Abstract
Specificity proteins (SPs) and Krüppel-like factors (KLFs) are C2H2-type Zinc finger transcription factors that play essential roles in differentiation, development, proliferation and cell death. SP/KLF proteins, similarly to Wilms tumor protein 1 (WT1), Early Growth Response (EGR), Huckebein, and Klumpfuss, prefer to bind GC-rich sequences such as GC-box and CACCC-box (GT-box). We searched various genomes and transcriptomes of metazoans and single-cell holozoans for members of these families. Seven groups of KLFs (KLFA–G) and three groups of SPs (SPA–C) were identified in the three lineages of Bilateria (Deuterostomia, Ecdysozoa, and Lophotrochozoa). The last ancestor of jawed vertebrates was inferred to have at least 18 KLFs (group A: KLF1/2/4/17, group B: KLF3/8/12; group C: KLF5/5l; group D: KLF6/7; group E: KLF9/13/16; group F: KLF10/KLF11; group G: KLF15/15l) and 10 SPs (group A: SP1/2/3/4; group B: SP5/5l; group C: SP6/7/8/9), since they were found in both cartilaginous and boned fishes. Placental mammals have added KLF14 (group E) and KLF18 (group A), and lost KLF5l (KLF5-like) and KLF15l (KLF15-like). Multiple KLF members were found in basal metazoans (Ctenophora, Porifera, Placozoa, and Cnidaria). Ctenophora has the least number of KLFs and no SPs, which could be attributed to its proposed sister group relationship to other metazoans or gene loss. While SP, EGR and Klumpfuss were only detected in metazoans, KLF, WT1, and Huckebein are present in nonmetazoan holozoans. Of the seven metazoan KLF groups, only KLFG, represented by KLF15 in human, was found in nonmetazoans. In addition, two nonmetazoan groups of KLFs are present in Choanoflagellatea and Filasterea. WT1 could be evolutionarily the earliest among these GC/GT-box-binding families due to its sole presence in Ichthyosporea.
Keywords: C2H2 zinc fingers, SP/KLF, WT1, EGR, Huckebein, Klumpfuss
1. Introduction
Specificity proteins (SPs) and Krüppel-like factors (KLFs) constitute a large family of transcription factors (SP/KLF family) with C2H2-type (also called Krüppel-type) zinc fingers (Znfs) (Turner and Crossley 1999; Bieker 2001; Kaczynski et al. 2003). SP/KLF members have three conserved Znfs responsible for binding GC-rich DNA motifs such as GC- box (GGGCGG) and CACCC-box (GT-box). The N-terminal regions before the Znfs are highly variable among SP/KLF subgroups and contain short sequence motifs for protein-protein interactions and post-translation modifications. These motifs are essential for the transcriptional activator/repressor function, sub-cellular localization and protein degradation. SP1 was the first cloned mammalian transcription factor with preferences for GC-rich sequences (Kadonaga et al. 1987). The first mammalian KLF gene, KLF1, was found to be highly expressed in erythroid (red blood) cells and thus named EKLF (Erythroid Krüppel-Like Factor) (Pei and Grishin 2013). Later discovered mammalian KLF genes were often initially named based on the abundance of their expression in certain tissues, such as LKLF (Lung KLF) (Dang et al. 2000), but the numerical naming of KLF proteins by Human Gene Nomenclature Committee (HGNC) (White et al. 1997) gained popularity and dominancy in literature (e.g., LKLF is named KLF2). Human has 17 KLF genes (KLF1–17) and nine SP genes (SP1–9) (van Vliet et al. 2006; Schaeper et al. 2010). Recently a new putative KLF gene (or pseudogene), KLF18, was discovered in placental mammals as a chromosomal neighbor, and likely a product of gene duplication of KLF17 (Pei and Grishin 2013). Compared with KLFs, SPs possess a unique CxCPxC motif (Buttonhead (BTD) box) in the N-terminal region outside the Znfs (Suske et al. 2005). SP/KLF family proteins are functionally diverse with key roles in differentiation, development, proliferation and cell death and are involved in diseases such as various cancers (Black et al. 2001; McConnell and Yang 2010).
Several other transcription factors also bind GC-box or CACCC-box, e.g., Wilms tumor protein 1 (WT1) (Call et al. 1990; Gessler et al. 1990), Early Growth Response proteins (EGRs) (Gomez-Martin et al. 2010), Huckebein (Weigel et al. 1990) and Klumpfuss (Klein and Campos-Ortega 1997; Yang et al. 1997). Like SP/KLF, they also have conserved residues in zinc finger regions responsible for the similar DNA-binding preferences. Most experimental studies focused on members from vertebrate and invertebrate model organisms such as mouse and fruit fly. While computational surveys of some of these families have been conducted for certain genomes (Materna et al. 2006; Shimeld 2008; Chen et al. 2010; Seetharam et al. 2010), the evolutionary history of the SP/KLF family and other related GC/GT-box-binding Znf families (WT1, EGR, Huckebein, and Klumpfuss) have not been fully explored in metazoans and beyond. High-throughput sequencing technology generated genomes and transcriptomes for a variety of eukaryotes from diverse lineages. These rich datasets offer a unique opportunity to investigate the distribution of SP/KLF and other GC/GT-box-binding Znf families.
2. Materials and methods
2.1. Genome and transcriptome analysis
We used BLAST (Altschul et al. 1997) to perform sequence similarity searches of the SP/KLF, WT1, EGR, Huckebein, and Klumpfuss proteins in the nr database (queries: human KLF proteins KLF1–KLF17). The BLAST e-value cutoff was set 1e-20 (manually selected) so that the majority of C2H2 zinc fingers that do not belong to these GC/GT-box binding families (SP/KLF, WT1, EGR, Huckebein, and Klumpfuss) were filtered out. Hits to protein sequences in the following organisms were selected and manually examined for the DNA-binding sequence motifs in these families: placental mammals – human (Homo sapiens) and mouse (Mus musculus); marsupial – opossum (Monodelphis domestica); birds – Gallus gallus and Pseudopodoces humilis; reptiles – lizard (Anolis carolinensis) and turtle (Chrysemys picta bellii); amphibian – Xenopus tropicalis; boned fishes – coelacanth (Latimeria chalumnae), spotted gar (Lepisosteus oculatus) and zebrafish (Danio rerio); cartilaginous fish – elephant shark (Callorhinchus milii); amphioxus – Branchiostoma floridae; urochordate – Ciona intestinalis; hemichordate – Saccoglossus kowalevskii; echinoderm – Strongylocentrotus purpuratus; lophotrochozoans – Lottia gigantea and Capitella teleta; ecdysozoans – Daphnia pulex, Tribolium castaneum, Drosophila melanogaster, Caenorhabditis elegans and Trichinella spiralis; cnidarians – Nematostella vectensis and Hydra vulgaris; placozoan – Trichoplax adhaerens; porifera – Amphimedon queenslandica, ctenophores – Mnemiopsis leidyi and Pleurobrachia bachei; choanoflagellates – Monosiga brevicollisi, Monosiga ovata, and Salpingoeca rosetta; Filasterea – Capsaspora owczarzaki. TBLASTN against NCBI genome and EST databases (hits with e-values worse than 0.001 were considered as false positives; other options were default: gap costs: 11 (existence) and 1 (extension); scoring matrix: BLOSUM62) and BLAT against genome sequences in the UCSC genome browser (Karolchik et al. 2014) were used to identify potential missing protein records. The predicted protein set of Oscarella carmela (in the Porifera group) was obtained from http://www.compagen.org. We also surveyed assembled transcriptomes of 11 Ctenophores (Moroz et al. 2014). De novo assembly of transcriptomes by Trinity (Grabherr et al. 2011) were performed using data available at the NCBI SRA (Sequence Read Archive) database (Leinonen et al. 2011) for the following species: Filasterea - Ministeria vibrans; Ichthyosporea - Abeoforma whisleri, Amoebidium parasiticum, Creolimax fragrantissima, Pirum gemmata and Sphaeroforma arctica. The SP/KLF, WT1, EGR, Huckebein, and Klumpfuss proteins in the above organisms are available in supplementary materials.
2.2. Assignment of SP/KLF members and groups
For member assignment of SP/KLFs in vertebrates (KLF1, KLF2, KLF3, KLF4, KLF5, KLF5-like (KLF5l), KLF6, KLF7, KLF8, KLF9, KLF10, KLF11, KLF12, KLF13, KLF14, KLF15, KLF15-like (KLF15l), KLF16, KLF17, KLF18, SP1, SP2, SP3, SP4, SP5, SP5-like (SP5l), SP6, SP7, SP8, and SP9), we mainly rely on overall sequence similarity in Znf regions. Most of the orthologous relationships were supported by reciprocal best BLAST hits. Some SP/KLF members have undergone divergent evolution, and their membership assignments were made by considering gene synteny, conservation in key positions in Znf regions, and presence/absence of short sequence motifs in the N-terminal regions before the Znfs. Gene synteny has been shown to be helpful in establishment of KLF17s in nonmammailan vertebrates (van Vliet et al. 2006; Antin et al. 2010).
The seven groups of bilaterian KLFs (KLFA–KLFG) were established mainly based on sequence similarity to groups of vertebrate KLFs. For example, top vertebrate BLAST hits of the lancelet KLFA are KLF1/2/4/17 proteins. The seven groups were also supported by their universal presence in a number of nonvertebrate bilaterian species such as lancelet, sea urchin, acorn worm (three nonvertebrate deuterostomes), Capitella teleta (a lophotrochozoan), and Daphnia pulex (an ecdysozoan). Definition of three groups of bilaterian SPs were based on a previous study (Schaeper et al. 2010).
We assigned SP/KLF proteins in basal metazoans to the seven KLF groups and three SP groups defined in bilaterians by considering a number of factors, including their sequence similarities in the Znf regions and full-length proteins to bilaterian SP/KLFs, residue conservation in key positions in the Znf regions and the presence/absence of group-specific motifs in the regions N-terminal to the Znfs.
2.3. Phylogenetic analysis
The MOLPHY package (Adachi and Hasegawa 1996) was used for phylogenetic reconstruction from the zinc finger regions of these proteins. First, the ProtML (Maximum Likelihood Inference of Protein Phylogeny) program of the MOLPHY package was used to derive a distance matrix for the sequences (-fD option, with “f” denoting the use of amino acid equilibrium frequencies estimated from input data). The NJdist (Neighbor Joining Phylogeny from Distance Matrix) program of the MOLPHY package was applied to build an initial tree based on the distance matrix. The ProtML program was then used to optimize the topology and branch lengths of the tree by the local rearrangement search (-R option) with the default JTT amino acid substitution model (Jones et al. 1992) (-j option) and the use of data-derived amino acid equilibrium frequencies (-f option). The local estimates of bootstrap percentages were obtained by the RELL method (-R option in the ProtML program of MOLPHY) (Hasegawa et al. 1991).
3. Results and discussion
3.1. Seven groups of KLFs and three groups of SPs are present in Bilateria
Sequence similarity searches against various genomes of deuterostomes including vertebrates, Urochordata, Cephalochordata, Hemichordata and Echinodermata revealed seven groups of KLFs with corresponding members of KLF1/2/4/17/18, KLF3/8/12, KLF5, KLF6/7, KLF9/13/14/16, KLF10/11, and KLF15 in the human genome. We name these groups KLFA, KLFB, KLFC, KLFD, KLFE, KLFF, and KLFG, respectively. Multiple genes in each group were found in vertebrates (Table 1, described below). In contrast, a single member in each of the seven groups was discovered in the three representative genomes of invertebrate deuterostomes – Branchiostoma floridae (a cephalochordate) (Putnam et al. 2008), Saccoglossus kowalevskii (a hemichordate) and Strongylocentrotus purpuratus (an echinoderm) (Cameron et al. 2009) (Table 2). Ciona intestinalis (a urochordate) (Satou et al. 2008) has one copy of KLFA/B/C/D/F/G and could have lost KLFE.
Table 1.
Distribution of KLF, SP, WT1 and EGR in selected jawed vertebrates.
| Organism | Krüppel-like factor (KLF) groups | Specificity protein (SP) groups | WT1 | EGR | ||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | A | B | C | |||||||||||||||||||||||
| 1 | 2 | 4 | 17 | 18 | 3 | 8 | 12 | 5 | 5l | 6 | 7 | 9 | 13 | 14 | 16 | 10 | 11 | 15 | 15l | 1 | 2 | 3 | 4 | 5 | 5l | 6 | 7 | 8 | 9 | |||
| Hom.sap | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | - | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | - | 1 | 1 | 1 | 1 | 1 | - | 1 | 1 | 1 | 1 | 1 | 4 |
| Mus.mus | 1 | 1 | 1 | 1 | 4 | 1 | 1 | 1 | 1 | - | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | - | 1 | 1 | 1 | 1 | 1 | - | 1 | 1 | 1 | 1 | 1 | 4 |
| Mon.del | 1 | 1 | 1 | - | - | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | - | - | 1 | 1 | 1 | - | - | 1 | 1 | 1 | 1 | - | 1 | - | 1 | 1 | 1 | 4 |
| Gal.gal | - | 1 | 1 | 1 | - | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | - | - | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | - | - | - | 1 | 1 | 1 | 4 |
| Pse.hum | - | 1 | 1 | 1 | - | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | - | - | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | - | 1 | 1 | 1 | 1 | 1 | 4 |
| Ano.car | 1 | 1 | 1 | 1 | - | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | - | - | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | - | - | 1 | 4 |
| Chr.pic | - | 1 | 1 | 1 | - | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | - | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 4 |
| Xen.tro | 1 | 1 | 1 | 1 | - | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | - | - | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 4 |
| Lat.cha | - | 1 | 1 | 1 | - | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | - | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 4 |
| Dan.rer | 1 | 2 | 1 | 1 | - | 1 | 1 | 2 | 2 | 1 | 2 | 2 | 1 | 1 | - | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 2 | 6 |
| Lep.ocu | 1 | 1 | 1 | 1 | - | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | - | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 4 |
| Cal.mil | 1 | 1 | 1 | 1 | - | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | - | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 3 |
KLF and SP groups are shown as letters A–G and A–C in the header, respectively. The KLF proteins belonging to a group are denoted by numbers, with “l” following the number as abbreviation of “-like”, e.g., KLF17 in KLF group A is shown as 17 and KLF5-like (KLF5l) in group C is denoted as 5l. The SP proteins are denoted the same way, e.g, SP5l in SP group B is denoted as 5l. Organism name abbreviations are as follows: Hom.sap - Homo sapiens; Mus.mus - Mus musculus; Mon.dom - Monodelphis domestica; Gal.gal - Gallus gallus; Pse.hum - Pseudopodoces humilis; Ano.car - Anolis carolinensis; Chr.pic - Chrysemys picta bellii; Xen.tro - Xenopus tropicalis; Lat.cha - Latimeria chalumnae; Dan.rer - Danio rerio; Lep.ocu - Lepisosteus oculatus; Cal.mil - Callorhinchus milii.
Table 2.
Distribution of KLF, SP, WT1, EGR, Klumpfuss, and Huckebein in selected nonvertebrate organisms.
| Lineage| | Organism | KLF | SP | WT1 | EGR | Klum-pfuss | Huck-ebein | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | A | B | C | ||||||
| Deuterostomia | Cio.int | 1 | 1 | 1 | 1 | - | 1 | 1 | - | - | 1 | 1 | 1 | - | 1 | - | - |
| Bra.flo | 1 | 1 | 1 | 1 | 1 | 1 | 1 | - | - | 1 | 1 | 1 | 1 | 1 | - | 1 | |
| Str.pur | 1 | 1 | 1 | 1 | 1 | 1 | 1 | - | - | 1 | 1 | 1 | - | 1 | - | - | |
| Sac.kow | 1 | 1 | 1 | 1 | 1 | 1 | 1 | - | - | 1 | 1 | 1 | - | 1 | - | 1 | |
| Lophotrochozoa | Cap.tel | 1 | 1 | 1 | 1 | 1 | 1 | 1 | - | - | 1 | 1 | 3 | - | 1 | 1 | - |
| Lot.gig | 1 | 1 | 1 | 1 | 1 | - | 1 | - | - | 1 | 1 | 2 | - | 1 | 1 | 1 | |
| Ecdysozoa | Dap.pul | 6 | 1 | 1 | 1 | 1 | 1 | 1 | - | - | 1 | 1 | 1 | - | 1 | 1 | 1 |
| Tri.cas | - | 1 | 1 | 1 | 1 | 1 | 1 | - | - | 1 | 1 | 1 | - | 1 | 1 | 1 | |
| Dro.mel | - | 1 | 1 | 1 | - | 1 | 1 | - | - | 1 | 1 | 2 | - | 1 | 1 | 1 | |
| Cae.ele | 1 | 1 | 1 | - | - | - | - | - | - | 1 | 1 | 1 | - | 3 | 2 | - | |
| Tri.spi | 1 | 1 | 1 | - | - | - | - | - | - | 1 | 1 | 1 | - | 1 | 1 | - | |
| Cnidaria | Nem.vec | - | 2 | 1 | 1 | 1 | 1 | 1 | - | - | 2 | 1 | 1 | - | 1 | - | 1 |
| Hyd.vul | - | 2 | 1 | 1 | 1 | 1 | - | - | - | 1 | - | 1 | - | 1 | - | - | |
| Placozoa | Tri.adh | - | 1 | 1 | 1 | 1 | 1 | - | - | - | 1 | - | 1 | - | 1 | - | 1 |
| Porifera | Amp.que | - | 1 | 1 | 1 | 1 | 1 | 1 | - | - | 2 | - | - | 1 | 1 | - | - |
| Osc.car | - | 1 | 1 | 1 | 1 | 1 | - | - | - | 2 | 1 | - | 2 | 1 | - | - | |
| Ctenophora | Mne.lei | - | 1 | - | 1 | 1 | - | - | - | - | - | - | - | 1 | 1 | - | 1 |
| Ple.bac | - | 1 | - | 1 | 1 | - | - | - | - | - | - | - | 1 | 1 | - | 1 | |
| Choanoflagellatea | Mon.bre | - | - | - | - | - | - | 1 | - | - | - | - | - | - | - | - | - |
| Sal.ros | - | - | - | - | - | - | 1 | 1 | - | - | - | - | - | - | - | - | |
| Mon.ova | - | - | - | - | - | - | 1 | 2 | - | - | - | - | - | - | - | ||
| Filasterea | Cap.owc | - | - | - | - | - | - | 1 | - | 2 | - | - | - | 1 | - | - | 1 |
| Min.vib | - | - | - | - | - | - | - | - | 2 | - | - | - | - | - | - | 1 | |
| Ichthyosporea | Amo.par | - | - | - | - | - | - | - | - | - | - | - | - | 3 | - | - | - |
| Cre.fra | - | - | - | - | - | - | - | - | - | - | - | - | 1 | - | - | - | |
| Sph.arc | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | - | |
Organism name abbreviations are as follows: Cio.int - Ciona intestinalis; Bra.flo - Branchiostoma floridae; Str.pur - Strongylocentrotus purpuratus; Sac.kow - Saccoglossus kowalevskii; Cap.tel - Capitella teleta; Lot.gig - Lottia gigantea; Dap.pul - Daphnia pulex; Tri.cas - Tribolium castaneum; Dro.mel - Drosophila melanogaster; Cae.ele - Caenorhabditis elegans; Tri.spi - Trichinella spiralis; Nem.vec - Nematostella vectensis; Hyd.vul - Hydra vulgaris; Tri.adh - Trichoplax adhaerens; Amp.que - Amphimedon queenslandica; Osc.car - Oscarella carmela; Mne.lei - Mnemiopsis leidyi; Ple.bac - Pleurobrachia bachei; Mon.bre - Monosiga brevicollisi; Sal.ros - Salpingoeca rosetta; Mon.ova - Monosiga ovata; Cap.owc - Capsaspora owczarzaki; Min.vib - Ministeria vibrans; Amo.par - Amoebidium parasiticum; Cre.fra - Creolimax fragrantissima; Sph.arc - Sphaeroforma arctica.
Besides Deuterostomia, members of the seven KLF groups were also identified in species of the other two major lineages of Bilateria – Ecdysozoa and Lophotrochozoa (Table 2). The model organism Drosophila melanogaster of the Ecdysozoa group has only five KLF proteins (KLFB: CG42741, KLFC: dar1, KLFD: luna, KLFF: cabut, and KLFG: bteb2) and lacks KLFA and KLFE. The other surveyed insect species, Tribolium castaneum (Tribolium Genome Sequencing et al. 2008), has six KLFs (KLFB/C/D/E/F/G). The arthropod Daphnia pulex possesses members of all seven KLF groups and has multiple KLFA copies (Colbourne et al. 2011). These data suggest that all seven KLF groups were present in the last common ancestor of ecdysozoans. The two nematodes Caenorhabditis elegans (Consortium 1998) and Trichinella spiralis (Mitreva et al. 2011) of the Ecdysozoa group have a smaller set of KLFs, missing members from four KLF groups (KLFD/E/F/G). Analysis of genomes of two lophotrochozoans, Capitella teleta (Simakov et al. 2013) and Lottia gigantean (Simakov et al. 2013), suggests that all seven KLF groups are present in the common ancestor of lophotrochozoans. The presence of the seven KLF groups in all three major groups of Bilateria suggests that the last common ancestor of Bilateria was equipped with all of them. Three groups of SPs have been identified in metazoans, corresponding to SP1/2/3/4, SP5, SP6/7/8/9 in the human genome (Schaeper et al. 2010). We name them SPA, SPB, and SPC, respectively. All surveyed bilaterian organisms contain at least one copy of each of the three SP groups (Table 2).
3.2. Lineage-specific gene expansion and loss of KLFs and SPs in jawed vertebrates
While invertebrate deuterostomes often contain one gene for each of the seven KLF groups (Table 2), multiple members of each group are commonly found in jawed vertebrates (Table 1). The last common ancestor of jawed vertebrates appears to possess at least 18 KLFs, as they are present in both cartilaginous fish (elephant shark (Callorhinchus milii) (Venkatesh et al. 2014)) and boned fish (such as zebrafish (Danio rerio) (Howe et al. 2013) and spotted gar (Lepisosteus oculatus) (Amores et al. 2011)). These 18 KLF members are KLF1/2/4/17 (group A), KLF3/8/12 (group B), KLF5/5l (group C), KLF6/7 (group D), KLF9/13/16 (group E), KLF10/11 (group F), and KLF15/15l (group G). Two other KLF members, KLF14 of group E and KLF18 of group A (possibly a pseudogene), are only found in placental mammals and might have originated subsequently in evolution. KLF14 and KLF18 are likely derived from KLF16 (by reverse transcription) and KLF17 (by local gene duplication), respectively. Although the lack of expression data suggests that KLF18 could be a pseudogene, several active genes such as zfp352 and zfp353 in mouse and rat could have been derived from KLF18 through reverse transcription and represent recent expansion of the KLF family in the murine lineage (Pei and Grishin 2013). KLF5l (KLF5-like) of group C was lost in placental mammals, but is still present in the genome of opossum, a marsupial. KLF15l (KLF15-like) of group G appears to be lost in all mammals, but is present in all surveyed nonmammalian species (Table 1). Highly similar KLF gene pairs in zebrafish should be the result of a recent whole genome duplication event in the ancestor of teleost fish (Meyer and Schartl 1999; Howe et al. 2013). In contrast, single copies of the 18 KLF members were found in spotted gar, a boned fish that diverged before the teleost genome duplication. KLF1 and KLF16 appear to be more prone to gene loss compared to other KLF members (Table 1).
Ten members of specificity proteins were found in cartilaginous fish and boned fish, suggesting that they were present in the last common ancestor of jawed vertebrates. These SP proteins form three groups – SP1/2/3/4 (group A), SP5/5l (group B), and SP6/7/8/9 (group C). Among them, SP5l (SP5-like) appears to have been lost in mammals and birds (Table 1).
3.3. SP/KLF proteins in basal metazoan groups
Recent genome sequencing efforts have shed light on the evolution of four basal metazoan groups – Cnidaria, Placozoa, Porifera, and Ctenophora (Sullivan et al. 2006; Srivastava et al. 2008; Chapman et al. 2010; Srivastava et al. 2010; Nichols et al. 2012; Ryan et al. 2013; Moroz et al. 2014). Our analysis shows that genomes of these groups also contain multiple copies of KLF members (Table 2). Compared with Bilateria, these basal metazoan groups lack KLFA members, suggesting that KLFA could be an invention in Bilateria. Each of the two recently sequenced Ctenophora genomes, Mnemiopsis leidyi (Ryan et al. 2013) and Pleurobrachia bachei (Moroz et al. 2014), contains three KLF genes (Table 2). Phylogenetic studies based on both genomes indicate that Ctenophora is the sister group to all other metazoans (Ryan et al. 2013; Moroz et al. 2014). The repertoire of KLFs and SPs appears to be consistent with this hypothesis, as Ctenophora contains fewer SP/KLFs (3 KLF members from groups B, D, E and no SPs) compared to the other three basal metazoan groups (Table 2). Alternatively, the smaller set of SP/KLF proteins in Ctenophora could be a result of gene loss. To investigate this possibility, we searched 11 Ctenophora transcriptomes (Moroz et al. 2014) and did not find other groups of SP/KLF proteins with one exception, which is a putative KLFF sequence fragment deduced from the transcriptome of Euplokamis dunlapae. Phylogenetic studies suggested that E. dunlapae may be the basal lineage among these 11 Ctenophora species (sister group to the other 10 species) (Moroz et al. 2014). It is thus likely that KLFF is present in the ancestor of Ctenophora, but is secondarily lost in most of its lineages. The absence of KLFG in Ctenophora could be attributed to gene loss as well, since KLFG is present in nonmetazoan lineages such as choanoflagellates and Filasterea (described below), suggesting its origination before the appearance of metazoans. KLFG was also not detected in Hydra vulgaris (a cnidarian) (Chapman et al. 2010), Trichoplax adhaerens (a placozoan) (Srivastava et al. 2008), and Oscarella carmela (a sponge) (Nichols et al. 2012), but is present in Nematostella vectensis (a cnidarian) (Sullivan et al. 2006) and Amphimedon queenslandica (a sponge) (Srivastava et al. 2010).
We assigned SP/KLF members in basal metazoans to the seven KLF groups and three SP groups defined in bilaterians (see Materials and methods). KLFs of group B/C/D/E/F appear to be present in all surveyed species in Cnidaria, Placozoa, and Porifera (Table 2) according to our manual assignment. To study the relationship of SP/KLF proteins in basal metazoans and bilaterians, we conducted a phylogenetic reconstruction using the Znf regions of SP/KLF members in basal metazoan groups and three bilaterians (B. floridae, S. kowalevskii, and S. purpuratus), with WT1 proteins in these organisms serving as the outgroup (WT1 proteins are discussed below) (supplementary Figure S1). KLFE/F and SPA/B/C appear to form a clade, within which the SPA/B/C subgroup received high support. Both the KLFG group and the WT1 out group received moderate support. A clade containing KLFA/B/C/D members received high support (supplementary Figure S1). However, KLFB, KLFC, and KLFD individually do not form monophyletic groups. For example, while KLFDs from Cnidarians (N. vectensis and H. vulgaris) and the placozoan (T. adhaerens) are grouped together with bilaterian KLFDs in the phylogenetic reconstruction, the two Ctenophora KLFDs are placed inside a group with mainly KLFB proteins (supplementary Figure S1). Using Ctenophora KLFDs as queries, the top BLAST hits to bilaterian SP/KLFs are mostly KLFD and KLFC proteins, instead of KLFB proteins. Ctenophora KLFDs possess the CxHCDRC motif in the third Znf that is conserved in most of bilaterian KLFDs, while the H and third C in this motif are not conserved in KLFBs (Figure 1(A) and supplementary Figure S2). Our assignments of Ctenophora KLDs were also based in part on motifs in the N-terminal regions (data not shown). The inconsistencies between the tree and our group assignment could be caused by the imprecision of our assignment or the limitations in tree reconstruction such as the limited number of positions (less than 100) for tree building and long-branch attraction artifacts of divergent sequences.
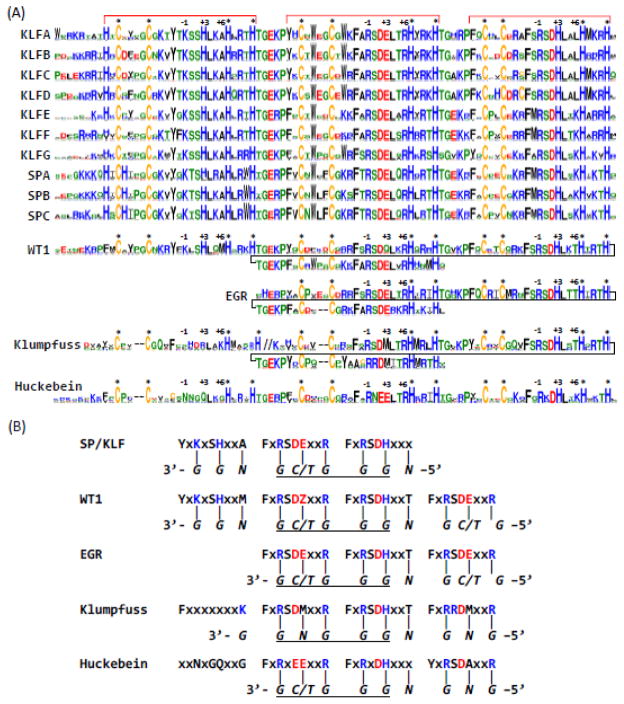
Figure 1. Sequence logos and DNA-binding preferences of Znfs of SP/KLF, WT1, EGR, Huckebein, and Klumpfuss.
(A) Sequence logos of SP/KLF groups, WT1, EGR, Huckebein and Klumpfuss. Znf regions are marked by red brackets on top. Up to eight positions before the first Znf were also shown as they provide some discrimination power among KLF and SP groups. Three key DNA-binding specificity-determining positions are marked by −1, +3 and +6 under the brackets. Zinc-binding positions with conserved cysteines and histidines are marked by stars (*). The last Znf of WT1, EGR, and Klumpfuss is aligned to the second Znf of SP/KLF and WT1. The coloring of amino acids is as follows: C – yellow; R,K,H – blue; D,E – red; W,F,Y,M,L,I,V,A – black; G,P,S,T, N, Q – green. The linker region between the first Znf and the second Znf of Klumpfuss is replaced with “//”, as it does not conform to the TGE[RK]P consensus in terms of length and amino acid composition. These sequence logos were obtained by using the WebLogo 3 server (Crooks et al. 2004). (B) Predicted DNA-binding preferences of SP/KLF, WT1, EGR, Huckebein and Klumpfuss. The motifs harboring key DNA recognition positions are shown for Znfs of each family. The preferred DNA sequences are shown in italic letters under the motifs with the direction marked by 5′ and 3′ at the ends. Each base was under its corresponding interaction position in the protein motif. The core GC/GT-box sequences are underlined. The last Znf of Huckebein is in grey letters as it is missing in many Huckebein proteins. The letter Z in the second Znf motif of WT1 denotes E or Q. Metazoan and nonmetazoan WT1s have Q and E in this position, respectively.
The cnidarian N. vectensis has members of all three SP groups (SPA–C), while the other surveyed basal metazoan organisms seem to miss SP members from one or more groups (Table 2). The absence of SPs in Ctenophora and its presence in the other three basal metazoan lineages (Porifera, Placozoa, and Cnidaria) and Bilateria suggest that SPs could have originated after the divergence of Ctenophora from other metazoans, if Ctenophora is indeed a sister group to all other metazoans (Ryan et al. 2013; Moroz et al. 2014). Alternatively, SPs could be present in the last common ancestor of all metazoans, but were secondarily lost in the Ctenophora lineage.
3.4. Sequence signatures of SP/KLF and the related EGR, WT1, Huckebein, and Klumpfuss families
C2H2-type Znfs have a consensus sequence of [FY]XCX2,5CX3[FY]X5ΨX2HX3,5[CH] (Xm,n: m to n residues; Ψ: a hydrophobic residue) (Klug 2010). Most of SP/KLF family members have three highly conserved C2H2-type Znfs that follow a cysteine-histidine pattern of “CX4CX12HX3HX7CX4CX12HX3HX7CX2CX12HX3H” (Xn: separation of n residues). The number of Znfs (three) coupled with the conserved residue separation numbers between zinc-coordinating cysteines and histidines appear to be a unique feature of mammalian SP/KLF members (Pei and Grishin 2013). The sequence logos of individual SP/KLF groups are shown in Figure 1(A) (Crooks et al. 2004).
Each Znf has two conserved cysteines and two conserved histidines functioning as zinc-coordinating ligands (marked by * in Figure 1(A)). For all metazoan SP/KLF groups, several positions between the second zinc-coordinating cysteine and the first zinc-coordinating histidine of each Znf exhibit high sequence conservation, forming motifs of YxKxSHxxA, FxRSDExxR, FxRSDHxxx (from the fourth position after the second zinc-binding cysteine to the position before the first zinc-coordinating histidine) in Znfs 1–3, respectively (Figure 1(B)). Each C2H2-type Znf mainly interacts with three consecutive DNA base pairs, and the third, sixth, and ninth positions in these motifs are key contributors to the binding specificity of the 3′ base, the middle base, and 5′ base of the primary interaction DNA strand, respectively (Turner and Crossley 1999; Wolfe et al. 2000; Klug 2010). These specificity-determining positions have been numbered as −1, +3, and +6 positions relative to the start of the helix in a C2H2-type Znf (Wolfe et al. 2000; Klug 2010) (marked on top of Figure 1(A)). For example, the N-terminal arginine, the glutamate, and the C-terminal arginine in FxRSDExxR of the second Znf of SP/KLF determine the preferences for G, C/T, and G, respectively (Figure 1(B)). The arginine in FxRSDHxxx of the third Znf of SP/KLF has preference for G (Figure 1(B)). Noticeably, the +3 positions of the first Znf and the third Znf contain conserved histidines in motifs of KxSH and RSDH, respectively (Figure 1(A)). These two conserved histidines serve as DNA-binding determinants with preference for G (Figure 1(B)) and play different functional roles than the zinc-coordinating histidines.
The sequence logos of SP/KLF groups revealed group(s)-specific conservation in some positions that could be used to support the SP/KLF grouping system and the relationships among the groups (supplementary Figure S2). For example, KLFA has a conserved arginine in the TGxRP linker between the second and third Znfs, while in all other KLF and SP groups, this linker has the consensus of TGxKP (Figure 1(A) and supplementary Figure S2). As a key difference from the KLF groups, the three SP groups contain the BTD box (CxCPxC) N-terminally to the Znfs (supplementary Figure S2). SP groups also share several positions with sequence conservation that differs from KLF groups (supplementary Figure S2). The three SP groups can be differentiated by varying compositional preferences in some positions before and after the BTD boxes (supplementary Figure S2). SP groups appear to be more closely related to KLFE and KLFF, reflected by several positions with common conservation in these groups but not in other KLF groups (supplementary Figure S2). Such an observation is consistent with the phylogenetic reconstruction in this work (supplementary Figure S1) and previous studies (Shimeld 2008; Pei and Grishin 2013).
Several other C2H2-type Znf families possess similar motifs and DNA-binding preferences (Figure 1). The Wilms tumor proteins (Wilm tumor 1, or WT1) have four Znfs with corresponding motifs of YxKxSHxxΨ (Ψ: a hydrophobic residue), FxRSDZxxR (Z: Q or E), FxRSDHxxT, and FxRSDExxR. The first three Znfs of WT1 proteins bear strong similarity to the three Znfs of SP/KLF proteins. The last Znf of WT1 exhibits the strongest similarity to the second Znf of WT1 and SP/KLF (Figure 1). The other related family of proteins, Early Growth Response proteins (EGRs), contain three Znfs with motifs of FxRSDExxR, FxRSDHxxT, and FxRSDExxR that are most similar to the last three Znfs of WT1 (Figure 1).
Two more related families, Huckebein and Klumpfuss, have similar DNA-binding motifs (Figure 1). Two Znfs of Huckebein proteins have FxRNEExxR and FxRKDHxx[KQ] motifs that contribute to the binding of GC/GT-boxes (Figure 1). Regarding DNA-binding residues, these two motifs of Huckebein exhibit the most similarity to the motifs of second and third Znfs of SP/KLF and WT1 proteins (Figure 1(B)). The Klumpfuss proteins often have four Znfs. The last three Znfs have motifs of FxRSDΦxxR, FxRSDHxxT, FxRRDΦxxR that show corresponding similarity to the three Znfs of EGRs and the last three Znfs of WT1s (Figure 1). However, the first Znf of Klumpfuss does not bear strong similarity to the Znfs of SP/KLF, EGR or WT1 proteins (Figure 1). In addition, the linker regions between the first and the second Znfs of Klumpfuss proteins are not conserved in length, in contrast to the highly conserved linker regions (five amino acids with a consensus of TGE[RK]P (Klug 2010)) between consecutive Znfs of SP/KLF, EGR, and WT1 proteins and many other C2H2-type Znfs. These observations suggest that the first Znf of Klumpfuss could have originated from a separate source by gene fusion. A recent large scale characterization of C2H2-type zinc finger proteins using a bacterial one-hybrid system indeed showed similar DNA-binding specificities of SP/KLF, EGR, Huckebein, and Klumpfuss from D. melanogaster (Enuameh et al. 2013).
3.5. Distribution of SP/KLF, EGR, WT1, Huckebein, and Klumpfuss proteins in Metazoa
Within Metazoa, these GC/GT-box-binding families exhibit different taxonomic distributions. KLFs and EGRs are present in all major groups of metazoans (Ctenophora, Porifera, Placozoa, Cnidaria, Ecdysozoa, Lophotrochozoa, and Deuterostomia) (Table 2). SPs were found in all major metazoan groups except Ctenophora. WT1 appears to be missing in many metazoan lineages as it is only found in Ctenophora, Porifera, and some deuterostomes such as B. floridae and vertebrates. Huckebein proteins are present in three basal metazoan lineages – Cnidaria, Placozoa, and Ctenophora, while missing in Porifera. Huckebein was also identified in all three groups of Bilateria – Ecdysozoa, Lophotrochozoa, and Deuterostomia. However, Huckebein is more prone to gene loss compared to SP/KLF and EGR, as it appears to be missing in C. elegans and T. spiralis (nematodes), L. gigantea (a lophotrochozoan), sea urchin, C. intestinalis (a tunicate), and all vertebrates (Table 2). Klumpfuss proteins were only identified in Ecdysozoa and Lophotrochozoa, two protostomian groups. Such a restricted distribution and the similarity of Klumpfuss’ Znf motifs (Figure 1) to EGR and WT1 suggest that Klumpfuss could be a product of gene duplication of EGR or WT1.
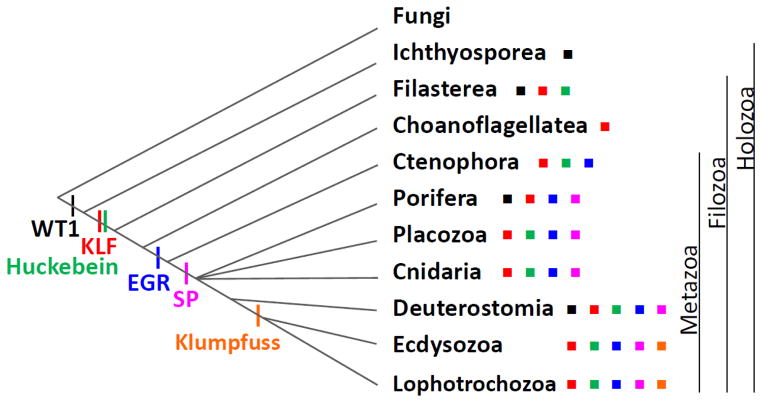
Among these proteins, SP, EGR and Klumpfuss are not found outside Metazoa (Table 2). Previous phylogenetic studies suggest that SPs form a clade together with KLF group E (such as vertebrate KLF9/13/14/16) and group F (such as vertebrate KLF10/11) (Pei and Grishin 2013), implying that SPs could have been derived from KLFs after the advent of metazoans, and possibly after the divergence of Ctenophora from other metazoans (Figure 2 and Table 2). The ubiquitous presence of EGR in all metazoans is consistent with its origin in the last ancestor of Metazoa, while the restricted distribution of Klumpfuss suggests its origin in the last ancestor of protostomes (Figure 2).
Figure 2. The distribution of KLF, SP, WT1, EGR, Huckebein and Klumpfuss in major groups of Holozoa.
The phylogenetic dendrogram assumes that Ctenophore is sister to other metazoans, and both Filozoa and Protostomia (Ecdysozoa+Lophotrochozoa) are monophyletic. Presence of WT1, KLF, Huckebein, EGR, SP, and Klumpfuss in a group is denoted by black, red, green, blue, magenta, and orange squares to the right of the group name, respectively. The inferred origins of these proteins were represented by vertical segments on the dendrogram and their names under the segments with corresponding colors.
3.6. Distribution of KLF, WT1 and Huckebein beyond Metazoa
KLF, WT1, and Huckebein are found outside Metazoa in some single-cell holozoans (Table 2 and Figure 2). Therefore, these proteins originated before the advent of metazoans. Nonmetazoan lineages of Holozoa include Ichthyosporea (also called Mesomycetozoa), Filasterea, and Choanoflagellatea (Shalchian-Tabrizi et al. 2008). Members of these families appear to be absent in lineages outside Holozoa such as Fungi.
Several KLF proteins were discovered in a few single-cell eukaryotes of the Choanoflagellatea and Filasterea groups (Table 2). Choanoflagellatea is likely the group closest to metazoans (Shalchian-Tabrizi et al. 2008). Both choanoflagellates with whole genome sequences, Monosiga brevicollis and Salpingoeca rosetta, contain a putative KLFG ortholog. S. rosetta also possesses an additional KLF protein. We searched EST sequences of another choanoflagellate, Monosiga ovata, and identified transcripts of three KLF proteins, including a putative KLFG ortholog. Capsaspora owczarzaki of the Filasterea group has three KLF proteins, one of which is a putative KLFG ortholog, as suggested by sequence similarity and phylogenetic analysis (Figure 3, described below). The other organism in the Filasterea group, Ministeria vibrans, has at least two KLF proteins according to transcripts from transcriptome assembly. KLFs appear to be absent in Ichthyosporea, another Holozoa lineage aside from Choanoflagellatea, Filasterea, and Metazoa (Figure 2). For example, KLFs were not detected in the whole genome sequences of the Ichthyosporea species Sphaeroforma arctica. We also did not find any KLF transcripts in the assembled transcriptomes of five species in the Ichthyosporea group (see Materials and methods). The presence of KLFs in all three lineages of Filozoa (Filasterea+Choanoflagellatea+Metazoa) (Shalchian-Tabrizi et al. 2008) suggests that they can be traced back to the ancestor of Filozoa (Figure 2).
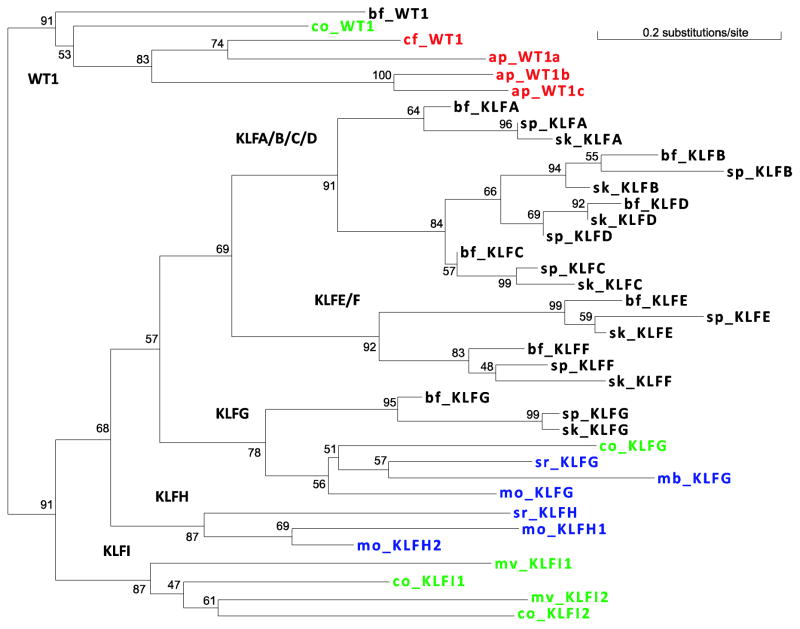
Figure 3. A phylogenetic tree of KLF and WT1 proteins in nonmetazoan organisms and three deuterostomes.
Each protein is denoted by species name abbreviation and protein name. Species name abbreviations are as follows: bf – Branchiostoma floridae; sk – Saccoglossus kowalevskii; sp – Strongylocentrotus purpuratus; mb – Monosiga brevicollisi; mo – Monosiga ovate; sr – Salpingoeca rosetta; co – Capsaspora owczarzaki; mv – Ministeria vibrans; ap – Amoebidium parasiticum; cf – Creolimax fragrantissima. Proteins of Metazoa (bf, sk, and sp), Choanoflagellatea (mb, mo, and sr), Filasterea (co and mv), and Ichthyosporea (ap and cf) are colored in black, blue, green, and red, respectively.
In addition to its presence in C. owczarzaki of the Filasterea group, WT1 was also found in two Ichthyosporea species (Amoebidium parasiticum and Creolimax fragrantissima) according to RNA-Seq-derived transcriptome data, suggesting its origin in the ancestor of Holozoa (Figure 2). In comparison, KLFs are present in Filozoa (Filasterea+Choanoflagellatea+Metazoa) but not in Ichthyosporea. As Ichthyosporea has been indicated to be the sister group of Filozoa (Shalchian-Tabrizi et al. 2008), it is likely that WT1 proteins appeared earlier in evolution than KLFs and KLFs were derived from WT1 proteins. Alternatively, KLFs could be present in the ancestor of Holozoa (Filozoa+Ichthyosporea), and WT1 proteins could have evolved from KLF proteins by adding a C-terminal fourth Znf. In this scenario, the lack of KLF proteins in Ichthyosporea should be due to gene loss. Huckebein was discovered in Filasterea, but not in Ichthyosporea, indicating an origin in the ancestor of Filozoa (Figure 2). Additional genome and transcriptome data could be helpful in clarifying the origins of these families.
To investigate the relationship of KLFs in metazoan and nonmetazoan organisms, we constructed a phylogenetic tree using KLFs in nonmetazoans and three invertebrate deuterostomes (S. kowalevskii, S. purpuratus, and B.floridae), with WT1 proteins in these organisms serving as an outgroup (Figure 3). The tree suggests that KLFG originated before the last common ancestor of metazoans. Nonmetazoan KLFG has Filozoa members from all three surveyed choanoflagellates (M. brevicollis, S. rosetta, and M. ovata) as well as C. owczarzaki of the Filasterea group. Two KLF groups formed by nonmetazoan members have moderate supports and are named KLFH and KLFI. KLFH and KLFI members are from choanoflagellates and Filasterea (two members each in C. owczarzaki and M. vibrans), respectively (Figure 3 and Table 2).
3.7. Variation of Znf numbers in SP/KLF, Huckebein, and Klumpfuss families
The number of Znfs is usually well conserved in the SP/KLF, WT1, EGR, and Klumpfuss families. However, a few exceptions were observed in divergent members. For example, SPC of the nematode C. elegans (encoded by the SPTF-1 gene) has lost two C-terminal Znfs. C. elegans contains two Klumpfuss proteins: one with four Znfs (GenBank accession: NP_493611) like Klumpfuss proteins in other organisms and a second, divergent Klumpfuss protein (GenBank accession: NP_491843) that appears to have lost the first Znf. D. melanogaster possesses a divergent SPC member (CG3065) with two additional Znfs in the C-terminus. These two additional Znfs have motifs FKRQDD and FVRSDH that are most similar to the second and third Znfs of the SP/KLF family, respectively, suggesting that they were derived from gene duplication and fusion. This divergent SP sequence also lacks the Btd box (CxCxxC N-terminally to Znfs) that is present in other SP proteins, but not in KLF proteins.
Huckebein proteins show varying number of Znfs in different lineages. Four Znfs were found in Huckebein proteins from the cnidarian N. vectensis, the hemichordate S. kowalevskii, the arthropod Stegodyphus mimosarum and two nonmetazoan species from the Filasteria group (Capsaspora owczarzaki and Ministeria vibrans). On the other hand, Huckebein proteins from the amphioxus B. floridae, three ecdysozoan species T. castaneum, D. pulex, and D. melanogaster, and the placozoan T. adhaerens appear to have lost the last Znf and only possess three Znfs. Moreover, Huckebeins of the two Ctenophora organisms M. leidyi and P. bachei appear to have lost the first and the last Znfs and only possess two Znfs.
Supplementary Material
Highlights.
Searches of SP/KLF, EGR, WT1, Huckebein and Klumpfuss proteins were performed.
Seven KLF groups (KLFA–G) and three SP groups (SPA–C) are found in bilaterians.
The ancestor of jawed vertebrates has at least 18 KLFs and 10 SPs.
KLF, WT1, and Huckebein proteins originated before the advent of metazoans.
KLFG (human representative: KLF15) was found in Choanoflagellatea and Filasterea.
Acknowledgments
We would like to thank Lisa Kinch for critical reading of the manuscript and discussions. We would like to thank Qian Cong for suggestions with transcriptome analysis. This work was supported by National Institutes of Health (GM094575 to NVG) and the Welch Foundation (I-1505 to NVG).
Abbreviations
- DNA
deoxyribonucleic acid
- EGR
Early Growth Response protein
- EKLF
Erythroid Krüppel-Like Factor
- HGNC
Human Gene Nomenclature Committee
- KLF
Krüppel-like factor
- LKLF
Lung Krüppel-Like Factor
- SP
Specificity protein
- WT1
Wilms tumor protein 1
- Znf
zinc finger
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi J, Hasegawa M. MOLPHY version 2.3, programs for molecular phylogenetics based on maximum likelihood. Computer Science Monographs (The Institute of Statistical Mathematics) 1996;28:1–150. [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amores A, Catchen J, Ferrara A, Fontenot Q, Postlethwait JH. Genome evolution and meiotic maps by massively parallel DNA sequencing: spotted gar, an outgroup for the teleost genome duplication. Genetics. 2011;188(4):799–808. doi: 10.1534/genetics.111.127324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antin PB, Pier M, Sesepasara T, Yatskievych TA, Darnell DK. Embryonic expression of the chicken Kruppel-like (KLF) transcription factor gene family. Dev Dyn. 2010;239(6):1879–1887. doi: 10.1002/dvdy.22318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieker JJ. Kruppel-like factors: three fingers in many pies. J Biol Chem. 2001;276(37):34355–34358. doi: 10.1074/jbc.R100043200. [DOI] [PubMed] [Google Scholar]
- Black AR, Black JD, Azizkhan-Clifford J. Sp1 and kruppel-like factor family of transcription factors in cell growth regulation and cancer. Journal of cellular physiology. 2001;188(2):143–160. doi: 10.1002/jcp.1111. [DOI] [PubMed] [Google Scholar]
- Call KM, Glaser T, Ito CY, Buckler AJ, Pelletier J, Haber DA, Rose EA, Kral A, Yeger H, Lewis WH, et al. Isolation and characterization of a zinc finger polypeptide gene at the human chromosome 11 Wilms’ tumor locus. Cell. 1990;60(3):509–520. doi: 10.1016/0092-8674(90)90601-a. [DOI] [PubMed] [Google Scholar]
- Cameron RA, Samanta M, Yuan A, He D, Davidson E. SpBase: the sea urchin genome database and web site. Nucleic Acids Res. 2009;37(Database issue):D750–754. doi: 10.1093/nar/gkn887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JA, Kirkness EF, Simakov O, Hampson SE, Mitros T, Weinmaier T, Rattei T, Balasubramanian PG, Borman J, Busam D, et al. The dynamic genome of Hydra. Nature. 2010;464(7288):592–596. doi: 10.1038/nature08830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Lei T, Chen X, Zhang J, Yu A, Long Q, Long H, Jin D, Gan L, Yang Z. Porcine KLF gene family: Structure, mapping, and phylogenetic analysis. Genomics. 2010;95(2):111–119. doi: 10.1016/j.ygeno.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Colbourne JK, Pfrender ME, Gilbert D, Thomas WK, Tucker A, Oakley TH, Tokishita S, Aerts A, Arnold GJ, Basu MK, et al. The ecoresponsive genome of Daphnia pulex. Science. 2011;331(6017):555–561. doi: 10.1126/science.1197761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium CeS. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282(5396):2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14(6):1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang DT, Pevsner J, Yang VW. The biology of the mammalian Kruppel-like family of transcription factors. Int J Biochem Cell Biol. 2000;32(11–12):1103–1121. doi: 10.1016/s1357-2725(00)00059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enuameh MS, Asriyan Y, Richards A, Christensen RG, Hall VL, Kazemian M, Zhu C, Pham H, Cheng Q, Blatti C, et al. Global analysis of Drosophila Cys(2)-His(2) zinc finger proteins reveals a multitude of novel recognition motifs and binding determinants. Genome Res. 2013;23(6):928–940. doi: 10.1101/gr.151472.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessler M, Poustka A, Cavenee W, Neve RL, Orkin SH, Bruns GA. Homozygous deletion in Wilms tumours of a zinc-finger gene identified by chromosome jumping. Nature. 1990;343(6260):774–778. doi: 10.1038/343774a0. [DOI] [PubMed] [Google Scholar]
- Gomez-Martin D, Diaz-Zamudio M, Galindo-Campos M, Alcocer-Varela J. Early growth response transcription factors and the modulation of immune response: implications towards autoimmunity. Autoimmunity reviews. 2010;9(6):454–458. doi: 10.1016/j.autrev.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature biotechnology. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M, Kishino H, Saitou N. On the maximum likelihood method in molecular phylogenetics. J Mol Evol. 1991;32(5):443–445. doi: 10.1007/BF02101285. [DOI] [PubMed] [Google Scholar]
- Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews L, et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496(7446):498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8(3):275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kaczynski J, Cook T, Urrutia R. Sp1- and Kruppel-like transcription factors. Genome Biol. 2003;4(2):206. doi: 10.1186/gb-2003-4-2-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga JT, Carner KR, Masiarz FR, Tjian R. Isolation of cDNA encoding transcription factor Sp1 and functional analysis of the DNA binding domain. Cell. 1987;51(6):1079–1090. doi: 10.1016/0092-8674(87)90594-0. [DOI] [PubMed] [Google Scholar]
- Karolchik D, Barber GP, Casper J, Clawson H, Cline MS, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 2014;42(Database issue):D764–770. doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein T, Campos-Ortega JA. klumpfuss, a Drosophila gene encoding a member of the EGR family of transcription factors, is involved in bristle and leg development. Development. 1997;124(16):3123–3134. doi: 10.1242/dev.124.16.3123. [DOI] [PubMed] [Google Scholar]
- Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annual review of biochemistry. 2010;79:213–231. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- Leinonen R, Sugawara H, Shumway M International Nucleotide Sequence Database C. The sequence read archive. Nucleic Acids Res. 2011;39(Database issue):D19–21. doi: 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Materna SC, Howard-Ashby M, Gray RF, Davidson EH. The C2H2 zinc finger genes of Strongylocentrotus purpuratus and their expression in embryonic development. Developmental biology. 2006;300(1):108–120. doi: 10.1016/j.ydbio.2006.08.032. [DOI] [PubMed] [Google Scholar]
- McConnell BB, Yang VW. Mammalian Kruppel-like factors in health and diseases. Physiol Rev. 2010;90(4):1337–1381. doi: 10.1152/physrev.00058.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A, Schartl M. Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr Opin Cell Biol. 1999;11(6):699–704. doi: 10.1016/s0955-0674(99)00039-3. [DOI] [PubMed] [Google Scholar]
- Mitreva M, Jasmer DP, Zarlenga DS, Wang Z, Abubucker S, Martin J, Taylor CM, Yin Y, Fulton L, Minx P, et al. The draft genome of the parasitic nematode Trichinella spiralis. Nature genetics. 2011;43(3):228–235. doi: 10.1038/ng.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroz LL, Kocot KM, Citarella MR, Dosung S, Norekian TP, Povolotskaya IS, Grigorenko AP, Dailey C, Berezikov E, Buckley KM, et al. The ctenophore genome and the evolutionary origins of neural systems. Nature. 2014;510(7503):109–114. doi: 10.1038/nature13400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols SA, Roberts BW, Richter DJ, Fairclough SR, King N. Origin of metazoan cadherin diversity and the antiquity of the classical cadherin/beta-catenin complex. Proc Natl Acad Sci U S A. 2012;109(32):13046–13051. doi: 10.1073/pnas.1120685109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Grishin NV. A new family of predicted Kruppel-like factor genes and pseudogenes in placental mammals. PLoS One. 2013;8(11):e81109. doi: 10.1371/journal.pone.0081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam NH, Butts T, Ferrier DE, Furlong RF, Hellsten U, Kawashima T, Robinson-Rechavi M, Shoguchi E, Terry A, Yu JK, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453(7198):1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- Ryan JF, Pang K, Schnitzler CE, Nguyen AD, Moreland RT, Simmons DK, Koch BJ, Francis WR, Havlak P, Program NCS, et al. The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science. 2013;342(6164):1242592. doi: 10.1126/science.1242592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satou Y, Mineta K, Ogasawara M, Sasakura Y, Shoguchi E, Ueno K, Yamada L, Matsumoto J, Wasserscheid J, Dewar K, et al. Improved genome assembly and evidence-based global gene model set for the chordate Ciona intestinalis: new insight into intron and operon populations. Genome Biol. 2008;9(10):R152. doi: 10.1186/gb-2008-9-10-r152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeper ND, Prpic NM, Wimmer EA. A clustered set of three Sp-family genes is ancestral in the Metazoa: evidence from sequence analysis, protein domain structure, developmental expression patterns and chromosomal location. BMC Evol Biol. 2010;10:88. doi: 10.1186/1471-2148-10-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seetharam A, Bai Y, Stuart GW. A survey of well conserved families of C2H2 zinc-finger genes in Daphnia. BMC Genomics. 2010;11:276. doi: 10.1186/1471-2164-11-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalchian-Tabrizi K, Minge MA, Espelund M, Orr R, Ruden T, Jakobsen KS, Cavalier-Smith T. Multigene phylogeny of choanozoa and the origin of animals. PLoS One. 2008;3(5):e2098. doi: 10.1371/journal.pone.0002098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimeld SM. C2H2 zinc finger genes of the Gli, Zic, KLF, SP, Wilms’ tumour, Huckebein, Snail, Ovo, Spalt, Odd, Blimp-1, Fez and related gene families from Branchiostoma floridae. Dev Genes Evol. 2008;218(11–12):639–649. doi: 10.1007/s00427-008-0248-6. [DOI] [PubMed] [Google Scholar]
- Simakov O, Marletaz F, Cho SJ, Edsinger-Gonzales E, Havlak P, Hellsten U, Kuo DH, Larsson T, Lv J, Arendt D, et al. Insights into bilaterian evolution from three spiralian genomes. Nature. 2013;493(7433):526–531. doi: 10.1038/nature11696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava M, Begovic E, Chapman J, Putnam NH, Hellsten U, Kawashima T, Kuo A, Mitros T, Salamov A, Carpenter ML, et al. The Trichoplax genome and the nature of placozoans. Nature. 2008;454(7207):955–960. doi: 10.1038/nature07191. [DOI] [PubMed] [Google Scholar]
- Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier ME, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten U, et al. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature. 2010;466(7307):720–726. doi: 10.1038/nature09201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan JC, Ryan JF, Watson JA, Webb J, Mullikin JC, Rokhsar D, Finnerty JR. StellaBase: the Nematostella vectensis Genomics Database. Nucleic Acids Res. 2006;34(Database issue):D495–499. doi: 10.1093/nar/gkj020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suske G, Bruford E, Philipsen S. Mammalian SP/KLF transcription factors: bring in the family. Genomics. 2005;85(5):551–556. doi: 10.1016/j.ygeno.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Tribolium Genome Sequencing C. Richards S, Gibbs RA, Weinstock GM, Brown SJ, Denell R, Beeman RW, Gibbs R, Beeman RW, Brown SJ, et al. The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452(7190):949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- Turner J, Crossley M. Mammalian Kruppel-like transcription factors: more than just a pretty finger. Trends in biochemical sciences. 1999;24(6):236–240. doi: 10.1016/s0968-0004(99)01406-1. [DOI] [PubMed] [Google Scholar]
- van Vliet J, Crofts LA, Quinlan KG, Czolij R, Perkins AC, Crossley M. Human KLF17 is a new member of the Sp/KLF family of transcription factors. Genomics. 2006;87(4):474–482. doi: 10.1016/j.ygeno.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Venkatesh B, Lee AP, Ravi V, Maurya AK, Lian MM, Swann JB, Ohta Y, Flajnik MF, Sutoh Y, Kasahara M, et al. Elephant shark genome provides unique insights into gnathostome evolution. Nature. 2014;505(7482):174–179. doi: 10.1038/nature12826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Jurgens G, Klingler M, Jackle H. Two gap genes mediate maternal terminal pattern information in Drosophila. Science. 1990;248(4954):495–498. doi: 10.1126/science.2158673. [DOI] [PubMed] [Google Scholar]
- White JA, McAlpine PJ, Antonarakis S, Cann H, Eppig JT, Frazer K, Frezal J, Lancet D, Nahmias J, Pearson P, et al. Guidelines for human gene nomenclature (1997). HUGO Nomenclature Committee. Genomics. 1997;45(2):468–471. doi: 10.1006/geno.1997.4979. [DOI] [PubMed] [Google Scholar]
- Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annual review of biophysics and biomolecular structure. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- Yang X, Bahri S, Klein T, Chia W. Klumpfuss, a putative Drosophila zinc finger transcription factor, acts to differentiate between the identities of two secondary precursor cells within one neuroblast lineage. Genes & development. 1997;11(11):1396–1408. doi: 10.1101/gad.11.11.1396. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.