 0.097 | 1.000 |
Normalized RNA-Seq gene expression data (RPKM) for each of the seven gene panel was downloaded from the cBioPortal for Cancer Genomics using the CGDS-R package http://www.cbioportal.org/cgds_r.jsp. One hundred eighty-six TCGA colon cancers had mRNA expression, BRAF mutation, methylation subtype and MLH1 methylation data available. Thirty one of 186 colon cancers (17%) were BRAF mutated, CIMP-H and/or MLH1 silenced. The majority of these cancers 20/31 (64%) had two or more of these DNA alterations highly suggestive of colon cancers developing via the serrated pathway. Panel A, The sensitivity and specificity of each of our seven gene panel, and two previously described SSA/P gene markers (ANXA10, CLDN1), in identifying BRAF mutant, CIMP-H and/or MLH1 silenced colon cancers. Panel B, The sensitivity and specificity of one or more genes from our seven gene panel showing a ≥ 2-fold increased expression in serrated pathway cancers compared to the average of all colon cancers.
Discussion
Sessile serrated adenoma/polyps (SSA/Ps) are now recognized as polyps with malignant potential, with SSA/Ps originating in the serrated polyposis syndrome having the highest risk for progression to colon cancer. Recent cancer surveillance guidelines recommend earlier follow up for patients with sporadic SSA/Ps almost at par with individuals with adenomatous polyps (22). Nevertheless, differentiating SSA/Ps from HPs by histopathology and identifying patients with SSA/Ps have some challenges in clinical practice. The RNA sequencing datasets we describe identifies 51 differentially expressed genes in SSA/Ps that molecularly distinguish them from HPs. These genes are also differentially expressed in sporadic microsatellite unstable (MSI-H) colon cancers. We further refined our panel to seven genes that also show frequent overexpression in BRAF mutant, CpG island methylator phenotype high (CIMP-H) and MLH1 silenced colon cancers. Our data provides clear evidence that RNA expression changes in BRAF mutant, CIMP-H and MLH1 silenced colon cancers are observed in early SSA/Ps and that these new gene expression markers may lead to improved diagnostics for SSA/Ps. Moreover, our data demonstrate similar gene expression profiles of SSA/Ps in the serrated polyposis syndrome and sporadic SSA/Ps indicating that common mechanisms of progression to cancer are operating in both.
Comparing the transcriptome of SSA/Ps and HPs produced findings that raise some critical questions about these two subtypes of serrated polyps with very different potentials for progression to colon cancer. It is unclear if serrated adenocarcinoma originates directly through SSA/Ps or if genetic alterations in certain hyperplastic polyps found in right colon lead to the development of SSA/Ps and eventually to colon cancer. SSA/Ps, especially in the serrated polyposis syndrome, have a significant risk for progression to cancer (9–11) whereas HPs have a negligible risk (32, 33). The finding that most of the genes found differentially expressed in HPs were also found in SSA/Ps at least partly explains why both types of polyps have a similar morphological appearance. On the other hand, there were many uniquely and highly differentially expressed genes in SSA/Ps compared to HPs. The unique SSA/Ps gene signature established in this study provides an opportunity to identify critical pathways that may explain these differences in cancer risk.
Our seven-gene panel (FSCN1, ZIC2, ZIC5, CRYBA2, MUC6, TRNP1 and SEMG1) identified BRAF mutant, CIMP-H and MLH1 silenced colon cancers with high sensitivity and specificity. In comparison with other gene markers described for SSA/Ps (ANXA10 and CLDN1) our seven-gene panel showed increased sensitivity and similar specificity. This increase in sensitivity might be related to the use of RNA-Seq versus microarray technology. RNA-Seq provides a more quantitative analysis of transcript abundance and is not dependent on previously defined gene annotation. Also, the analysis of SSA/Ps from serrated polyposis (SPS) patients, known to have high colon cancer risk, may have further increased our ability to identify a gene signature more closely associated with sporadic colon cancer developing from the serrated pathway.
Three genes (FSCN1, TRNP1, ZIC2) of our seven-gene panel were previously identified to be overexpressed in BRAF positive colon cancers in a European patient cohort (34). These genes were part of a 64-gene expression classifier for BRAF positive colon cancers with poor prognosis. Another study classifying colon cancers into four consensus molecular subtypes with subtype 1 (CMS1) consisting of microsatellite unstable, CIMP-H and BRAF positive tumors identified one of our seven-gene panel (ZIC2) as a marker of serrated cancers (35,36). ZIC proteins play a role in regulating the sonic hedgehog and Wnt/B-catenin signaling pathways (37, 38). ZIC2 expression has been associated with multiple cancers including brain, ovarian and cervical cancer (39, 40). FSCN1 is an actin-binding protein frequently overexpressed in a variety of cancers including colon cancer and predicts poor prognosis (41). FSCN1 is also highly expressed in serrated colon cancers (42). TMF-regulated nuclear protein (TRNP1) is a nuclear protein that plays a role in mammalian brain cortex development (43). The significance of TRNP1 overexpression in colon cancer remains unknown. Our study reinforces the importance of these genes in serrated colon cancers providing the first evidence that these mRNA changes occur early in the cancer process in pre-neoplastic serrated lesions (SSA/Ps).
Other genes described in our seven-gene panel may also participate in colon cancer progression. MUC6 is a gastric mucin protein shown to have increased expression in SSA/Ps compared to HPs (44). Increased expression of MUC6 has been documented in hypermethylated colon cancers suggesting its possible role in serrated pathway (45). Data lacks about the role of SEMG1 and CRYBA2 in colon cancer. SEMG1 is a seminal vesical protein that has been studied as a biomarker for detection of prostate cancer (46). CRYBA2 belongs to beta/gamma-crystallin family of genes and is found to be hypermethylated in CIMP-H neuroblastoma tumors (47). Further mechanistic studies will be needed to understand the functions of these key genes in the serrated pathway.
A significant number of the genes that were differently expressed in the uninvolved colonic mucosa of patients with syndromic (SPS) and sporadic SSA/Ps, relative to normal colon (patients with no polyps), overlapped and suggest a field effect may be present in the colonic mucosa of patients with SSA/Ps. These genes were different from those found common to syndromic and sporadic SSA/Ps and had smaller fold changes relative to controls. A ‘field cancerization’ effect has been reported in studies of sporadic colon cancer (48, 49). There are also limited studies investigating possible field effects in patients with colon polyps, particularly SSA/Ps (50). Our data raise important questions regarding the origin of such changes. The question of predictive value of field effect will require studies with larger number of patients, which are underway at this time.
Microsatellite instability, CpG island methylation (CIMP), inactivation of MLH1 and BRAF mutations have all been implicated as underlying events in the serrated pathway to colon cancer (14–18,51). A recent study showed MLH1 silencing in a subgroup of hypermutated colon cancers that had increased BRAF and decreased APC and KRAS mutations. The authors concluded that MLH1 silencing occurred through a different pathway, suggestive of the serrated pathway (52). However, not all SSA/Ps have these changes, and it remains uncertain if they are absolute requirements for progression to cancer. A recent large serrated polyp study only identified MLH1 methylation in 11% of SSA/Ps (53). We report a new set of 51 genes that are differently expressed in most SSA/Ps and sporadic MSI-high cancers in the TCGA cancer database. A smaller seven-gene panel identified BRAF mutant, CIMP-H and MLH1 silenced colon cancers with both high sensitivity and specificity. Our findings provide novel molecular markers for SSA/Ps that may play a role in the development of serrated colon cancers.
Limitations of our study include a small sample size in each individual patient cohort (n=9 to12). This is, in part, due to colon biopsies being collected prospectively and the low prevalence of sporadic SSA/Ps and the serrated polyposis syndrome in the general population. Even with this limitation, this is the largest RNA-sequencing study performed to characterize the transcriptome of SSA/Ps. Finally, our gene panel was not validated in a separate RNA-Seq study of serrated polyps because these datasets are not publically available. However, our gene signature did accurately classify SSA/Ps from MVHPs using expression data from a previous microarray study. Future validation studies are currently being designed and are beyond the scope of this study.
In summary, this report provides a comprehensive gene expression comparison of SSA/Ps with HPs, which share many histopathological similarities but differ markedly in risk of progression to colon cancer. Despite many similarities in gene expression in SSA/Ps and HPs, both sporadic and syndromic SSA/Ps have a unique gene signature with a number of highly differentially expressed genes of interest relative to oncogenesis. The identification of a set of novel genes uniquely differentially expressed in SSA/Ps and BRAF mutant, CIMP-H and MLH1 silenced colon cancers provides additional leads for further understanding the molecular pathways leading to cancer progression via the serrated pathway. This may lead to the development of a gene panel that can be used in clinical practice to stratify patients with increased colon cancer risk from serrated polyps. This could be especially helpful in identifying patients with serrated polyposis syndrome in who no currently recognized genetic mutation has been identified.
Supplementary Material
Acknowledgments
Funding: This study was supported by a pilot clinical research award from American College of Gastroenterology (P Kanth), University of Utah Personalized Medicine Program Seed Grant (CH Hagedorn), and National Institutes of Health (NIH) CA176130 and CA148068 (CH Hagedorn), plus CA073992 and CA146329 (RW Burt). Cancer Center Support Grant P30-CA42014. National Center for Advancing Translational Sciences of the NIH Award 1ULTR001067.
We thank: Kathleen Boynton and Michelle Done for their assistance in sample collection and Mark Hazel for qPCR validation experiments.
Footnotes
Conflict of Interest: None
References
- 1.Department of Health and Human Services, Centers for Disease Control and Prevention, and National Cancer Institute. 2010 [Google Scholar]
- 2.Burgess NG, Pellise M, Nanda KS, Hourigan LF, Zanati SA, Brown GJ, et al. Clinical and endoscopic predictors of cytological dysplasia or cancer in a prospective multicentre study of large sessile serrated adenomas/polyps. Gut. 2015 doi: 10.1136/gutjnl-2014-308603. [DOI] [PubMed] [Google Scholar]
- 3.Lieberman DA, Weiss DG, Bond JH, Ahnen DJ, Garewal H, Chejfec G. Use of colonoscopy to screen asymptomatic adults for colorectal cancer. Veterans Affairs Cooperative Study Group 380. N Engl J Med. 2000;343:162–168. doi: 10.1056/NEJM200007203430301. [DOI] [PubMed] [Google Scholar]
- 4.Kahi CJ, Hewett DG, Norton DL, Eckert GJ, Rex DK. Prevalence and variable detection of proximal colon serrated polyps during screening colonoscopy. Clin Gastroenterol Hepatol. 2011;9:42–46. doi: 10.1016/j.cgh.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 5.Bosman FT, Carneiro F, Hruban RH, Theise ND, editors. WHO Classification of Tumors of the Digestive System. Lyon: IARC; 2010. pp. 160–165. [Google Scholar]
- 6.Torlakovic E, Skovlund E, Snover DC, Torlakovic G, Nesland JM. Morphologic reappraisal of serrated colorectal polyps. Am J Surg Pathol. 2003;27:65–81. doi: 10.1097/00000478-200301000-00008. [DOI] [PubMed] [Google Scholar]
- 7.Wong NA, Hunt LP, Novelli MR, Shepherd NA, Warren BF. Observer agreement in the diagnosis of serrated polyps of the large bowel. Histopathology. 2009;55:63–66. doi: 10.1111/j.1365-2559.2009.03329.x. [DOI] [PubMed] [Google Scholar]
- 8.Khalid O, Radaideh S, Cummings OW, O'Brien MJ, Goldblum JR, Rex DK. Reinterpretation of histology of proximal colon polyps called hyperplastic in 2001. World J Gastroenterol. 2009;15:3767–3770. doi: 10.3748/wjg.15.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jasperson KW, Kanth P, Kirchhoff AC, Huismann D, Gammon A, Kohlmann W, et al. Serrated polyposis: colonic phenotype, extracolonic features, and familial risk in a large cohort. Dis Colon Rectum. 2013;56:1211–1216. doi: 10.1097/DCR.0b013e3182a11cca. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rashid A, Houlihan PS, Booker S, Petersen GM, Giardiello FM, Hamilton SR. Phenotypic and molecular characteristics of hyperplastic polyposis. Gastroenterology. 2000;119:323–332. doi: 10.1053/gast.2000.9361. [DOI] [PubMed] [Google Scholar]
- 11.Boparai KS, Mathus-Vliegen EM, Koornstra JJ, Nagengast FM, van Leerdam M, van Noesel CJ, et al. Increased colorectal cancer risk during follow-up in patients with hyperplastic polyposis syndrome: a multicentre cohort study. Gut. 2010;59:1094–1100. doi: 10.1136/gut.2009.185884. [DOI] [PubMed] [Google Scholar]
- 12.Holme O, Bretthauer M, Eide TJ, Loberg EM, Grzyb K, Loberg M, et al. Long-term risk of colorectal cancer in individuals with serrated polyps. Gut. 2015;64:929–936. doi: 10.1136/gutjnl-2014-307793. [DOI] [PubMed] [Google Scholar]
- 13.Mäkinen MJ. Colorectal serrated adenocarcinoma. Histopathology. 2007;50:131–150. doi: 10.1111/j.1365-2559.2006.02548.x. [DOI] [PubMed] [Google Scholar]
- 14.O'Brien MJ. Hyperplastic and serrated polyps of the colorectum. Gastroenterol Clin North Am. 2007;36:947–968. doi: 10.1016/j.gtc.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 15.O'Brien MJ, Zhao Q, Yang S. Colorectal serrated pathway cancers and precursors. Histopathology. 2015;66:49–65. doi: 10.1111/his.12564. [DOI] [PubMed] [Google Scholar]
- 16.Iino H, Jass JR, Simms LA, Young J, Leggett B, Ajioka Y, et al. DNA microsatellite instability in hyperplastic polyps, serrated adenomas, and mixed polyps: a mild mutator pathway for colorectal cancer? J Clin Pathol. 1999;52:5–9. doi: 10.1136/jcp.52.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guarinos C, Sánchez-Fortún C, Rodríguez-Soler M, Perez-Carbonell L, Egoavil C, Juarez M, et al. Clinical subtypes and molecular characteristics of serrated polyposis syndrome. Clin Gastroenterol Hepatol. 2013;11:705–711. doi: 10.1016/j.cgh.2012.12.045. [DOI] [PubMed] [Google Scholar]
- 18.Samowitz WS, Albertsen H, Herrick J, Levin TR, Sweeney C, Murtaugh MA, et al. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology. 2005;129:837–845. doi: 10.1053/j.gastro.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalo DH, Lai KK, Shadrach B, Goldblum JR, Bennett AE, Downs-Kelly E, et al. Gene expression profiling of serrated polyps identifies annexin A10 as a marker of a sessile serrated adenoma/polyp. J Pathol. 2013;230:420–429. doi: 10.1002/path.4200. [DOI] [PubMed] [Google Scholar]
- 20.Caruso M, Moore J, Goodall GJ, Thomas M, Phillis S, Tyskin A, et al. Over-expression of cathepsin E and trefoil factor 1 in sessile serrated adenomas of the colorectum identified by gene expression analysis. Virchows Archive: an international journal of pathology. 2009;454:291–302. doi: 10.1007/s00428-009-0731-0. [DOI] [PubMed] [Google Scholar]
- 21.Delker DA, McGettigan BM, Kanth P, Pop S, Neklason DW, Bronner MP, et al. RNA sequencing of sessile serrated colon polyps identifies differentially expressed genes and immunohistochemical markers. PLoS One. 2014;9:e88367. doi: 10.1371/journal.pone.0088367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lieberman DA, Rex DK, Winawer SJ, Giardiello FM, Johnson DA, Levin TR, et al. United States Multi-Society Task Force on Colorectal Cancer. Guidelines for colonoscopy surveillance after screening and polypectomy: a consensus update by the US Multi-Society Task Force on Colorectal Cancer. Gastroenterology. 2012;143:844–857. doi: 10.1053/j.gastro.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 23.Folkers ME, Delker DA, Maxwell CI, Nelson CA, Schwartz JJ, Nix DA, et al. ENCODE tiling array analysis identifies differentially expressed annotated and novel 5' capped RNAs in hepatitis C infected liver. PLoS One. 2011;6:e14697. doi: 10.1371/journal.pone.0014697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papic N, Maxwell CI, Delker DA, Liu S, Heale BS, Hagedorn CH. RNA-sequencing analysis of 5' capped RNAs identifies many new differentially expressed genes in acute hepatitis C virus infection. Viruses. 2012;4:581–612. doi: 10.3390/v4040581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Comprehensive molecular characterization of human colon and rectal cancer. Cancer Genome Atlas Network. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. https://tcga-data.nci.nih.gov/tcga/tcgaHome2.jsp.
- 28.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science Signaling. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discovery. 2012 May;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ciriello G, Cerami E, Sander C, Schultz N. Mutual exclusivity analysis identifies oncogenic network modules. Genome Res. 2012;22:398–406. doi: 10.1101/gr.125567.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caruso M1, Fung KY2, Moore J3, Brierley GV, Cosgrove LJ, Thomas M, et al. Claudin-1 Expression Is Elevated in Colorectal Cancer Precursor Lesions Harboring the BRAF V600E. Mutation. Transl Oncol. 2014;7:456–463. doi: 10.1016/j.tranon.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DiSario JA, Foutch PG, Mai HD, Pardy K, Manne RK. Prevalence and malignant potential of colorectal polyps in asymptomatic, average-risk men. Am J Gastroenterol. 1991;86:941–945. [PubMed] [Google Scholar]
- 33.Weston AP, Campbell DR. Diminutive colonic polyps: histopathology, spatial distribution, concomitant significant lesions, and treatment complications. Am J Gastroenterol. 1995;90:24–28. [PubMed] [Google Scholar]
- 34.Popovici V, Budinska E, Tejpar S, Weinrich S, Estrella H, Hodgson G, et al. Identification of a poor-prognosis BRAF-mutant-like population of patients with colon cancer. J Clin Oncol. 2012;30:1288–1295. doi: 10.1200/JCO.2011.39.5814. [DOI] [PubMed] [Google Scholar]
- 35.Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C, et al. The consensus molecular subtypes of colorectal cancer. Nature Medicine. 2015;21:1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Sousa E Melo F, Wang X, Jansen M, Fessler E, Trinh A, de Rooij LP, et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nature Medicine. 2013;19:614–618. doi: 10.1038/nm.3174. [DOI] [PubMed] [Google Scholar]
- 37.Merzdorf CS. Emerging roles for zic genes in early development. Dev Dyn. 2007;236:922–940. doi: 10.1002/dvdy.21098. [DOI] [PubMed] [Google Scholar]
- 38.Sanek NA, Taylor AA, Nyholm MK, Grinblat Y. Zebrafish zic2a patterns the forebrain through modulation of Hedgehog-activated gene expression. Development. 2009;136:3791–3800. doi: 10.1242/dev.037820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marchini S, Poynor E, Barakat RR, Clivio L, Cinquini M, Fruscio R, et al. The zinc finger gene ZIC2 has features of an oncogene and its overexpression correlates strongly with the clinical course of epithelial ovarian cancer. Clin Cancer Res. 2012;18:4313–4324. doi: 10.1158/1078-0432.CCR-12-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chan DW, Liu VW, Leung LY, Yao KM, Chan KK, Cheung AN, et al. Zic2 synergistically enhances Hedgehog signalling through nuclear retention of Gli1 in cervical cancer cells. J Pathol. 2011;225:525–534. doi: 10.1002/path.2901. [DOI] [PubMed] [Google Scholar]
- 41.Ma Y, Machesky LM. Fascin1 in carcinomas: Its regulation and prognostic value. Int J Cancer. 2015;137:2534–2544. doi: 10.1002/ijc.29260. [DOI] [PubMed] [Google Scholar]
- 42.Conesa-Zamora P, García-Solano J, García-García F, Turpin Mdel C, Trujillo-Santos J, Torres-Moreno D, et al. Expression profiling shows differential molecular pathways and provides potential new diagnostic biomarkers for colorectal serrated adenocarcinoma. Int J Cancer. 2013;132:297–307. doi: 10.1002/ijc.27674. [DOI] [PubMed] [Google Scholar]
- 43.Stahl R, Walcher T, De Juan Romero C, Pilz GA, Cappello S, Irmler M, et al. Trnp1 regulates expansion and folding of the mammalian cerebral cortex by control of radial glial fate. Cell. 2013;153:535–549. doi: 10.1016/j.cell.2013.03.027. [DOI] [PubMed] [Google Scholar]
- 44.Owens SR, Chiosea SI, Kuan SF. Selective expression of gastric mucin MUC6 in colonic sessile serrated adenoma but not in hyperplastic polyp aids in morphological diagnosis of serrated polyps. Mod Pathol. 2008;21:660–669. doi: 10.1038/modpathol.2008.55. [DOI] [PubMed] [Google Scholar]
- 45.Walsh MD, Clendenning M, Williamson E, Pearson SA, Walters RJ, Nagler B, et al. Expression of MUC2, MUC5AC, MUC5B, and MUC6 mucins in colorectal cancers and their association with the CpG island methylator phenotype. Mod Pathol. 2013;26:1642–1656. doi: 10.1038/modpathol.2013.101. [DOI] [PubMed] [Google Scholar]
- 46.Neuhaus J, Schiffer E, von Wilcke P, Bauer HW, Leung H, Siwy J, et al. Seminal plasma as a source of prostate cancer peptide biomarker candidates for detection of indolent and advanced disease. PLoS One. 2013;8(6):e67514. doi: 10.1371/journal.pone.0067514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abe M, Watanabe N, McDonell N, Takato T, Ohira M, Nakagawara A, et al. Identification of genes targeted by CpG island methylator phenotype in neuroblastomas, and their possible integrative involvement in poor prognosis. Oncology. 2008;74:50–60. doi: 10.1159/000139124. [DOI] [PubMed] [Google Scholar]
- 48.Hawthorn L, Lan L, Mojica W. Evidence for field effect cancerization in colorectal cancer. Genomics. 2014;103:211–221. doi: 10.1016/j.ygeno.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 49.Lochhead P, Chan AT, Nishihara R, Fuchs CS, Beck AH, Giovannucci E, et al. Etiologic field effect: reappraisal of the field effect concept in cancer predisposition and progression. Mod Pathol. 2015;28:14–29. doi: 10.1038/modpathol.2014.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen LC, Hao CY, Chiu YS, Wong P, Melnick JS, Brotman M, et al. Alteration of gene expression in normal-appearing colon mucosa of APC(min) mice and human cancer patients. Cancer Res. 2004;64:3694–3700. doi: 10.1158/0008-5472.CAN-03-3264. [DOI] [PubMed] [Google Scholar]
- 51.Huang CS, Farraye FA, Yang S, O'Brien MJ. The clinical significance of serrated polyps. Am J Gastroenterol. 2011;106:229–240. doi: 10.1038/ajg.2010.429. [DOI] [PubMed] [Google Scholar]
- 52.Donehower LA, Creighton CJ, Schultz N, Shinbrot E, Chang K, Gunaratne PH, et al. MLH1-silenced and non-silenced subgroups of hypermutated colorectal carcinomas have distinct mutational landscapes. J Pathol. 2013;229:99–110. doi: 10.1002/path.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burnett-Hartman AN, Newcomb PA, Potter JD, Passarelli MN, Phipps AI, Wurscher MA, et al. Genomic aberrations occurring in subsets of serrated colorectal lesions but not conventional adenomas. Cancer Res. 2013;73:2863–2872. doi: 10.1158/0008-5472.CAN-12-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.