Abstract
The sequestrate false truffles Elaphomyces favosus, E. iuppitercellus, and E. labyrinthinus spp. nov. are described as new to science from the Dja Biosphere Reserve, Cameroon. Elaphomyces adamizans sp. nov. is described as new from the Pakaraima Mountains of Guyana. The Cameroonian species are the first Elaphomyces taxa to be formally described from Africa, occurring in lowland Guineo-Congolian tropical rainforests dominated by the ectomycorrhizal (ECM) canopy tree Gilbertiodendron dewevrei (Fabaceae subfam. Caesalpinioideae). The Guyanese species is the third to be discovered in lowland tropical South America, occurring in forests dominated by the ECM trees Pakaraimaea dipterocarpacea (Dipterocarpaceae) and Dicymbe jenmanii (Fabaceae subfam. Caesalpinioideae). Macromorphological, micromorphological, habitat, and DNA sequence data are provided for each new species. Molecular and morphological data place these fungi in Elaphomycetaceae (Eurotiales, Ascomycota). Unique morphological features are congruent with molecular delimitation of each of the new species based on a phylogenetic analysis of the rDNA ITS and 28S loci across the Elaphomycetaceae. The phylogenetic analysis also suggests that a common ancestor is shared between some Elaphomyces species from Africa and South America, and that species of the stalked, volvate genus Pseudotulostoma may be nested in Elaphomyces.
Keywords: biogeography, ectomycorrhizal fungi, Gilbertiodendron, Guiana Shield, Guineo-Congolian rainforest, Pakaraimaea, sequestrate fungi, Uapaca
INTRODUCTION
Elaphomyces Nees 1820 (Elaphomycetaceae, Eurotiales, Ascomycota) is a sequestrate, ectomycorrhizal (ECM) fungal genus that associates with a broad range of primarily north or south temperate angiosperm and gymnosperm hosts (Trappe et al. 2009, Castellano et al. 2011, Quandt et al. 2015). Elaphomyces species generally fruit hypogeously and have relatively large cleistothecial ascomata with a thick peridium, a powdery, hydrophobic gleba, and dark, globose, ornamented ascospores (Trappe 1979). Aside from new tropical Australian species recently described by Castellano et al. (2011), there is a paucity of published Elaphomyces records from the tropics (e.g. Corner & Hawker 1955, Castellano et al. 2012). Unpublished and currently undescribed Elaphomyces collections have been reported from Costa Rica, Java, Papua New Guinea, and Thailand (Reynolds 2011, Nampia Sukarno pers. comm., T.F.E., unpubl.data).
Castellano et al. (2012) provided the first report of Elaphomyces from the lowland South American tropics, describing two new species associated with ECM Fabaceae hosts in Guyana. Subsequently, our continued collecting efforts in the tropics of Africa and South America have uncovered four additional new Elaphomyces species. Here we describe Elaphomyces favosus, E. iuppitercellus, and E. labyrinthinus spp. nov. from the Dja Biosphere Reserve in Cameroon, and E. adamizans sp. nov. from the Pakaraima Mountains of Guyana. The Cameroonian species are the first to be formally described from Africa, although Elaphomyces partial ITS root tip sequences have been reported from the African tropics (e.g. Tedersoo et al. 2010, 2011) and as yet undescribed Elaphomyces ascomata have been collected in Madagascar (Bart Buyck, pers. comm.). The Cameroonian species are currently only known from primary Guineo-Congolian tropical rainforests dominated by the ECM canopy tree Gilbertiodendron dewevrei (Fabaceae subfam. Caesalpinioideae) with additional scattered trees of the ECM genus Uapaca (Phyllanthaceae). The Guyanese species occurs in primary forests co-dominated by the ECM trees Pakaraimaea dipterocarpacea (Dipterocarpaceae) and Dicymbe jenmanii (Fabaceae subfam. Caesalpinioideae). Macromorphological, micromorphological, habitat, and DNA sequence data are provided for each new species. A molecular phylogenetic analysis assesses the relationships of the new species within Elaphomycetaceae, and suggests that a common ancestor is shared between some species from Africa and South America, and that stalked, volvate species of Pseudotulostoma may be nested within Elaphomyces.
MATERIALS AND METHODS
Collections
In Guyana, ascomata were collected during the June rainy season of 2012 from Pakaraima Mountains, Upper Mazaruni River Basin, near a base camp at 5o26’21.3” N 60o4’43.1” W, 800 m a.s.l., in savanna fringing forest dominated by P. dipterocarpacea and D. jenmanii (Smith et al. 2013). In Cameroon, ascomata and ECM root tips were collected during the Aug.–Sep. early rainy season of 2014 from the Dja Biosphere Reserve, Northwest Sector near the village of Somalomo, Upper Dja River Basin, within a two km radius of a base camp located at 3o21’29.8” N 12o43’46.9” W, 650 m a.s.l., in forests dominated by G. dewevrei (Peh et al. 2014).
Descriptions of macromorphological features were made from fresh material in the field. Colours were compared with colour plates from Kornerup & Wanscher (1978) and are cited in parentheses (e.g. 5A4). Fresh collections were dried with silica gel. Preserved specimens were later examined in 3 % KOH, Melzer’s reagent, and Cotton blue. Microscopic descriptions are based on 3 % KOH mounts unless specified otherwise. Twenty ascospores were measured from each type collection; dimensions reported include ornamentation. Dried ascospores were mounted on aluminum pegs with double-sided tape and coated with gold for scanning electron microscopy (SEM) with an AmRay 3300 FE field emission scanning electron microscope. Type and additional specimens are deposited in the following herbaria: BRG, University of Guyana; YA, Cameroon National Herbarium; HSC, Humboldt State University; OSC, Oregon State University; K(M), Fungarium, Royal Botanic Gardens, Kew.
DNA extraction, PCR amplification, and sequencing
All DNA work was carried out in the Jodrell Laboratory, Royal Botanic Gardens, Kew. DNA extractions were performed on ascoma tissue from specimens and ECM root tips using the Extract-N-Amp Plant PCR kit (SIGMA-ALDRICH, Saint Louis, MO), followed or not by plate filtration (Dentinger et al. 2010), or using a Plant DNeasy mini kit (QIAGEN, Valencia, CA). Full internal transcribed spacers 1 and 2, along with the 5.8S rDNA (ITS), were PCR-amplified with primers ITS1F and ITS4 (White et al. 1990, Gardes & Bruns 1993), and the nuclear 28S rDNA D1–D2 domains (28S) were PCR-amplified with LR0R/LR5 (Vilgalys & Hester 1990) following the cycling conditions in Dentinger et al. (2010). PCR products were visualized by UV fluorescence after running out 2 μL PCR products in a 1 % agarose gel containing 0.005 % ethidium bromide. Prior to sequencing, amplicons were cleaned of unincorporated dNTPs and excess primers by adding 1 μL ExoSAP-IT (USB, Cleveland, OH) to 5 μL PCR reaction mix and incubating for 15 min at 37 °C followed by 15 min at 80 °C. Unidirectional dye-terminator sequencing used BigDye3.1 (Applied Biosystems, Foster City, CA), by adding 2 μL of cleaned PCR template to 3 μL of solution containing 0.2 μL BigDye, 1 μL sequencing buffer, 0.15 μL 50mM MgCl2, 0.15 μL of 10 μM primer, and 1.5 μL of Milli-Q (Merck Millipore, Darmstadt, Germany) purified water. Sequencing was performed with 60 cycles of 95 °C denaturation for 10 sec, 50 °C annealing for 10 sec, and 60 °C extension for 2 min. Sequencing reactions were cleaned using ethanol precipitation and resuspended in purified water before loading into an ABI 3730 DNA Analyzer (Applied Biosystems, Foster City, CA). Complementary unidirectional sequence reads were aligned and edited in Sequencher v. 4.2 (Gene Codes, Ann Arbor, MI) and deposited in GenBank (Table 1).
Table 1.
Elaphomycetaceae taxa, voucher numbers, collection locales, and GenBank accession numbers for ITS and 28S nuc rDNA used in the phylogenetic analysis. Taxa described here and newly generated sequences are in bold at the top. Sequences on the complementary strand are indicated by an asterisk (*).
| Taxon | Voucher | Collection locale as indicated in GenBank | ITS | 28S |
|---|---|---|---|---|
| Elaphomyces adamizans | TH9660 (type) | Region 7 Cuyuni-Mazaruni, Guyana | KT694133 | KT694144 |
| Elaphomyces favosus | TH10015 | East Province, Cameroon | KT694134 | KT694145 |
| Elaphomyces favosus | TH9859 (type) | East Province, Cameroon | KT694138 | KY694149 |
| Elaphomyces favosus | TH9874 | East Province, Cameroon | KT694135 | KT694147 |
| Elaphomyces favosus | TH9897 | East Province, Cameroon | KT694136 | KT694146 |
| Elaphomyces iupperticellus | M3 (root tip) | East Province, Cameroon | KT694140 | |
| Elaphomyces iupperticellus | TH9934 | East Province, Cameroon | KT694141 | KT694142 |
| Elaphomyces iupperticellus | THDJA 39 (type) | East Province, Cameroon | KT694139 | KT694143 |
| Elaphomyces labryinthinus | TH9918 (type) | East Province, Cameroon | KT694137 | KT694148 |
| Elaphomyces aculeatus | 16952 | Italy | JF907985 | |
| Elaphomyces aff. decipiens | GO-2009-211 | Mexico | KC152093 | |
| Elaphomyces citrinus | 16955 | Spain | JF907986 | |
| Elaphomyces compleximurus | TH8880 | Guyana | JN711441 | JN711441 |
| Elaphomyces compleximurus | TH8880 | Guyana | NR_121522 | |
| Elaphomyces decipiens | Trappe 12436 | USA | EU837229 | |
| Elaphomyces decipiens | Trappe 28269 | USA | EU846311 | |
| Elaphomyces digitatus | MCA1512 | Guyana | JN713147 | |
| Elaphomyces digitatus | TH8887 | Guyana | JQ657705 | |
| Elaphomyces digitatus | MCA1923 | Guyana | JN713148 | |
| Elaphomyces granulatus | K(M)47712 | UK | EU784197 | |
| Elaphomyces guangdongensis | KH-TW09-030 | Taiwan | HM357249 | |
| Elaphomyces guangdongensis | KH-TW09-031 | Taiwan | HM357250 | HM357248 |
| Elaphomyces leveillei | 16960 | Italy | JF907987 | |
| Elaphomyces maculatus | 16961 | Italy | JF907988 | |
| Elaphomyces muricatus | src641 | USA | DQ974740 | |
| Elaphomyces muricatus | K(M)121442 | UK | EU784198 | |
| Elaphomyces muricatus | Hy14 (root tip) | Finland | GU550112 | |
| Elaphomyces muricatus | n.a. | Poland | JF834198 | |
| Elaphomyces muricatus | HA38 (root tip) | Latvia | KR019869 | |
| Elaphomyces sp. | YM144 (root tip) | Japan | AB848482* | |
| Elaphomyces sp. | HB1 | Indonesia | LC010285* | |
| Elaphomyces sp. | HB3 | Indonesia | LC010287* | |
| Elaphomyces sp. | HB2 | Indonesia | LC010286* | |
| Elaphomyces sp. | 7381.2 (root tip) | UK | FJ876187 | |
| Elaphomyces sp. | fc1639 (root tip) | UK | FJ876188 | |
| Elaphomyces sp. | AM3GA3A4 | USA | JQ272414 | |
| Elaphomyces sp. | GM 13-32 (root) | USA | KF359559 | |
| Elaphomyces sp. | M6ELA | Poland | KJ524537 | |
| Elaphomyces sp. | GO-2009-040 | Mexico | KJ594995 | |
| Elaphomyces sp. | GO-2009-028 | Mexico | KJ594996 | |
| Elaphomyces sp. | LM2779 (root tip) | Romania | KM576390 | |
| Elaphomyces sp. | LM5570B (root tip) | Hungary | KM576391 | |
| Elaphomyces sp. | LM1606 (root tip) | UK | KM576392 | |
| Elaphomyces sp. | LM1612 (root tip) | UK | KM576393 | |
| Elaphomyces sp. | LM2195 (root tip) | UK | KM576394 | |
| Elaphomyces sp. | LM34 (root tip) | Spain | KM576395 | |
| Elaphomyces sp. | ITS-545ek (root tip) | Latvia | KP753307 | |
| Elaphomyces sp. | PA5 (root tip) | Latvia | KR019785 | |
| Pseudotulostoma volvatum | TH8975 | Guyana | JN168735 | JN168735 |
| Uncultured Elaphomyces | O17 (root tip) | Estonia | AJ534702 | |
| Uncultured Elaphomyces | L503Z_E1 (root tip) | Estonia | AJ893252 | |
| Uncultured Elaphomyces | UBCOGTR184 (root tip) | Canada | EU597039 | |
| Uncultured Elaphomyces | SLUBC46 (environmental sample) | Canada | FJ152544 | FJ152544 |
| Uncultured Elaphomyces | SDL33 (root tip) | USA | FJ769549 | |
| Uncultured Elaphomyces | BJP93T_102 (root tip) | UK | GU289427 | |
| Uncultured Elaphomyces | root tip | Poland | HM015499 | |
| Uncultured Elaphomyces | 4174-1205 (root tip) | UK | HM146813 | |
| Uncultured Elaphomyces | 4115-1205 (root tip) | UK | HM146814 | |
| Uncultured Elaphomyces | 5237-1201 (root tip) | UK | HM146815 | |
| Uncultured Elaphomyces | 1Bart526S (soil) | USA | HQ021767 | |
| Uncultured Elaphomyces | Bart1760S (soil) | USA | HQ021973 | |
| Uncultured Elaphomyces | 4Bart240R (root tip) | USA | HQ021974 | |
| Uncultured Elaphomyces | 4Bart24S (soil) | USA | HQ022116 | |
| Uncultured Elaphomyces | 1Bart34R (root tip) | USA | HQ022117 | |
| Uncultured Elaphomyces | 4Bart309S (soil) | USA | HQ022186 | |
| Uncultured Elaphomyces | Ref_306 (root tip) | USA | HQ285385 | |
| Uncultured Elaphomyces | Brg_333 (root tip) | USA | HQ285386 | |
| Uncultured Elaphomyces | LMAS17c-09 (soil) | France | JF506754 | |
| Uncultured Elaphomyces | T566 | Tasmania | JF960758 | |
| Uncultured Elaphomyces | ecm1108 (root tip) | Guyana | JN168718 | JN168718 |
| Uncultured Elaphomyces | 1_28M5 (root tip) | USA | JQ393049 | |
| Uncultured Elaphomyces | ECM92 (root tip) | China | JQ991717 | |
| Uncultured Elaphomyces | ECM93 (root tip) | China | JQ991718 | |
| Uncultured Elaphomyces | ECM94 (root tip) | China | JQ991719 | |
| Uncultured Elaphomyces | ECM95 (root tip) | China | JQ991720 | |
| Uncultured Elaphomyces | SJ-LM318 (root tip) | UK | KC412512 | |
| Uncultured Elaphomyces | B4pos3.4_35 (clone) | Canada | KC702640 | |
| Uncultured Elaphomyces | F4pos1.1_43 (clone) | Canada | KC702648 | |
| Uncultured Elaphomyces | F4pos1.2_49 (clone) | Canada | KC702649 | |
| Uncultured Elaphomyces | 15 (root tip) | Poland | KF484416 | |
| Uncultured Elaphomyces | HVM21 (root tip) | USA | KF879472 | |
| Uncultured Elaphomyces | ecm62 (root tip) | Latvia | KF954066 | |
| Uncultured Elaphomyces | 141A (root) | Canada | KM403019 | KM403019 |
| Uncultured Elaphomycetaceae | jj046 (root tip) | Sweden | AY839226 |
Taxa used, sequence alignment, and phylogenetic analysis
All Elaphomycetaceae (e.g. Elaphomyces, Pseudotulostoma) ITS and 28S sequences derived from ascomata and ECM root tips available in GenBank were downloaded. Each gene region was aligned separately with the sequences from our new species using the RNA structure-based algorithm Q-INS-i implemented in MAFFT v7.023b (Katoh et al. 2002, Katoh & Toh 2008, Katoh & Standley 2013). After correcting the orientations of four ITS sequences, removing one short sequence (GenBank accession AM087442) and one sequence with substantial numbers of ambiguous bases (GenBank accession AB161194), the uneven ends were trimmed and alignments refined with a second round of alignment in MAFFT, as above, and refined alignments concatenated into a single dataset. Phylogenetic analysis under the maximum likelihood criterion was performed using the Pthreads parallelised version of RAxML v7.0.3 (Stamatakis 2006, Ott et al. 2007) with a GTRGAMMA model, allowing model parameters to be estimated for each gene partition separately. Branch support was assessed using nonparametric bootstrapping with the autoMRE option. Geographic sources of sequences used from GenBank were determined primarily from locality information in GenBank records. The alignment and tree have been accessioned in TreeBase at http://purl.org/phylo/treebase/phylows/study/TB2:S18165.
RESULTS
Final alignments consisted of 82 sequences and 832 positions for the ITS (381 parsimony informative, 361 constant, 90 autapomorphic), and of 18 sequences and 887 positions for the 28S (145 parsimony informative, 712 constant, 30 autapomorphic). All 28S sequences had corresponding ITS sequences derived from the same source, except for Elaphomyces digitatus where they were derived from two separate conspecific sources and subsequently combined in the dataset. All characters were included in the analysis. RAxML rapid bootstrapping terminated after 550 replicates (WRF average of 100 random splits = 2.319227) and the best ML tree had a likelihood score of –10065.206922 (Fig. 1). Analysis of a data set consisting only of ITS sequences recovered a best ML tree that differed only in the placement of a few unsupported branches (Fig. 2).
Fig. 1.
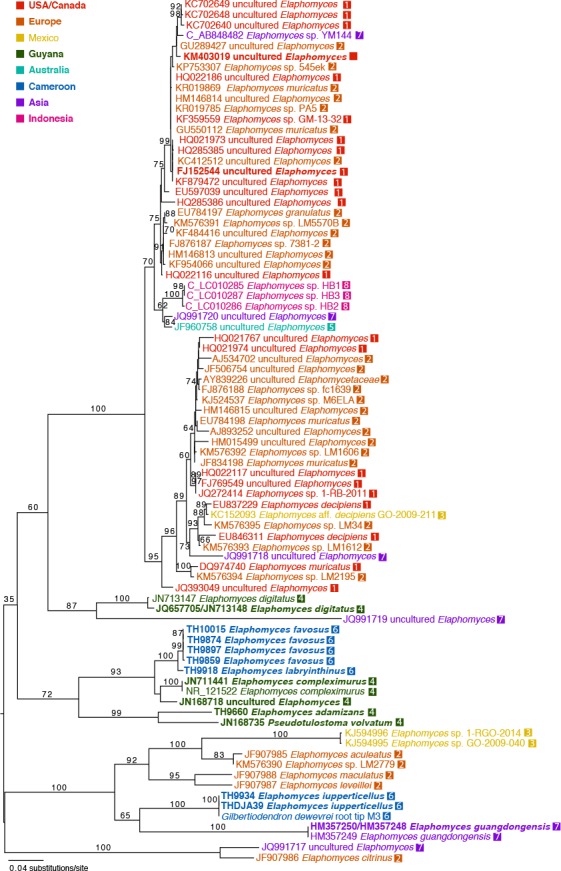
Best maximum likelihood phylogram (–ln = 10065.206922) of a combined analysis of ITS and 28S sequences of Elaphomycetaceae taxa in RAxML using a GTRGAMMA substitution model. Tree is midpoint rooted. Numbers on or next to branches are nonparametric bootstrap supports >70 % from 550 bootstrap replicates. For terminals downloaded from GenBank, labels begin with GenBank accession number and are colour-coded by geographic origin of the sources of the sequences as determined by GenBank records. Terminal labels for taxa generated in this study begin with the collection number. Terminals in bold are represented by both ITS and 28S sequences. Those beginning with a ‘C’ had their sequence orientation corrected for phylogenetic analysis.
Fig. 2.
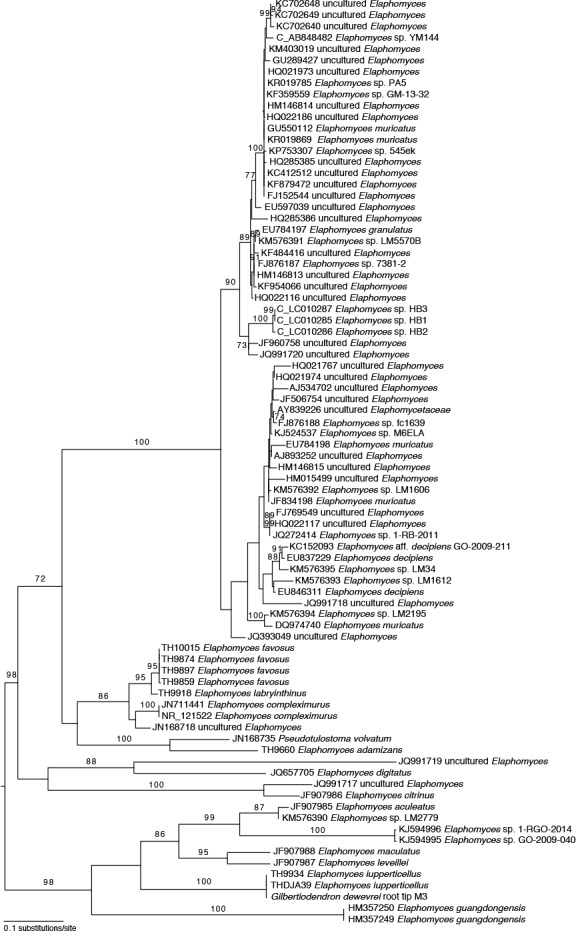
Best maximum likelihood phylogram of an analysis of ITS sequences in RAxML using a GTRGAMMA substitution model. Tree is midpoint rooted. Numbers on or next to branches are nonparametric bootstrap supports >70 % from 550 bootstrap replicates. Terminal labels for taxa generated in this study begin with the collection number.
The new species described here were resolved in strongly supported lineages at the 93–100 % bootstrap level within Elaphomyces (Fig. 1). Amongst the Cameroonian species, E. iuppitercellus was recovered in a strongly supported clade (100 % bootstrap) containing the European E. aculeatus, E. leveillei, and E. maculatus, an unidentified Elaphomyces species, and the east Asian E. guangdongensis. Ascoma-derived sequences of E. iuppitercellus were identical to those of a sympatric ECM G. dewevrei root tip, confirming the ECM status of the species. Elaphomyces favosus and E. labyrinthinus were strongly supported (100 % bootstrap) as sisters within a well supported (93 % bootstrap) clade that includes E. compleximuris and an ECM root tip from Guyana, suggesting that these taxa share a common ancestor within the genus (Fig. 1). The new Guyanese species, E. adamizans, was strongly supported (99 % bootstrap) as sister with the stalked, volvate Pseudotulostoma volvatum from Guyana; together these sympatric taxa formed a larger moderately supported (72 % bootstrap) clade with E. favosus, E. labryinthinus, and E. compleximurus (Fig. 1). These phylogenetic results, along with unique morphological features, warrant the description of the Cameroonian and Guyanese species as new to science, and suggest that Pseudotulostoma and Elaphomyces may not be reciprocally monophyletic.
TAXONOMY
Elaphomyces favosus Castellano & T.W. Henkel, sp. nov.
Index Fungorum IF551318
(Fig. 3)
Fig. 3.
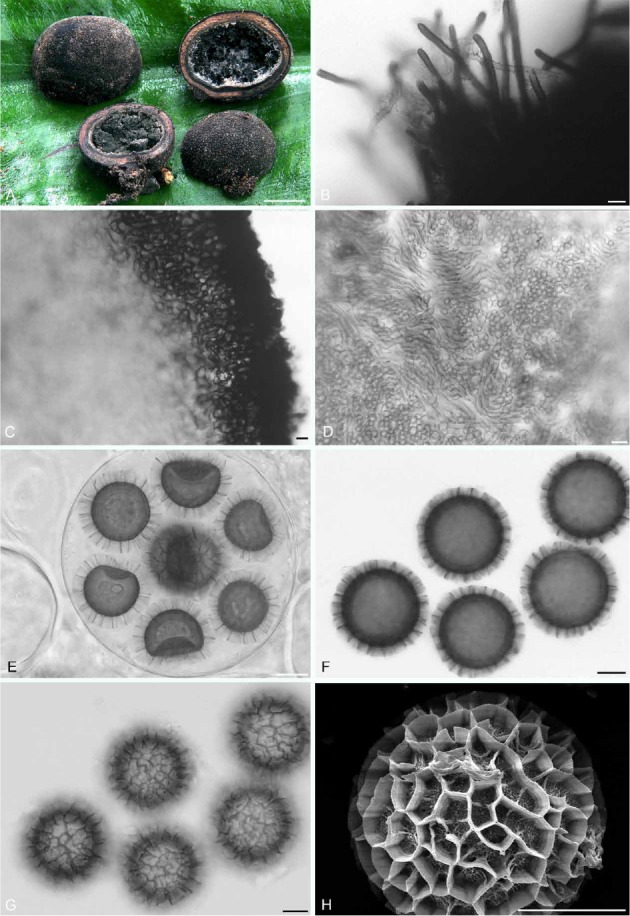
Elaphomyces favosus (holotype; Henkel 9859). A. Ascomata showing peridial surface, gleba, and peridium in section. B. Erect peridial hairs found in patches on peridial surface. C. Microscopic view of peridium in section. D. Third layer of peridium with cross-hatched, bundled hyphae. E. Ascus with eight ascospores. F. Ascospores with ornamentation in outline. G. Ascospores with ornamentation in surface view. H. Scanning electron micrograph of an ascospore. Bars A = 10 mm, B, D–H = 10 μm, C = 20 μm.
Etymology: favosus (L. adj. A) = honey-combed; referring to the distinctive reticulate-alveolate ascospore ornamentation.
Diagnosis: Similar to the neotropical E. compleximurus in ascospore ornamentation and colours of the outer peridium and gleba, but differing in its distinctly larger ascospores (mean diameter with ornamentation = 35.7 μm vs. 23.2 μm), and grey (vs. white) inner peridium.
Type: Cameroon: East Province: Dja Biosphere Reserve, Northwest Sector near the village of Somalomo, Upper Dja River Basin, within 2 km radius of Dja base camp at 3°21’29.8” N 12°43’46.9” W, ~400 m west of base camp on edge of Gilbertiodendron dewevrei monodominant plot 1, 16 Aug 2014, Henkel 9859 (YA 0063174 – holotype; HSC G1174, OSC 149785, K(M) 200223 – isotypes). GenBank accession numbers ITS: KT694138; 28S: KY694149.
Description: Ascomata 6–20 mm tall (without basal attachment) × 7–27 mm broad, subglobose to ovate or somewhat lobed, black overall, with a distinct subturbinate base encompassing dark brown to black ectomycorrhizas, dense extramatrical mycelium, and sand; peridial surface nearly smooth on immature ascomata, on larger, mature ascomata verrucose throughout; warts 0.1–0.2 mm tall and 0.6–0.8 mm broad, polygonal, 4–5–6-sided, with flattened apices. Peridium in section subcartilaginous, three-layered; outer layer black, carbonaceous, < 0.25 mm thick, underlain by a greyish tan second layer with occasional reddish tones, to 0.5 mm thick, with embedded, black ectomycorrhizas; inner third layer dark grey to black, to 0.75 mm thick. Gleba initially off-white to pale grey, greyish black at maturity, somewhat powdery but mostly arranged in irregular moist masses, with fine, grey hyphae particularly near gleba-peridium interface. Odour none. Taste mild with a hint of sweetness.
In microscopic section outer first peridium layer carbonaceous, 65–90 μm thick, composed of a palisade-like tier of nearly black, globose to irregularly-shaped cells, these to 9.5 × 17.5 μm; walls 1–2 μm broad; surface with occasional scattered patches of hair-like projecting hyphae, these erect, pale brown to dark brown with obtuse apices, 4.5–6.5 μm broad with walls 2–3 μm thick; underlying second layer 460–500 μm thick, composed of a textura epidermoidea of pale brown, irregularly-shaped to elongate, occasionally branched hyphae, to 8.5 μm broad with walls ±1 μm thick, grading into the third layer that is to 750 μm thick, composed of a textura obita of bundles of up to 10 hyphae arranged in a cross-hatched arrangement; individual hyphae hyaline, somewhat sinuous, 5.0–5.5 μm broad with walls 0.5 μm thick. Gleba of ascospores and sinuous, hyaline, septate, loosely interwoven hyphae, these 2.5–4.5 μm broad with walls < 0.5 μm thick. Asci globose, 90–95 μm diam, hyaline, walls 2–2.5 μm thick, eight-spored. Ascospores globose, dark brown, (30–)34–38.5(–40.5) μm diam (mean = 35.5 μm) including the reticulate-alveolate ornamentation; alveolae well-defined, 4.5–5.5 μm broad and to 4.5 μm tall, with irregular to wavy walls; under SEM the individual alveolar wall is a composite of densely spaced vertical ribs, these with numerous ends emerging from the wall margin; ascospore surface exposed inside the alveolae with an irregular, extremely roughened, subreticulate texture with occasional ridged projections onto the surrounding alveolar wall.
Habit, habitat, and distribution: Solitary or in small groups, hypogeous in lateritic mineral soil or semi-emergent in leaf litter of forest floor in Gilbertiodendron dewevrei monodominant forest with nearby stands of Uapaca species; known only from the type locality in the Dja River Basin of southern Cameroon.
Additional specimens examined: Cameroon: East Province: Dja Biosphere Reserve, Northwest Sector near the village of Somalomo, Upper Dja River Basin, within 2 km radius of Dja base camp located at 3°21’29.8” N 12°43’46.9” W, ~1.4 km WNW of base camp on trail between Gilbertiodendron plots 1 & 2, in semi-inundated G. dewevrei monodominant forest, 20 Aug. 2014, Henkel 9874 (YA, HSC G1175, OSC 149786, K(M) 200224; GenBank accession numbers: ITS KT694135; 28S KT694147); 28 Aug. 2014, Henkel 9897 (YA, HSC G1176, OSC 149788, K(M) 200219; GenBank accession numbers ITS: KT694136; 28S: KT694146); ~2 km WNW of base camp in vicinity of Gilbertiodendron plot 3, in G. dewevrei monodominant forest, 26 Sep. 2014, Henkel 10015 (YA, HSC G1177, OSC 149787, K(M) 200220, GenBank accession numbers ITS: KT694134; 28S: KT694145).
Commentary: The molecular phylogenetic analysis strongly supported E. favosus as sister to, but distinct from, the sympatric E. labyrinthus described here, and showed that, within the genus Elaphomyces, these two African species share a common ancestor with E. compleximuris from Guyana (Fig. 1). Both Elaphomyces favosus and E. labrynthinus have a warty, black ascoma with a tapered base, but the peridial warts of E. favosus are both taller and broader than those of E. labyrinthinus. Additionally, the ascospore ornamentation of E. favosus is distinctly reticulate-aveolate while that of E. labyrinthinus is labyrinthine. The Guyanese E. compleximurus has similar overall ascoma morphology and ascospore ornamentation to those of E. favosus, but has smaller ascospores and a white inner peridium (vs. grey in E. favosus).
Amongst other Elaphomyces species worldwide, only two European species, E. cyanosporus and E. persoonii, combine the features of reticulate ascospores and a black, warty peridium. Elaphomyces persoonii has a tapered base like E. favosus but its peridial warts are to 1.5 mm broad, twice as wide as those of E. favosus. Also, the globose ascospores of E. persoonii are somewhat smaller with a mean diameter of 31.3 μm (vs. 35.7 μm for E. favosus). The ascospores of E. cyanosporus, with a mean diameter of 28.0 μm, are much smaller than those of E. favosus.
Elaphomyces iuppitercellus Castellano & T.W. Henkel, sp. nov.
Index Fungorum IF551320
(Fig. 4)
Fig. 4.
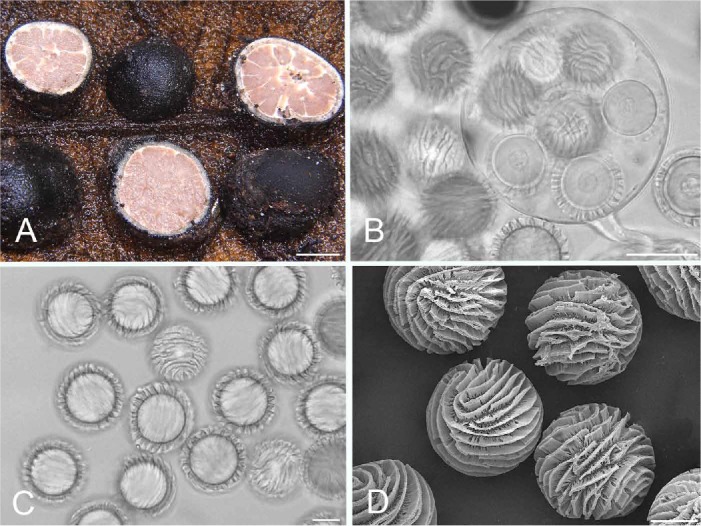
Elaphomyces iuppitercellus (holotype; Henkel THDJA 39). A. Ascomata showing peridial surface, gleba, and peridium in section. B. Ascus showing seven (of eight) ascospores. C. Ascospores with ornamentation in outline and surface views. D. Scanning electron micrograph of ascospores. Bars A = 5 mm, B = 25 μm, C–D = 10 μm.
Etymology: iuppiter (L.) = Jupiter and –cellus (L. adj. suf.) = diminutive for small, hence “small Jupiter”, in reference to the ascospore ornamentation resembling the swirling atmospheric patterns of the planet Jupiter.
Diagnosis: Similar to the European E. virgatosporus in peridium characteristics and ascospore ornamentation but differs in its pinkish brown gleba and larger ascospores (mean diameter = 24.7 μm vs. 20.2 μm in E. virgatosporus).
Type: Cameroon: East Province: Dja Biosphere Reserve, Northwest Sector near the village of Somalomo, Upper Dja River Basin, within 2 km radius of Dja base camp at 3° 21’ 29.8” N 12° 43’ 46.9” W, ~1 km WNW of base camp on trail between Gilbertiodendron plots 1 & 2, in semi-inundated G. dewevrei monodominant forest, 25 Aug. 2014, Henkel THDJA 39 (YA 0063175 – holotype; HSC G1178, OSC 149782, K(M) 200226). GenBank accession numbers ITS: KT694139; 28S: KT694143.
Description: Ascomata 4–8 mm tall and 6–12 mm broad, subglobose to ovate, without a distinct base, dark brown to black overall under scattered adherent soil, debris, and ectomycorrhizas; peridial surface somewhat smooth macroscopically but close inspection reveals low warts covering the entire surface; warts subcircular, to 150 μm tall and 400 μm broad, blunt at apex. Peridium in section subcartilaginous, with two distinct layers; outer first layer ~ 0.15 mm thick, black, carbonaceous; underlying second layer to 0.5 mm thick, greyish to grey-brown, with an apparent “third” inner layer appearing as a thin band of white cottony hyphae emanating from the outer gleba and contiguous at irregular intervals with white glebal veins. Gleba white to off-white and arranged in irregular moist masses when immature, at maturity pinkish brown (7C4–7D4) with white veins and eventually powdery, in larger specimens hollow in the center. Odour indistinct to mild. Taste mild to mealy.
Peridium in microscopic section two-layered; outer layer carbonaceous, 130–140 μm thick, composed of dark brown, polygonal cells, to 2.0–4.5 μm broad with walls 1 μm thick; surface with adhering debris but lacking erect hyphae; underlying second layer to 450 μm thick and composed of a textura intricata of hyaline hyphae, to 9 μm broad with walls 2 μm thick; hyphae closest to gleba with clavate ends to 15.5 μm broad. Gleba of ascospores and sinuous, hyaline, septate, loosely interwoven hyphae, these 3.5–4.5 μm broad with walls < 0.5 μm thick. Asci globose, 75–80 μm diam, hyaline, walls 1 μm broad, with an elongate, stipe-like base, eight-spored. Ascospores globose, hyaline, 23.5–25.5(–26.5) μm diam (mean = 24.7 μm) including the striate ornamentation that is to 2.5 μm tall; ornamentation irregular to wavy; under SEM the individual walls consist of a lattice-work of anastomosed rods and spines, with individual ridges varying somewhat in thickness.
Habit, habitat and distribution: Solitary or in small groups, hypogeous or semi-emergent in leaf litter of forest floor in Gilbertiodendron dewevrei monodominant forest with nearby stands of Uapaca spp.; known only from the type locality in the Dja River Basin of southern Cameroon.
Additional specimens examined: Cameroon: East Province: Dja Biosphere Reserve, Northwest Sector near Somalomo Village, Upper Dja River Basin, within 2 km radius of Dja base camp located at 3°21’29.8” N 12°43’46.9” W, ~1.5 km WNW of base camp in Gilbertiodendron plot 2, in G. dewevrei monodominant forest, 4 Sep. 2014, Henkel 9934 (YA, HSC G1179, OSC 149783, K(M) 200221; GenBank accession numbers ITS: KT694141; 28S: KT694142).
Commentary: The striate ascospore ornamentation seen in E. iuppitercellus is uncommon within Elaphomyces, found previously only in the European E. spirosporus, E. striatosporus, and E. virgatosporus, and in the Asian E. guangdongensis. The phylogenetic relationships, if any, between these species could not be assessed here as ITS and 28S sequence data for E. spirosporus, E. striatosporus, and E. virgatosporus are lacking in GenBank. Elaphomyces iuppitercellus has larger ascospores (mean diameter = 24.7 μm) than E. guangdongensis (mean diameter 17.8 μm), E. spirosporus (mean diameter 20.5 μm), E. striatosporus (mean diameter = 17.5 μm), and E. virgatosporus (mean diameter = 20.2 μm); additionally, each of the European species has a grey-toned gleba, which contrasts with the pinkish brown gleba of E. iuppitercellus. Also, the striate ornamentation walls of E. iuppitercellus are much thinner than those of all other striate-spored Elaphomyces species. Elaphomyces iuppitercellus ascomata had an identical ITS sequence with that obtained from a G. dewevrei ECM root tip collected at the Dja site, confirming its ECM status (Fig. 1).
Elaphomyces labyrinthinus Castellano & T.W. Henkel, sp. nov.
Index Fungorum IF551319
(Fig. 5)
Fig. 5.
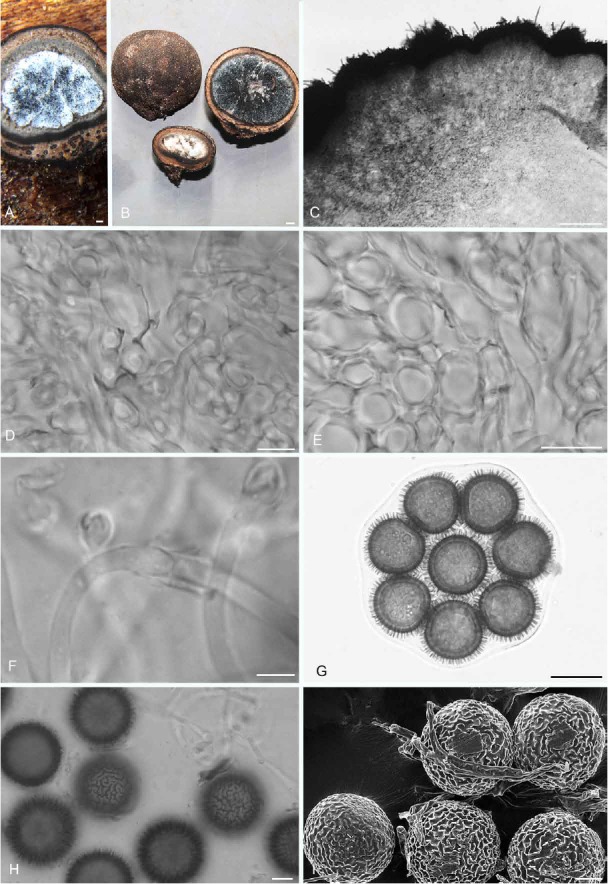
Elaphomyces labrynthinus (holotype; Henkel 9918). A. Sectioned ascoma showing the embedded black-mantled ectomycorrhizas within the inner peridial layer. B. Ascomata showing peridial surface, gleba, and peridium in section. C. Microscopic view of sectioned peridium. D. Thick-walled, interwoven hyphae from the fourth peridial layer. E. Thick-walled, somewhat swollen, interwoven hyphae from the fifth peridial layer. F. Collared septa on hyphae within the gleba. G. Ascus with eight ascospores in transverse planar view. H. Ascospores with ornamentation in outline and surface views. I. Scanning electron micrograph of ascospores. Bars A = 0.5 mm, B = 1 mm, C = 100 μm, D = 5 μm, E = 15 μm, F = 5 μm, G = 20 μm, H–I = 10 μm.
Etymology: labyrinthinus (L. adj. A) = labyrinthine, referring to the labyrinthine structure of the ascospore ornamentation.
Diagnosis: Similar to the Cameroonian E. favosus in overall macromorphology but differing in having peridial warts that are shorter and narrower than those of E. favosus, and ascospore ornamentation that is labyrinthine while that of E. favosus is reticulate-alveolate.
Type: Cameroon: East Province: Dja Biosphere Reserve, Northwest Sector near the village of Somalomo, Upper Dja River Basin, within 2 km radius of Dja base camp located at 3o21’29.8 “ N 12o43’46.9” W, ~1.5 km WNW of base camp in Gilbertiodendron plot 2, in G. dewevrei monodominant forest, 1 Sep. 2014, Henkel 9918 (YA 0063176 – holotype; HSC G1180, OSC 149781, K(M) 200225 – isotypes). GenBank accession numbers ITS: KT694137; 28S: KT694148.
Description: Ascomata to 13 mm tall and 20 mm broad, broadly ovate to depressed ovate, with a distinct, slightly tapered base composed of ectomycorrhizas, sand, and dense, dark brown to nearly black mycelium; peridium slightly thickened in this area; peridial surface verrucose beneath a turf of erect, dark brown hyphae; warts black, polygonal, 4–6-sided, with uneven side lengths, 100 μm tall and ± 300 μm broad, flattened, on close inspection appearing finely ridged. Peridium in section subcartilaginous, five-layered; outer layer ± 0.05 mm thick, dark brown; second layer ± 0.1 mm thick, black, carbonaceous; third layer 0.35–0.60 mm thick, pale tan, with scattered embedded, black-mantled ectomycorrhizas across the lower portion of the ascoma, these more dense near the basal attachment; fourth layer ± 0.1 mm thick, dark brown; fifth layer ± 0.20 mm thick, grey; all layers most distinct in young specimens; inner layers obscured with age. Gleba off-white to pale grey, cottony when immature, becoming greyish black, powdery, with fine, off-white to grey hyphae concentrated near the peridium. Odour and taste not recorded.
Peridium in microscopic section five-layered; outer layer ± 50 μm thick, composed of a turf of erect, dark brown, septate, capitate hyphae, 6–7(–9.5) μm broad to 17.5 μm long with walls 1.5–2.0 μm thick; surface with scattered patches of erect hair-like hyphae, these pale brown to dark brown with obtuse apices, 4.5–6.5 μm broad with walls 2–3 μm thick; underlying second layer ± 100 μm thick, a textura epidermoidea of dark brown, tangled, irregularly-shaped hyphae that are densely packed near the surface and less so towards the gleba; hyphal cells to 7 × 13.5 μm with walls ±1 μm thick; third layer 350–600 μm thick, a compact textura intricata of pale brown hyphae, ± 5.5 μm broad with walls 1.0–2.0 μm thick, with amorphous dark brown particles scattered throughout; fourth layer ± 100 μm thick, a textura intricata of hyaline, loosely interwoven hyphae, 2.5–5.0 μm broad with walls 1.0–1.5 μm thick; innermost fifth layer ± 200 μm thick, a textura intricata of pale brown, interwoven hyphae, somewhat swollen, 5.5–15.0 μm broad, with occasional dark particles scattered through the layer. Gleba consisting of ascospores and sinuous, hyaline, septate, loosely tangled hyphae, ± 3.5 μm broad with walls < 0.5 μm thick, with a collar-like thickening at the septa. Asci globose, 66–88 μm diam, hyaline, walls 1.5–2.0 μm thick, eight-spored. Ascospores globose, dark brown, 33.5–37.5 μm diam (mean = 35.2 μm), including the labyrinthine-like ornamentation ± 3.5 μm tall; ornamentation with irregular to angular walls, appearing as short, variously shaped, unconnected lines in surface view, spiny in outline view; under SEM individual walls slightly variable in thickness and formed into semi-circles, lines, or irregular shapes.
Habit, habitat, and distribution: In small groups semi-emergent in leaf litter of the forest floor in Gilbertiodendron dewevrei monodominant forest, with nearby stands of Uapaca spp.; known only from the type locality in the Dja River Basin of southern Cameroon.
Commentary: See above for differences of E. labrynthinus from the morphologically and phylogenetically similar E. favosus, and the close phylogenetic relationship between these two species and E. compleximuris from Guyana. The labyrinthine ascospore ornamentation of E. labyrinthinus is similar to that of E. digitatus from Guyana, but the distinctly orange peridial surface and much smaller ascospores allow easy separation of the latter species from E. labyrinthinus. The European E. citrinus has labyrinthine ascospore ornamentation but its ascospores are half the size (mean diameter = 15.8 μm) than those of E. labyrinthinus (mean diameter = 35.2 μm).
Elaphomyces adamizans Castellano & T.W. Henkel, sp. nov.
Index Fungorum IF551682
(Fig. 6)
Fig. 6.
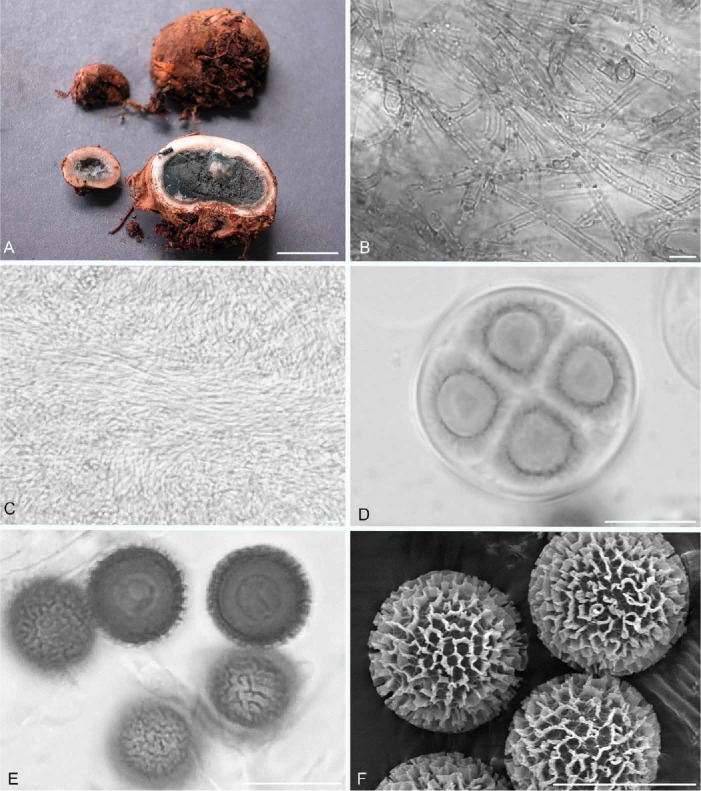
Elaphomyces adamizans (holotype; Henkel 9660). A. Ascomata showing peridial surface, gleba, and peridium with embedded ectomycorrhizas in section. B. Interwoven hyphae with numerous, adherent, dark granules in the first peridial layer. C. Cross-hatched, interwoven, bundled hyphae in the third peridial layer. D. Immature ascus in focal plane showing four (of eight) developing ascospores. E. Ascospores with ornamentation in outline and surface views. F. Scanning electron micrograph of ascospores. Bars A = 10 mm, B–F = 10 μm.
Etymology: adamas (L.) = diamond and –izans (L. adj. suf.) = “becoming like” or “resembling”; in reference to the alluvial diamonds originally found in the Upper Mazaruni River Basin of Guyana, the type locality of the fungus.
Diagnosis: Similar to the Australian E. rugosisporus in peridium structure and ascospore size but differs in having a labryinthine ascospore ornamentation (vs. finely reticulate for E. rugosisporus) and lack of a carbonaceous outer peridial layer.
Type: Guyana: Region 9 Cuyuni-Mazaruni: Pakaraima Mountains, Upper Mazaruni River Basin, ~10 km west of Mt Ayanganna, within 0.5 km of a base camp at 5° 26’ 21.3” N 60° 04’ 43.1” W, 100 m north of base camp in savanna fringing forest dominated by Pakaraimaea dipterocarpacea and Dicymbe jenmanii, 2 Jun. 2012, Henkel 9660 (BRG 41125 – holotype; HSC G1181, OSC 149784, K(M) 200222 – isotypes). GenBank accession numbers ITS: KT694133; 28S: KT694144.
Description: Ascomata 7–14 mm tall and 10–22 mm broad, ovoid-flattened, with a dense layer of ectomycorrhizal roots, mycelium, humic particles, and soil covering the lower quarter, earthen brown (5E7–6E7), unchanging; peridial surface a tightly appressed, brown, tomentose mat, occasionally organized into cord-like fibrils. Peridium in section cartilaginous, three-layered; outer layer 0.18–0.22 mm thick, brown; underlying second layer p to 0.13 mm thick, off-white to pale orange-tan; inner third layer overall 1.5–2.0 mm thick but this varying across entire section, off-white, with numerous orange-brown, embedded ectomycorrhizas along the lower half of the ascoma, and there darkening to pale brown to grey-brown near the gleba. Gleba hollow, grey when immature, at maturity of ascospores that are dark olivaceous blue-grey (4F2–5F2) in mass, powdery, with scattered narrow hyphae. Odour indistinct to slightly earthy. Taste slightly sweet.
Peridium in microscopic section three-layered; outer layer 175–220 μm thick, composed of a pale brown, somewhat loose textura intricata, not carbonaceous; hyphae 3.5–4.5 μm broad with walls 1 μm thick, with numerous adhering dark small granules; second layer ± 130 μm thick, similarly structured as the first but hyphae lacking adherent granules; inner third layer 1500-2000 μm thick, composed of compact, agglutinated, hyaline hyphae, 3.5–4.5 μm broad, arranged in bundles that are occasionally cross-hatched. Gleba consisting of ascospores and sinuous, hyaline, septate, irregularly swollen hyphae, 2.0–3.5 μm broad with walls < 0.5 μm thick. Asci irregularly globose, 26–27 μm broad, hyaline, with walls to 2.0 μm thick, eight-spored. Ascospores globose, pale brown to brown, 10.5–12.0(–12.5) μm diam (mean = 11.1 μm) including the labyrinthine ornamentation that is 1.5–2.0 μm tall; ornamentation of irregular to wavy walls, appearing as short, variously shaped, unconnected lines; under SEM the individual walls are slightly variable in thickness and formed into semi-circles, lines, or irregular shapes, often with small pits at the tips.
Habit, habitat, and distribution: In group of two, semi-emergent in leaf litter in forest dominated by Pakaraimaea dipterocarpacea and Dicymbe jenmanii; known only from the type locality in the Upper Mazaruni River Basin of Guyana.
Commentary: In the field, the brown, tomentose peridial surface of E. adamizans allows it to be easily distinguished from the two other Elaphomyces known from Guyana (Castellano et al. 2012). The very small ascospores (mean diameter = 11.1 μm) contrast with the larger ascospores of the Guyanese E. compleximurus (mean = 23.2 μm broad) and E. digitatus (mean = 21.9 μm broad). There are a number of recently described Australian Elaphomyces species with mean ascospore diameter ranging from 9–12 μm, including E. chlorocarpus, E. symeae, E. timgroveii, E. cooloolanus, E. pedicellaris, and E. rugosisporus (Castellano et al. 2011). Each of these species differs from E. adamizans in peridial characteristics and ascospore ornamentation, and all are associated with ECM Myrtaceae hosts (Castellano et al. 2011).
Molecular phylogenetic analysis places E. adamizans as sister to the stalked, volvate Pseudotulostoma volvatum. While E. adamizans and P. volvatum have highly dissimilar macromorphologies at maturity, the ascospore morphologies E. adamizans and P. volvatum are very similar. Ascospores in each are between 7–12.5 μm diam with a labyrinthine ornamentation. The stalked, epigeous habit with an exposed ascospore mass in P. volvatum allows the species to be easily separated from the fully sequestrate E. adamizans. Additionally, SEM images reveal differences in fine detail of the ascospore ornamentation of these taxa that under light microscopy appear similar (Miller et al. 2001, Asai et al. 2004).
DISCUSSION
In addition to supporting the recognition of the new species of Elaphomyces reported here, the phylogenetic analysis suggests that the stalked, volvate Pseudotulostoma volvatum may be nested within the genus Elaphomyces. Pseudotulostoma volvatum was described as a new taxon by Miller et al. (2001; as “volvata”) from Guyana with a macromorphology resembling a basidiomycete stalked puffball but micromorphology consistent with Elaphomyces. At maturity this fungus exhibits a powdery ascospore and pseudocapillitium mass exposed on the apex of a woody stalk, having expanded upward through the peridium, which remains as a volva-like basal structure (Fig. 7). The 18S rDNA phylogenetic analysis presented by Miller et al. (2001) placed P. volvatum within the Eurotiales and sister to Elaphomyces within Elaphomycetaceae. Pseudotulostoma was therefore recognized as a new genus related to, but outside of Elaphomyces, supported morphologically by the radically different form of the mature ascoma. The close relationship of P. volvatum to Elaphomyces was corroborated by its thick, tough, multi-layered peridium with embedded ectomycorrhizas, and gleba of hydrophobic, thick-walled, ornamented ascospores with Elaphomyces-like ultrastructure. The ECM nutritional status typical of Elaphomyces species was also demonstrated for P. volvatum based on morphological and molecular analysis of ECM roots of Dicymbe corymbosa found in proximity to the ascomata (Henkel et al. 2006).
Fig. 7.

Ascomata of Pseudotulostoma volvatum (Henkel 9786) showing developmental stages. Bar = 10 mm.
Masuya & Asai (2004) subsequently placed P. volvatum and the Japanese P. japonicum (as “japonica”) in Elaphomycetaceae as sister to Elaphomyces based on a SSU rDNA phylogenetic analysis. It is clear from the detailed descriptions and illustrations of P. japonicum from Kawamura (1954), Otani (1960), and Asai et al. (2004) that the species shares the unusual macromorphological structure with P. volvatum, and both species have key micromorphological features shared with Elaphomyces. Masuya & Asai (2004) stated “…the fact that unopened ascomata of P. japonica are highly similar to the fruit-body found in the genus Elaphomyces suggests that this species, which we believe should be treated as Pseudotulostoma, may also exist as a species of Elaphomyces”. It should be noted that during the time of both Miller et al. (2001) and Masuya & Asai (2004) very few Elaphomyces sequences were available in GenBank, so taxon sampling for the genus was low in both studies. Subsequently, Reynolds (2011) provided unpublished data suggesting a congeneric relationship of Pseudotulostoma and Elaphomyces, a relationship also suggested by our phylogenetic analysis (Fig. 1). Although our results suggest that Pseudotulostoma and Elaphomyces are not reciprocally monophyletic and may need to be treated as a single genus, more taxon-extensive sampling with multi-locus DNA sequence data is needed to better understand the relationship between them before formal taxonomic changes can be proposed.
ACKNOWLEDGEMENTS
We thank the following funding sources: The National Geographic Society’s Committee for Research and Exploration grant 9235-13 and National Science Foundation (NSF) DEB-0918591 and DEB-1556338 to T.W.H, and a grant to C.T. from the Basler Stiftung für Biologische Forschung. In Cameroon the Ministry of Research and Scientific Innovation issued research permits. Jean Michel Onana, Head of The National Herbarium of Cameroon (Institute of Agricultural Research for Development, IRAD), provided much logistical assistance. The Conservator of the Dja Biosphere Reserve, Mengamenya Goue Achille, and his staff greatly assisted the fieldwork in the Dja. Additional research permits were granted by the Guyana Environmental Protection Agency. Field assistance in Cameroon was provided by Alamane Gabriel (a.k.a. Sikiro), Abate Jackson, Mamane Jean-Pierre, and Mei Lin Chin, and in Guyana by Dillon Husbands, Christopher Andrew, Peter Joseph, Francino Edmund, and Luciano Edmund. Caitlin Winterbottom and Connor Adams assisted with scanning electron microscopy. Laura Martínez-Suz isolated the ECM G. dewevrei root tip and generated its fungal ITS sequence. James Trappe assisted with deriving the Latin species names. Two anonymous reviewers provided very helpful comments on an earlier version of the manuscript. This paper is number 213 in the Smithsonian Institution’s Biological Diversity of the Guiana Shield Program publication series.
Footnotes
*This paper was prepared by USA Govt. employees on official time, and is therefore in the public domain and not subject to copyright.
REFERENCES
- Asai I, Sato H, Nara T. (2004) Pseudotulostoma japonica, comb. nov. (=Battarrea japonica), a species of the Eurotiales, Ascomycota. Bulletin of the National Science Museum Tokyo, Series B 30: 1–7. [Google Scholar]
- Castellano MA, Henkel TW, Miller SL, Smith ME, Aime MC. (2012) New Elaphomyces species (Elaphomycetaceae, Eurotiales, Ascomycota) from Guyana. Mycologia 104: 1244–1249. [DOI] [PubMed] [Google Scholar]
- Castellano MA, Trappe JM, Vernes K. (2011) Australian species of Elaphomyces (Elaphomycetaceae, Eurotiales, Ascomycota). Australian Systematic Botany 24: 32–57. [Google Scholar]
- Corner EJH, Hawker LE. (1953) Hypogeous fungi from Malaya. Transactions of the British Mycological Society 36: 125–137. [Google Scholar]
- Dentinger BTM, Margaritescu S, Moncalvo J-M. (2010) Rapid and reliable high-throughput methods of DNA extraction for use in barcoding and molecular systematics of mushrooms. Molecular Ecology Resources 10: 628–633. [DOI] [PubMed] [Google Scholar]
- Gardes M, Bruns TD. (1993) ITS primers with enhanced specificity for basidiomycetes –application to the identification of mycorrhizae and rusts. Molecular Ecology 2: 113–118. [DOI] [PubMed] [Google Scholar]
- Henkel TW, James TY, Miller SL, Aime MC, Miller OK., Jr (2006) The mycorrhizal status of Pseudotulostoma volvata (Elaphomycetaceae, Eurotiales, Ascomycota). Mycorrhiza 16: 241–244. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research 30: 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Toh H. (2008) Improved accuracy of multiple ncRNA alignment by incorporating structural information into a MAFFT-based framework. BMC Bioinformatics 9: 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura S. (1954) Icones of Japanese Fungi. Vol. 7 Tokyo: Kazama Shobo. [Google Scholar]
- Kornerup A, Wanscher JH. (1978) Methuen Handbook of Colour. 3rd edn. London: Eyre Methuen. [Google Scholar]
- Masuya H, Asai I. (2004) Phylogenetic position of Battarrea japonica (Kawam.) Otani. Bulletin of the National Science Museum Tokyo, Series B 30: 9–13. [Google Scholar]
- Miller OK, Jr, Henkel TW, James TY, Miller SL. (2001) Pseudotulostoma, a remarkable new volvate genus in the Elaphomycetaceae from Guyana. Mycological Research 105: 1268–1272. [Google Scholar]
- Otani Y. (1960) On Battarrea japonica (Kawam.) Otani, comb. nov. Transactions of the Mycological Society of Japan 2: 11–13. [Google Scholar]
- Ott M, Zola J, Aluru S, Stamatakis A. (2007) Large-scale Maximum Likelihood-based Phylogenetic Analysis on the IBM BlueGene/L. Proceedings of ACM/IEEE Conference on Supercomputing Article No. 4. New York: Association for Computing Machinery. [Google Scholar]
- Peh KS, Sonke B, Séné O, Djuikouo MN, Nguembou CK, et al. (2014) Mixed-forest species establishment in a monodominant forest in central Africa: implications for tropical forest invasibility. PLoS One 9: e97585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt CA, Kohler A, Hesse CN, Sharpton TJ, Martin F, Spatafora JW. (2015) Metagenome sequence of Elaphomyces granulatus from sporocarp tissue reveals Ascomycota ectomycorrhizal fingerprints of genome expansion and a Proteobacteria-rich microbiome. Environmental Microbiology 17: 2952–2968. [DOI] [PubMed] [Google Scholar]
- Reynolds HT. (2011) Systematics, phylogeography, and ecology of Elaphomycetaceae. PhD dissertation, Department of Biology, Duke University. [Google Scholar]
- Smith ME, Henkel TW, Uehling JK, Fremier AK, Clarke HD, Vilgalys R. (2013) The ectomycorrhizal fungal community in a Neotropical forest dominated by the endemic dipterocarp Pakaraimaea dipterocarpacea. PLoS One 8: e55160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- Tedersoo L, Nilsson RH, Abarenkov K, Jairus T, Sadam A, Saar I, Kõljalg U. (2010) 454 pyrosequencing and Sanger sequencing of tropical mycorrhizal fungi provide similar results but reveal substantial methodological biases. New Phytologist 188: 291–301. [DOI] [PubMed] [Google Scholar]
- Tedersoo L, Bahram M, Jairus T, Bechem E, Chinoya S, Mpumba R, Koljalg U. (2011) Spatial structure and the effects of host and soil environments on communities of ectomycorrhizal fungi in wooded savannas and rain forests of Continental Africa and Madagascar. Molecular Ecology 20: 3071–3080. [DOI] [PubMed] [Google Scholar]
- Trappe JM. (1979) The orders, families, and genera of hypogeous Ascomycotina (truffles and their relatives). Mycotaxon 9: 297–340. [Google Scholar]
- Trappe JM, Molina R, Luoma DL, Cázares E, Pilz D, et al. (2009) Diversity, ecology and conservation of truffle fungi in forests of the Pacific Northwest. USDA Forest Service [General Technical Report PNW-GTR-772.] Portland: Pacific Northwest Research Station. [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR Protocols: a guide to methods and applications (Innis M, Gelfand D, Sninsky J, White T, eds): 315–322. San Diego: Academic Press. [Google Scholar]


