Abstract
STUDY HYPOTHESIS
We hypothesized that a better discrimination between follicles containing oocytes with high developmental competence and those containing oocytes with low competence, based on a combination of a follicle's size and transcriptomic signature, will provide a reliable method to predict embryonic outcome of IVF.
STUDY FINDING
This study provides new insights on the impact of follicular size on oocyte quality as measured by embryonic development and demonstrates that medium follicles yield a better percentage of transferable embryos.
WHAT IS KNOWN ALREADY
Although it is generally accepted that large ovarian follicles contain better eggs, other studies report that a better follicular size subdivision and a better characterization are needed.
STUDY DESIGN, SAMPLES/MATERIALS, METHODS
Individual follicles (n = 136), from a total of 33 women undergoing IVF, were aspirated and categorized on the basis of their follicular liquid volume (small, medium or large) and the embryonic outcome of the enclosed oocyte: poor or good development. Comprehensive gene expression analysis between cells from the different sized follicles was performed using microarrays and quantitative RT–PCR to find molecular markers associated with follicular maturity and oocyte developmental competence.
MAIN RESULTS AND THE ROLE OF CHANCE
The analysis of embryonic outcome in relation to follicular size indicates that the medium-sized follicles category yielded more transferable embryos (35%) compared with the largest follicles (30%) (NS). Gene expression analysis revealed expression markers with significant (P < 0.05) discrimination between the poor development groups for all three follicle sizes, and good development medium-size follicles, including up-regulation of thrombomodulin, transforming growth factor, beta receptor II and chondrolecti, and those associated with hyaluronan synthesis, coagulation and hepatocyte growth factor signalling.
LIMITATIONS, REASONS FOR CAUTION
These analyses were performed in a single cohort of patients coming from a single clinic and the biomarkers generated will require validation in different geographical and biological contexts to ensure their global applicability.
WIDER IMPLICATIONS OF THE FINDINGS
Medium-size follicles seem to be the optimal size for a positive embryonic outcome and are associated with competence markers that may help in understanding the ideal differentiation status during late folliculogenesis.
LARGE SCALE DATA
The data discussed in this publication have been deposited in The National Center for Biotechnology Information Gene Expression Omnibus database and are accessible through GEO Series accession number GSE52851.
STUDY FUNDING AND COMPETING INTEREST(S)
This study was supported by Canadian Institutes of Health Research (CIHR) and Natural Sciences and Engineering Research Council of Canada (NSERC) to M.A.S. There are no competing interests to declare.
Keywords: follicular cells, granulosa cells, follicular size, oocyte competence, gene expression, hepatocyte growth factor
Introduction
In order to improve the success rate of assisted reproductive technologies (ART), the analysis of the reasons for failure is a key component. Nearly 64% of ART cycles using fresh own oocytes fail to produce a pregnancy (CDC report USA national survey http://www.cdc.gov/art/pdf/2013-report/art_2013_national_summary_report.pdf#page=21), raising medical, scientific, public health, economic and social issues for patients and society. The most important research question is why some oocytes have higher developmental competence potential than others for a given cycle and patient, or across patients.
In the past years, much evidence has demonstrated a relationship between oocyte developmental acquisition and the surrounding somatic cell environment acquired through folliculogenesis (Sugiura and Eppig, 2005; Li and Albertini, 2013; Dumesic et al., 2015). Moreover, efforts have been made to link follicle size to developmental competence using different evaluation criteria (Nataprawira et al., 1992; Wittmaack et al., 1994; Arnot et al., 1995; Dubey et al., 1995; Miller et al., 1996; Ectors et al., 1997; Bergh et al., 1998; Salha et al., 1998; Teissier et al., 2000; Trounson et al., 2001; Triwitayakorn et al., 2003; Rosen et al., 2008; Farhi et al., 2010; Lee et al., 2010; Mehri et al., 2014). Most studies have compared two populations of follicle sizes (variable threshold) and reached the conclusion that larger follicles are superior in terms of developmental competence (a list of relevant publications is summarized in Supplementary Table SI). Using the likelihood of cleavage as a criterion for evaluating developmental competence, Ectors and collaborators (Ectors et al., 1997), defined follicles in the 16–23 mm diameter size range as superior to the <16 mm and >23 mm categories. Based on the early cleavage timing, Lee and co-authors (Lee et al., 2010), determined that follicles with a volume of 3–5 ml (18–21 mm follicles) were superior to follicles >5 ml (>21 mm). More recently, Farhi et al. (2010) showed that when the size of the largest follicle in a patient is 18–22 mm, the likelihood of pregnancy is greater than when it is 26 mm (Farhi et al., 2010).
In a previous study, we have found molecular markers of oocyte competence associated with individual oocytes collected from individual follicles (Hamel et al., 2008, 2010a, b). Since a correlation between follicular physiological status and embryo outcome has been demonstrated, we want to explore if a classification of these follicles based on their size and embryonic outcome could explain the reasons for success or failure. Therefore, this study has two main goals. The first one is to determine whether or not the sizes of human follicles are associated with oocyte competence for embryonic development. The second one is to attempt to understand the impact of follicular status by associating a differential gene expression list to each follicular size category. This study thus allowed identification of markers of follicular maturity and embryo transferability and highlighted a new physiological pathway, namely hepatocyte growth factor (HGF) signalling, unknown until now in follicular physiology.
Materials and Methods
This work utilized the same database and samples as used previously (Hamel et al., 2008, 2010a, b), and the institutional review board approval was obtained for those studies. The specific tissues and procedures for this analysis are outlined below as well as all the information related to sample preparation.
Follicular cells recovery
Follicular cells (FC) composed principally of granulosa cells but containing contaminating cumulus cells from individual follicles (n = 136) were collected from 33 consenting patients who were undergoing IVF treatment at the Ottawa Fertility Centre (Ottawa, ON, Canada). Samples were collected irrespective of hormonal treatment (FSH or hMG) or age. Patients with polycystic ovary syndrome, severe male factor and ovarian failure were excluded from the study.
FCs were collected from individual follicular aspiration and the cumulus-oocyte complexes were used for IVF/ICSI procedures, as previously described (Hamel et al., 2008, 2010a, b). Only follicles containing a mature oocyte were kept for the analysis. Cells collected from the aspirated follicular fluid are mainly granulosa cells. Ovulation was triggered when the 2 largest follicles reached 17 mm (measured by ultrasound) in a given cycle and oocyte retrieval was carried out 34 h later. Patient follicular population repartition was normal, with a mean of 5.7 ± 2.3 (SEM) follicles >15 mm diameter and 12.1 ± 6.3 (SEM) follicles <15 mm diameter at trigger. Although no particular treatment was used to remove contaminating blood cells, the fact that individual aspiration was used has minimized the contaminant and we believe that the contamination would be similar across follicular sizes and therefore masked by the contrast analysis. Data from individual follicles, including follicular fluid volume, oocyte maturity, embryonic development and treatment outcomes, were collected. Depending on the aspirated follicular volume, the samples were categorized on the basis of size (small (S): 0.5–1.5 ml, medium (M): 2–3 ml or large (L): >3 ml) and embryo outcome: poor development/blocked (−), or transferable (transferred or cryopreserved) embryos. A positive embryo development (+) refers to a transferable embryo, at Day 3 (67%) or Day 5 (33%) and these embryos have been cryopreserved for potential transfer later.
From the dataset associated with the 33 patients, a subset of samples with specific follicular size and enclosed oocyte competence outcome were selected for gene expression analysis with microarrays and quantitative RT–PCR (qRT–PCT) (Supplementary Table SII).
Experimental design
For the microarray and qRT–PCR analyses, the four categories (S−, M−, M+ and L−) each containing four different individual samples were included and three comparisons were performed in triplicate. The medium-sized follicles associated with transferable embryos (M+) were compared with all follicle size categories correlated with poor or blocked embryonic development (S−, M− and L−). Schematic overview of the general experimental design for microarrays and qRT–PCR is provided in Supplementary Fig. S1. Considering the limited individual tissues, the same samples utilized for the microarray experiment have also been used before amplification for the qRT–PCR assays.
RNA extraction and amplification
Total RNA was extracted using the RNeasy mini kit (Qiagen, Mississauga, Canada) following the manufacturer's protocol and including a DNAse digestion with the RNase-free DNase Set (Qiagen), directly on the extraction column. Total RNA integrity and concentration were evaluated on a 2100-Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) using the RNA 6000 Pico Kit (Agilent Technologies) and the RNA integrity number threshold for usable samples was set as >7. To generate enough material for hybridization, the samples were amplified using the RiboAmp HS RNA amplification kit (Life Technologies Inc., Burlington, ON, Canada). After two amplification rounds of 6 h each, the amplified RNA (aRNA) output was quantified using the NanoDrop ND-1000 system (NanoDrop Technologies, Wilmington, DE, USA).
Sample labelling and microarray hybridization
For each sample, 2 µg of aRNA were labelled using the ULS Fluorescent Labelling Kit for Agilent arrays (with Cy3 and Cy5) (Kreatech Diagnostics, Amsterdam, Netherlands). The labelled product was then purified with the PicoPure RNA Isolation Kit but without DNase treatment. Labelling efficiency was measured using the Nano-Drop ND-1000 (NanoDrop Technologies). Samples were hybridized on the human gene expression 4 × 44K v2 microarray (Agilent).
Overall, 18 hybridizations were performed, corresponding to M+ versus S−, M+ versus M− and M+ versus L− with three biological replicates samples for each group and including a technical dye-swap replicate. A total of 825 ng of each labelled sample (Cy3 and Cy5) was incubated in a solution containing 2× blocking agent and 5× fragmentation buffer in a volume of 55 µl at 60°C for 15 min and put on ice immediately afterwards. 2X HI-RPM hybridization buffer (Agilent) was added (55 µl of 2× GEx) for a total volume of 110 µl. The hybridization mix was added onto the array and hybridization was performed at 65°C for 17 h using an Agilent Hybridization chamber in a rotating oven. Slides were then washed with gene expression wash buffer 1 (containing 0.005% Triton X-102) for 3 min at room temperature and then transferred to gene expression wash buffer 2 (also containing 0.005% Triton X-102) for 3 min at 42°C. Final washes at room temperature with acetonitrile (for 10 s at room temperature) and with drying and stabilization solution (for 30 s at room temperature) were performed before air-drying slides. The slides were then scanned using the Tecan PowerScanner microarray scanner (Tecan Group Ltd, Männerdorf, Switzerland) and features were extracted using ArrayPro 6.4 (Media Cybernetics, Bethesda, MD, USA).
Microarray data analysis
Microarray data were processed with Flexarray 1.6.1 (Efron and Tibshirani, 2002) using a background correction (simple subtraction method) performed on raw intensities, Loess within-array normalization, and between-array Quantile normalization based on intensities. To improve the power and gain a more stable inference with our small number of arrays, the Limma package in Bioconductor was used in FlexArray. Limma uses linear models to analyse the microarray data and an empirical Bayes method to assess differential expression by moderating the standard errors of the estimated log-fold changes (Smyth, 2004). Differences between treatments were considered significant when the Limma P-value was less than 0.05. The data discussed in this publication have been deposited in The National Center for Biotechnology Information Gene Expression Omnibus database and are accessible through GEO Series accession number GSE52851.
Ingenuity pathway analysis
Data were also analysed through the use of Ingenuity Pathway Analysis (IPA, Ingenuity Systems, www.ingenuity.com). IPA is able to construct networks showing the known potential direct or indirect interactions between molecules, and to overlay our microarray data on it.
Complementary DNA (cDNA) preparation and qRT–PCR
Total RNA from 12 individual follicles (10 ng) (three biological replicates of M+, S−, M− and L−) was reverse transcribed using a qScript Flex cDNA Synthesis Kit (Quanta Biosciences, Gaithersburg, MD, USA) with oligo dT(20) primers following the manufacturer's recommendations. Based on the P-values and fold changes observed in the microarray analysis, in combination with a known gene function, eight genes were selected for validation by qPCR. The primers used in qRT–PCR are listed in Supplementary Table SII and were designed using the IDT PrimerQuest tool (http://www.idtdna.com/Scitools/Applications/Primerquest/, accessed May 2012). To confirm the specificity of each pair of primers, electrophoresis on a standard 1.2% agarose gel was performed for each amplified fragment. The PCR product was then purified using the QIAquick Gel Extraction kit (Qiagen), quantified using the NanoDrop ND-1000, and then sequenced. The products were used to create the standard curve for the quantification experiment, with dilutions ranging from 2 × 10−4 to 2 × 10−8 ng µl−1. qRT–PCR was performed on a LightCycler 480 (Roche Diagnostics, Laval, QC, Canada) using SYBR incorporation. Each reaction mix (in a final volume of 20 µl) contained 2 µl (0.5 ng) of the cDNA product, 0.25 mmol L−1 of each primer and 1× SYBR mix (LightCycler 480 SYBR Green I Master, Roche Diagnostics). The PCR conditions used for all genes were as follows: denaturing cycle for 10 min at 95°C, 50 PCR cycles (denaturing, 95°C for 1 s; annealing, for 5 s; extension, 72°C for 5 s), a melting curve (94°C for 5 s, 72°C for 30 s and a step cycle starting at 72°C up to 94°C at 0.2°C/s), and a final cooling step at 40°C. cDNA quantification was performed using LightCycler 480 Software Version 1.5 (Roche Diagnostics) by comparison with the standard curve. PCR specificity was confirmed by melting-curve analysis.
Statistical analysis of qRT–PCR results
Analysis of gene expression stability over the different follicles was performed using the GeNorm VBA applet software as described by Vandesompele et al. (2002). The most stable reference genes were identified by the stepwise exclusion of the least stable gene and recalculating the M values. Following GeNorm analysis, beta actin (ACTB) and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were the most stable genes among 4 tested (ACTB, GAPDH, tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta (YWHAZ), cyclophilin A (PPIA)), with M values of 0.545 (i.e. <1.5, as recommended by the software developer). Evaluation of mRNA differences was performed using Mann–Whitney test for the comparisons of two conditions with GraphPad Prism Version 5.0 (GraphPad Software Inc., CA, USA). Differences were considered statistically significant at the 95% confidence level (P < 0.05). Data are presented as mean ± SEM.
Results
Follicle volume analysis and its association with embryonic outcome
The complete dataset, including 136 follicles from 33 patients, was analysed and follicles were grouped into three categories based on volume. Small (S) correspond to follicles with a volume between 0.5 and 1.5 ml, medium (M) follicles have a volume of 2–3 and large (L) have >3 ml volume. Also, the follicles were also classified in two categories based on the oocyte developmental competence from them: (i) poor development (blocked) and (ii) transferred/cryopreserved. The relative mean frequency of each embryonic developmental category per follicular size is illustrated in Fig. 1. The ‘no development’ group was not used since it can be indicative of an IVF or sperm issue instead of considering the level of oocyte competence. Additional information regarding data and relative proportions of each follicular size groups per development category is shown in Table I.
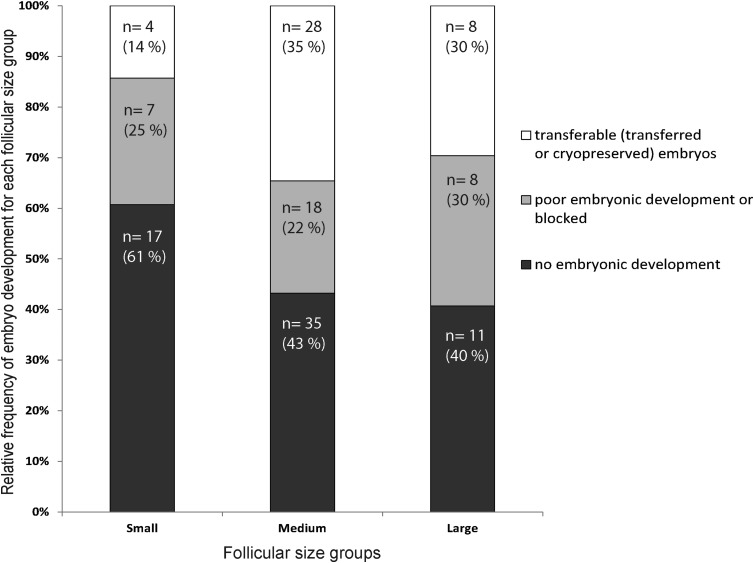
Figure 1.
Individual human follicle and embryo development dataset analysis. Relative mean frequency of embryo development category per follicular size group. The number of embryos in each developmental category from each follicular size range and their percentage (%) of the total is indicated.
Table I.
Information regarding human follicular cells and embryonic developmental stage used in microarrays experiments.
| Small follicle | Medium follicle | Large follicle | Medium follicle | |
|---|---|---|---|---|
| Poor or blocked embryonic development |
Transferable (transferred or cryopreserved) embryos | |||
| Follicular fluid | 0.5–1.5 ml | 2–3 ml | >3 ml | 2–3 ml |
| Equivalent diameter based on fluid volume | 9.8–14.2 mm | 15.6–17.9 mm | >18 mm | 15.6–17.9 mm |
| Embryonic developmental stages associated with enclosed oocytes | Zygote to up to 10 cells | Zygote to up to 10 cells | Zygote to up to 10 cells | Blastocyst stage at Day 5 or 6 |
The largest proportion of transferable embryos is associated with medium-size follicles (2–3 ml of follicular fluid, Fig. 1) and also associated with the smallest proportion of blocked embryos. Consequently three times more follicles were available for this category. Consistent with previous studies (see references in Supplementary Table SI), the smallest follicles are correlated with the smallest proportion of transferable embryos. Conversely, the poor development (blocked) embryos are more likely to come from large follicles (>3 ml) than medium-sized follicles (2–3 ml) but the majority of them are from the smallest size category (0.5–1.5 ml). Using two-way analysis of variance, the follicular volume has a significant effect on embryo development (P-value <0.0001) and the correlation (r) of follicle size and embryonic development is also significant (P-value = 0.0086) showing that the embryo development is influenced by follicular size.
Based on our analyses, the best oocyte developmental competence was associated with the medium-size category. Using regression analysis, it was found that within the small and medium categories, a bigger follicle volume is associated with higher occurrence of a transferable embryo (P-value = 0.009, R2 = 0.99, data not shown). However, this is not the case for the large follicle group (P-value = 0.163). Although our data set was small (136 follicles), each follicle was tracked individually, thus providing much higher resolution of the outcome.
Gene expression analysis
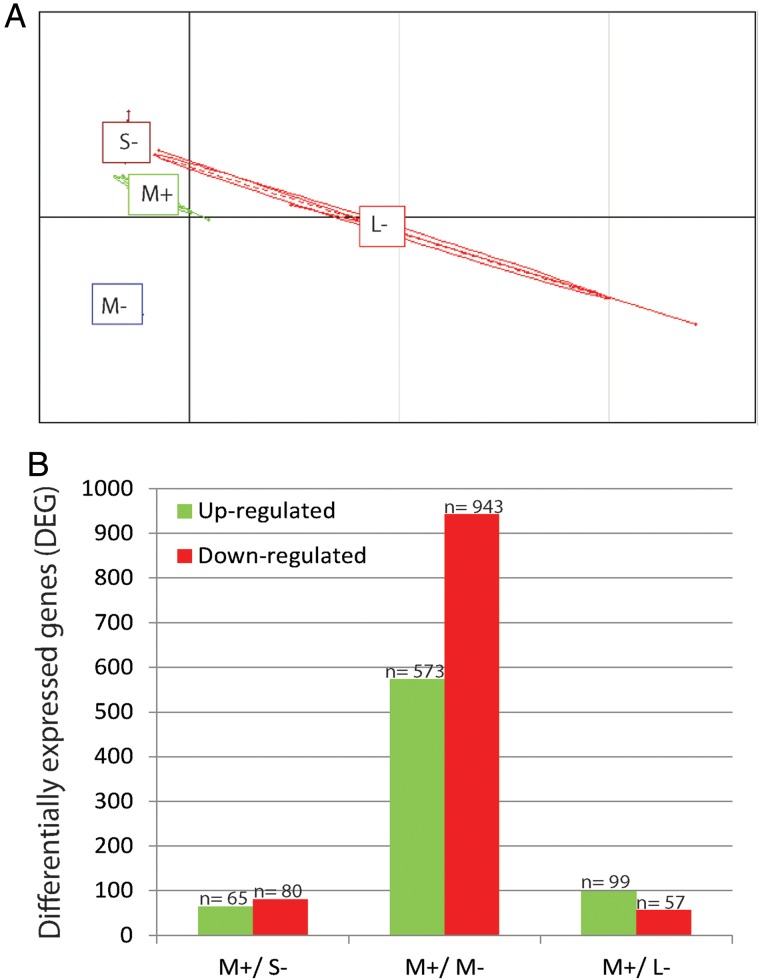
The microarray experiment was performed using the medium volume follicles associated with transferred or cryopreserved embryos groups (M+) as the group of interest since they have been shown to have a higher proportion of transferable embryos. The M+ follicles were compared to the poor or blocked embryonic development follicular groups of small, medium and large follicular volumes (S−, M− and L−). The experimental microarray design is illustrated in Supplementary Fig. S1. Principal component analysis (PCA), which groups the categories or samples based on their gene expression, shows that the small negative (S−), medium positive (M+), and medium negative (M−) follicles genes have a more homogenous expression pattern, and much less dispersed, compared to the large negative population (Fig. 2A). PCA also indicates that biological replicates tightly co-segregate, which adds value to our gene analysis by showing homogeneity within categories.
Figure 2.
Characterization of the three different sizes of follicle by microarrays. (A) Principal component analysis of follicle group comparisons using biological triplicate microarray data. (B) Changes in gene expression profile among the different follicular sizes. Unigenes up-regulated (green) and down-regulated (red) between the different microarrays. M+, medium follicles associated with transferable (transferred or cryopreserved) embryos; S− small, M− medium and L− large follicles linked to a poor or blocked embryo development.
A total of 1817 targets/probes were detected to be significantly changed between M+/ S−, M+/ M− and M+/ L− follicular groups (Fig. 2B). Details regarding gene ID and fold-change (>1.5) of the differentially expressed genes (DEG) among the three comparisons can be found in Supplementary Tables SIII–SV.
Quantitative RT–PCR validation
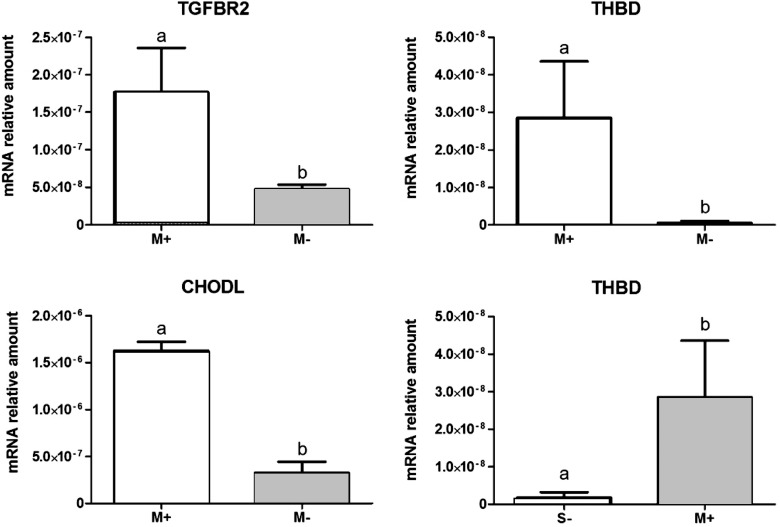
Validation of the microarray results was performed using qRT–PCR (Fig. 3). Candidate selection for qRT–PCR validation was based on different expression between the M+ and the M− groups transforming growth factor, beta receptor II (TGFBR2), thrombomodulin (THBD) and chondrolectin (CHODL) transcripts) and also compared across S− and M+ follicles (THBD). Moreover, Supplementary Fig. S2 illustrates the gene expression comparison of the qRT–PCR validation and the microarray results for the four different follicular groups (S−, M−, L− and M+) and shows a similar profile for both methods.
Figure 3.
Validation by quantitative RT–PCR of the potential markers of follicles associated with transferable embryos. M+: medium-sized follicles associated with developmental competence (transferable embryos); M−: medium-sized follicles associated with poor or blocked embryonic development; S−: small follicles associated with poor development or blocked embryonic development; TGFBR2: transforming growth factor receptor 2; CHODL: chondrolectin; THBD: thrombomodulin. Different superscripts represent significant differences (Mann–Whitney test, P = 0.05).
Ingenuity pathway analysis
Among the tools available in Ingenuity software, it is possible to select extracellular proteins as a way to explore the possible secretion or interaction of molecules between granulosa or cumulus cells. The extracellular molecules represented, respectively, 9.6, 6.4, and 5.3% of the DEG (P-value <0.05 and 1.5-fold change) in the comparisons M+/S−, M+/M− and M+/L− (Supplementary Tables SIII–SV). There were six molecules in common between the M+/S− and M+/M− groups: epidermal growth factor containing fibulin-like extracellular matrix protein 1 (EFEMP1), lectin, galactoside-binding, soluble, 12 (LGALS12), lectin, galactoside-binding, soluble, 3 (LGALS3), nidogen 1 (NID1), PLBD1 (phospholipase B domain containing 1), vitronectin (VTN). Three molecules are shared between M+/ M− and M+/ L− groups: (apolipoprotein C-I (APOC1), proprotein convertase subtilisin/kexin type 9 (PCSK9), metallopeptidase inhibitor 1 (TIMP1)) (Supplementary Tables SIV and SV).
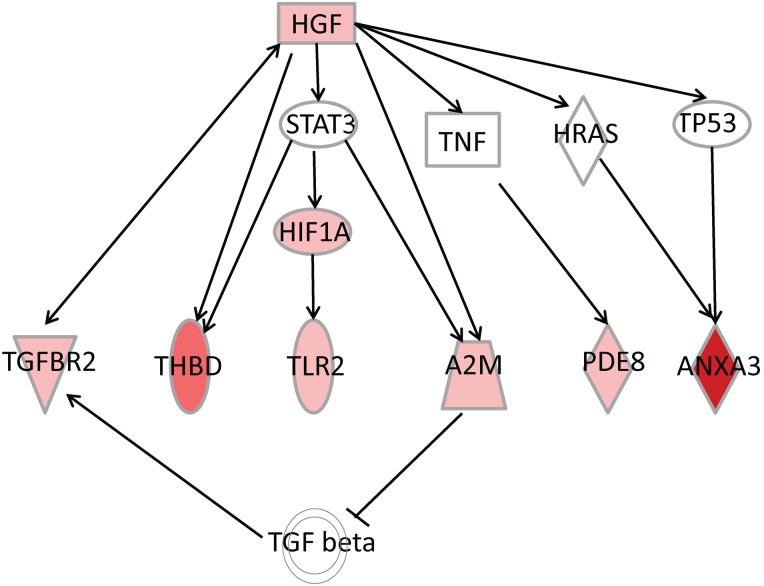
Ingenuity software has a function to identify upstream regulators by looking for pathways and genes that can have increased activity based on expression levels of their downstream targets. Among the different upstream regulators associated with oocyte competence, the HGF pathway has been identified (Fig. 4). Indeed, many DEG in M+ follicles seem to be driven by the upstream regulator HGF. HGF is also up-regulated when M+ and M− groups are compared (Supplementary Table SIV). The majority of DEG (Z score of 2.8, where 2 is considered to be a very significant threshold) under this influence is also activated in M+/M− (Supplementary Table SIV). However, we note with interest that HGF is upstream of TGFBR2, THBD, toll-like receptor 2 (TLR2), alpha-2-macroglobulin (A2M), PDE8B and annexin A3 (ANXA3), which are all up-regulated in M+/M− (Fig. 4) and hence associated with developmental competence.
Figure 4.
Ingenuity pathway analysis revealed hepatocyte growth factor (HGF) as an upstream regulator. Various downstream targets found among over-expressed genes in medium follicular cells associated with developmental competence (M+) compared to medium-sized follicles associated with poor or blocked embryonic development (M−) are also shown. Gene symbol colour (red to pink) indicates degrees of expression. Full arrow line indicates ‘acts on’ and arrow dotted line: ‘acts on indirectly’. TLR2: toll-like receptor 2; ANXA3: annexin-3; A2M: alpha 2 macroglobulin; PDE8B: phosphodiesterase 8B; HIF1A: hypoxia induced factor 1A; TNF: tumour necrosis factor; STAT3: signal transducer and activator of transcription 3; HRAS: v-Ha-ras Harvey rat sarcoma viral oncogene homologue; TP53: tumour protein P53; TGF beta: transforming growth factor beta.
Discussion
To our knowledge, this is the first study where FC samples from individually punctured follicles are compared in relation to follicular volume and embryonic outcome. Indeed, no gene analysis has ever been done in conjunction with such precise knowledge of follicle volume and the developmental competence of the enclosed oocyte.
The present study included an analysis of a dataset of 136 follicles to correlate follicular volume to embryonic development outcomes. The results show that the medium follicles yielded more transferable embryos compared with the largest and the smallest ones. These results are consistent with studies by Ectors (Ectors et al., 1997) and Lee (Lee et al., 2010), who linked medium-sized follicles to a superior cleavage rate compared with the largest follicles. Our study goes further by examining embryo transferability, a more advanced criterion of embryo development evaluation.
The first conclusion emerging from the volume-outcome analysis is that oocyte quality seems to be associated with specific follicular conditions, since follicles that are too small or too large contain oocytes that are less likely to become transferable embryos. However, follicle size does not explain oocyte quality completely, since both competent and incompetent oocytes are observed in all categories. This intra-size diversity supports our hypothesis of the existence of a potential relationship between granulosa cell gene expression and the developmental competence of the enclosed oocyte. Based on PCA, the volume-outcome groupings have reduced overlap and M+ follicles (the best quality) distinguish themselves from M− follicles and relatively well from S− but with more difficulty from L−, which corroborates the results obtained using qRT–PCR. Gene expression analysis using the microarray revealed particularities of each of the four follicle categories, which may hold the key to explaining the variations in oocyte potential.
Distinctive gene expression patterns associated with small incompetent follicles (S−) were related principally to follicle growth, for example, small follicles overexpress angiotensin I converting enzyme 2 (ACE2) (fc: 2.7) (Goncalves et al., 2012), glutathione S-transferase theta 3 (GSTA3) (fc: 2.3), and fascin homologue 1 (FSCN1) (fc: 2.3) (Kostopoulou et al., 2008). It is noted with interest that secreted phosphoprotein 1 (SPP1), a gene associated with antral follicle growth in bovine (Skinner et al., 2008; Hayashi et al., 2010), is down-regulated (fc: −3.9) in S− follicles and therefore could be related to low follicular competence (Supplementary Table SIII).
Among the differentially overexpressed genes in the largest incompetent follicles uromodulin (UMODL1) (fc: 37.3) is particularly interesting. Umodl1 is regulated by gonadotrophins. Mice carrying extra copies of functional Umodl1 were generated by bacterial artificial chromosome transgenesis showing reduced or diminished fertility (Wang et al., 2012). Among the multilayered pre-antral follicles, elevated apoptosis was observed in both the oocytes and surrounding granulosa cells (Wang et al., 2012). Genes associated with cellular proliferation, such as filamin A interacting protein 1-like (FILIP1L) (fc: 17,3) (Burton et al., 2011), Y box binding protein 1 (YBX1) (fc: 2.1) (Raffetseder et al., 2009) and TGFBR2 (fc: 1.9) (Kingsley, 1994), and with apoptosis, for example NOD-like receptors (NLR) family, pyrin domain containing 12 (NLRP12) (fc: 5.1) (Wang et al., 2002) and B-cell lymphoma 2 (BCL2)-like 13 (BCL2L13) (fc: 1.9) an apoptotic facilitator (Kataoka et al., 2001), have been up-regulated (Supplementary Table SV).
In the M+/ M− DEG list (Supplementary Table SIV), and also identified as a major upstream regulator, HGF has been shown to act positively on the reduction of apoptosis in granulosa cells (Uzumcu et al., 2006). It corroborates with the down-regulation in M+ follicles of molecules associated with hypoxia, like spectrin alpha non-erythrocytic 1 (SPTAN1) (fc: 9.4) (Weigand et al., 2012), oxidative stress with dual oxidase 2 (DUOX2) (fc: 3.4) (Ameziane-El-Hassani et al., 2005) and apoptosis, epithelial membrane protein 3 (EMP3) (fc: 3.1) (Taylor and Suter, 1996). HGF acts positively on angiogenesis, which is consistent with ANXA3 downstream over-expression (Park et al., 2005). Among the genes associated with M+ and downstream HGF, TLR2 (fc: 1.7) is of potential interest from a physiological perspective. Toll-like receptors are well known to be associated with antigen detection but also have endogenous ligands (Zhang and Schluesener, 2006). TLR2 and TLR4 recognize extracellular matrix (ECM) degradation products, such as fragments of hyaluronan and heparin sulphate, a function that represents a key element in our study (Supplementary Fig. S3). Supplementary Figure S3 illustrates how the medium follicles associated with oocyte competence prepare for ovulation by programming the right context for blood coagulation, an important feature to prevent intra-abdominal bleeding and for corpus luteum formation. Also TLR2 acts positively on the production of cytokines interleukin 6 and tumour necrosis factor-alpha, which promote tissue regeneration. This positive feedback constitutes a system in which post-inflammatory wound defence and repair processes are promoted. TLR2 expression might be associated with follicles that have responded adequately to stimuli associated with ECM degradation, and hence with an appropriate inflammatory response.
With respect to inflammation, TGFBR2 (also downstream from HGF) is over-expressed in M+ compared with M− (fc: 1.8) and down-regulated in L− (fc: 1.9) (Supplementary Tables SIV and SV). The tissue repair process involving the TLR2 gene also involves hyaluronic acid and is positively regulated by members of the TGF-beta family. Some of these are produced by ovarian follicles (anti-Mullerian hormone, bone morphogenetic protein 2 (BMP2), BMP4, BMP6) and others are secreted by oocytes, for example BMP6, BMP15 and growth differentiation factors (GDFs) (Pangas, 2007). TGFBR1 acts positively on hyaluronic acid via its action on hyaluronan synthase, the enzyme necessary for hyaluronic acid synthesis. Hyaluronic acid is incorporated into the plasma membrane or into the ECM (Weigel et al., 1997), acting on specific receptors such as CD44 and hyaluronan-mediated motility receptor. Hyaluronic acid is involved in ECM regulation, cellular motility, and has a positive impact on cellular proliferation, preparing the luteal cells for differentiation (Stern, 2004).
THBD, downstream from HGF, is overexpressed in follicles above a certain size and associated with developmental competence. THBD forms a complex with thrombin, which produces activated protein C by cleavage of the inactive form. Activated protein C binds to endothelial protein C receptor, which stimulates Prader–Willi/ Angelman gene region 1 (PAR1) and PAR4 receptors and G protein signalling (Riewald et al., 2003; Griffin et al., 2006). This system is activated in granulosa cells from pre-ovulatory follicles following the LH surge (Cheng et al., 2012). Furthermore, PAR1 and PAR4 act negatively on cyclic AMP production and inhibit progesterone production (Cheng et al., 2012). THBD could therefore regulate the luteinization process in M+ follicles and could be representative of an optimal pre-luteinization state 34 h hCG as in this case.
Finally, expression of another molecule downstream of HGF, the enzyme PDE8B which inactivates cyclic AMP, would have a transitory expression associated with the oocyte's developmental competence in terms of capacity to reach the morulae/blastocyst stage. The PDE8B transcript has been detected in human ovaries (Hayashi et al., 1998), and found under-expressed in granulosa cells in patients with diminished ovarian reserve (Skiadas et al., 2012). It appears that PDE8B could also modulate the level of luteinization in concert with THBD.
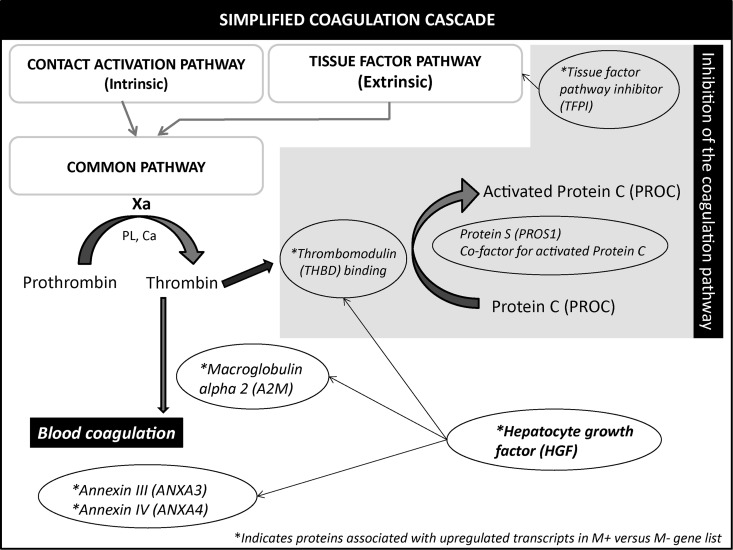
Also the role of THBD in the coagulation cascade might be important (Fig. 5). It is clear that the preparation of the follicles towards ovulation requires the appropriate anti-coagulant factors to be present and ready to be activated to promote optimal blood clotting for corpus luteum formation.
Figure 5.
Schematic illustration of the coagulation pathway and its inhibitor systems. Three pathways are implicated in the classical blood coagulation: (i) intrinsic pathway (ii) extrinsic and (iii) the common pathway. The majority of the molecules identified in this study is in the common pathway. * represents transcripts over-expressed in medium follicular cells associated with developmental competence (M+/ M−). The grey box contains proteins associated with anticoagulation. Xa, factor Xa; PL, phospholipids; Ca, calcium.
HGF downstream signalling seems to play a key role in follicle developmental competence and acts at various levels, including apoptosis reduction, ECM organization and inflammatory processes, luteinization and angiogenesis. The potential cross signalling of HGF also involves the theca cells, as HGF increased expression of the SCF (Stem cell factor c-kit) gene in granulosa cells, and SCF reciprocally increased expression of the HGF gene in theca cells (Ito et al., 2001). Several molecules associated with negative regulation of the coagulation canonic pathway, notably tissue factor pathway inhibitor (TFPI) (fc: 1.6)), THBD (fc: 3.7), alpha 2 macroglobulin (A2M) (fc: 1.6), ANXA3 (fc: 36.0) and ANXA4 (fc: 1.6), are up-regulated in M+/M− follicles (Fig. 4). The anticoagulation process could facilitate the ovulation process of medium-sized follicles associated with competent oocytes. This study has allowed us to identify several potential new indicators (THBD, TLR2, PDE8B, UMODL1L, TGFBR2 and ANXA3) associated with embryo transferability. The markers identified herein may be useful for identifying the best follicles but may also serve to help assess whether or not a given cycle has created permissive conditions for oocyte competence.
Conclusions
Based on three gene expression comparisons between follicles of different volumes, this study brings important new information on the impact of follicular size on oocyte quality and the potential explanation(s) for such differences. Several biomarkers have been identified with positive and negative associations with the outcome. Moreover, a key modulator of important downstream targets, HGF, has been identified and singled out for further characterization.
Supplementary data
Supplementary data are available at http://molehr.oxfordjournals.org/.
Authors' roles
A.L.N. participated in the conception and design of the study, analysed the complete dataset, processed all the samples (RNA extraction, microarrays experiment and real-time PCR) and analysed, performed the statistical analysis, interpreted the data, and drafted the manuscript. M.C.L. participated in the conception and design of the study, participated in the coordination of the sample collection and the dataset acquisition, and revised the manuscript. A.L. participated in the conception and design of the study, participated in the coordination of the sample collection and the dataset acquisition, and revised the manuscript. M.A.S. participated in the conception and design of the study, the analysis and the interpretation of the data, and improved the manuscript. All the authors read and approved the final manuscript.
Funding
This study was supported by grants from Natural Sciences and Engineering Research Council of Canada (NSERC) and Canadian Institutes of Health Research (CIHR).
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary Material
Acknowledgements
The authors would like to express a special tribute to the invaluable contribution of Melanie Hamel (1974–2012), who died shortly after defending her thesis. Melanie was involved in the early stages of this research and published the initial papers on the follicle transcriptome and pregnancy outcome. The assistance of Drs Claman, Kotarba and Haebe of the Ottawa Fertility Centre in this study is gratefully acknowledged. The authors thank the embryologists of the Fertility Centre for the collection of the human material and scoring of the embryos.
References
- Ameziane-El-Hassani R, Morand S, Boucher JL, Frapart YM, Apostolou D, Agnandji D, Gnidehou S, Ohayon R, Noel-Hudson MS, Francon J et al. Dual oxidase-2 has an intrinsic Ca2+-dependent H2O2-generating activity. J Biol Chem 2005;280:30046–30054. [DOI] [PubMed] [Google Scholar]
- Arnot AM, Vandekerckhove P, DeBono MA, Rutherford AJ. Follicular volume and number during in-vitro fertilization: association with oocyte developmental capacity and pregnancy rate. Hum Reprod 1995;10:256–261. [DOI] [PubMed] [Google Scholar]
- Bergh C, Broden H, Lundin K, Hamberger L. Comparison of fertilization, cleavage and pregnancy rates of oocytes from large and small follicles. Hum Reprod 1998;13:1912–1915. [DOI] [PubMed] [Google Scholar]
- Burton ER, Gaffar A, Lee SJ, Adeshuko F, Whitney KD, Chung JY, Hewitt SM, Huang GS, Goldberg GL, Libutti SK et al. Downregulation of Filamin A interacting protein 1-like is associated with promoter methylation and induces an invasive phenotype in ovarian cancer. Mol Cancer Res 2011;9:1126–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Kawamura K, Deguchi M, Takae S, Mulders SM, Hsueh AJ. Intraovarian thrombin and activated protein C signaling system regulates steroidogenesis during the periovulatory period. Mol Endocrinol 2012;26:331–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey AK, Wang HA, Duffy P, Penzias AS. The correlation between follicular measurements, oocyte morphology, and fertilization rates in an in vitro fertilization program. Fertil Steril 1995;64:787–790. [DOI] [PubMed] [Google Scholar]
- Dumesic DA, Meldrum DR, Katz-Jaffe MG, Krisher RL, Schoolcraft WB. Oocyte environment: follicular fluid and cumulus cells are critical for oocyte health. Fertil Steril 2015;103:303–316. [DOI] [PubMed] [Google Scholar]
- Ectors FJ, Vanderzwalmen P, Van Hoeck J, Nijs M, Verhaegen G, Delvigne A, Schoysman R, Leroy F. Relationship of human follicular diameter with oocyte fertilization and development after in-vitro fertilization or intracytoplasmic sperm injection. Hum Reprod 1997;12:2002–2005. [DOI] [PubMed] [Google Scholar]
- Efron B, Tibshirani R. Empirical bayes methods and false discovery rates for microarrays. Genet Epidemiol 2002;23:70–86. [DOI] [PubMed] [Google Scholar]
- Farhi J, Orvieto R, Gavish O, Homburg R. The association between follicular size on human chorionic gonadotropin day and pregnancy rate in clomiphene citrate treated polycystic ovary syndrome patients. Gynecol Endocrinol 2010;26:546–548. [DOI] [PubMed] [Google Scholar]
- Goncalves PB, Ferreira R, Gasperin B, Oliveira JF. Role of angiotensin in ovarian follicular development and ovulation in mammals: a review of recent advances. Reproduction 2012;143:11–20. [DOI] [PubMed] [Google Scholar]
- Griffin JH, Fernandez JA, Mosnier LO, Liu D, Cheng T, Guo H, Zlokovic BV. The promise of protein C. Blood Cells Mol Dis 2006;36:211–216. [DOI] [PubMed] [Google Scholar]
- Hamel M, Dufort I, Robert C, Gravel C, Leveille MC, Leader A, Sirard MA. Identification of differentially expressed markers in human follicular cells associated with competent oocytes. Hum Reprod 2008;23:1118–1127. [DOI] [PubMed] [Google Scholar]
- Hamel M, Dufort I, Robert C, Leveille MC, Leader A, Sirard MA. Genomic assessment of follicular marker genes as pregnancy predictors for human IVF. Mol Hum Reprod 2010a;16:87–96. [DOI] [PubMed] [Google Scholar]
- Hamel M, Dufort I, Robert C, Leveille MC, Leader A, Sirard MA. Identification of follicular marker genes as pregnancy predictors for human IVF: new evidence for the involvement of luteinization process. Mol Hum Reprod 2010b;16:548–556. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Matsushima K, Ohashi H, Tsunoda H, Murase S, Kawarada Y, Tanaka T. Molecular cloning and characterization of human PDE8B, a novel thyroid-specific isozyme of 3′,5’-cyclic nucleotide phosphodiesterase. Biochem Biophys Res Commun 1998;250:751–756. [DOI] [PubMed] [Google Scholar]
- Hayashi KG, Ushizawa K, Hosoe M, Takahashi T. Differential genome-wide gene expression profiling of bovine largest and second-largest follicles: identification of genes associated with growth of dominant follicles. Reprod Biol Endocrinol 2010;8:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Harada T, Tanikawa M, Fujii A, Shiota G, Terakawa N. Hepatocyte growth factor and stem cell factor involvement in paracrine interplays of theca and granulosa cells in the human ovary. Fertil Steril 2001;75:973–979. [DOI] [PubMed] [Google Scholar]
- Kataoka T, Holler N, Micheau O, Martinon F, Tinel A, Hofmann K, Tschopp J. Bcl-rambo, a novel Bcl-2 homologue that induces apoptosis via its unique C-terminal extension. J Biol Chem 2001;276:19548–19554. [DOI] [PubMed] [Google Scholar]
- Kingsley DM. The TGF-beta superfamily: new members, new receptors, and new genetic tests of function in different organisms. Genes Dev 1994;8:133–146. [DOI] [PubMed] [Google Scholar]
- Kostopoulou E, Angelidou S, Daponte A, Galani C, Chiotoglou I, Terzis A, Koukoulis G. Fascin can be an auxiliary immunomarker of ovarian granulosa cell tumors: comparison with calretinin and inhibin-alpha. Eur J Gynaecol Oncol 2008;29:638–642. [PubMed] [Google Scholar]
- Lee TF, Lee RK, Hwu YM, Chih YF, Tsai YC, Su JT. Relationship of follicular size to the development of intracytoplasmic sperm injection-derived human embryos. Taiwan J Obstet Gynecol 2010;49:302–305. [DOI] [PubMed] [Google Scholar]
- Li R, Albertini DF. The road to maturation: somatic cell interaction and self-organization of the mammalian oocyte. Nat Rev Mol Cell Biol 2013;14:141–152. [DOI] [PubMed] [Google Scholar]
- Mehri S, Levi Setti PE, Greco K, Sakkas D, Martinez G, Patrizio P. Correlation between follicular diameters and flushing versus no flushing on oocyte maturity, fertilization rate and embryo quality. J Assist Reprod Genet 2014;31:73–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KF, Goldberg JM, Falcone T. Follicle size and implantation of embryos from in vitro fertilization. Obstet Gynecol 1996;88:583–586. [DOI] [PubMed] [Google Scholar]
- Nataprawira DS, Harada T, Sekijima A, Mio Y, Terakawa N. Assessment of follicular maturity by follicular diameter and fluid volume in a program of in vitro fertilization and embryo transfer. Asia Oceania J Obstet Gynaecol 1992;18:225–230. [PubMed] [Google Scholar]
- Pangas SA. Growth factors in ovarian development. Semin Reprod Med 2007;25:225–234. [DOI] [PubMed] [Google Scholar]
- Park JE, Lee DH, Lee JA, Park SG, Kim NS, Park BC, Cho S. Annexin A3 is a potential angiogenic mediator. Biochem Biophys Res Commun 2005;337:1283–1287. [DOI] [PubMed] [Google Scholar]
- Raffetseder U, Rauen T, Djudjaj S, Kretzler M, En-Nia A, Tacke F, Zimmermann HW, Nelson PJ, Frye BC, Floege J et al. Differential regulation of chemokine CCL5 expression in monocytes/macrophages and renal cells by Y-box protein-1. Kidney Int 2009;75:185–196. [DOI] [PubMed] [Google Scholar]
- Riewald M, Petrovan RJ, Donner A, Ruf W. Activated protein C signals through the thrombin receptor PAR1 in endothelial cells. J Endotoxin Res 2003;9:317–321. [DOI] [PubMed] [Google Scholar]
- Rosen MP, Shen S, Dobson AT, Rinaudo PF, McCulloch CE, Cedars MI. A quantitative assessment of follicle size on oocyte developmental competence. Fertil Steril 2008;90:684–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salha O, Nugent D, Dada T, Kaufmann S, Levett S, Jenner L, Lui S, Sharma V. The relationship between follicular fluid aspirate volume and oocyte maturity in in-vitro fertilization cycles. Hum Reprod 1998;13:1901–1906. [DOI] [PubMed] [Google Scholar]
- Skiadas CC, Duan S, Correll M, Rubio R, Karaca N, Ginsburg ES, Quackenbush J, Racowsky C. Ovarian reserve status in young women is associated with altered gene expression in membrana granulosa cells. Mol Hum Reprod 2012;18:362–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner MK, Schmidt M, Savenkova MI, Sadler-Riggleman I, Nilsson EE. Regulation of granulosa and theca cell transcriptomes during ovarian antral follicle development. Mol Reprod Dev 2008;75:1457–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004;3:Article3. [DOI] [PubMed] [Google Scholar]
- Stern R. Hyaluronan catabolism: a new metabolic pathway. Eur J Cell Biol 2004;83:317–325. [DOI] [PubMed] [Google Scholar]
- Sugiura K, Eppig JJ. Society for Reproductive Biology Founders’ Lecture 2005. Control of metabolic cooperativity between oocytes and their companion granulosa cells by mouse oocytes. Reprod Fertil Dev 2005;17:667–674. [DOI] [PubMed] [Google Scholar]
- Taylor V, Suter U. Epithelial membrane protein-2 and epithelial membrane protein-3: two novel members of the peripheral myelin protein 22 gene family. Gene 1996;175:115–120. [DOI] [PubMed] [Google Scholar]
- Teissier MP, Chable H, Paulhac S, Aubard Y. Comparison of follicle steroidogenesis from normal and polycystic ovaries in women undergoing IVF: relationship between steroid concentrations, follicle size, oocyte quality and fecundability. Hum Reprod 2000;15:2471–2477. [DOI] [PubMed] [Google Scholar]
- Triwitayakorn A, Suwajanakorn S, Pruksananonda K, Sereepapong W, Ahnonkitpanit V. Correlation between human follicular diameter and oocyte outcomes in an ICSI program. J Assist Reprod Genet 2003;20:143–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trounson A, Anderiesz C, Jones G. Maturation of human oocytes in vitro and their developmental competence. Reproduction 2001;121:51–75. [DOI] [PubMed] [Google Scholar]
- Uzumcu M, Pan Z, Chu Y, Kuhn PE, Zachow R. Immunolocalization of the hepatocyte growth factor (HGF) system in the rat ovary and the anti-apoptotic effect of HGF in rat ovarian granulosa cells in vitro. Reproduction 2006;132:291–299. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002;3:RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Manji GA, Grenier JM, Al-Garawi A, Merriam S, Lora JM, Geddes BJ, Briskin M, DiStefano PS, Bertin J. PYPAF7, a novel PYRIN-containing Apaf1-like protein that regulates activation of NF-kappa B and caspase-1-dependent cytokine processing. J Biol Chem 2002;277:29874–29880. [DOI] [PubMed] [Google Scholar]
- Wang W, Tang Y, Ni L, Kim E, Jongwutiwes T, Hourvitz A, Zhang R, Xiong H, Liu HC, Rosenwaks Z. Overexpression of Uromodulin-like1 accelerates follicle depletion and subsequent ovarian degeneration. Cell Death Dis 2012;3:e433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigand JE, Boeckel JN, Gellert P, Dimmeler S. Hypoxia-induced alternative splicing in endothelial cells. PLoS One 2012;7:e42697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel PH, Hascall VC, Tammi M. Hyaluronan synthases. J Biol Chem 1997;272:13997–14000. [DOI] [PubMed] [Google Scholar]
- Wittmaack FM, Kreger DO, Blasco L, Tureck RW, Mastroianni L Jr, Lessey BA. Effect of follicular size on oocyte retrieval, fertilization, cleavage, and embryo quality in in vitro fertilization cycles: a 6-year data collection. Fertil Steril 1994;62:1205–1210. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Schluesener HJ. Mammalian toll-like receptors: from endogenous ligands to tissue regeneration. Cell Mol Life Sci 2006;63:2901–2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.