Abstract
At present, there are no studies that have established a microRNA (miRNA)-based signature profile in patients with radiographic axial spondyloarthritis (rad-axial SpA), and we hypothesized that these patients may have aberrantly expressed circulating miRNAs reflective of underlying disease and inflammation. This study aims to determine the expression profile of miRNAs in plasma of patients with rad-axial SpA and compare it with healthy, age, and sex-matched controls. Fifteen subjects with rad-axial SpA based on ASAS classification criteria and 5 controls were recruited from our local SpA registry. Demographic data were collected and disease activity was measured using Bath Ankylosing Spondylitis Disease Activity Index (BASDI). Peripheral blood samples (5 ml) were obtained from eligible consenting patients and controls. RNA from the plasma was prepared using miRNeasy kit (Qiagen) by a modified protocol. Expression of 175 miRNAs was screened in the plasma of all 15 patients and 5 controls using serum/plasma miRNA PCR arrays (Exiqon Inc. Woburn, MA) essentially following the manufacturer’s instructions. Real-time PCR was carried out on StepOne Plus (Applied Biosystems) and the data was extracted and analyzed using ExiGen Enterprise software (MultiD, Göteborg, Sweden). Potential miRNA targets were identified using bioinformatics. ESR and CRP levels were measured by standard laboratory methods. We identified 7 differentially expressed miRNAs (2 upregulated and 5 downregulated). miR-34a, which was overexpressed in patients with rad-axial SpA, was predicted to target BMP-3 mRNA by TargetscanS and PicTar miRNA target algorithms. miR-150 was downregulated in all of the samples analyzed by us using the TaqMan Gene Expression assay. The most repressed miRNA was miR-16 and is predicted to regulate the expression of activin A receptor (ACVR2B), a receptor for growth, and differentiation factor-5 (GDF-5). Our data indicates that (1) patients with axial SpA, as compared to controls, have dysregulated expression of selected miRNAs in the plasma; and (2) the differentially expressed miRNAs are predicted to target genes that play a role in bone morphogenesis, growth, and immune response.
Keywords: Ankylosing spondylitis, microRNAs, Radiographic axial spondyloarthritis, Spondyloarthritis
Background
The main challenges in the management of spondyloarthritis (SpA) [1] are related to the lack of complete understanding of the pathogenesis of the disease, availability of biomarkers associated with disease activity as well as the inability to predict response to treatment. MicroRNAs (miRNAs), endogenous small noncoding RNAs regulating the activities of target mRNAs and cellular processes [2], are present in human plasma in a stable form and have emerged as potential biomarkers for disease activity, pathogenesis, and prognosis of the disease. At present, there are no studies that have established a miRNA-based signature profile in patients with axial SpA.
The hypothesis guiding our proposed study was that patients with radiographic axial SpA have aberrantly expressed circulating miRNAs reflective of underlying disease and inflammation and these dysregulated miRNAs can be detected through miRNA expression profiling. This was tested in our proof of the concept pilot study. The aim of our study was to determine the expression profile of miRNAs in plasma of patients with rad-axial SpA and compare it with healthy, age, and sex-matched controls.
Methods
Study was approved by our institutional review board (IRB). Fifteen subjects with rad-axial SpA based on Assessment of Spondyloarthritis International Society criteria [3] and 5 normal controls were recruited from the Arthritis Clinic. Subjects with (i) active malignancy in last 5 years, (ii) subjects with rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), and other rheumatic diseases, and (iii) subjects with evidence of HIV or chronic hepatitis B or C were excluded from the study. Patients and controls were screened and consent was obtained.
Demographic data were collected and disease activity was measured using Bath Ankylosing Spondylitis Disease Activity Index (BASDAI) questionnaire. ESR and CRP were measure using standard methods.
Peripheral blood samples (5 ml) were obtained from eligible consenting patients and controls. The blood samples were drawn into EDTA-containing tubes and centrifuged at 400 × g for 7 min, and 1 ml aliquots of plasma were transferred into nuclease-free Eppendorf tubes and stored at −20 °C until analyses.
RNA extraction
RNA from the plasma was prepared using miRNeasy kit (Qiagen) by a modified protocol. Briefly, 1 ml of plasma was denatured in 10 ml of Qiazol reagent to ensure effective denaturation of high protein content present in plasma. Phase separation was carried out by adding 2.2 ml of chloroform followed by centrifugation at 12,000 g for 15 min at 4 °C. RNA in the aqueous layer was precipitated by adding 100 % ethanol. This mixture was loaded on to the kit supplied column and washed 3 times with buffer and the RNA was eluted in 50 μl RNase-free water.
Screening of miRNA expression using Exiqon human plasma/serum miRNA PCR array
Expression of 175 miRNAs was screened in the plasma of all 15 patients and 5 controls using serum/plasma miRNA PCR arrays (Exiqon Inc. Woburn, MA) essentially following the manufacturer’s instructions. Briefly, 4 μl of purified plasma RNA was used to synthesize single-stranded complementary DNA (cDNA) and 20 μl of the cDNA reaction mix was added to a SYBR Green master mix. Ten microliters of the cDNA and SYBR Green mix was aliquoted in each well of the two separate 96-well plates containing specific primers for individual human miRNAs. Real-time PCR was carried out on StepOne Plus (Applied Biosystems) and the data was extracted and analyzed using ExiGen Enterprise software (MultiD, Göteborg, Sweden). Potential miRNA gene targets were identified using bioinformatics [4]. Comparisons between patients and controls were performed using two-sample t test. Fold changes in scores were compared to a baseline value set at 1. miRNAs with twofold difference in expression were considered differentially expressed and labeled. Comparison of differentially expressed miRNAs was also made between HLA-B27-positive and -negative patients, respectively.
Results
The patient demographics and characteristics are shown in Table 1. The mean age of the patients was 46 years (range 22–66), with 60 % being males and 26 % African-Americans. About 60 % of the patients were HLAB-B27 positive, mean BASDAI of 5.34 (range 1–8.1), and 40 % on anti-tumor necrosis factor therapy. Mean ESR and CRP were 18.2 mm/h (range 1–53) and 0.8 mg/dl (0.5–2.2). The demographics of the controls are shown in Table 2.
Table 1.
Patient demographics and disease activity measures
| Patient number | Sex | Race | Age (Years) | HLA-B27 | BASDAI | ESR (mm/h) | CRP (mg/dl) | Anti-TNF therapy |
|---|---|---|---|---|---|---|---|---|
| 1 | F | W | 48 | + | 1 | 14 | 0.5 | + |
| 2 | M | H | 52 | − | 3.7 | 7 | 0.5 | + |
| 3 | F | W | 47 | + | 3.85 | 53 | 0.5 | |
| 4 | M | W | 62 | − | 7.6 | — | — | + |
| 5 | M | W | 66 | + | 1 | 1 | 0.6 | |
| 6 | M | W | 53 | − | 4.5 | 19 | 0.5 | |
| 7 | M | B | 53 | + | 6.8 | 10 | 2.2 | |
| 8 | M | W | 56 | + | 4.6 | 23 | 1.4 | |
| 9 | M | B | 44 | − | 7.9 | 47 | 1.2 | |
| 10 | M | W | 33 | + | 8.1 | 25 | 1.1 | + |
| 11 | F | W | 22 | + | 5.8 | 1 | 0.5 | |
| 12 | F | B | 59 | + | 7.4 | 48 | 0.5 | + |
| 13 | F | B | 44 | + | 5.7 | 31 | 0.5 | |
| 14 | M | W | 42 | − | 7.1 | 11 | 0.5 | |
| 15 | F | W | 53 | − | 5.1 | 7 | 0.6 | + |
Table 2.
Demographics of controls
| Sex | Race | Age in years | |
|---|---|---|---|
| 1 | F | W | 54 |
| 2 | M | B | 26 |
| 3 | M | W | 24 |
| 4 | F | B | 39 |
| 5 | M | W | 48 |
We identified 7 differentially expressed miRNAs (2 upregulated and 5 downregulated) as shown in Fig. 1. miR-34a and miR-32 were both overexpressed and miR-16, miR-150, miR-10b, miR-30a, and miR-154 were underexpressed as shown in Table 3. miR-150 was downregulated in all of the samples analyzed by us using the TaqMan Gene Expression assay and miR-16 was the most repressed.
Fig. 1.
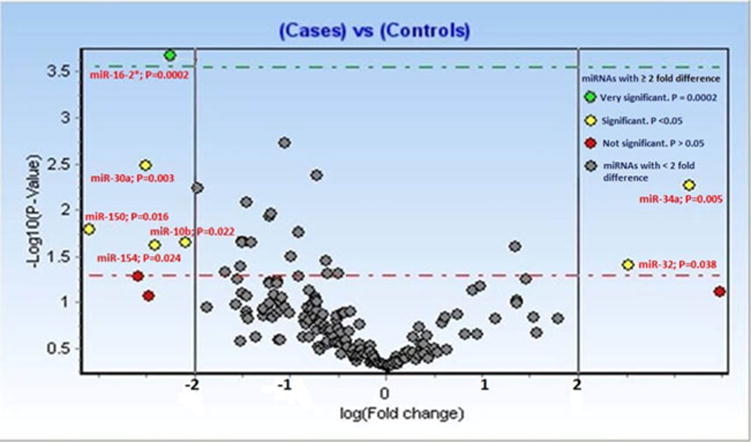
Several miRNAs are differentially expressed in the plasma of axial SpA patients. Human serum/plasma miRNA PCR array profiling was performed using RNA extracted from the plasma of 15 axial SpA patients and 5 healthy controls. miRNAs with at least twofold difference in expression level were considered differentially expressed and are labeled
Table 3.
miRNAs with at least twofold difference in expression level were considered differentially expressed (P < 0.05)
| Under expressed miRNAs | Fold change | Overexpressed miRNAs | Fold change |
|---|---|---|---|
| Hsa-miR-150 | −2.55 | Has-miR-34a | +2.74 |
| Hsa-miR-16 | −3.13 | Has-miR-32 | +2.26 |
| Hsa-miR-10b | −2.05 | ||
| Hsa-miR-30a | −2.26 | ||
| Hsa-miR-154 | −2.20 |
When miRNA expression profile was compared between HLA-B27-positive and HLA-B27-negative patients, miR-32 (which was also highly expressed in total patients vs control pool) was highly expressed in HLA-B27 positive patients and was not detected in the plasma of HLA-B27 negative patients.
Discussion
miRNAs are short (22 nucleotides) non-coding RNA molecules that regulate the expression of target genes at the post-transcriptional level by translational repression or degradation of messenger RNAs (mRNAs) [5–7]. A single miRNA regulates translation of multitude of distinct target genes involved in a certain function and the expression of a single coding gene can be regulated by several miRNAs. They play a role in multiple cellular processes, including various immune pathways, and regulate the function of both the innate and the adaptive immune system. More than 700 miRNAs have been identified in the human genome. Since miRNA expression pattern is believed to be reflective of underlying pathophysiologic processes and is specific to various disease states, it may help us to understand the pathophysiology of this disease. They are present in human plasma in a stable form and have emerged as potential biomarkers for disease activity, pathogenesis, and prognosis of the disease.
Deregulation of miRNA expression has been implicated in the pathogenesis of human diseases [7–10]. Altered miRNA expression has also been reported in autoimmune diseases like SLE, RA, and multiple sclerosis [11–17]. Studies also suggest that circulating miRNAs are promising as candidate biomarkers of diagnosis, prognosis, disease activity, and severity in several rheumatic diseases and in many cancers [17–21]. Recent study has shown that plasma concentrations of miR-24 and miR-125a-5p and ePRAM are potential diagnostic markers of RA even if patients were anti-citrullinated peptide antibody negative [19]. A pilot study has shown that miRNA expression profile can clearly separate healthy controls from Sjögren’s syndrome patients [21].
Our study is the first study to establish a miRNA-based signature profile in patients with rad-axial SpA. miR-34a and miR-32 were both overexpressed and miR-16, miR-150, miR-10b, miR-30a, and miR-154 were underexpressed as shown in Table 2. Some of the predicted biologic targets of selected differentially expressed microRNAs were associated with bone morphogenesis and growth. For example, miR-34a, which was overexpressed in patients with rad-axial SpA, was predicted to target bone morphogenic protein 3 (BMP3) mRNA by TargetscanS and PicTar miRNA target algorithms (data not shown). Its role in regulating the expression of BMP-3 can be tested in future studies. Similarly, miR-150 has recently been shown to be a negative regulator of the transcription factor c-Myb and its downregulation correlated with the metastasis of pancreatic cancer [22]. Although miR-150 has been shown to be upregulated in other diseases like but consistent with the array data, miR-150 was downregulated in all of the samples analyzed by us using the TaqMan Gene Expression assay (data not shown). The most repressed miRNA was miR-16 and is predicted to regulate the expression of activin A receptor (ACVR2B), a receptor for growth, and differentiation factor-5 (GDF-5) [23]. Expression or role of GDF-5 in axial SpA is currently not known.
Our study is a pilot study with a small sample size. A study with larger number of patients with rad-axial SpA for better validity of the results is needed. Also, we need to test whether the presence of identified miRNAs in the plasma correlates with the disease activity measures like validated patient-reported index, BASDAI, and ASDAS, as well as with ESR and CRP levels in patients with axial SpA.
In summary, the our study indicates that (1) patients with rad-axial SpA, as compared to controls, have dysregulated expression of selected miRNAs in the plasma; and (2) the differentially expressed miRNAs are predicted to target genes that play a role in bone morphogenesis, growth, and immune response.
Acknowledgments
We would likely to acknowledge Dianne Morus for her role as research coordinator.
Footnotes
Disclosures None.
Contributor Information
Marina N. Magrey, Email: mmagrey@metrohealth.org.
Tariq Haqqi, Email: thaqqi@neomed.edu.
Abdul Haseeb, Email: ahaseeb@neomed.edu.
References
- 1.Dougados M, Baeten D. Spondyloarthritis. Lancet. 2011;377:2127–37. doi: 10.1016/S0140-6736(11)60071-8. [DOI] [PubMed] [Google Scholar]
- 2.Dai R, Ahmed SA. MicroRNA, a new paradigm for understanding immunoregulation, inflammation, and autoimmune diseases. Transl Res. 2011;157:163–79. doi: 10.1016/j.trsl.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rudwaleit M, van der Heijde D, Landewé R, Listing J, Akkoc N, Brandt J, et al. The development of assessment of spondyloarthritis international society classification criteria for axial spondyloarthritis (part II): validation and final selection. Ann Rheum Dis. 2009;68:777–83. doi: 10.1136/ard.2009.108233. [DOI] [PubMed] [Google Scholar]
- 4.Kuhn DE, Martin MM, Feldman DS, Terry AV, Jr, Nuovo GJ, Elton TS. Experimental validation of miRNA targets. Methods. 2008;44:47–54. doi: 10.1016/j.ymeth.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pauley KM, Chan EK. MicroRNAs and their emerging roles in immunology. Ann N Y Acad Sci. 2008;1143:226–39. doi: 10.1196/annals.1443.009. [DOI] [PubMed] [Google Scholar]
- 6.Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet. 2008;9:831–42. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9:839–45. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 8.Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9:405–14. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du C, et al. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the Pathogenesis of multiple sclerosis. Nat Immunol. 2009;10:1252–1259. doi: 10.1038/ni.1798. [DOI] [PubMed] [Google Scholar]
- 10.Otaegui D, Baranzini SE, Armañanzas R, Calvo B, Muñoz-Culla M, Khankhanian P. Differential micro RNA expression in PBMC from multiple sclerosis patients. PLoS ONE. 2009;4:e6309. doi: 10.1371/journal.pone.0006309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duroux-Richard I, Jorgensen C, Apparailly F. What do microRNAs mean for rheumatoid arthritis? Arthritis Rheum. 2011;26 doi: 10.1002/art.30651. [DOI] [PubMed] [Google Scholar]
- 12.Pauley KM, Satoh M, Chan AL, Bubb MR, Reeves WH, Chan EK, et al. Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis Res Ther. 2008;10:R101. doi: 10.1186/ar2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nakasa T, Shibuya H, Nagata Y, Niimoto T, Ochi M. The inhibitory effect of microRNA-146a expression on bone destruction in collagen-induced arthritis. Arthritis Rheum. 2011;63:1582–1590. doi: 10.1002/art.30321. [DOI] [PubMed] [Google Scholar]
- 14.Dai Y, Huang YS, Tang M, Lv TY, Hu CX, Tan YH, et al. Microarray analysis of microRNA expression in peripheral blood cells of systemic lupus erythematosus patients. Lupus. 2007;16:939–946. doi: 10.1177/0961203307084158. [DOI] [PubMed] [Google Scholar]
- 15.Te JL, Dozmorov IM, Guthridge JM, Nguyen KL, Cavett JW, Kelly JA, et al. Identification of unique microRNA signature associated with lupus nephritis. PLoS ONE. 2010;5:e10344. doi: 10.1371/journal.pone.0010344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chatzikyriakidou A, Voulgari PV, Georgiou I, Drosos AA. The role of microRNA-146a (miR-146a) and its target IL-1R-associated kinase (IRAK1) in psoriatic arthritis susceptibility. Scand J Immunol. 2010;71:382–5. doi: 10.1111/j.1365-3083.2010.02381.x. [DOI] [PubMed] [Google Scholar]
- 17.Murata K, Yoshitomi H, Tanida S, Ishikawa M, Nishitani K, Ito H. Plasma and synovial fluid microRNAs as potential biomarkers of rheumatoid arthritis and osteoarthritis. Arthritis Res Ther. 2010;12:R86. doi: 10.1186/ar3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alevizos I, Illei GG. MicroRNAs as biomarkers in rheumatic diseases. Nat Rev Rheumatol. 2010;6(7):391–8. doi: 10.1038/nrrheum.2010.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murata K, Furu M, Yoshitomi H, Ishikawa M, Shibuya H, Hashimoto M. Comprehensive microRNA analysis identifies miR-24 and miR-125a-5p as plasma biomarkers for rheumatoid arthritis. PLoS ONE. 2013;8(7):e69118. doi: 10.1371/journal.pone.0069118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Wan Y, Guo Q, Zou L, Zhang J, Fang Y, et al. Altered microRNA expression profile with miR-146a upregulation in CD4+ T cells from patients with rheumatoid arthritis. Arthritis Res Ther. 2010;12:R81. doi: 10.1186/ar3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alevizos I, Alexander S, Turner RJ, Illei GG. MicroRNA expression profiles as biomarkers of minor salivary gland inflammation and dysfunction in Sjögren’s syndrome. Arthritis Rheum. 2011;63:535–44. doi: 10.1002/art.30131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Srivastava SK, Bhardwaj A, Singh S, Arora S, Wang B, Grizzle WE, et al. MicroRNA-150 directly targets MUC4 and suppresses growth and malignant behavior of pancreatic cancer cells. Carcinogenesis. 2011;32:1832–9. doi: 10.1093/carcin/bgr223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishitoh H, Ichijo H, Kimura M, Matsumoto T, Makishima F, Yamaguchi A, et al. Identification of type I and type II serine/threonine kinase receptors for growth/differentiation factor-5. J Biol Chem. 1996;271:21345–52. doi: 10.1074/jbc.271.35.21345. [DOI] [PubMed] [Google Scholar]


