Abstract
Please cite this paper as: Fry et al. (2012) The first cases of 2009 pandemic influenza A (H1N1) virus infection in the United States: a serologic investigation demonstrating early transmission. Influenza and Other Respiratory Viruses 6(3), e48–e53.
Background The first two laboratory‐confirmed cases of 2009 pandemic influenza A (H1N1) virus (H1N1pdm09) infection were detected in San Diego (SD) and Imperial County (IC) in southern California, April 2009.
Objectives To describe H1N1pdm09 infections and transmission early in the 2009 H1N1 pandemic.
Patients/Methods We identified index case‐patients from SD and IC with polymerase chain reaction (PCR)‐confirmed H1N1pdm09 infections and investigated close contacts for a subset of case‐patients from April 17–May 6, 2009. Acute and convalescent serum was collected. Serologic evidence for H1N1pdm09 infection was determined by microneutralization and hemagglutination inhibition assays.
Results Among 75 close contacts of seven index case‐patients, three reported illness onset prior to patient A or B, including two patient B contacts and a third with no links to patient A or B. Among the 69 close contacts with serum collected >14 days after the onset of index case symptoms, 23 (33%) were seropositive for H1N1pdm09, and 8 (35%) had no fever, cough, or sore throat. Among 15 household contacts, 8 (53%) were seropositive for H1N1pdm09. The proportion of contacts seropositive for H1N1pdm09 was highest in persons aged 5–24 years (50%) and lowest in persons aged ≥50 years (13%) (P = 0·07).
Conclusions By the end of April 2009, before H1N1pdm09 was circulating widely in the community, a third of persons with close contact to confirmed H1N1pdm09 cases had H1N1pdm09 infection in SD and IC. Three unrelated clusters during March 21–30 suggest that transmission of H1N1pdm09 had begun earlier in southern California.
Keywords: 2009 pandemic influenza A (H1N1), influenza, serology, Southern California, transmission
Background
The first two laboratory‐confirmed cases of what is now known as the 2009 pandemic influenza A (H1N1) virus (H1N1pdm09) were reported in April 17, 2009, from southern California. 1 Patient A, a 10‐year‐old resident of San Diego County (SD), had onset of symptoms on March 30, 2009. Patient B, a 9‐year‐old resident of Imperial County (IC), had onset of symptoms on March 28, 2009. Shortly after the identification of these two cases, enhanced surveillance for H1N1pdm09 infections was initiated in California and early case‐patients and their close contacts were actively investigated. To better characterize the infections caused by the new virus and investigate secondary transmission rates, we collected sera from polymerase chain reaction (PCR)‐confirmed H1N1pdm09 case‐patients and their close contacts identified early in the outbreak in SD and IC for serologic testing.
Methods
We conducted active surveillance of PCR‐confirmed H1N1pdm09 case‐patients and their close contacts between April 17 and May 6, 2009. Convalescent serum samples were collected on April 18, 2009, for patient A and April 17 and May 5, 2009, for patient B. For all other PCR‐confirmed H1N1pdm09 index case‐patients, we attempted to collect well‐timed paired sera, for example, an acute serum sample within 7 days of symptom onset and a convalescent sample at least 14 days later. Single serum specimens collected >14 days since symptom onset were considered convalescent. 2 , 3 , 4 Close contacts were defined as persons within six feet of a case‐patient for at least 1 hour or in contact with an index case‐patient for a shorter period of time but with direct contact with case‐patient droplets, such as being coughed on by the case‐patient. Close contacts of a convenience sample of PCR‐confirmed H1N1pdm09 index case‐patients from SD and IC had blood collected within 2 days of blood specimen collection on the index case‐patient and again at least 14 days later. Single serum specimens collected >14 days since the index case‐patient symptom onset were considered convalescent. A standardized form was used to collect information from PCR‐confirmed H1N1pdm09 patients and their contacts, including demographic information and the presence of symptoms at each blood draw. Symptoms were defined as any of the following: cough or sore throat. The presence of subjective fever was not required but was present in some symptomatic patients. Patients were considered to have subclinical infections if they did not report cough, sore throat, or subjective fever. Information on use of antiviral agents was not systematically collected.
Serum samples were tested by microneutralization (MN) and hemagglutination inhibition (HI) assays using an A/California/07/2009‐like virus. 4 , 5 , 6 , 7 The MN assay was performed with twofold dilutions of serum samples and a standard infectious dose of virus. After 18–20 hours, non‐neutralized virus was detected in Madin‐Darby kidney cells (MDKC) cells by an ELISA targeting viral nucleoprotein. A 50% endpoint was used for neutralization. The HI assay was performed with 0·5% turkey RBCs using twofold dilutions of sera and a standard amount of hemagglutination units of virus. Serum samples for the HI assay were treated with receptor‐destroying enzyme and adsorbed with RBCs if non‐specific agglutinins were detected prior to use in the assay. A 100% endpoint was used for hemagglutination inhibition.
Individuals with paired serum samples that demonstrated greater than or equal to fourfold rise in titer by either assay were considered to have seroconverted. In addition, because specimen collection was not optimal for all participants (e.g., poor timing of acute specimens or lack of convalescent specimen), individuals who did not seroconvert but who had serum antibody titers of ≥40 by MN and ≥20 by HI in any serum sample were considered seropositive. This combination of H1N1pdm09‐specific antibody titers was shown to provide 90% sensitivity and 96% specificity for detection of H1N1pdm09 infection in individuals <60 years of age and 92% specificity in those aged 60–79 years. 4 We defined serologic evidence of H1N1pdm09 infection as detection of either seroconversion or seropositivity. Because of the lack of adequate specificity of serologic testing for individuals aged ≥80 years, in this age‐group, we defined seroconversion as seropositive, a convalescent serum antibody titers of ≥40 by MN and ≥20 by HI as an indeterminate serologic test result, and those without this combination of HI and MN results to be negative. We compared log2‐transformed MN and HI titer values and differences between categorical and continuous variables with Student’s t‐test, chi‐square, and rank sum tests, respectively. P ≤ 0·05 was considered statistically significant.
The Centers for Disease Control and Prevention determined that this investigation represented public health response not requiring institutional review board authorization.
Results
A total of 19 PCR‐confirmed H1N1pdm09 case‐patients were identified, and 16 had serum collected; however, only ten case‐patients had at least one serum specimen collected >14 days post‐symptom onset, including those with either well‐timed paired sera. Of the four case‐patients with well‐timed paired sera, three demonstrated seroconversion, and the fourth was seropositive in both samples. Six case‐patients with at least one specimen collected >14 days post‐symptom onset were all seropositive. In addition, six case‐patients had suboptimal timing for serum collection, single sera collected 7–13 days post‐symptom onset, and of those, four were seropositive. The median age of the 16 case‐patients with serum collected was 23 years [interquartile range (IQR): 8–39 years]. Serologic responses and the highest titers were best detected at >14 days post‐symptom onset (Figure 1). 4
Figure 1.
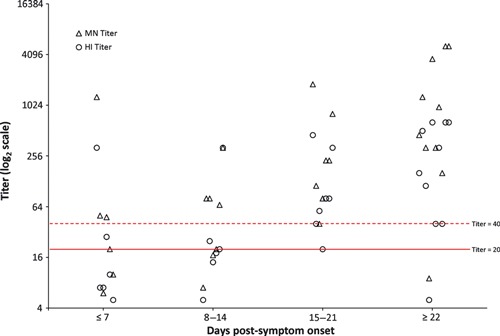
Kinetics of antibody response to H1N1pdm09 infection among PCR‐confirmed index cases. The Microneutralization (MN) and hemagglutination inhibition (HI) titers of 30 serum samples from 16 PCR‐confirmed index cases and three PCR‐confirmed close contacts are grouped by number of days after symptom onset in 7‐day intervals. Random staggering of paired data points (MN and HI titers) was used to optimize data presentation.
Seven (37%) of the 19 PCR‐confirmed H1N1pdm09 index case‐patients had close contacts that agreed to participate in the investigation. Among 80 identified close contacts, 75 (94%) participated. Three close contacts with serologic evidence of H1N1pdm09 infection reported symptoms prior to the onset of symptoms of patient A (March 30) and B (March 28) (Figure 2). One extended family contact of patient B had a reported onset of illness of March 21, and one household contact of patient B reported onset on March 25; all had extensive contact before and after illness dates. The third person was a SD resident with no epidemiologic links to patient A or B, a close contact of a PCR‐confirmed case identified from surveillance, with a probable onset date of March 23. None of these persons reported travel to Mexico during the week prior to illness onset. These three persons were reclassified as index case‐patients for the remaining analyses.
Figure 2.
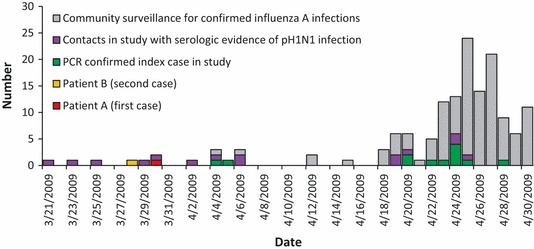
The number of patients infected with H1N1pdm09 in the serologic survey or patients infected with H1N1pdm09 or influenza A virus from community surveillance from San Diego and Imperial counties, California, by date of onset of illness from March 21, 2009–April 30, 2009.
Most close contacts were either household [16/75 (22%)] or extended family [43/75 (57%)] contacts (Figure 3); the remaining were occupational (n = 7), neighbor (n = 2), or school or daycare (n = 3) contacts. Among 69 close contacts with at least one specimen collected >14 days since the onset of index case‐patient symptoms, 23 (33%) had serologic evidence of H1N1pdm09 infection (eight seroconversions and 15 seropositive), and 8 (35%) reported no symptoms. Four contacts aged ≥80 years had indeterminate serology and were excluded from subsequent analyses. The proportion of contacts with serologically confirmed H1N1pdm09 infection was highest in persons aged 5–24 years [12/24 (50%)], followed by persons aged 25–49 years [8/23 (35%)], and lowest in persons aged ≥50 years [2/15 (2%)], the difference was of borderline statistical significance (P = 0·07). Household contacts had a statistically higher frequency of serologic evidence of H1N1pdm09 infection [8/15 (53%)] compared to extended family contacts [14/40 (35%)] and other contacts [occupational, school or daycare and neighbor contacts: 1/14 (7·1%)] (P = 0·03).
Figure 3.
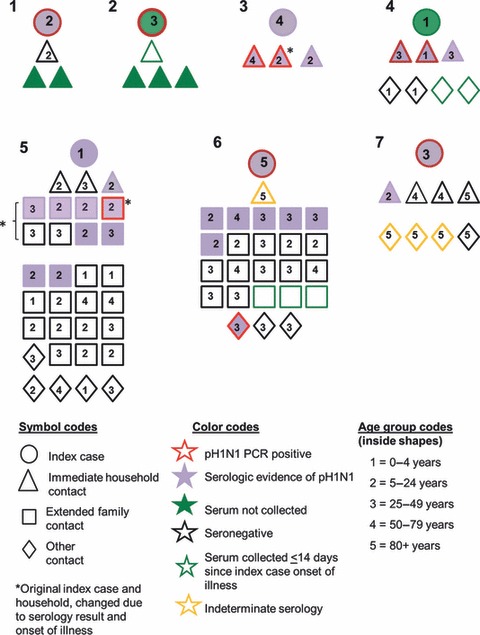
Graphic depiction of seven index case‐patients and their 80 close contacts included in the serologic survey, San Diego and Imperial counties, California, USA, April 2009 (2b). The clusters are not numbered in any specific order.
Overall, 24 (35%) close contacts reported symptoms; however, only 15 (63%) of these symptomatic contacts had serologic evidence for H1N1pdm09 infection. All symptomatic contacts had serum collected >14 days after the onset of illness. The median age of contacts with and without serologic evidence of H1N1pdm09 infection was 22 (IQR: 13–38) and 36 (IQR: 11–53) years, respectively (P = 0·22).
The geometric mean titers (GMT) for MN and HI among contacts with subclinical infection and symptomatic close contacts with serologic evidence of H1N1pdm09 infection appeared similar (P = 0·36, P = 0·33, respectively) (Table 1). The MN and HI GMT values for 10 index cases with serum specimen >14 days after symptom onset (including four with well‐timed paired sera) were statistically higher than the GMT values from contacts with subclinical infections (P = 0·01, P = 0·04, respectively) and not different than the GMT values from symptomatic contacts (P = 0·20, P = 0·16, respectively).
Table 1.
Microneutralization (MN) and hemagglutination inhibition (HI) geometric mean titers (GMT) of selected pH1N1 index cases* and of the close contacts* of six index cases
| Index cases (n = 10) | Seropositive contacts with symptoms (n = 12) | Seropositive contacts withsubclinical infection (n = 11) | |
|---|---|---|---|
| MN GMT (95% CI) | 707 (414, 1209) | 375 (178, 788) | 241 (144, 404) |
| HI GMT (95% CI) | 221 (111, 326) | 118 (64, 154) | 83 (51, 135) |
| Time from onset of illness to convalescent serum, median days (IQR) | 25 (22–31) | 36 (19–39) | 23 (22–35) |
| Age, median years (IQR) | 23 (7, 39) | 16 (11, 24) | 38 (29, 44) |
| Reported fever, No. (%) | 9 (100)** | 10 (83) | 0 |
CI, confidence interval; IQR, interquartile range.
*Index cases with a convalescent specimen >14 days after symptom onset, including those with well‐timed paired sera. Contacts with blood draw >14 days after symptom onset or after onset of index patient symptoms.
**Information missing on one index case.
Discussion
We provide serological evidence of early community transmission of H1N1pdm09 in southern California in March 2009. Also, the H1N1pdm09 outbreak in the United States began slightly earlier than previously documented. The detection of three unrelated clusters of H1N1pdm09 illness in southern California during March 21–30 suggests that community transmission of H1N1pdm09 had begun prior to this time. The lack of detection of H1N1pdm09 viruses in routine surveillance 8 or reports of respiratory outbreaks (Michele Ginsberg and Paula Kriner, personal communication) implies that transmission was of limited scope or occurred in populations not easily captured by surveillance, such as persons that do not access standard healthcare providers.
While our sample size was small, this very early serologic investigation during the U.S H1N1 pdm09 pandemic provided some important information that has been corroborated by other studies. Persons aged 5–24 years who were close contacts of PCR‐confirmed H1N1pdm09 index cases had the highest secondary attack rates (SAR), similar to other reports. 2 , 9 , 10 However, unlike other studies, the difference between age‐groups was of borderline statistical significance, probably secondary to our small sample size. Household contacts had higher SAR compared to other contact types. Also, the household SAR that we report was higher than other reports that used syndromic case definitions or PCR to detect H1N1pdm09 infection, and not serology. 9 , 10 However, a third of the close contacts with serologically confirmed H1N1pdm09 infection in our investigation had subclinical infections. Also, the household SAR is consistent with a report from Canada that also used serology to estimate household SAR (SAR = 45%). 11 Other published studies have reported between 9% and 45% subclinical H1N1 pdm09 infections among those seropositive. 11 , 12 , 13
Interestingly, the proportion of contacts with serological evidence of H1N1pdm09 infection at the very beginning of the pandemic in the United States was similar to serology‐based estimates of community H1N1pdm09 infection determined later in the pandemic with larger serosurveys. 2 , 13 , 14 A third of the contacts of PCR‐confirmed case‐patients from SD and IC had evidence of infection with H1N1pdm09 virus by serology. We did not limit the contacts in this investigation to household members but included several large extended families, school classmates, and persons with possible occupational exposure. Thus, transmission in our cohort from early in the pandemic prior to widespread community transmission appeared similar to community transmission at the peak of virus circulation. This study demonstrates how early field investigations that utilize serology may provide useful estimates to help characterize the transmission of pandemic viruses and can supplement syndrome‐based evidence.
We used a lower HI cutoff value for seropositivity than other studies (e.g., ≥20 versus either ≥32 or ≥40 used in other studies). 2 , 13 , 14 The use of both MN and HI improved our sensitivity to detect H1N1pdm09 infection, and by day 14, most persons with PCR‐confirmed H1N1pdm09 infection had MN and HI titers at or above our cutoff values. 4 However, all close contacts with serologic evidence of H1N1pdm09 infection had HI titers ≥40 except for one person who had HI = 20 and MN titers = 80. Thus, our results are comparable to other reports; the improved sensitivity of both HI and MN added few H1N1pdm09 infections.
Our conclusions are limited because of the suboptimal timing of blood draws for all participants and small sample size. While we initiated the investigation at the time the two‐first patients were detected with a novel virus, the timing of our investigation, at the beginning of detection of the 2009 pandemic, made optimal timing for paired serum collection difficult and once the pandemic was declared other public health duties were prioritized. Also, some close contacts reported illnesses several weeks prior to interviews and were not tested for influenza virus infection at the time of illness. Thus, despite serologic evidence for H1N1pdm09 infection, we cannot be certain that the illnesses they recalled were because of H1N1pdm09 infection. Finally, we did not collect standard information on antiviral use; however, we have no evidence that any contacts received antiviral chemoprophylaxis prior to, or in between, their blood draws (M. Patel and M. Gladden, personal communication).
Despite shortcomings, our findings are noteworthy as they demonstrate early community transmission of H1N1pdm09 in southern California in March 2009 and establish a slightly earlier onset of the H1N1pdm09 outbreak in the United States.
Addendum
Alicia M. Fry participated in data cleaning and analysis, interpretation of analysis, and critical writing and revising the intellectual content. Kathy Hancock, Vic Veguilla, Xiuhua Lu, Heather Noland, Yaohui Bai and Jacqueline M. Katz participated in study concept and design, serologic assay development, interpretation of analysis, and revising the intellectual content. Minal Patel, Matthew Gladden and Dianna M. Blau participated in study concept and design, data collection and cleaning, and revising the intellectual content. Saumil Doshi, David Sugerman, Azarnoush Maroufi, Darlene Sunega, Annie Kao, Paula Kriner, Karla Lopez and Michele Ginsberg participated in study concept and design, data collection, and revising the intellectual content. Seema Jain and Sonja J. Olsen participated in study concept and design and revising the intellectual content. All co‐authors approve this version of the manuscript for publication.
Conflicts of interest and financial disclosures
None of the authors have any affiliation, financial agreement, or involvement with a company whose product figures prominently in this manuscript.
Funding
No external funding was received for this project.
Disclaimer
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Acknowledgement
We acknowledge Dr Paul Garguillo from Centers for Disease Control and Prevention, Atlanta, GA, for his statistical help.
Working Group members: Jarad Schiffer, Darbi Aranio, Alicia Branch, Libo Dong, Crystal Holiday, Feng Liu, Evelene Steward‐Clark, Hong Sun, Byron Tsang, David Wang, Melissa Whaley, Yaohui Bai, Li Cronin, Peter Browning, Hanan Dababneh, Leilani Thomas, Lydia Foster, Conrad Quinn, and Stephen Soroka from Centers for Disease Control and Prevention, Atlanta, GA.
Previous presentation: These results were previously presented at Options for the Control of Influenza VII, September 3–7, 2010, Hong Kong China, abstract #P‐334.
References
- 1. Centers for Disease Control and Prevention . Swine Influenza A (H1N1) Infection in Two Children – Southern California, March–April 2009. Morb Mortal Wkly Rep 2009; 58:400–402. [PubMed] [Google Scholar]
- 2. Miller E, Hoschler K, Hardelid P, Stanford E, Andrews N, Zambon M. Incidence of 2009 pandemic influenza A H1N1 infection in England: a cross‐sectional serological study. Lancet 2010; 375:1100–1108. [DOI] [PubMed] [Google Scholar]
- 3. Chen MI, Barr IG, Koh GCH et al. Serological response in RT‐PCR confirmed H1N2‐2009 influenza A by hemagglutination inhibition and virus neutralization assays: an observational study. PLoS ONE 2010; 5:e12474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Veguilla V, Hancock K, Schiffer J et al. Sensitivity and Specificity of Serologic Assays for the Detection of Human Infection with 2009 Pandemic H1N1 Virus in U.S. Populations. J Clin Microbiol 2011; 49:2210–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kendal AP, Pereira MS, Skehel JJ. Concepts and Procedures for Laboratory‐Based Influenza Surveillance; 1982. Atlanta GA: United States Department of Health and Human Services, Public Health Service Centers for Disease Control, 1982. [Google Scholar]
- 6. Rowe T, Abernathy RA, Hu‐Primmer J et al. Detection of antibody to avian influenza A (H5N1) virus in human serum by using a combination of serologic assays. J Clin Microbiol 1999; 37:937–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. World Health Organization (2010) Serological diagnosis of influenza by microneutralization assay. Available at http://www.who.int/csr/disease/influenza/influenzanetwork/en/index.html (Accessed 12 December 2010).
- 8. Centers for Disease Control and Prevention . FluView. Available at http://www.cdc.gov/flu/weekly/. (Accessed 4 January 2011)
- 9. Cauchemez S, Donnelly CA, Reed C et al. Household transmission of 2009 pandemic influenza A (H1N1) virus in the United States. N Engl J Med 2009; 361:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cowling BJ, Chan KH, Fang VJ et al. Comparative epidemiology of pandemic and seasonal influenza A in households. N Engl J Med 2010; 362:2175–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Papenburg J, Baz M, Hamelin MÈ et al. Household transmission of the 2009 pandemic A/H1N1 influenza virus: elevated laboratory‐confirmed secondary attack rates and evidence of asymptomatic infections. Clin Infect Dis 2010; 51:1033–1041. [DOI] [PubMed] [Google Scholar]
- 12. Lau LL, Cowling BJ, Fang VJ et al. Viral shedding and clinical illness in naturally acquired influenza virus infections. J Infect Dis 2010; 201:1509–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bandaranayake D, Huang QS, Bissielo A et al. Risk factors and immunity in a nationally representative population following the 2009 influenza A(H1N1) pandemic. PLoS ONE 2010; 5:e13211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen MIC, Lee VLM, Lim W et al. 2009 Influenza A(H1N1) 2, and Risk Factors Among Distinct Adult Cohorts in Singapore. JAMA 2010; 303:1383–1391. [DOI] [PubMed] [Google Scholar]


