Abstract
Please cite this paper as: Friesema et al. (2012). Course of pandemic influenza A(H1N1) 2009 virus infection in Dutch patients. Influenza and Other Respiratory Viruses 6(3), e16–e20.
The clinical dynamics of influenza A(H1N1) 2009 infections in 61 laboratory‐confirmed Dutch cases were examined. An episode lasted a median of 7·5 days of which 2 days included fever. Respiratory symptoms resolved slowly, while systemic symptoms peaked early in the episode and disappeared quickly. Severity of each symptom was rated highest in the first few days. Furthermore, diarrhoea was negatively associated with viral load, but not with faecal excretion of influenza virus. Cases with comorbidities appeared to have higher viral loads than the cases without, suggesting a less effective immune response. These results complement information obtained through traditional surveillance.
Keywords: Clinical dynamics, influenza A(H1N1) 2009, pandemic, symptoms, viral load
Introduction
In 2009, a novel influenza A(H1N1) virus caused the first influenza pandemic of the 21st century. 1 , 2 In the Netherlands, the first case of infection with the pandemic virus strain was reported on 30 April, and 1622 cases were registered until 15 August 2009. 3 On this date, the notification criteria were narrowed to hospitalized and deceased laboratory‐confirmed cases only. 4 Little is known about the clinical dynamics of symptoms during an influenza infection, despite this information being of potential use for control efforts, policy and communication. We investigated the dynamics of symptoms of pandemic influenza A(H1N1)pdm09 infections in community cases in the Netherlands, and studied possible relationships between symptoms and viral load.
Methods
A core research protocol was designed to enable collection of relevant data in case of an epidemic of an unusual human influenza virus. This protocol and the adapted 2009 version were approved by the Medical Ethical Review Committee of the University Medical Centre, Utrecht.
Laboratory‐confirmed cases were recruited via municipal health services, general practitioners and academic hospitals. Between 30 April and 14 August 2009, cases could be drawn from notification data. As from 15 August, recruitment was continued in collaboration with the Sentinel General Practice Network of NIVEL, the Netherlands Institute for Health Services Research. 5 Twelve of the 42 GP practices were willing to participate requiring the GP to collect a nose and throat swab and finger prick sample at time of consultation. These samples had to be taken within 4 days of onset of illness. In November and December 2009, patients with a laboratory‐confirmed infection hospitalized in one of three university hospitals were also approached.
Consenting patients agreed to sequential sampling of nose‐throat swabs and blood specimens (Table 1). Furthermore, cases completed a questionnaire about demographics, medical history, vaccinations, symptoms and the use of antivirals. The cases were also asked to keep a daily diary recording symptoms and the use of antivirals each day for a maximum of 14 days. The cases rated severity of each symptom on a scale ranging from 0 (none experienced) to 10 (symptom was present throughout the day) by the case.
Table 1.
Research scheme of sampling and questionnaires per house visit
| Visit 1* = Day 0 | Visit 2* = Day 4–6 | Visit 3 = Day 9–11 | Visit 4 = Day 28–35 | |
|---|---|---|---|---|
| Informed consent | X | |||
| Swabs | ||||
| Throat/nose | X | X | ||
| Rectal | X | |||
| Venous blood | X | X | X | |
| Questionnaire | X | |||
| Symptoms diary | X | X | X | X |
*For cases recruited through the GP, visit 1 and 2 were combined.
Real‐time RT‐PCR for detection of A(H1N1)pdm09 virus in combined nose and throat swabs and in rectal swabs was performed as described previously. 6 To determine of the exact viral load in virus particles per ml, an electron microscopy (EM)‐counted standard of human influenza virus A/PuertoRico/8/1934 (H1N1) was used (provided by M. Schutten, Erasmus MC, Rotterdam, NL). All clinical specimens and a dilution series of the EM‐counted control were retrospectively subjected batch‐wise to RNA isolation and one‐step matrix gene‐based real‐time RT‐PCR, and the viral loads of the clinical specimens were estimated based on the batch calibration curve.
The evolution of symptoms was examined by calculating the percentage of cases experiencing each symptom per day, together with the median severity score per symptom per day. The diary could be completed for a maximum of 14 days, which led to a censoring of duration of illness for cases who had symptoms for a longer period. Associations between the highest estimated viral load in the first 5 days of illness and age, gender, underlying disease, duration of illness, and reported symptoms on the day of sampling were examined using the Kruskal–Wallis test or Spearman correlation. Secondly, associations between all estimated viral loads and severity scores per symptom on the day of sampling were calculated using Spearman correlations. All analyses were carried out for all cases, stratifying by the use of antivirals.
Results
A total of 120 laboratory‐confirmed cases were approached; 76 (63%) were willing to participate of whom 61 (51%) cases reported their symptoms either in the questionnaire (46 cases) and/or in the diary (38 cases). Table 2 shows the characteristics of the cases. Forty‐three per cent of the cases had one or more underlying diseases. Thirty‐four cases were treated with oseltamivir, and about two‐thirds started treatment within 3 days of the first symptoms occurring (range 1–7 days). The cases reported an overall median duration of illness of 7·5 days (range 2–21 days). The admitted cases were hospitalized for 3–9 days, and had a median duration of illness of 10 days (range 7–13 days).
Table 2.
Characteristics of cases with a laboratory‐confirmed influenza A(H1N1) 2009 virus infection (n = 61)
| n (%) | |
|---|---|
| 30 April–14 August 2009 | |
| Via municipal health services | 36 (59) |
| 15 August–31 December 2009 | |
| Via GPs | 19 (31) |
| Via academic centres | 6 (10) |
| Age, in years | |
| 4–18 | 16 (26) |
| 20–39 | 26 (43) |
| 40–59 | 12 (20) |
| 60–69 | 7 (11) |
| Gender | |
| Male | 26 (43) |
| Female | 35 (57) |
| Underlying disease | |
| No severe or chronic disease | 20 (33) |
| Severe or chronic disease | 26 (43) |
| Lung disease | 13 (21) |
| Other disease | 7 (11) |
| More than one disease | 6 (10) |
| Unknown | 15 (25) |
| Antiviral medication | |
| Yes | 36 (59) |
| No | 23 (38) |
| Unknown | 2 (3) |
Cases with fever reported a maximum of 6 days of fever with a median of 2 days (Figure 1). Other symptoms lasted longer. Complaints of coughing were still present in more than 10% of the cases at day 14. Men appeared to have fever for a longer period (4 days; 1–6 days) than women [1·5 days; 1–5 days (P = 0·09)].
Figure 1.
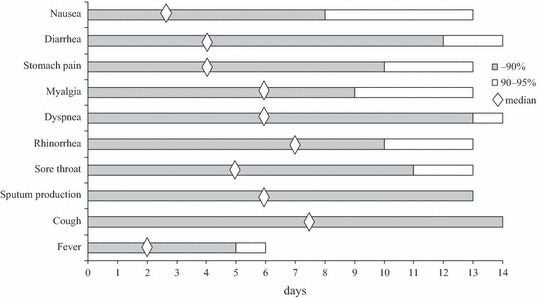
Duration of clinical symptoms, showing median, 90‐percentile and 95‐percentile per symptom.
All cases coughed in the first few days of illness and these complaints persisted (Figure 2). Dyspnoea (data not shown) followed a comparable pattern as rhinorrhoea. Myalgia peaked at day 2–4 (71–75% of the cases). Diarrhoea was reported most frequently between day 4 and 6 (almost half of the cases). Nausea was reported by one‐third of the cases on day 1, and then declined. The severity of complaints was highest in the first 4 days for all symptoms (Figure 3). A notable result is the unexplained rise in score for sputum production after day 9.
Figure 2.
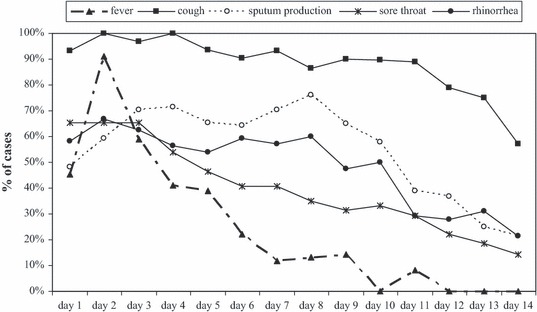
Percentage of cases reporting the experience of each symptom per day.
Figure 3.
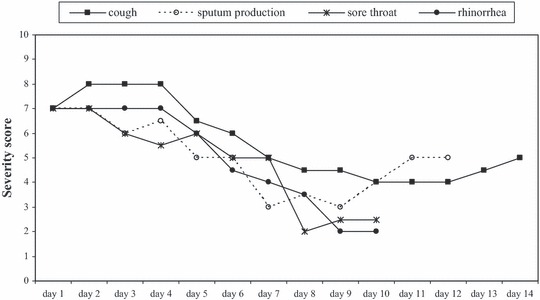
Median severity score (1–10) per day per symptom as reported by the cases (when reported by at least six cases per day).
Ninety‐two per cent of the nose‐throat swabs taken within 1 week after onset of illness were positive (46/50) compared to 24% (9/37) in the second and 18% (4/22) in the third week. Two samples from one case were still positive at day 24 and 39. Five of the six positive samples taken more than 14 days after day of onset were from three hospitalized cases. Four cases, with a period of illness of 3–12 days, had a positive swab (<3000 viral particles/ml) at 1–3 days after clinical recovery.
Viral load data were available for 73 samples of 42 cases. Median load was 8130 viral particles/ml (range: 20–117 000 000). No difference in load was seen between samples taken at day 1–2 versus day 3–5 post onset of illness or use of antivirals. Cases with underlying disease had a higher viral load than the cases without. No difference in prescription of antivirals was seen between cases with and without underlying disease. A negative correlation was found between number of days with diarrhoea and highest available viral load [−0·39 (P = 0·02)], but no association between viral load and severity score for diarrhoea on the day of sampling was seen.
A rectal swab, taken between day 4 and day 13, was available for 31 cases. Four specimens were positive, two cases had diarrhoea somewhere during their episode of influenza, but none on the day of sampling. Of the 27 cases with a negative rectal sample, 13 had reported at least 1 day of diarrhoea.
Discussion
Cases with comorbidities had on average a higher viral load than cases without, but without difference in the duration of illness. The opposite has been reported for seasonal influenza, but no association was found for A(H1N1)pdm09 influenza. 7 Previous comparisons of cases of varying severity did not reveal differences in initial viral loads, but patients with more severe disease or with comorbidities cleared the virus more slowly. 8 , 9 A negative association was only found between viral load and diarrhoea in the present study, whereas Li et al 10 found higher viral loads in patients with fever and in patients who developed pneumonia. Faecal excretion of A(H1N1)pdm09 influenza virus could not be linked to diarrhoea in the current study, which has also been reported elsewhere. 11
Initially, the course of the 2009 pandemic resulted in staff resource challenges, which made it difficult to include more cases. Subsequently, the mandatory notification in the Netherlands was restricted, and a new route for inclusions had to be found. The relatively low numbers reduce the power of the study. Seventy per cent of the participants reported underlying diseases, mainly lung diseases (39%), compared to 16·8% of all notified laboratory‐confirmed cases in the Netherlands (9·1% lung diseases), 3 indicating that selection bias is likely to have occurred. Cases with comorbidities may be more prone to participate. Furthermore, people with comorbidities will visit their GP faster in the case of influenza‐like illness (ILI) than healthy persons experiencing ILI, and thus have a higher chance to be included. However, no indications were found for a different course of the disease in the cases with comorbidities.
In conclusion, the pandemic offered a unique opportunity to investigate the clinical dynamics of influenza, complementing the information obtained through traditional public health surveillance. Systemic symptoms dominated during the first 4 days, whereas respiratory symptoms peaked in the first few days and resolved slowly. The severity of each symptom was rated highest in the first few days and then declined. Diarrhoea appeared to be negatively associated with viral load, but not with faecal excretion of influenza virus. Patients with underlying disease seemed to have higher viral loads, suggesting a less effective immune response, although they were not significantly more severely ill.
Authors’ contribution
IF participated in the design and coordination of the study, performed the statistical analyses and drafted the manuscript. AM participated in the design and coordination of the study, drafting the manuscript and was responsible for the virological assays. AvG participated in the design and coordination of the study, in performing the statistical analyses and drafting the manuscript. MvdL participated in the design and coordination of the study, and was responsible for the virological assays. JvB participated in the design and coordination of the study, and processing the laboratory results. GD participated in the coordination of the study, and headed the network of general practices. JP and MdJ participated in the design and coordination of the study with respect to the academic centres. SB participated in the implementation and processing of the virological tests. LI participated in the design and coordination of the study. MK participated in the design and coordination of the study as project leader of the Dutch ZonMw Influenza A(H1N1) 2009 consortium. MvdS participated in the design and coordination of the study in performing the statistical analyses and drafting the manuscript. All authors read and approved the final manuscript.
Acknowledgments
We thank all the municipal health services, hospitals, general practitioners of the sentinel network and laboratories for their cooperation in the data collection. We thank the employees from SALTRO for visiting the participants and taking specimens. Furthermore, we acknowledge the participants in this study for providing specimens and completing the questionnaires. We thank Dr LSB Gelinck, Erasmus MC Rotterdam, and Dr JJ Oosterheert, UMCU Utrecht, who enrolled the hospitalized patients. We thank Mrs. M. Heshusius‐Valen for valuable support in data collection of the Sentinel General Practice Network of NIVEL. We also thank the technicians Shireen Jenny, Yaobi Hu, Ngoc Hoa Chung, Cheraine Paulsen and Pieter Overduin of the Laboratory for Infectious Diseases and Perinatal Screening of the RIVM for their technical assistance. Finally, we thank Georgia Ladburry for reading and correcting the text.
This work was supported by the Ministry of Health, Welfare and Sport (the Netherlands) and by The Netherlands Organisation for Health Research and Development Funding (ZonMw) [50‐50800‐98‐105].
Appendix: Dutch ZonMw Influenza A(H1N1) 2009 consortium
RIVM, Bilthoven: L van Asten, D Baas, D Beaujean, J van Beek, R van Binnendijk, M van Boven, R Coutinho, F Dijkstra, T Donker, IHM Friesema, AB van Gageldonk‐Lafeber, F Heijningen, W van der Hoek, L Isken, F van der Klis, T van ‘t Klooster, M Koopmans, M Kretzschmar, IM van der Lubben, M Mak, A Meijer, J Reimerink, R Riesmeijer, MAB van der Sande, J van Steenbergen, A Steens, P Teunis, A Timen, L Vinck, J Wallinga, A Westerhof, L Wielders, CC van den Wijngaard; NIVEL, Utrecht: GA Donker, M Hooiveld, J IJzermans, F Schellevis, R Verheij; Erasmus Medical Centre, Rotterdam: C Boucher, R Fouchier, P de Fraaij, A Osterhaus, G Rimmelzwaan, M Schutten; Academical Medical Centre – University Amsterdam: M de Jong, J Prins; Utrecht University Medical Centre, Utrecht: C Kesmir, JJ van Oosterheert; Radboud University Nijmegen Medical Centre, Nijmegen: JC Braspenning, M Bults, R Grol, M Hulscher, J van der Meer, MAJB Tacken; Leiden University Medical Centre, Leiden: J van Dissel; Public Health Service of Amsterdam and LOI, Amsterdam: A van den Hoek; Rotterdam‐Rijnmond Public Health Service, Rotterdam: O de Zwart; iResearch, Berg en Dal: M van der Velden.
References
- 1. Cohen J. Swine flu outbreak. Out of Mexico? Scientists ponder swine flu’s origins. Science 2009; 324:700–702. [DOI] [PubMed] [Google Scholar]
- 2. Fraser C, Donnelly CA, Cauchemez S et al. Pandemic potential of a strain of influenza A (H1N1): early findings. Science 2009; 324:1557–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vriend HJ, Hahné SJM, Donker T et al. Epidemiologie van Nieuwe Influenza A(H1N1) in Nederland, 30 April–15 augustus 2009. [Epidemiology of the new influenza A(H1N1) in the Netherlands, 30 April–15 August 2009.]. Ned Tijdschr Geneeskd 2009; 153:A969. [Google Scholar]
- 4. van’t Klooster TM, Wielders CC, Donker T et al. Surveillance of hospitalisations for 2009 pandemic influenza a(H1N1) in the Netherlands, 5 June–31 December 2009. Euro Surveill 2010; 15:9–16. [DOI] [PubMed] [Google Scholar]
- 5. Donker GA. Continuous Morbidity Registration at Dutch Sentinel General Practice Network 2009. Utrecht: Nivel, 2011. [Google Scholar]
- 6. Meijer A, Beerens A, Claas E et al. Preparing the outbreak assistance laboratory network in the Netherlands for the detection of the influenza virus A(H1N1) variant. J Clin Virol 2009; 45:179–184. [DOI] [PubMed] [Google Scholar]
- 7. Lee CK, Lee HK, Loh TP et al. Comparison of pandemic (H1N1) 2009 and seasonal influenza viral loads, Singapore. Emerg Infect Dis 2011; 17:287–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. To KKW, Hung IFN, Li IWS et al. Delayed clearance of viral load and marked cytokine activation in severe cases of pandemic H1N1 2009 influenza virus infection. Clin Infect Dis 2010; 50:850–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Na S, Chong YP, Kim MN et al. Duration of viral shedding in patients admitted to hospital with pandemic influenza A/H1N1 2009 infection. J Med Virol 2011; 83:5–9. [DOI] [PubMed] [Google Scholar]
- 10. Li CC, Wang L, Eng HL et al. Correlation of pandemic (H1N1) 2009 viral load with disease severity and prolonged viral shedding in children. Emerg Infect Dis 2010; 16:1265–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yoo SJ, Moon SJ, Kuak EY et al. Frequent detection of pandemic (H1N1) 2009 virus in stools of hospitalized patients. J Clin Microbiol 2010; 48:2314–2315. [DOI] [PMC free article] [PubMed] [Google Scholar]


