Summary
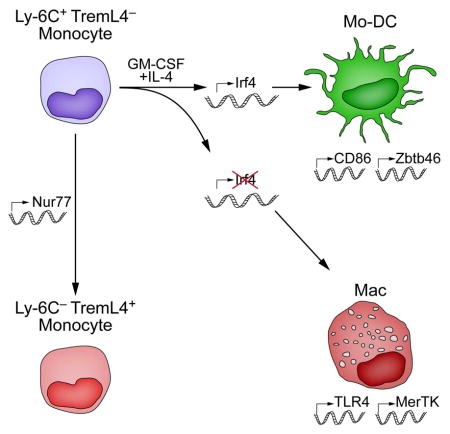
Both classical DCs (cDCs) and monocyte-derived DCs (Mo-DCs) are capable of cross-priming CD8+ T cells in response to cell-associated antigens. We found that Ly-6ChiTREML4− monocytes can differentiate into Zbtb46+ Mo-DCs in response to GM-CSF and IL-4, but that Ly-6ChiTREML4+ monocytes were committed to differentiate into Ly-6CloTREML4+ monocytes. Differentiation of Zbtb46+ Mo-DCs capable of efficient cross-priming required both GM-CSF and IL-4, and was accompanied by induction of Batf3 and Irf4. However, monocytes require IRF4, but not BATF3, to differentiate into Zbtb46+ Mo-DCs capable of cross-priming CD8+ T cells. Instead, Irf4−/− monocytes differentiate into macrophages in response to GM-CSF and IL-4. Thus, cDCs and Mo-DCs require distinct transcriptional programs of differentiation in acquiring the capacity to prime CD8+ T cells. These differences may be of consideration in the use of therapeutic DC vaccines based on Mo-DCs.
In Brief
The transcriptional programs required for differentiation of cross-priming APCs from various lineages are unknown. Briseño et al. show that Mo-DCs use a distinct program than cDCs, requiring IRF4 but not Batf3. These differences may impact the design of vaccines based on Mo-DCs that would require efficient cross-priming of T cells.
Introduction
Cross-presentation functions in initiating cytolytic CD8+ T cell responses during viral infections (Joffre et al., 2012) and is mediated by classical dendritic cells (cDCs) derived from the common dendritic cell progenitor (Naik et al., 2007; Liu et al., 2007) and by monocyte-derived dendritic cells (Mo-DCs) (Nierkens et al., 2013). Efficient cross-presentation is carried out in vivo by a CD24+ cDC subset requiring IRF8 and BATF3 (Briseno et al., 2014; Satpathy et al., 2012b), but the transcriptional requirements for Mo-DCs are undefined. In mice, monocytes can produce DCs under inflammatory conditions in vivo (Auffray et al., 2009; Cheong et al., 2010) or upon ex vivo treatment with granulocyte-macrophage colony stimulating factor (GM-CSF) (Inaba et al., 1992; Inaba et al., 1993; Caux et al., 1992). Human monocytes treated ex vivo with GM-CSF and IL-4 also acquire DC characteristics (Sallusto and Lanzavecchia, 1994; Romani et al., 1994). Mo-DCs express CD11c and MHC-II (Leon et al., 2004) and the DC-specific transcription factors Zbtb46 and Mycl1 (Satpathy et al., 2012a; KC et al., 2014). However, monocytes differentiated with GM-CSF alone generate a heterogeneous population of CD11c+ cells (Helft et al., 2015), resembling either macrophages (GM-Macs, CD11b+MHC-IIlo) or DCs (GM-DCs, CD11b+MHC-IIhi). GM-DCs cross-present soluble antigen more efficiently than GM-Macs (Helft et al., 2015).
Mo-DCs can promote TH1 and CD8+ T cell responses (Leon et al., 2007; Aldridge, Jr. et al., 2009; Ji et al., 2013), but differ in the antigen processing pathways they employ (Segura et al., 2009) and the phases of infection they are involved in compared to cDCs (Ballesteros-Tato et al., 2010). Mo-DCs react distinctly from cDCs in response to adjuvant (Langlet et al., 2012) and unlike cDCs, act independently of GM-CSF signaling in vivo during steady state and immunization (Greter et al., 2012). Human Mo-DCs generated ex vivo with GM-CSF and IL-4 can elicit CD8+ T cell responses against tumor antigens (Nestle et al., 1998; Holtl et al., 1999; Timmerman et al., 2002; Thurner et al., 1999) and subdominant neoantigens (Carreno et al., 2015), and have been used in cancer vaccines (Palucka and Banchereau, 2013; Carreno et al., 2015). Although CDPs have been suggested as sources of DC vaccines (Guilliams and Malissen, 2015), the abundance and practical value of monocytes motivates understanding their cross-presentation capacity for use in future vaccine design.
How IL-4 regulates Mo-DC differentiation is still unclear. In macrophages, IL-4 signaling induces M2 polarization (El Chartouni et al., 2010) by Stat6 activation and induction of Jumonji domain-containing-3 (Jmjd3). JMJD3 functions as a demethylase of histone 3 lysine 27 (Ishii et al., 2009) and promotes M2 polarization by regulating IRF4 expression (Satoh et al., 2010). Loss of either JMJD3 or IRF4 impairs expression of M2 macrophage genes, such as Arg1, IL13, and Fizz1 (Satoh et al., 2010). Whether similar actions of IL-4 and IRF4 occur during Mo-DC differentiation has not been examined. In CD11b+ cDCs, IRF4 is required for migration (Bajana et al., 2012), survival in mucosal tissues (Schlitzer et al., 2013; Persson et al., 2013), and capacity to induce TH17 and TH2 responses (Gao et al., 2013; Williams et al., 2013; Schlitzer et al., 2013; Persson et al., 2013). Human Mo-DCs induce IRF4 in response to GM-CSF and IL-4 (Lehtonen et al., 2005), but its function there is undefined. In this study, we compared the transcriptional programs between cDCs and Mo-DCs for their ability to prime T cells in response to cell-associated antigens, finding that Mo-DCs do not require IRF8 and BATF3 like cDCs do, but instead require IRF4.
Results
IL-4 is required for optimal cross-priming by GM-CSF induced Mo-DCs
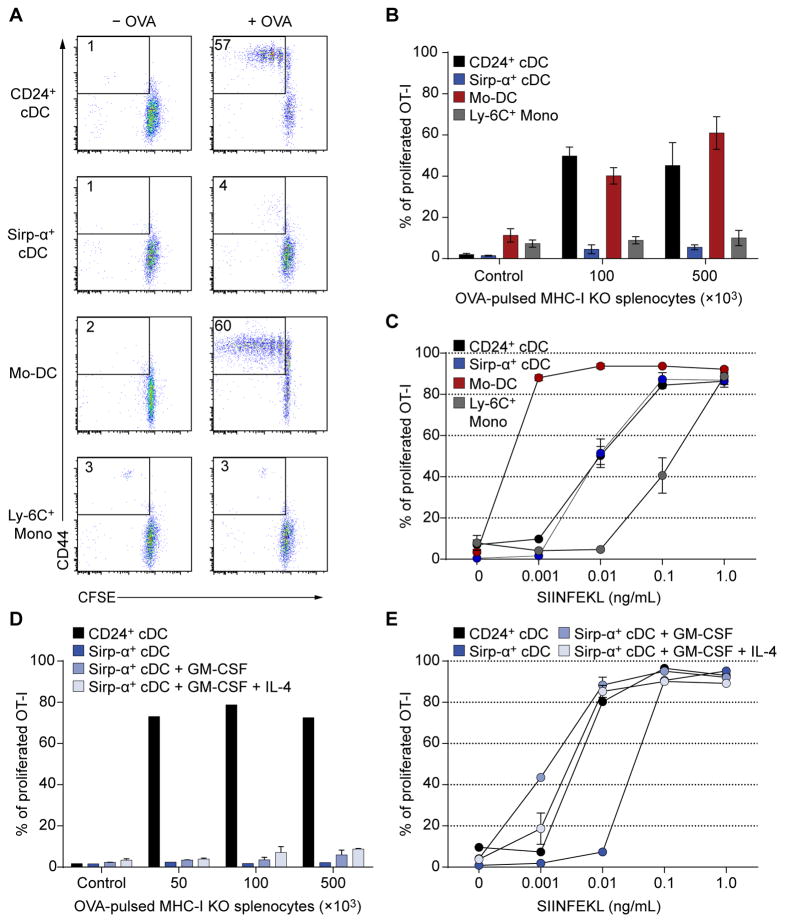
Splenic CD24+ cDCs, but not Sirp-α+ cDCs, efficiently cross-primed T cells with cell-associated antigen (Fig. 1A, B), as reported (den Haan et al., 2000; Becker et al., 2014). As control, both cDC subsets presented SIINFEKL peptide (Fig. 1C). Mo-DCs generated with GM-CSF and IL-4 efficiently activated T cells in response to cell-associated antigen and SIINFEKL peptide, in contrast to Ly-6C+ monocytes (Fig. 1A–C), as reported (Cheong et al., 2010). Unlike Mo-DCs, sorted splenic Sirp-α+ DCs cultured in GM-CSF with or without IL-4 did not cross-prime T cells to cell-associated antigen (Fig. 1D), but presented SIINFEKL peptide (Fig. 1E). Thus, Mo-DCs, but not Sirp-α+ cDCs, are able to cross-prime T cells to cell-associated antigens.
Figure 1. Mo-DCs, but not Sirp-α+ cDCs, cross-present cell-associated antigen as efficiently as CD24+ cDCs.
(A, B) Splenic CD24+ and Sirp-α+ cDCs, BM Ly-6Chi monocytes and Mo-DCs cultured in GM-CSF + IL-4 were purified by cell sorting. APCs were co-cultured with CFSE-labeled OT-1 cells and the indicated number of OVA-loaded γ-irradiated Kb−/−Db−/−β2m−/− (MHC-I TKO) splenocytes. OT-I cells were analyzed after three days by flow cytometry. (A) Representative flow cytometry analysis of OT-I proliferation after cross-presentation assay. (B) Summary of OT-I proliferation after cell-associated cross-presentation assay determined as the percentage of CD44+ OT-I cells that had at least one CFSE dilution. n=3 biological replicates per group; control: 1×105 γ-irradiated MHC-I TKO splenocytes without OVA. (C) SIINFEKL peptide presentation by sorted splenic CD24+ and Sirp-α+ cDCs, BM Ly-6Chi monocytes and Mo-DCs. OT-I cell proliferation was analyzed by flow cytometry three days after culture. n=2 biological replicates per group. (D, E) Sorted splenic Sirp-α+ cDCs were cultured in GM-CSF with or without IL-4 for two days and tested for cross-presentation (D) as in A and for SIINFEKL peptide presentation (E) as in C. Sorted splenic CD24+ and Sirp-α+ cDCs without treatment were used as positive and negative controls respectively; n=2 biological replicates per group.
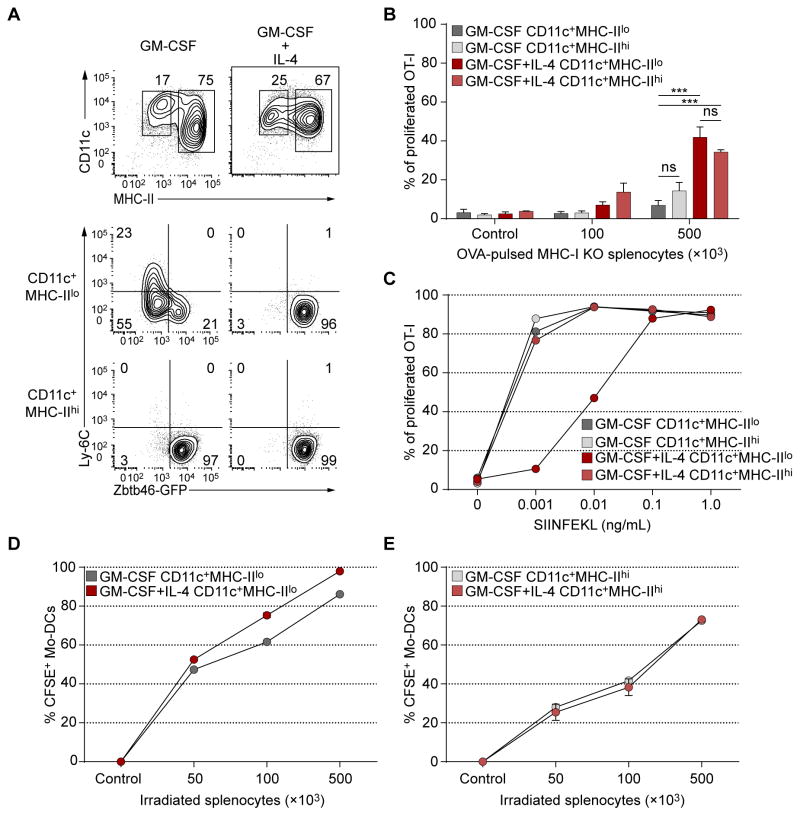
Monocytes cultured in GM-CSF produce a heterogeneous population of MHC-IIhi GM-DCs and MHC-IIlo GM-Macs in (Fig. 2A), in agreement with a recent study (Helft et al., 2015). MHC-IIhi GM-DCs expressed Zbtb46gfp (Satpathy et al., 2012a), but MHC-IIlo GM-Macs did not (Fig. 2A), consistent with specific Zbtb46 expression in cDCs but not macrophages (Meredith et al., 2012; Satpathy et al., 2012a). Addition of IL-4 with GM-CSF induced uniform Zbtb46gfp expression in both MHC-IIhi and MHC-IIlo populations of CD11c+ cells (Fig. 2A). Both MHC-IIhi and MHC-IIlo cells that developed in GM-CSF alone were weak cross-primers of cell-associated antigen, but addition of IL-4 significantly enhanced their activity (Fig. 2B) to levels similar to CD24+ cDCs (Fig. 1A, B). All populations presented SIINFEKL peptide (Fig. 2C). MHC-IIlo (Fig. 2D) and MHC-IIhi (Fig. 2E) Mo-DCs differentiated with GM-CSF alone or with IL-4 showed similar uptake of apoptotic cells. Thus, IL-4 signaling during GM-CSF-induced monocyte differentiation induces Zbtb46 expression in MHC-IIlo cells and increases cross-priming in both MHC-IIhi and MHC-IIlo cell populations.
Figure 2. Mo-DCs require IL-4 treatment during differentiation for optimal cross-priming.
(A) Ly-6Chi BM monocytes from Zbtb46gfp/+ mice were sorted and cultured in GM-CSF with or without IL-4 for 4 days, and analyzed by flow cytometry for expression of Zbtb46-GFP. Data are representative of three independent experiments. (B) WT Mo-DCs were generated as in A. CD11c+ Mo-DCs were then sorted as MHC-II− or MHC-II+ and co-cultured with CFSE-labeled OT-I cells and OVA-loaded γ-irradiated MHC-I TKO splenocytes. OT-I proliferation was analyzed by flow cytometry after three days. Data are pooled from three independent experiments, with at least 4 biological replicates per group. Two-way analysis of variance (ANOVA) with Sidak’s multiple comparison test; n.s. not significant; ***P<0.001. (C) SIINFEKL peptide presentation by Mo-DCs to CFSE labelled OT-I cells. OT-I proliferation was analyzed on the third day as in B. n=2 biological replicates per group. (D, E) Mo-DCs were sorted as in B, and co-cultured with γ-irradiated CFSE labelled CD45.1+ splenocytes for 16 hours. Uptake of apoptotic cells was determined as the percentage of CD45.2+CD45.1−CD11c+ Mo-DCs that were CFSE+. n=2 biological replicates per group.
Expression of TREML4 and NUR77 identifies monocytes lacking Mo-DC potential
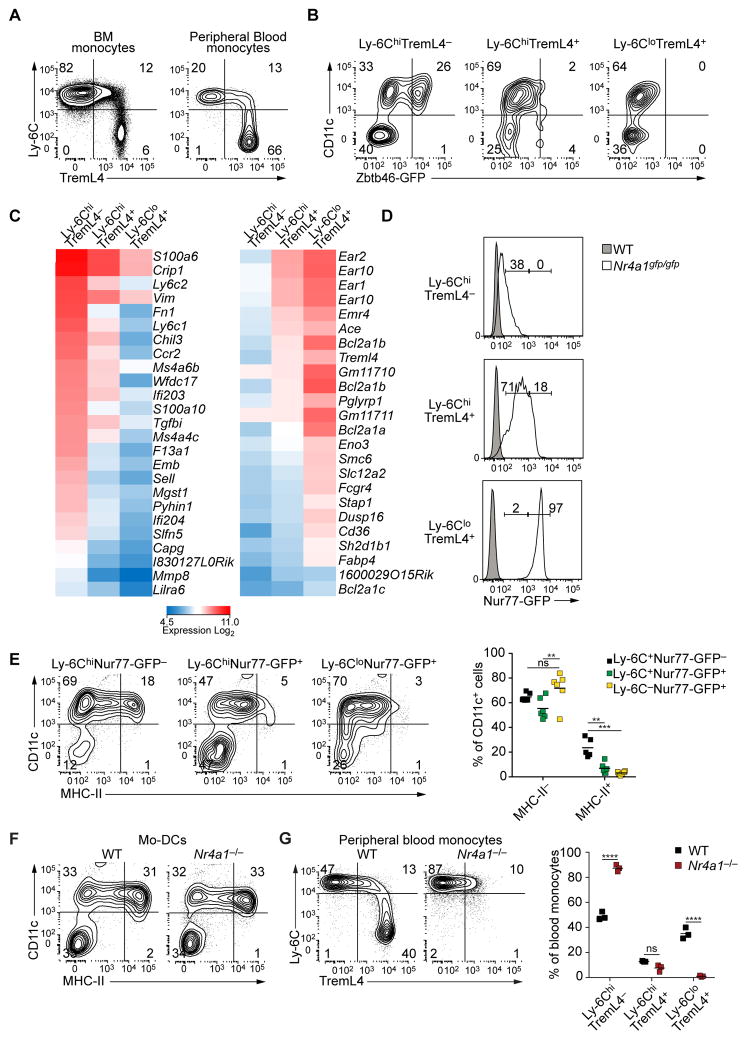
TREML4, a triggering receptor family member expressed on myeloid cells-like (Ford and McVicar, 2009), is induced during heme-mediated differentiation of macrophages from monocytes and BM progenitors (Haldar et al., 2014). TREML4 is expressed on CD24+ cDCs, monocytes (Hemmi et al., 2012) and macrophages, where it regulates TLR7 signaling amplification (Ramirez-Ortiz et al., 2015). Ly-6Chi monocytes were heterogeneous for TREML4 expression, but Ly-6Clo monocytes were uniformly TREML4 positive (Fig. 3A). Only LY-6Chi TREML4− monocytes were able to induce Zbtb46gfp expression in response to GM-CSF and IL-4, whereas Ly-6Chi TREML4+ monocytes and Ly-6Clo TREML4+ monocytes could not (Fig. 3B). Thus, TREML4 may mark the commitment of monocytes to the Ly-6Clo monocyte and macrophage lineages. Gene expression profiling suggested that Ly-6Chi TREML4+ monocytes were an intermediate stage of differentiation between Ly-6Chi TREML4− and Ly-6Clo monocytes (Fig. 3C). In Ly-6Chi TREML4− monocytes, expression of Ccr2 was 3-fold higher and 10-fold higher compared to Ly-6C+ TREML4+ and Ly-6Clo TREML4+ monocytes, respectively, while Treml4 expression was about 4-fold higher in Ly-6Chi TREML4+ monocytes and 6-fold higher in Ly-6Clo TREML4+ monocytes relative to Ly-6Chi TREML4− monocytes (Fig. 3C).
Figure 3. TREML4 identifies a subset of Ly-6Chi monocytes committed to macrophage-lineage differentiation.
(A) Flow cytometry of bone marrow (BM) and peripheral blood (PB) cells from WT mice. BM monocytes were gated as Ter-119−B220−Ly-6G−CD117−CD135−CD11c−MHC-II−CD115+CD11b+ live cells. PB monocytes were gated as CD45.2+B220−Ter-119−MHC-II−Ly-6G−CD115+CD11b+ live cells. Data is representative of three independent experiments. (B) Ly-6ChiTREML4−, Ly-6ChiTREML4+ and Ly-6CloTREML4+ monocytes were sorted from PB of Zbtb46gfp/+ mice, cultured in GM-CSF and IL-4 for 3 days and analyzed by flow cytometry for Zbtb46-GFP expression. Data are representative of three independent experiments. (C) Gene expression microarray analysis of sorted Ly-6ChiTREML4−, Ly-6ChiTREML4+, Ly-6CloTREML4+ PB monocytes. Shown are genes that were at least 3-fold different between Ly-6ChiTREML4− and Ly-6CloTREML4+ monocytes. (D) Flow cytometry analysis of Nur77-GFP expression in the indicated monocyte populations from peripheral blood of Nr4a1gfp/+ mice. Monocytes were pre-gated as in A. (E) Sorted Ly-6ChiNur77-GFP−, Ly-6ChiNur-77GFP+, Ly-6CloNur77-GFP+ peripheral blood monocytes from Nr4a1gfp/gfp mice were cultured as in B. Left panels show representative two-color histograms for CD11c and MHC-II expression. Right panel shows summarized data; each dot represents a biological replicate. n=6 biological replicates from two independent experiments. (F) Sorted Ly-6ChiTREML4− PB monocytes from Nr4a1−/− and WT littermate controls were cultured as in B and analyzed by flow cytometry. Data are representative of three independent experiments. (G) Flow cytometry analysis of PB from WT and Nr4a1−/− mice. Cells were gated as in A. Right panel shows summarized data; each dot represents a biological replicate; two-way analysis of variance (ANOVA) with Sidak’s multiple comparison test; n.s. not significant, **P<0.01; ***P<0.001; ****P<0.0001. See also Figure S1.
NUR77 (Nr4a1) is required for development of Ly-6Clo monocytes (Martinez-Gonzalez and Badimon, 2005; Hanna et al., 2011). Analysis of NUR77-GFP reporter mice (Moran et al., 2011) shows that TREML4 expression increased along with Nur77 (Fig. 3D). NUR77-GFP was absent in Ly-6Chi TREML4− monocytes, but expressed at intermediate levels in all Ly-6Chi TREML4+ monocytes and at high levels in all Ly-6Clo TREML4+ monocytes (Fig. 3D). We then tested the DC potential of monocytes expressing different levels of NUR77 (Fig. 3E). Ly-6C+ NUR77-GFP− monocytes differentiated into Mo-DCs in response to GM-CSF and IL-4 (Fig. 3E). In contrast, Ly-6C+ Nur77-GFP+ monocytes and Ly-6Clo NUR77-GFP+ monocytes were unable to differentiate into CD11c+MHC-II+ Mo-DCs (Fig. 3E). NUR77-deficient monocytes could not develop into Ly-6Clo monocytes, as reported (Moran et al., 2011), but could develop into Mo-DCs (Fig. 3F). Unsupervised analysis using SPADE (Qiu et al., 2011) reconstituted the successive steps of monocyte differentiation in vivo (Fig. S1, related to Figure 3). Thus, Ly-6Chi TREML4− Nur77-GFP− monocytes are the last stage of monocyte differentiation that retains potential for Mo-DC development.
IL-4 induces BATF3 and IRF4 during Mo-DC differentiation
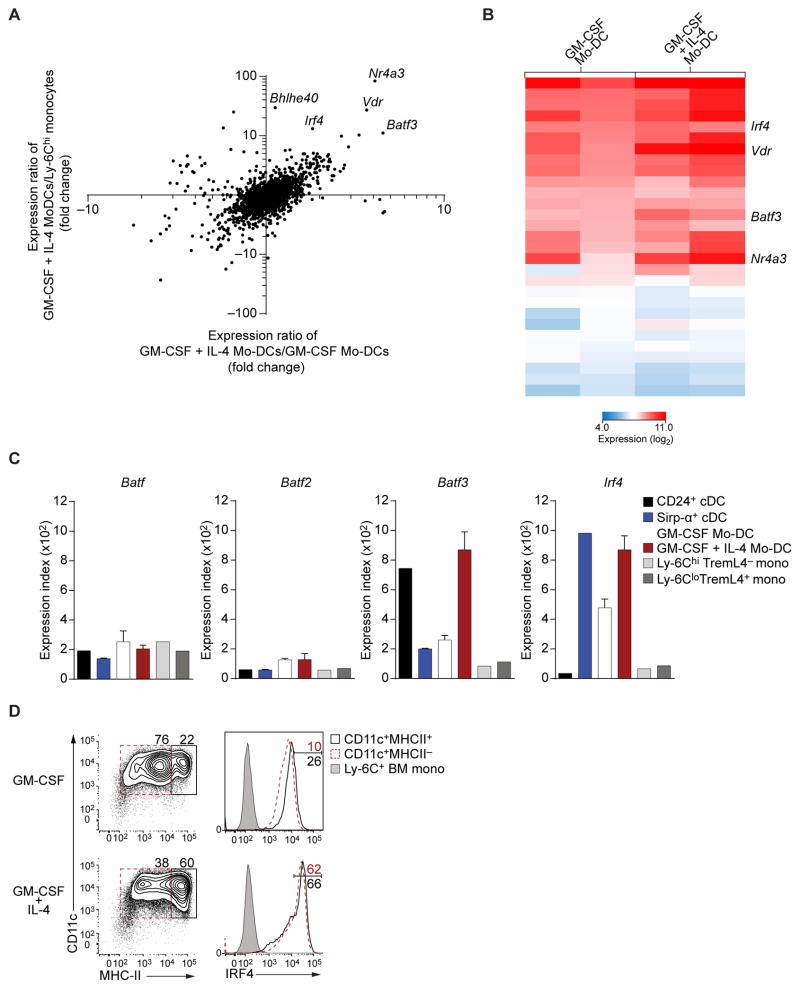
We examined gene expression microarrays of Ly-6Chi TREML4− and Ly-6Clo monocytes, Mo-DCs cultured with or without IL-4, and splenic CD24+ and Sirp-α+ cDCs. Several transcription factors were increased when Mo-DCs were differentiated with GM-CSF and IL-4, compared to monocytes or Mo-DCs cultured in GM-CSF alone (Fig. 4A). Specifically, Batf3 was induced by GM-CSF and IL-4 by 10-fold and 4-fold relative to monocytes and Mo-DCs cultured with GM-CSF alone, respectively. In addition, Irf4 was induced more than 25-fold relative to monocytes and 2-fold relative to Mo-DCs cultured with GM-CSF (Fig. 4A, B), as reported in human Mo-DCs (Lehtonen et al., 2005). Two other factors, Nr4a3 (DeYoung et al., 2003) and Vdr (Yoshizawa et al., 1997; Li et al., 1997), were induced, but have not been associated with antigen presentation. In contrast, Batf3 is required for the development of cDCs capable of cross-presentation (Hildner et al., 2008; Torti et al., 2011) and Irf4 was shown to be required for MHC-II expression in GM-DCs (Vander et al., 2014). Mo-DCs induced Batf3, but not Batf or Batf2, to levels equivalent to both splenic CD24+ and Sirp-α+ cDCs (Fig. 4C). Likewise, Mo-DCs expressed Irf4 to levels similar to Sirp-α+ cDCs (Fig. 4C). Also, IL-4 increased IRF4 expression in Mo-DCs (Fig. 4D). In summary, IL-4 induced both BATF3 and IRF4 during Mo-DC differentiation.
Figure 4. Mo-DCs induce expression of Batf3 and Irf4 in response to IL-4.
(A) Gene expression microarray analysis of Ly-6Chi monocytes and Mo-DCs differentiated with GM-CSF alone or GM-CSF and IL-4. Shown is the ratio of expression in Mo-DCs generated with GM-CSF and IL-4 versus that of Mo-DCs generated with GM-CSF alone (horizontal axis) plotted against the ratio of expression in Mo-DCs generated with GM-CSF and IL-4 versus that in monocytes (vertical axis) for all transcription factor-encoding genes. (B) Gene expression of transcription factors induced at least 2-fold in Mo-DCs cultured with IL-4 relative to Mo-DCs cultured with GM-CSF alone. Shown are biological replicates for each cell lineage. (C) Relative expression of Batf, Batf2, Batf3 and Irf4 from microarrays of the indicated cell type. (D) Representative intracellular flow cytometry analysis of sorted Ly-6ChiTREML4− BM monocytes cultured in GM-CSF with or without IL-4. Ly-6C+ BM monocytes are shown as control. Data is representative of three independent experiments.
Cross-priming by Mo-DCs is independent of BATF3
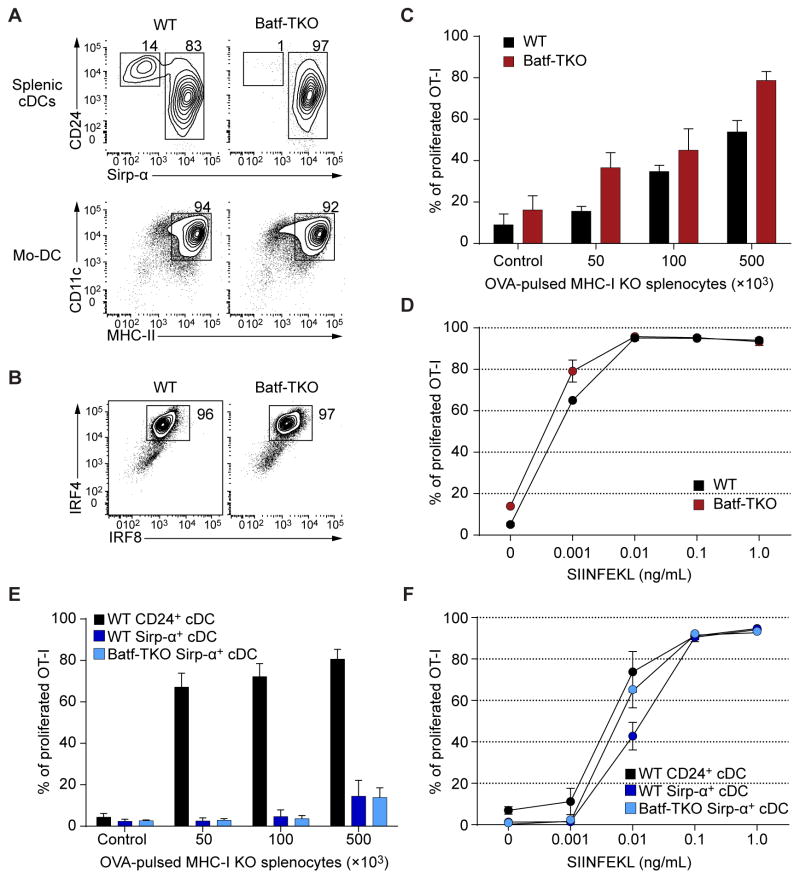
To examine Mo-DC differentiation and function, we used monocytes from Batf, Batf2 and Batf3 triple knockout mice (Batf-TKO), since Batf and Batf2 can compensate for Batf3 in CD24+ cDC development (Tussiwand et al., 2012). Mo-DCs developed normally from Batf-TKO monocytes (Fig, 5A), with normal expression of IRF4 and IRF8 (Fig. 5B). As reported (Tussiwand et al., 2012), Batf-TKO lacked splenic CD24+ cDCs but retained Sirp-α+ cDCs (Fig. 5A). We found no difference in cross-priming between WT and Batf-TKO Mo-DCs over a range of antigen concentrations or in presentation of SIINFEKL peptide (Fig. 5C, D). Splenic Batf-TKO Sirp-α+ DCs did not cross-prime but could present SIINFEKL peptide (Fig. 5E, F). Thus, development and cross-priming of Mo-DCs was independent of BATF3.
Figure 5. Mo-DCs do not require BATF3 for differentation into APCs capable of cross-priming.
(A) Flow cytometry analysis of splenocytes and Mo-DCs generated with GM-CSF and IL-4 from WT and Batf−/−Batf2−/−Batf3−/− (Batf-TKO) mice. Splenic cDCs are pre-gated as B220−CD11c+MHC-II+ cells. Mo-DCs are gated as Ly-6C− cells. Data are representative of two independent experiments. (B) Intracellular flow cytometry analysis for IRF4 and IRF8 in WT and Batf-TKO Mo-DCs. Data is representative of three independent experiments. (C) Cross-presentation of cell-associated antigen by WT and Batf-TKO Mo-DCs. Percent proliferation was determined as the percentage of CD44+ OT-I cells that had undergone at least one CFSE dilution. n=3 biological replicates per group; control: 1×105 γ-irradiated MHC-I TKO splenocytes without OVA. (D) SIINFEKL peptide presentation by WT and Batf-TKO Mo-DCs. OT-I proliferation was analyzed by flow cytometry as in C after three days of culture. n=2 biological replicates per group. (E) Cell-associated cross-presentation assay by Batf-TKO Sirp-α+ cDCs as in C.Splenic WT CD24+ and Sirp-α+ cDCs were used as controls. (F) SIINFEKL peptide presentation by WT CD24+ and Sirp-α+ cDCs and Batf-TKO Sirp-α+ cDCs as in D, n=2 biological replicates per group.
IRF4 is required for development of in vitro derived Mo-DCs but not for Sirp-α+ cDCs
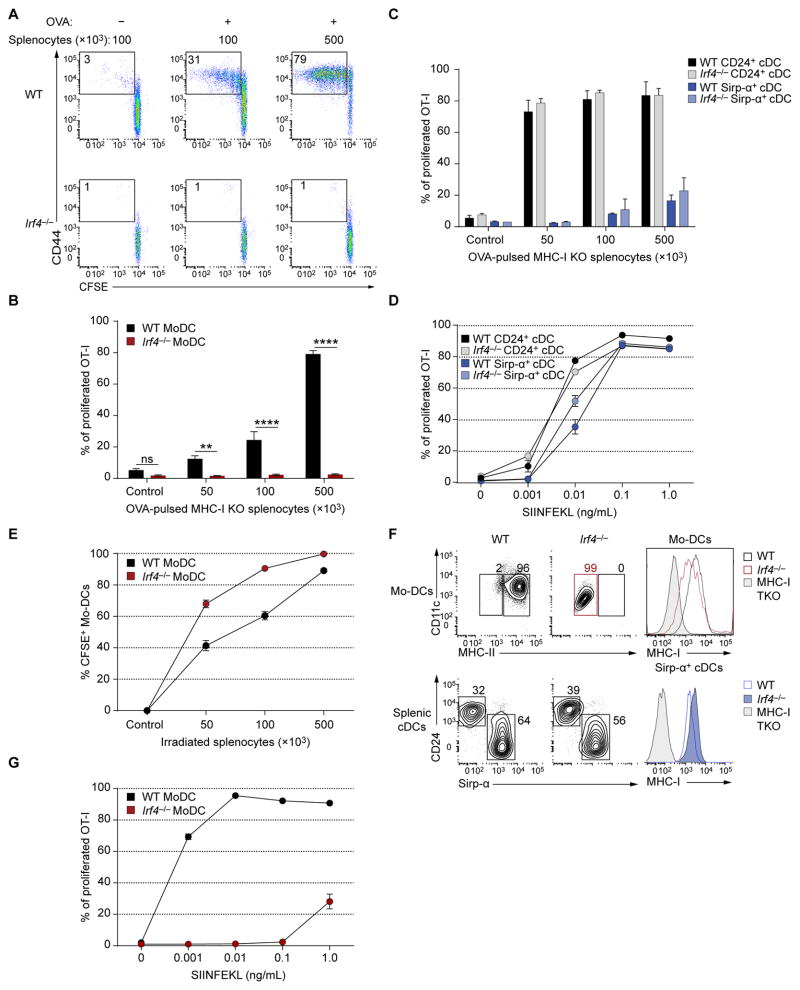
IRF4 is required for migration and homeostasis of Sirp-α+ cDCs (Bajana et al., 2012; Schlitzer et al., 2013; Persson et al., 2013) and promotes MHC-II expression by bone marrow-derived GM-DCs (Vander et al., 2014) but its role in priming of CD8+ T cells by Mo-DCs is unknown. Mo-DCs derived from Irf4−/− Ly-6Chi TREML4− monocytes were inactive for cross-priming (Fig. 6A, B). In contrast, Irf4−/− splenic CD24+ cDCs were as efficient as WT CD24+ DCs in cross-priming OT-I cells (Fig. 6C, D). Uptake of apoptotic cells was similar between WT and Irf4−/− Mo-DCs (Fig. 6E). Mo-DCs lacking IRF4 did not express MHC-II, as reported (Vander et al., 2014), but expressed normal MHC-I levels (Fig. 6F). However, they were unable to induce OT-I proliferation with SIINFEKL peptide (Fig. 6G).
Figure 6. Mo-DCs require IRF4 for cross-priming CD8+ T cells to cell-associated antigen.
(A, B) Cross-presentation of cell-associated antigen by WT and Irf4−/− Mo-DCs. OT-I cell proliferation was analyzed by flow cytometry three days after culture. (A) Representative two color histograms of OT-I cell proliferation after cross-presentation assay. (B) Summary of cell-associated cross-presentation by WT and Irf4−/− Mo-DCs. Percent proliferation of OT-I cells was determined as CD44+ OT-I cells that had undergone at least one CFSE dilution. Data are pooled from 6 biological replicates per group; control: 1×105 γ-irradiated MHC-I TKO splenocytes without OVA. (C) Cross-presentation of cell-associated antigen by sorted CD24+ and Sirp-α+ DCs from spleens of WT and Irf4−/− mice as in A; n=2 biological replicates per group. (D) SIINFEKL peptide presentation by splenic CD24+ and Sirp-α+ DCs from WT and Irf4−/− mice; n=2 biological replicates per group. (E) Apoptotic cell uptake after by WT and Irf4−/− Mo-DCs after 16 hours of culture. Cells were analyzed by flow cytometry, pre-gated as CD45.2+CD45.1−CD11c+; n=2 biological replicates per group. (F) Flow cytometry analysis of WT and Irf4−/− ex vivo derived Mo-DCs and splenic cDCs. Splenic cDCs were gated as B220−SiglecH−CD11c+MHC-II+. One-color histograms show MHC-I expression for the indicated populations. Cells from MHC-I TKO mice are shown as control. Data are representative of three biological replicates. (G) SIINFEKL peptide presentation assay by WT and Irf4−/− Mo-DCs. OT-I proliferation was measured as in A; n=3 biological replicates per group. Two-way analysis of variance (ANOVA) with Sidak’s multiple comparison test; n.s. not significant, **P<0.01, ****P<0.0001.
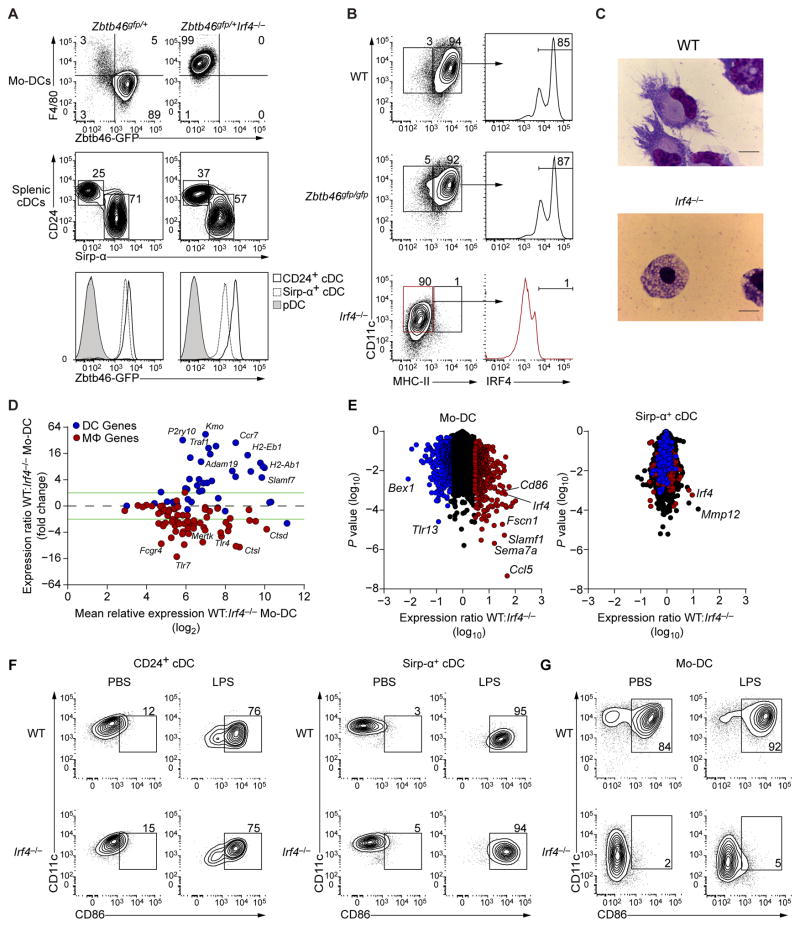
Unlike WT Mo-DCs, Irf4−/− monocytes failed to induce Zbtb46-GFP, and instead acquired expression of F4/80 following treatment with GM-CSF and IL-4 (Fig. 7A). IRF4 was not required for ZBTB46 expression in CD24+ or Sirp-α+ splenic cDCs (Fig. 7A). By contrast, Zbtb46-deficient Mo-DCs expressed normal levels of MHC-II and IRF4 (Fig. 7B). Consistent with the lack of MHC-II and Zbtb46 expression, the normal dendritic cell morphology of Mo-DCs was not seen in Irf4−/− Mo-DCs, which instead had the appearance of macrophages (Fig. 7C), suggesting that IRF4 may be required for induction of a broader DC transcriptional program in Mo-DCs beyond MHC-II gene expression.
Figure 7. Irf4−/− monocytes divert to macrophages upon GM-CSF and IL-4 signaling.
(A) Flow cytometry analysis of Zbtb46 expression in Mo-DCs generated with GM-CSF and IL-4 and splenic cDCs from Zbtb46gfp/+ and Zbtb46gfp/+Irf4−/− mice. Splenic cDCs were gated as B220−CD11c+MHC-II+, pDCs are shown as negative control. Data are representative of three independent experiments. (B) Flow cytometry analysis of WT, Zbtb46gfp/gfp and Irf4−/− Mo-DCs generated as in A. Expression of IRF4 in the indicated gates is shown in right panels. Data are representative of three biological replicates per group. (C) Microscopy of WT and Irf4−/− Mo-DCs stained with Wright-Giemsa stain. Scale bars: 10μm. (D) MA plot of the expression ratio of DC and macrophage (MΦ) specific genes from (Miller et al., 2012; Gautier et al., 2012) in WT and Irf4−/− Mo-DCs. (E) Gene expression analysis of Mo-DCs and splenic Sirp-α+ cDCs from WT and Irf4−/− mice. Colors indicate expression three fold higher (red) or lower (blue) in WT MoDCs than in Irf4−/− Mo-DCs. Welch’s t test, P value (vertical axis). (F, G) Flow cytometry analysis of sorted CD24+ and Sirp-α+ splenic DCs (F) and Mo-DCs (G) from WT or Irf4−/− mice treated with LPS for 16 hours. Data is representative of two independent experiments.
To determine the identity of cells originating from IRF4-deficient monocytes cultured with GM-CSF and IL-4, we performed microarray analysis of WT and Irf4−/− Mo-DCs cells (Figs. 7D, E). Consistent with the macrophage identity observed by flow cytometry and microscopy, Irf4−/− monocytes cultured in GM-CSF and IL-4 induced high expression of macrophage-specific genes such as Mertk, Tlr4 and Tlr7 (Gautier et al., 2012), and unlike WT Mo-DCs, failed to induce DC-associated genes such as Kmo, Traf1 and Slamf7 (Miller et al., 2012) (Fig. 7D). Since, IRF4 has been previously implicated in the development of splenic Sirp-α+ cDCs (Suzuki et al., 2004) we asked if IRF4 regulated a similar genetic program in both Mo-DCs and splenic Sirp-α+ cDCs. Comparison of the microarrays of WT and Irf4−/− Mo-DCs showed 747 genes to be differentially expressed by at least 3-fold between these two populations (Fig. 7E). However, only 49 of those targets were also at least 3-fold different between WT and Irf4−/− Sirp-α+ cDCs (Fig. 7E), suggesting Mo-DCs but not splenic Sirp-α+ cDCs require IRF4 for their development. We identified CD86 to be specifically downregulated in Mo-DCs, but not Sirp-α+ cDCs, lacking Irf4. We confirmed this result by assaying the expression of CD86 on WT and Irf4−/− splenic cDCs and Mo-DCs activated with LPS. Only MoDCs, and neither CD24+ nor Sirp-α+ cDCs, required IRF4 for CD86 expression (Fig. 7F, G). Altogether, these results indicate a specific requirement for IRF4 by monocytes for their differentiation into DC-like cells.
Discussion
Vaccines based on Mo-DCs can enhance immune responses against human melanoma (Carreno et al., 2015). Mo-DCs have been generated either in culture of GM-CSF alone or with IL-4 (Linette and Carreno, 2013). We show that IL-4 augments expression of Zbtb46 and Irf4, and that Irf4 is required for monocytes to differentiate into DCs. Mo-DCs can cross-prime CD8+ T cells for cell-associated antigen as efficiently as CD24+ cDCs. We show Mo-DCs rely on a distinct transcriptional program compared with cDCs in acquiring the ability to prime CD8+ T cells. Cross-presenting Mo-DCs require IRF4 but not BATF3, while cross-presenting cDCs require BATF3 but not IRF4.
Circulating Ly6Chi monocytes can differentiate either into Mo-Macs, Mo-DCs, or Ly6Clo ‘patrolling’ monocytes. Nur77 is required for differentiation of Ly6Chi monocytes into patrolling monocytes (Hanna et al., 2011), but not into Mo-DCs (Fig. 3F). We find that Ly6Chi monocytes that express Nur77 or TREML4, lack Mo-DC potential. In CD8+ T cells, Nur77 may inhibit IRF4 expression (Nowyhed et al., 2015), suggesting it may act similarly in Ly6Chi TREML4+ monocytes to repress IRF4 and thus Mo-DC development.
The biochemical basis for cross-presentation by different cells remains incompletely understood. Several proteins implicated in cross-presentation have been analyzed only in cells generated from BM cells treated with GM-CSF alone (Joffre et al., 2012; Segura and Amigorena, 2015). In this setting, NOX2 (Savina et al., 2006; Savina et al., 2009), Rac2 (Savina et al., 2009) and VAMP8 (Matheoud et al., 2013) were shown to regulate acidification of phagosomes in GM-DCs, suggesting that they act to preserving antigens from complete degradation. While NOX2 and Rac2 also regulate phagosomal acidification in CD8+ cDCs (Savina et al., 2009), only NOX2, but not Rac2, deficiency reduced CD8+ cDC cross-presentation of soluble antigen. Rab11a (Nair-Gupta et al., 2014), Rab3b (Zou et al., 2009) and Sec22b (Cebrian et al., 2011), which regulate vesicular trafficking, were shown to promote cross-presentation, but were studied using BM cultures treated with GM-CSF or in the DC2.4 cell line. In our studies, Mo-DCs generated with GM-CSF alone were relatively inefficient in cross-priming of cell-associated antigen compared with CD8+ cDCs and Mo-DCs generated with both GM-CSF and IL-4 (Fig. 2B).
Other known proteins such as ERAP1 (Firat et al., 2007) and IRAP (Segura et al., 2009; Saveanu et al., 2009) may be also be involved in cross-presentation. ERAP1 was required in vivo but not in GM-CSF BM-derived cells (Firat et al., 2007), while IRAP was required for both in vivo and in vitro cross-priming of CD8+ T cells to cell-associated antigen (Saveanu et al., 2009). IRAP was required for cross-presentation of soluble antigen only in inflammatory Mo-DCs generated in vivo, and not in CD24+ DCs (Segura et al., 2009). Alternately, unknown proteins may remain unidentified that differentially act in cross-presentation.
Experimental Procedures
Mice
Zbtb46gfp/+ mice (Satpathy et al., 2012a) were backcrossed to C57BL/6J for at least 8 generations. Batf−/−Batf2−/−Batf3−/−(Batf-TKO), Irf8−/− and Irf4−/− mice have been described mice (Tussiwand et al., 2012; Grajales-Reyes et al., 2015). The following mice were purchased from Jackson Laboratories: Nr4a1−/− (B6;129S2-Nr4a1tm1Jmi/J), OT-I (C57BL/6-Tg(TcraTcrb)1100Mjb/J), CD45.1+ (B6.SJL-Ptprca Pepcb/BoyJ). Nr4a1gfp/+ mice were a gift from Chyi-Song Hsieh and Kb−/−Db−/−β2m−/− mice (MHCI-TKO, Lybarger et al., 2003) were a gift from Herbert W. Virgin IV and Ted Hansen, Washington University in St. Louis. Mice, except Batf-TKO (129/SvEvTac), were maintained on the C57BL/6 background. All mice were housed in a specific pathogen-free animal facility following institutional guidelines with protocols approved by the Animal Studies Committee at Washington University in St. Louis. Experiments were performed with mice 8–12 weeks of age using sex-matched littermates.
Antibodies and flow cytometry
Cells were stained at 4°C in MACS buffer (PBS with 0.5% BSA and 2 mM EDTA) with CD16/32 Fc block (BD clone 2.4G2).
These antibodies were purchased from Becton Dickinson (BD): CD11b (M1/70); CD45.2 (104); CD135 (A2F10.1); MHC-II (M5/114.15.2); Ly-6C (AL-21); from eBioscience: CD4 (GK1.5); CD8α (53-6.7); CD11b (M1/70); CD45.1 (A20); CD44 (IM7); CD117 (2B8); CD115 (AFS98); CD11c (N418); CD24 (M1/69); CD172a (P84); Ly-6C (HK1.4); Ly-6A/E (D7); Ly-6G (IA8); Siglec-H (eBio440C); Ter-119 (Ter-119); CD105 (MJ7/18); Irf8 (V3GYWCH); CD45R (RA3-6B2); NK1.1 (PK136); Irf4 (3E4); 7AAD viability staining solution; from Tonbo Biosciences CD45.1 (A20); CD11c (N418); from BioLegend CD8α (53-6.7); CD45.2 (104); CD115 (ASF98); Ly-6G (IA8); TCR Vα2 (B20.1); TREML4 (16E5); from ThermoFisher Scientific: TCR Vα2 (B20.1), Live/dead Fixable Aqua Dead cell Stain kit. Carboxyfluorescein succinimidyl ester (CFSE) was purchased from Sigma.
Anti-Biotin and anti-B220 microbeads were purchased from Miltenyi. Cells were fixed and permeabilized for intracellular staining of IRF4 and IRF8 using the FoxP3/Transcription Buffer Set (eBioscience). Cells were sorted on a FACS Aria Fusion flow cytometers (BD) and with FlowJo software (Tree Star).
Isolation and culture of BM cells and splenic DCs
Femurs, pelvis and tibias were crushed using mortar and pestle in MACS buffer and filtered through a 70-μm strainer, and purified on Histopaque-119 gradient and depleted of Ly-6G and B220-expressing cells with biotinylated anti-Ly-6G and B220 antibodies and anti-biotin microbeads (Miltenyi). BM monocytes were identified as Lin−SiglecH−Ly-6G−MHCIIloCD11c−CD117−CD135−CD115+ and sorted as Ly-6ChiTREML4− or Ly-6CloTREML4+ for microarray analysis. Lin includes B220, CD105, NK1.1, and Ter-119. Blood monocytes were defined as Ter-119−CD45.2+MHC IIloLy-6G−CD115+ and segregated based on Ly-6C and TREML4 expression. Cells were sorted into Iscove’s modified Dulbecco’s medium + 10% FCS kept at 4°C. Spleens were minced and digested for 45 minutes at 37°C with stirring in 5 ml’s complete media with 250 μg/ml collagenase B (Roche) and 30 U/mL DNase I (Sigma-Aldrich). Red blood cells were lysed with ACK lysis buffer and splenocytes were passed through a 70-μm strainer. CD24+ cDCs were defined as B220−CD11c+MHC-II+CD24+CD172a−. Sirp-α+ cDCs were defined as B220−CD11c+MHCII+CD24−CD172a+. For Mo-DC differentiation, sorted Ly-6C+ TREML4− monocytes from BM or peripheral blood were cultured (0.25×105-0.5×105 cells/mL) at 37°C in complete media with GM-CSF and IL-4 (20ng/mL, Peprotech) for 3–4 days. Loosely adherent Mo-DCs were harvested by gentle pipetting. Sorted Sirp-α+ cDCs were cultured in 20ng/mL of GM-CSF and IL-4 for 48 hours. For induction of CD86, sorted CD24+ and Sirp-α+ cDCs, and Mo-DCs were cultured with LPS (1ng/mL) for 16 hours.
Microscopy
Cytospins of sorted Mo-DCs generated from GM-CSF and IL-4 culture of Ly-6C+TREML4− monocytes were stained with Wright-Giemsa stain using Hema 3 kit (Fisher Scientific). Images were acquired at room temperature with an Axioskop microscope (Objective: 100x, 1.25, oil) using an Axiocam ICc3 camera (Zeiss).
Gene expression microarray analysis
Total RNA was extracted from purified splenic cDCs, Mo-DCs, and monocytes from BM and peripheral blood using the RNAqueous-Micro Kit (Ambion). RNA was amplified using the Ovation Pico WTA Sytem (NuGEN) and hybridized to GeneChip Mouse Gene 1.0 ST microarrays (Affymetrix). Data was processed using robust multiarray average summarization and quartile normalization using ArrayStar software version 5 (DNASTAR). Sirp-α+ cDC expression values from WT and Irf4−/− mice were averaged from biological duplicates; all other expression values were from one biological sample.
Antigen presentation assays
Splenic OT-I cells were sorted as B220−CD11c−CD45.1+TCR-Vα2+CD4−CD8α+ to >95% purity, labeled with CFSE and plated at a density of 12.5×105 cells/mL. Splenocytes from MHC-I TKO mice were processed as described above. OVA loading of MHC-I TKO splenocytes has been described before (Carbone and Bevan, 1990). Splenocytes (2.5×107/mL) were incubated in hypertonic medium (RPMI 1640, 0.5M sucrose, 10% w/v poly-ethylyne glycol, 10 mM HEPES pH 7.2) with 5 mg OVA (Worthington) for 10 minutes at 37°C. Cells were diluted 10-fold with hypotonic media (60% FBS, 40% Sterile water) and incubated for 2 minutes at 37°C. Cells were washed with PBS and irradiated (13.5 Gy). Sorted splenic CD24+ and Sirp-α+ cDCs, and Mo-DCs (12.5×105 cells/mL) were co-cultured with CFSE labeled OT-I’s (12.5×105 cells/mL) and OVA-loaded MHC-I TKO cells (2.5×105-25.0×106 cells/mL). For peptide presentation, 2.5×104 APCs were cultured with SIINFEKL peptide (1.0×10−3-1.0 ng/mL) for 45 minutes in complete media at 37°C, washed twice, and cultured with 2.5×104 CFSE labeled OT-I cells. Cells were cultured at 37°C for 3 days and analyzed by flow cytometry. OT-I proliferation was determined as the percent of CD45.1+CD8α+TCR-Vα2+CD44+ cells that had undergone at least one CFSE dilution.
Phagocytosis assay
To prepare target cells, CD45.1+ splenocytes were harvested as described above, loaded with CFSE and γ-irradiated (13.5 Gy). Sorted CD45.2+ Mo-DCs (12.5×105 cells/mL) were co-cultured with CFSE labeled irradiated splenocytes (2.5×105-25.0×105 cells/200uL) for 16 hours at 37° C. After culture, Mo-DCs were washed, and stained for CD45.2, CD45.1, CD11c, Ly-6C, Aqua, and MHC-II. Percent phagocytosis was determined as the percentage of live (Aqua−) singlet Mo-DCs (CD45.1−CD45.2+Ly-6C−CD11c+) that were CFSE positive.
Statistical analysis
Error bars indicate standard error of mean. Statistical analyses were performed using two-way analysis of variance (ANOVA) with Sidak’s multiple comparison test unless otherwise specified. All statistical analyses were performed using Prism (GraphPad Software).
Supplementary Material
Highlights.
GM-CSF derived Mo-DCs require IL-4 to cross-present cell-associated antigen.
Monocytes expressing TremL4 lose potential to differentiate into DCs.
Monocytes require IRF4 but not Batf3 to become APCs that can prime CD8 T cells.
Acknowledgments
We thank C.S. Hsieh for Nr4a1gfp/+ mice; the Alvin J. Siteman Cancer Center at Washington University School of Medicine for use of the Center for Biomedical Informatics and Multiplex Gene Analysis Genechip Core Facility; and Ansuman T. Satpathy for helpful discussions. This work was supported by the Howard Hughes Medical Institute (K.M.M.), the US National Institutes of Health (F30DK108498 to V.D., 1F31CA189491-01 to G.E.G.-R., 1K08AI106953 to M.H.), the American Heart Association (12PRE12050419 to W.K.), and the Burroughs Wellcome Fund Career Award for Medical Scientists (M.H.)
Footnotes
Accession Numbers.
Author Contributions.
C.G.B., M.H., T.L.M. and K.M.M. designed the study; C.G.B., X.W. and W.K. performed microarray experiments with advice from A.I.; C.G.B., N.M.K. and D.J.T. performed cross-presentation assays; C.G.B. and M.H. performed experiments related to cell sorting, culture and flow cytometry with advice from V.D., G.E.G.-R., D.J.T. and P.B.; C.G.B., M.H. and K.M.M. wrote the manuscript with contributions from all authors.
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aldridge JR, Jr, Moseley CE, Boltz DA, Negovetich NJ, Reynolds C, Franks J, Brown SA, Doherty PC, Webster RG, Thomas PG. TNF/iNOS-producing dendritic cells are the necessary evil of lethal influenza virus infection. Proc Natl Acad Sci U S A. 2009;106:5306–5311. doi: 10.1073/pnas.0900655106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auffray C, Fogg DK, Narni-Mancinelli E, Senechal B, Trouillet C, Saederup N, Leemput J, Bigot K, Campisi L, Abitbol M, et al. CX3CR1+ CD115+ CD135+ common macrophage/DC precursors and the role of CX3CR1 in their response to inflammation. J Exp Med. 2009;206:595–606. doi: 10.1084/jem.20081385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajana S, Roach K, Turner S, Paul J, Kovats S. IRF4 promotes cutaneous dendritic cell migration to lymph nodes during homeostasis and inflammation. J Immunol. 2012;189:3368–3377. doi: 10.4049/jimmunol.1102613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros-Tato A, Leon B, Lund FE, Randall TD. Temporal changes in dendritic cell subsets, cross-priming and costimulation via CD70 control CD8(+) T cell responses to influenza. Nat Immunol. 2010 doi: 10.1038/ni.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker M, Guttler S, Bachem A, Hartung E, Mora A, Jakel A, Hutloff A, Henn V, Mages HW, Gurka S, Kroczek RA. Ontogenic, Phenotypic, and Functional Characterization of XCR1(+) Dendritic Cells Leads to a Consistent Classification of Intestinal Dendritic Cells Based on the Expression of XCR1 and SIRPalpha. Front Immunol. 2014;5:326. doi: 10.3389/fimmu.2014.00326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briseno CG, Murphy TL, Murphy KM. Complementary diversification of dendritic cells and innate lymphoid cells. Curr Opin Immunol. 2014;29C:69–78. doi: 10.1016/j.coi.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbone FR, Bevan MJ. Class I-restricted processing and presentation of exogenous cell-associated antigen in vivo. J Exp Med. 1990;171:377–387. doi: 10.1084/jem.171.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreno BM, Magrini V, Becker-Hapak M, Kaabinejadian S, Hundal J, Petti AA, Ly A, Lie WR, Hildebrand WH, Mardis ER, Linette GP. Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science. 2015;348:803–808. doi: 10.1126/science.aaa3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caux C, Dezutter-Dambuyant C, Schmitt D, Banchereau J. GM-CSF and TNF-alpha cooperate in the generation of dendritic Langerhans cells. Nature. 1992;360:258–261. doi: 10.1038/360258a0. [DOI] [PubMed] [Google Scholar]
- Cebrian I, Visentin G, Blanchard N, Jouve M, Bobard A, Moita C, Enninga J, Moita LF, Amigorena S, Savina A. Sec22b regulates phagosomal maturation and antigen crosspresentation by dendritic cells. Cell. 2011;147:1355–1368. doi: 10.1016/j.cell.2011.11.021. [DOI] [PubMed] [Google Scholar]
- Cheong C, Matos I, Choi JH, Dandamudi DB, Shrestha E, Longhi MP, Jeffrey KL, Anthony RM, Kluger C, Nchinda G, et al. Microbial stimulation fully differentiates monocytes to DC-SIGN/CD209(+) dendritic cells for immune T cell areas. Cell. 2010;143:416–429. doi: 10.1016/j.cell.2010.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Haan JM, Lehar SM, Bevan MJ. CD8(+) but not CD8(−) dendritic cells cross-prime cytotoxic T cells in vivo. J Exp Med. 2000;192:1685–1696. doi: 10.1084/jem.192.12.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung RA, Baker JC, Cado D, Winoto A. The orphan steroid receptor Nur77 family member Nor-1 is essential for early mouse embryogenesis. J Biol Chem. 2003;278:47104–47109. doi: 10.1074/jbc.M307496200. [DOI] [PubMed] [Google Scholar]
- El Chartouni C, Schwarzfischer L, Rehli M. Interleukin-4 induced interferon regulatory factor (Irf) 4 participates in the regulation of alternative macrophage priming. Immunobiology. 2010;215:821–825. doi: 10.1016/j.imbio.2010.05.031. [DOI] [PubMed] [Google Scholar]
- Firat E, Saveanu L, Aichele P, Staeheli P, Huai J, Gaedicke S, Nil A, Besin G, Kanzler B, Van Endert P, Niedermann G. The role of endoplasmic reticulum-associated aminopeptidase 1 in immunity to infection and in cross-presentation. J Immunol. 2007;178:2241–2248. doi: 10.4049/jimmunol.178.4.2241. [DOI] [PubMed] [Google Scholar]
- Ford JW, McVicar DW. TREM and TREM-like receptors in inflammation and disease. Curr Opin Immunol. 2009;21:38–46. doi: 10.1016/j.coi.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Nish SA, Jiang R, Hou L, Licona-Limon P, Weinstein JS, Zhao H, Medzhitov R. Control of T helper 2 responses by transcription factor IRF4-dependent dendritic cells. Immunity. 2013;39:722–732. doi: 10.1016/j.immuni.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grajales-Reyes GE, Iwata A, Albring J, Wu X, Tussiwand R, KCW, Kretzer NM, Briseno CG, Durai V, Bagadia P, et al. Batf3 maintains autoactivation of Irf8 for commitment of a CD8alpha(+) conventional DC clonogenic progenitor. Nat Immunol. 2015;16:708–717. doi: 10.1038/ni.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greter M, Helft J, Chow A, Hashimoto D, Mortha A, Agudo-Cantero J, Bogunovic M, Gautier EL, Miller J, Leboeuf M, et al. GM-CSF controls nonlymphoid tissue dendritic cell homeostasis but is dispensable for the differentiation of inflammatory dendritic cells. Immunity. 2012;36:1031–1046. doi: 10.1016/j.immuni.2012.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilliams M, Malissen B. A Death Notice for In-Vitro-Generated GM-CSF Dendritic Cells? Immunity. 2015;42:988–990. doi: 10.1016/j.immuni.2015.05.020. [DOI] [PubMed] [Google Scholar]
- Haldar M, Kohyama M, So AY, KCW, Wu X, Briseno CG, Satpathy AT, Kretzer NM, Arase H, Rajasekaran NS, et al. Heme-mediated SPI-C induction promotes monocyte differentiation into iron-recycling macrophages. Cell. 2014;156:1223–1234. doi: 10.1016/j.cell.2014.01.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna RN, Carlin LM, Hubbeling HG, Nackiewicz D, Green AM, Punt JA, Geissmann F, Hedrick CC. The transcription factor NR4A1 (Nur77) controls bone marrow differentiation and the survival of Ly6C- monocytes. Nat Immunol. 2011;12:778–785. doi: 10.1038/ni.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helft J, Bottcher J, Chakravarty P, Zelenay S, Huotari J, Schraml BU, Goubau D, Sousa Reis E. GM-CSF Mouse Bone Marrow Cultures Comprise a Heterogeneous Population of CD11c(+)MHCII(+) Macrophages and Dendritic Cells. Immunity. 2015;42:1197–1211. doi: 10.1016/j.immuni.2015.05.018. [DOI] [PubMed] [Google Scholar]
- Hemmi H, Zaidi N, Wang B, Matos I, Fiorese C, Lubkin A, Zbytnuik L, Suda K, Zhang K, Noda M, et al. TREML4, an Ig superfamily member, mediates presentation of several antigens to T cells in vivo, including protective immunity to HER2 protein. J Immunol. 2012;188:1147–1155. doi: 10.4049/jimmunol.1102541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildner K, Edelson BT, Purtha WE, Diamond M, Matsushita H, Kohyama M, Calderon B, Schraml BU, Unanue ER, Diamond MS, et al. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 2008;322:1097–1100. doi: 10.1126/science.1164206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtl L, Rieser C, Papesh C, Ramoner R, Herold M, Klocker H, Radmayr C, Stenzl A, Bartsch G, Thurnher M. Cellular and humoral immune responses in patients with metastatic renal cell carcinoma after vaccination with antigen pulsed dendritic cells. J Urol. 1999;161:777–782. [PubMed] [Google Scholar]
- Inaba K, Inaba M, Deguchi M, Hagi K, Yasumizu R, Ikehara S, Muramatsu S, Steinman RM. Granulocytes, macrophages, and dendritic cells arise from a common major histocompatibility complex class II-negative progenitor in mouse bone marrow. Proc Natl Acad Sci U S A. 1993;90:3038–3042. doi: 10.1073/pnas.90.7.3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba K, Inaba M, Romani N, Aya H, Deguchi M, Ikehara S, Muramatsu S, Steinman RM. Generation of large numbers of dendritic cells from mouse bone marrow cultures supplemented with granulocyte/macrophage colony-stimulating factor. J Exp Med. 1992;176:1693–1702. doi: 10.1084/jem.176.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii M, Wen H, Corsa CA, Liu T, Coelho AL, Allen RM, Carson WF, Cavassani KA, Li X, Lukacs NW, et al. Epigenetic regulation of the alternatively activated macrophage phenotype. Blood. 2009;114:3244–3254. doi: 10.1182/blood-2009-04-217620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Q, Castelli L, Goverman JM. MHC class I-restricted myelin epitopes are cross-presented by Tip-DCs that promote determinant spreading to CD8(+) T cells. Nat Immunol. 2013;14:254–261. doi: 10.1038/ni.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joffre OP, Segura E, Savina A, Amigorena S. Cross-presentation by dendritic cells. Nat Rev Immunol. 2012;12:557–569. doi: 10.1038/nri3254. [DOI] [PubMed] [Google Scholar]
- KCW, Satpathy AT, Rapaport AS, Briseno CG, Wu X, Albring JC, Russler-Germain EV, Kretzer NM, Durai V, Persaud SP, et al. L-Myc expression by dendritic cells is required for optimal T-cell priming. Nature. 2014;507:243–247. doi: 10.1038/nature12967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlet C, Tamoutounour S, Henri S, Luche H, Ardouin L, Gregoire C, Malissen B, Guilliams M. CD64 expression distinguishes monocyte-derived and conventional dendritic cells and reveals their distinct role during intramuscular immunization. J Immunol. 2012;188:1751–1760. doi: 10.4049/jimmunol.1102744. [DOI] [PubMed] [Google Scholar]
- Lehtonen A, Veckman V, Nikula T, Lahesmaa R, Kinnunen L, Matikainen S, Julkunen I. Differential expression of IFN regulatory factor 4 gene in human monocyte-derived dendritic cells and macrophages. J Immunol. 2005;175:6570–6579. doi: 10.4049/jimmunol.175.10.6570. [DOI] [PubMed] [Google Scholar]
- Leon B, Lopez-Bravo M, Ardavin C. Monocyte-derived dendritic cells formed at the infection site control the induction of protective T helper 1 responses against Leishmania. Immunity. 2007;26:519–531. doi: 10.1016/j.immuni.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Leon B, Martinez dH, Parrillas V, Vargas HH, Sanchez-Mateos P, Longo N, Lopez-Bravo M, Ardavin C. Dendritic cell differentiation potential of mouse monocytes: monocytes represent immediate precursors of CD8- and CD8+ splenic dendritic cells. Blood. 2004;103:2668–2676. doi: 10.1182/blood-2003-01-0286. [DOI] [PubMed] [Google Scholar]
- Li YC, Pirro AE, Amling M, Delling G, Baron R, Bronson R, Demay MB. Targeted ablation of the vitamin D receptor: an animal model of vitamin D-dependent rickets type II with alopecia. Proc Natl Acad Sci U S A. 1997;94:9831–9835. doi: 10.1073/pnas.94.18.9831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linette GP, Carreno BM. Dendritic cell-based vaccines: Shining the spotlight on signal 3. Oncoimmunology. 2013;2:e26512. doi: 10.4161/onci.26512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Waskow C, Liu X, Yao K, Hoh J, Nussenzweig M. Origin of dendritic cells in peripheral lymphoid organs of mice. Nat Immunol. 2007;8:578–583. doi: 10.1038/ni1462. [DOI] [PubMed] [Google Scholar]
- Lybarger L, Yu YY, Miley MJ, Fremont DH, Myers N, Primeau T, Truscott SM, Connolly JM, Hansen TH. Enhanced immune presentation of a single-chain major histocompatibility complex class I molecule engineered to optimize linkage of a C-terminally extended peptide. J Biol Chem. 2003;278:27105–27111. doi: 10.1074/jbc.M303716200. [DOI] [PubMed] [Google Scholar]
- Martinez-Gonzalez J, Badimon L. The NR4A subfamily of nuclear receptors: new early genes regulated by growth factors in vascular cells. Cardiovasc Res. 2005;65:609–618. doi: 10.1016/j.cardiores.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Matheoud D, Moradin N, Bellemare-Pelletier A, Shio MT, Hong WJ, Olivier M, Gagnon E, Desjardins M, Descoteaux A. Leishmania evades host immunity by inhibiting antigen cross-presentation through direct cleavage of the SNARE VAMP8. Cell Host Microbe. 2013;14:15–25. doi: 10.1016/j.chom.2013.06.003. [DOI] [PubMed] [Google Scholar]
- Meredith MM, Liu K, Darrasse-Jeze G, Kamphorst AO, Schreiber HA, Guermonprez P, Idoyaga J, Cheong C, Yao KH, Niec RE, Nussenzweig MC. Expression of the zinc finger transcription factor zDC (Zbtb46, Btbd4) defines the classical dendritic cell lineage. J Exp Med. 2012a;209:1153–1165. doi: 10.1084/jem.20112675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Brown BD, Shay T, Gautier EL, Jojic V, Cohain A, Pandey G, Leboeuf M, Elpek KG, Helft J, et al. Deciphering the transcriptional network of the dendritic cell lineage. Nat Immunol. 2012;13:888–899. doi: 10.1038/ni.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran AE, Holzapfel KL, Xing Y, Cunningham NR, Maltzman JS, Punt J, Hogquist KA. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med. 2011;208:1279–1289. doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik SH, Sathe P, Park HY, Metcalf D, Proietto AI, Dakic A, Carotta S, O’Keeffe M, Bahlo M, Papenfuss A, et al. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat Immunol. 2007;8:1217–1226. doi: 10.1038/ni1522. [DOI] [PubMed] [Google Scholar]
- Nair-Gupta P, Baccarini A, Tung N, Seyffer F, Florey O, Huang Y, Banerjee M, Overholtzer M, Roche PA, Tampe R, et al. TLR signals induce phagosomal MHC-I delivery from the endosomal recycling compartment to allow cross-presentation. Cell. 2014;158:506–521. doi: 10.1016/j.cell.2014.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestle FO, Alijagic S, Gilliet M, Sun Y, Grabbe S, Dummer R, Burg G, Schadendorf D. Vaccination of melanoma patients with peptide- or tumor lysate-pulsed dendritic cells. Nat Med. 1998;4:328–332. doi: 10.1038/nm0398-328. [DOI] [PubMed] [Google Scholar]
- Nierkens S, Tel J, Janssen E, Adema GJ. Antigen cross-presentation by dendritic cell subsets: one general or all sergeants? Trends Immunol. 2013;34:361–370. doi: 10.1016/j.it.2013.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowyhed HN, Huynh TR, Thomas GD, Blatchley A, Hedrick CC. Cutting Edge: The Orphan Nuclear Receptor Nr4a1 Regulates CD8+ T Cell Expansion and Effector Function through Direct Repression of Irf4. J Immunol. 2015;195:3515–3519. doi: 10.4049/jimmunol.1403027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palucka K, Banchereau J. Dendritic-cell-based therapeutic cancer vaccines. Immunity. 2013;39:38–48. doi: 10.1016/j.immuni.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson EK, Uronen-Hansson H, Semmrich M, Rivollier A, Hagerbrand K, Marsal J, Gudjonsson S, Hakansson U, Reizis B, Kotarsky K, Agace WW. IRF4 Transcription-Factor-Dependent CD103(+)CD11b(+) Dendritic Cells Drive Mucosal T Helper 17 Cell Differentiation. Immunity. 2013;38:958–969. doi: 10.1016/j.immuni.2013.03.009. [DOI] [PubMed] [Google Scholar]
- Qiu P, Simonds EF, Bendall SC, Gibbs KD, Jr, Bruggner RV, Linderman MD, Sachs K, Nolan GP, Plevritis SK. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol. 2011;29:886–891. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Ortiz ZG, Prasad A, Griffith JW, Pendergraft WF, III, Cowley GS, Root DE, Tai M, Luster AD, El Khoury J, Hacohen N, Means TK. The receptor TREML4 amplifies TLR7-mediated signaling during antiviral responses and autoimmunity. Nat Immunol. 2015;16:495–504. doi: 10.1038/ni.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romani N, Gruner S, Brang D, Kampgen E, Lenz A, Trockenbacher B, Konwalinka G, Fritsch PO, Steinman RM, Schuler G. Proliferating dendritic cell progenitors in human blood. J Exp Med. 1994;180:83–93. doi: 10.1084/jem.180.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto F, Lanzavecchia A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J Exp Med. 1994;179:1109–1118. doi: 10.1084/jem.179.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Takeuchi O, Vandenbon A, Yasuda K, Tanaka Y, Kumagai Y, Miyake T, Matsushita K, Okazaki T, Saitoh T, et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol. 2010;11:936–944. doi: 10.1038/ni.1920. [DOI] [PubMed] [Google Scholar]
- Satpathy AT, KCW, Albring JC, Edelson BT, Kretzer NM, Bhattacharya D, Murphy TL, Murphy KM. Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J Exp Med. 2012a;209:1135–1152. doi: 10.1084/jem.20120030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpathy AT, Wu X, Albring JC, Murphy KM. Re(de)fining the dendritic cell lineage. Nat Immunol. 2012b;13:1145–1154. doi: 10.1038/ni.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saveanu L, Carroll O, Weimershaus M, Guermonprez P, Firat E, Lindo V, Greer F, Davoust J, Kratzer R, Keller SR, Niedermann G, Van Endert P. IRAP identifies an endosomal compartment required for MHC class I cross-presentation. Science. 2009;325:213–217. doi: 10.1126/science.1172845. [DOI] [PubMed] [Google Scholar]
- Savina A, Jancic C, Hugues S, Guermonprez P, Vargas P, Moura IC, Lennon-Dumenil AM, Seabra MC, Raposo G, Amigorena S. NOX2 controls phagosomal pH to regulate antigen processing during crosspresentation by dendritic cells. Cell. 2006;126:205–218. doi: 10.1016/j.cell.2006.05.035. [DOI] [PubMed] [Google Scholar]
- Savina A, Peres A, Cebrian I, Carmo N, Moita C, Hacohen N, Moita LF, Amigorena S. The small GTPase Rac2 controls phagosomal alkalinization and antigen crosspresentation selectively in CD8(+) dendritic cells. Immunity. 2009;30:544–555. doi: 10.1016/j.immuni.2009.01.013. [DOI] [PubMed] [Google Scholar]
- Schlitzer A, McGovern N, Teo P, Zelante T, Atarashi K, Low D, Ho AW, See P, Shin A, Wasan PS, et al. IRF4 Transcription Factor-Dependent CD11b(+) Dendritic Cells in Human and Mouse Control Mucosal IL-17 Cytokine Responses. Immunity. 2013;38:970–983. doi: 10.1016/j.immuni.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura E, Albiston AL, Wicks IP, Chai SY, Villadangos JA. Different cross-presentation pathways in steady-state and inflammatory dendritic cells. Proc Natl Acad Sci U S A. 2009;106:20377–20381. doi: 10.1073/pnas.0910295106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura E, Amigorena S. Cross-Presentation in Mouse and Human Dendritic Cells. Adv Immunol. 2015;127:1–31. doi: 10.1016/bs.ai.2015.03.002. [DOI] [PubMed] [Google Scholar]
- Suzuki S, Honma K, Matsuyama T, Suzuki K, Toriyama K, Akitoyo I, Yamamoto K, Suematsu T, Nakamura M, Yui K, Kumatori A. Critical roles of interferon regulatory factor 4 in CD11bhighCD8alpha- dendritic cell development. Proc Natl Acad Sci U S A. 2004;101:8981–8986. doi: 10.1073/pnas.0402139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurner B, Haendle I, Roder C, Dieckmann D, Keikavoussi P, Jonuleit H, Bender A, Maczek C, Schreiner D, von den DP, et al. Vaccination with mage-3A1 peptide-pulsed mature, monocyte-derived dendritic cells expands specific cytotoxic T cells and induces regression of some metastases in advanced stage IV melanoma. J Exp Med. 1999;190:1669–1678. doi: 10.1084/jem.190.11.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerman JM, Czerwinski DK, Davis TA, Hsu FJ, Benike C, Hao ZM, Taidi B, Rajapaksa R, Caspar CB, Okada CY, et al. Idiotype-pulsed dendritic cell vaccination for B-cell lymphoma: clinical and immune responses in 35 patients. Blood. 2002;99:1517–1526. doi: 10.1182/blood.v99.5.1517. [DOI] [PubMed] [Google Scholar]
- Torti N, Walton SM, Murphy KM, Oxenius A. Batf3 transcription factor-dependent DC subsets in murine CMV infection: differential impact on T-cell priming and memory inflation. Eur J Immunol. 2011;41:2612–2618. doi: 10.1002/eji.201041075. [DOI] [PubMed] [Google Scholar]
- Tussiwand R, Lee WL, Murphy TL, Mashayekhi M, Wumesh KC, Albring JC, Satpathy AT, Rotondo JA, Edelson BT, Kretzer NM, et al. Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature. 2012;490:502–507. doi: 10.1038/nature11531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander LB, Khan AA, Hackney JA, Agrawal S, Lesch J, Zhou M, Lee WP, Park S, Xu M, DeVoss J, et al. Transcriptional programming of dendritic cells for enhanced MHC class II antigen presentation. Nat Immunol. 2014;15:161–167. doi: 10.1038/ni.2795. [DOI] [PubMed] [Google Scholar]
- Williams JW, Tjota MY, Clay BS, Vander LB, Bandukwala HS, Hrusch CL, Decker DC, Blaine KM, Fixsen BR, Singh H, et al. Transcription factor IRF4 drives dendritic cells to promote Th2 differentiation. Nat Commun. 2013;4:2990. doi: 10.1038/ncomms3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizawa T, Handa Y, Uematsu Y, Takeda S, Sekine K, Yoshihara Y, Kawakami T, Arioka K, Sato H, Uchiyama Y, et al. Mice lacking the vitamin D receptor exhibit impaired bone formation, uterine hypoplasia and growth retardation after weaning. Nat Genet. 1997;16:391–396. doi: 10.1038/ng0897-391. [DOI] [PubMed] [Google Scholar]
- Zou L, Zhou J, Zhang J, Li J, Liu N, Chai L, Li N, Liu T, Li L, Xie Z, Liu H, Wan Y, Wu Y. The GTPase Rab3b/3c-positive recycling vesicles are involved in cross-presentation in dendritic cells. Proc Natl Acad Sci U S A. 2009;106:15801–15806. doi: 10.1073/pnas.0905684106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.