Abstract
We recently reported a 2-aminoimidazole-based antibiotic adjuvant that reverses colistin resistance in two species of Gram-negative bacteria. Mechanistic studies in Acinetobacter baumannii demonstrated that this compound downregulated the PmrAB two-component system and abolished a lipid A modification that is required for colistin resistance. We now report the synthesis and evaluation of two separate libraries of substituted 2-aminoimidazole analogues based on this parent compound. From these libraries, a new small molecule was identified that lowers the minimum inhibitory concentration of colistin by up to 32-fold greater than the parent compound while also displaying less inherent bacterial effect, thereby minimizing the likelihood of resistance evolution.
Keywords: Antibiotic adjuvant, 2-aminoimidazole, colistin, ESKAPE pathogens, Acinetobacter baumannii, Pseudomonas aeruginosa
Introduction
Antibiotic resistant organisms represent a major threat to global health. The Center for Disease Control and Prevention (CDC) estimates that 2 million people acquire antibiotic-resistant infections each year, of which 23,000 are fatal.1 The main culprits behind these infections are referred to as the ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species).2 The severity of the problem of multi-drug resistant (MDR) bacteria has been significantly exacerbated by the fact that there have only been two new classes of antibiotics introduced to the clinic in the past two decades, daptomycin and linezolid.3 More concerning, these two classes of antibiotics are exclusively active towards Gram-positive bacteria, which leaves four of the ESKAPE pathogens untreated. As the well of clinically relevant antibiotics runs dry, the polymyxin antibiotic colistin has become a last line of defense against MDR Gram-negative infections.4
Unfortunately the frequency of colistin resistant strains of Gram-negative bacteria that have been observed in the clinic has increased as reliance upon colistin therapy has escalated.5 The mechanistic basis of colistin resistance is thought to occur predominantly through modification of lipid A6; however, the two-component system (TCS) signaling that drives these modifications has recently been shown to activate additional mechanisms that are also required for resistance.7 Our group has been focused on combating the inevitable development of antibiotic resistant bacteria by developing compounds capable of disrupting the mechanisms through which these organisms express resistance8–12. We recently established that the 2-aminoimidazole (2-AI) compound 1 is capable of reversing colistin resistance in multiple primary clinical isolates of two of the four Gram-negative ESKAPE pathogens: K. pneumoniae, and A. baumannii.11 Against several strains of both bacteria, the minimum inhibitory concentration (MIC) of colistin was lowered from 512 to ≤4 (in some cases as low as 0.25) µg/mL in the presence of 30 µM (8.4 µg/mL) 1. Mechanistic studies revealed that treatment of colistin-resistant A. baumannii with 1 led to downregulation of the PmrAB two-component system while mass spectrometry demonstrated reversal of the phosphoethanolamine modification of lipid A responsible for colistin resistance in A. baumannii.
Despite this unprecedented activity, we noted that compound 1 itself harbored some inherent toxicity to the bacteria in the absence of colistin. Given that this toxicity may lead to an accelerated rate of resistance evolution, we wondered whether we could augment the activity of 1 while decreasing inherent toxicity through analogue synthesis. In this regard, our group has also recently developed several 2-AIs based upon compound 2 that are capable of reversing β-lactam resistance in methicillin-resistant S. aureus (MRSA).10,13,14 In these studies, we were able to modify adjuvant activity through imprinting either a 1,4- or 1,5-substitution pattern on the 2-AI ring. Specifically, compound 2 was able to lower the MIC of oxacillin against MRSA fourfold at 25 µM, while from the library of 1,5 substituted derivatives of compound 2, a compound emerged that is capable of lowering the MIC of oxacillin against MRSA up to 512-fold at 5 µM.10 Inspired by these results, we set out to determine whether imparting either a 1,4- or 1,5-substitution pattern upon the 2-AI of 1 would deliver compounds with augmented activity and reduced inherent toxicity. Herein we report the synthesis of both 1,5- and 1,4-substituted analogues of 1, as well as the evaluation of their biological activity in terms of colistin resistance suppression. Moreover, we report a compound capable of lowering the MIC of colistin against resistant strains of both A. baumannii and P. aeruginosa to a greater degree than compound 1.
Results and Discussion
Synthesis and Biological Evaluation of 1,5-2-AIs
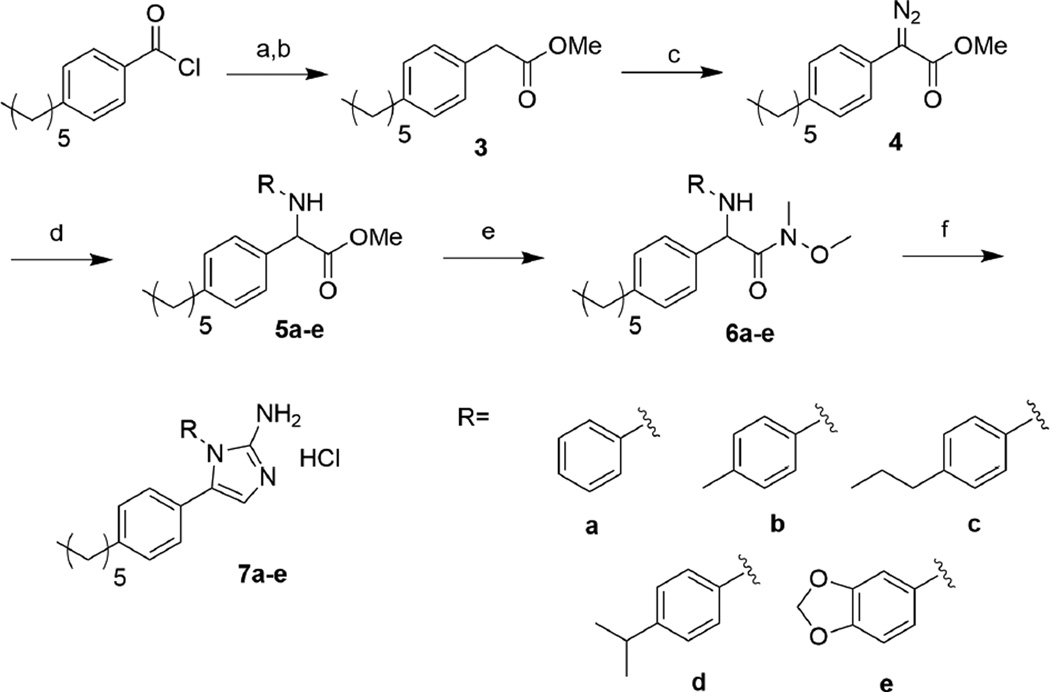
As we had the most success in our previous MRSA studies with the 1,5-substitution pattern where the introduced appendage was an aromatic substituent, we chose to initially evaluate a pilot library of five aryl-1,5-substituted 2-AIs that were synthesized according to Scheme 1. Briefly, commercially available 4-hexylbenzoyl chloride was reacted with diazomethane, and the resulting diazoketone was subjected to standard Arndt-Eistert conditions (silver benzoate in methanol).15 The homologated ester, 3 underwent diazotransfer reaction, accomplished by reaction with para-acetamidobenzenesulfonyl azide (p-ABSA) in the presence of DBU, to yield diazoketone 4.16 Analog diversity was then introduced via a Ru-catalyzed N-H insertion reaction.17 Conversion of the ester to the N-methoxy-N-methylamide (Weinreb amide) proceeded without the need for protection of the newly installed amine. Finally, reduction of the Weinreb amide to the corresponding aldehyde using diisobutylaluminum hydride (DIBAL-H), followed by cyclization with cyanamide afforded the 1,5-2AI derivatives 7a–e.10
Scheme 1.
Synthesis of 1,5-substituted 2-aminoimidazoles. a) i. CH2N2, rt, 1h. ii. AcOH, rt, 1h. b) AgOBz, Et3N, MeOH, rt, 16h. c) p-ABSA, DBU, MeCN, rt, 16h. d) [Ru(p-cymene)Cl2]2, R-NH2, CH2Cl2, rt, 1h. e) HN(OCH3)CH3, i-PrMgCl, THF, −40 °C, 8h. f) i. DIBAL-H, THF −78 °C, 1h. ii. EtOH/H2O, pH 4.3, H2NCN, 95 °C, 2h. iii. MeOH/HCl
Our pilot library of 1,5-2-AIs was evaluated for the ability to break resistance to colistin against the colistin-resistant strains of A. baumannii that we employed in our previous study.11 These strains, obtained from the Walter Reed Army Institute of Research (WRAIR), have colistin MICs significantly higher (512–1024 µg/mL) than the Clinical and Laboratory Standards Institute (CLSI) defined threshold for resistance for A. baumannii (≥4 µg/mL).18 As is common practice for evaluating adjuvant activity of our 2-AIs, we first established the intrinsic antibiotic activity of our library alone. Whereas the parent compound 1 has an MIC of 100 µM against all strains, all members of our library had MICs of ≥ 200 µM. We then determined the MIC of colistin against two strains of A. baumannii in the presence of 30 and 60 µM of each compound (Table 1). Surprisingly, the 1,5 substitution pattern essentially eradicated activity against A. baumannii in the context of colistin resensitization. At 30 µM compounds 7a–e only reduced the colistin MIC four-fold, from 512 to 128 µg/mL, whereas the parent compound at the same concentration was able to lower the MIC to 4 µg/mL.
Table 1.
Antibiotic activity and antibiotic resensitization activity of 1,5 2-AI library against two strains of A. baumannii
| A. baumannii strain | ||||
|---|---|---|---|---|
| Compound | MIC (µM) | Concentration Tested (µM) |
3941 | 4112 |
| Colistin MIC (µg/mL) | ||||
| 1 | 100 | 30 | 4 | 4 |
| 7a | >200 | 60 30 |
128 512 |
128 512 |
| 7b | >200 | 60 30 |
128 512 |
128 512 |
| 7c | >200 | 60 30 |
128 512 |
128 512 |
| 7d | >200 | 60 30 |
128 512 |
128 512 |
| 7e | 200 | 60 30 |
128 512 |
128 512 |
Synthesis and Biological Evaluation of 1,4-2-AIs
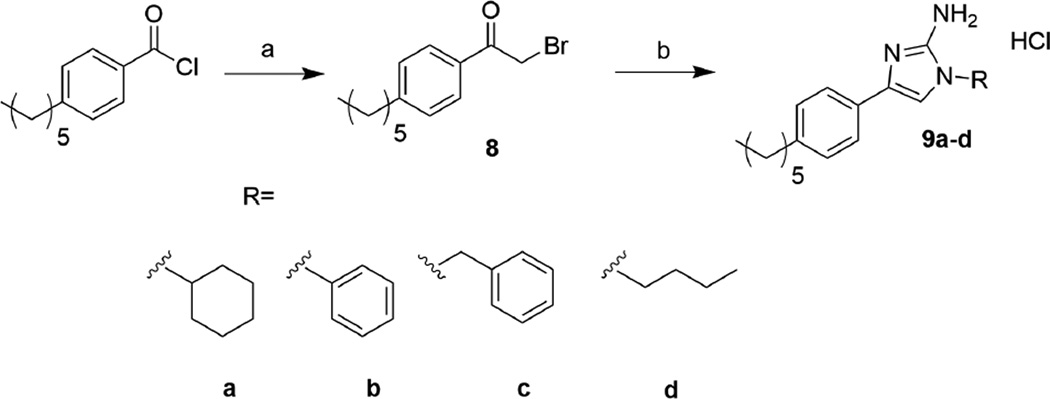
Given the failure of the 1,5-substitution pattern to enhance activity, we elected to evaluate the potential of 1,4-substitutions in the context of compound 1. Similar to the approach taken for the 1,5 2-AI derivatives, a pilot library of four 1,4 2-AIs was synthesized according to previous methods developed in our group,13 and is summarized in Scheme 2. Briefly, commercially available 4-hexylbenzoyl chloride was reacted with diazomethane and quenched with concentrated hydrobromic acid to yield the corresponding α-bromoketone, 8. This intermediate was reacted with various primary amines, the pH was then lowered to 4.3 through addition of 1M HCl, at which point cyanamide was added and the reaction was heated to reflux for 3 h to yield the 1,4 2-AI derivatives 9a–d.13
Scheme 2.
Synthesis of 1,4-substituted 2-aminoimidazoles. a) i. CH2N2, 0 °C, 1h. ii. HBr, 0 °C, 1h. b) i. RNH2, EtOH, rt, 30 min. ii. NH2CN, pH 4.3, EtOH, 95 °C. iii. MeOH/HCl
We evaluated this initial library of 1,4-substituted 2-AIs in the same manner as our 1,5 library, first assessing their intrinsic antibiotic activity alone, and then their ability to restore activity of colistin towards A. baumannii at sub-MIC levels. As observed with the 1,5-substituted 2 AIs, the 1,4 substitution pattern drastically lowered antibiotic activity as compared to the parent compound, with the MICs all being above 200 µM. Once again, the MIC of colistin against two strains of A. baumannii was determined in the presence of 60 and 30 µM of each compound (Table 2). In the case of compounds 9a–c the substitution led to a decrease in activity, however in the case of 9d at 30 µM, the MIC of colistin was in line with the parent compound. Encouraged by these initial results, the pilot library was screened against three additional strains of colistin resistant A. baumannii (Supplemental Information). Against all five strains, the initial lead 9d was able to lower colistin MICs to 0.5 – 4 µg/mL at 30 µM.
Table 2.
Antibiotic activity and antibiotic resensitization activity of 1,4 2-AI library against two strains of A. baumannii
| A. baumannii strain | ||||
|---|---|---|---|---|
| Compound | MIC (µM) | Concentration Tested (µM) |
3941 | 4112 |
| Colistin MIC (µg/mL) | ||||
| 1 | 100 | 30 | 4 | 4 |
| 9a | >200 | 60 30 |
8 16 |
8 16 |
| 9b | >200 | 60 30 |
8 16 |
8 32 |
| 9c | >200 | 60 30 |
8 64 |
16 64 |
| 9d | >200 | 60 30 |
1 2 |
2 4 |
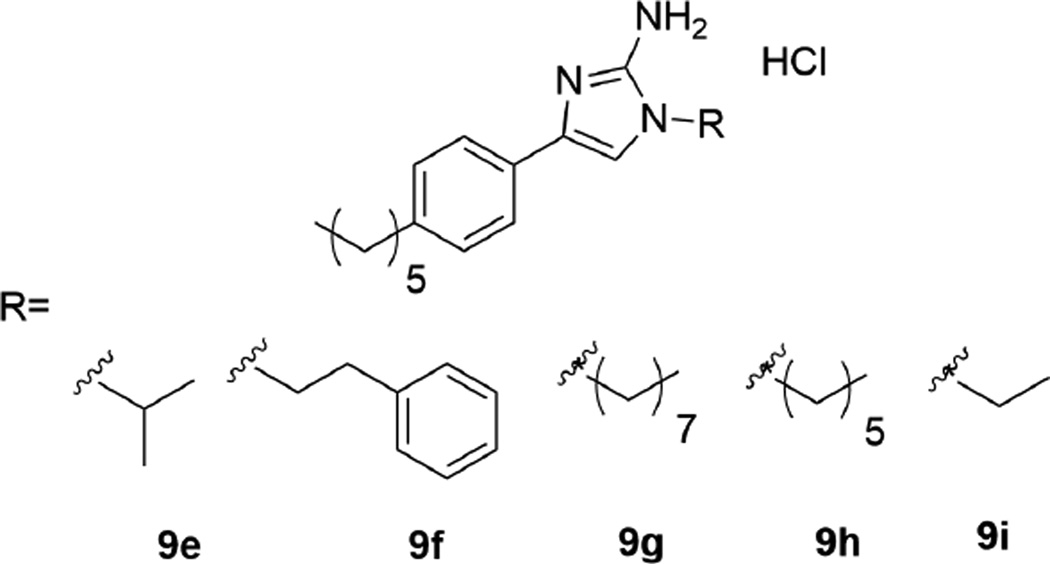
With the initial library results indicating that an alkyl group as opposed to an aryl group was better tolerated, five more compounds primarily focused on alkyl groups were synthesized, with the structures being shown in Figure 2. Compounds 9g–i along with the original 9d lead delivered compounds with side chains of various lengths while 9e afforded a branched alkyl side chain. In addition, since compounds 9b and 9c have a phenyl ring with an increasing number of methylene units, compound 9f was synthesized to further quantify the activity trend in this series of compounds. Compounds 9e–i were evaluated identically to the other 1,4 2-AIs, and the colistin resensitization results against A. baumannii strains 3941 and 4112 are shown in Table 3, while results against all strains can be found in Supplemental information.
Figure 2.
Structure of second library of 1,4 2-aminoimidazoles
Table 3.
Antibiotic activity and antibiotic resensitization activity of second-generation 1,4 2-AI library against two strains of A. baumannii
| A. baumannii strain | ||||
|---|---|---|---|---|
| Compound | MIC (µM) | Concentration Tested (µM) |
3941 | 4112 |
| Colistin MIC (µg/mL) | ||||
| 9e | >200 | 60 30 |
0.5 0.5 |
0.5 0.5 |
| 9f | >200 | 60 30 |
2 2 |
1 2 |
| 9g | >200 | 60 30 |
64 128 |
128 128 |
| 9h | >200 | 60 30 |
4 16 |
4 8 |
| 9i | >200 | 60 30 |
0.5 0.5 |
0.5 0.5 |
From the second generation library, the isopropyl and ethyl analogues 9e and 9i, emerged as the most active compounds, exceeding the activity of compound 1 and any members of the initial library and lowering the MIC of colistin against all five strains of A. baumannii to 0.5 µg/mL, constituting up to a 1024-fold reduction in MIC. Both compounds lowered the MIC of colistin further than compound 1 while remaining significantly less toxic alone. In addition, compound 9f was nearly as active as 9e and 9i, and still more active than the parent. Following the series of 9b, and 9c, compound 9f has a two-methylene spacer between the 2-AI and the benzene ring. With compound 9c being less active than 9b, we expected 9f to follow the trend, so it was surprising when it was one of our most active compounds.
With more active compounds in hand, we next wanted to verify that, as we observed with compound 1, we were reversing lipid A modification. To this end, A. baumannii 4106 was grown in the presence of 30 µM 9e for 16 h. Bacteria were then collected and subjected to mass spectrometry-based analysis of their lipid A fraction. As we observed with compound 1, 2-AI 9e reversed the phosphoethanolamine modification that drives colistin resistance in A. baumannii.
Encouraged by the results against A. baumannii, we then evaluated the spectrum of activity of our library by testing whether we could reverse colistin resistance in both clinical and laboratory strains of P. aeruginosa.19,20 These strains have MICs ranging from 64 to > 1024 µg/mL (Table 4), all of which are above the CLSI threshold for colistin resistance for P. aeruginosa (≥8 µg/mL).18 The MICs of our entire library of 1,4 2-AIs as well as the parent compound were then determined against all six strains, and in all cases the MICs were above the highest concentration tested (200 µM). All nine compounds were then evaluated for their ability to lower the MIC of colistin across the six strains of P. aeruginosa at an initial concentration of 50 µM. Select results from the initial screening are shown in Table 4 (full results in Supplemental Information).
Table 4.
Select resensitization activity of 1,4 library at 50 µM against colistin resistant strains of P. aeruginosa
|
P. aeruginosa strain |
Colistin MIC (µg/mL) |
Colistin MIC + 1 (µg/mL) |
Colistin MIC + 9d (µg/mL) |
Colistin MIC + 9e (µg/mL) |
Colistin MIC + 9i (µg/mL) |
|---|---|---|---|---|---|
| 1016 | >1024 | 128 | 64 | 8 | 32 |
| 1018 | 1024 | 8 | 16 | 4 | 8 |
| 1029 | 64 | 8 | 8 | 1 | 2 |
| 1033 | 64 | 8 | 16 | 8 | 8 |
| 1109 | 512 | 64 | 16 | 2 | 4 |
| 1133 | >1024 | 8 | 128 | 8 | 8 |
As was the case for A. baumannii, compound 9e emerged as the lead compound, lowering the colistin MIC to at or below the resistance threshold for all six strains, while compound 9i was also more active than the parent against several strains. In addition, at 50 µM, 9e lowered the colistin MIC further than the parent compound in four of the six strains, and had identical levels of resensitization against the other two strains.
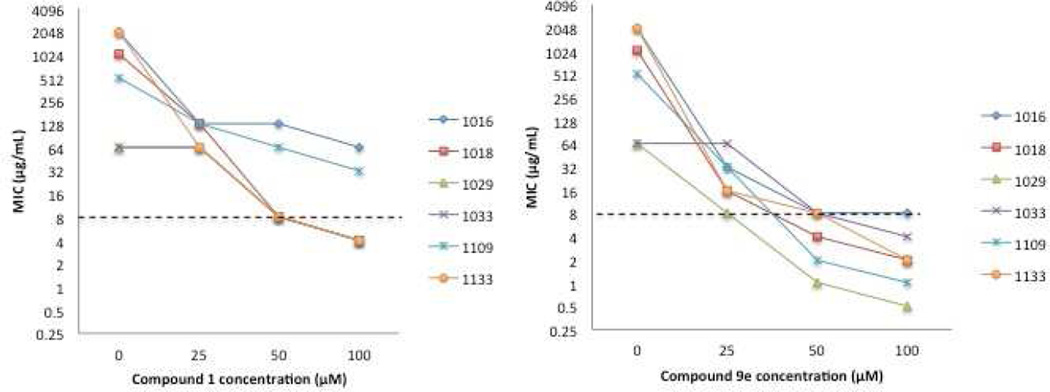
After identifying 9e as the lead compound, we investigated the dose-response activity of the most active compound, 9e in comparison to compound 1 (Figure 3). This was accomplished by determining the colistin MIC against each strain in the presence of 25 or 100 µM of either compound. In both cases, increasing compound concentration to 100 µM led to a decrease in colistin MIC compared to 50 µM. Lowering the concentration to 25 µM led to a dramatic decrease in activity for compound 1, which was unable to lower the MIC of colistin below the CLSI threshold in any of the strains tested. On the other hand, compound 9e was able to effect significant levels of antibiotic repotentiation at 25 µM.
Figure 3.
Dose response colistin resensitization data for compounds 1 and 9e against colistin resistant strains of P. aeruginosa. Dashed line represents the CLSI breakpoint.
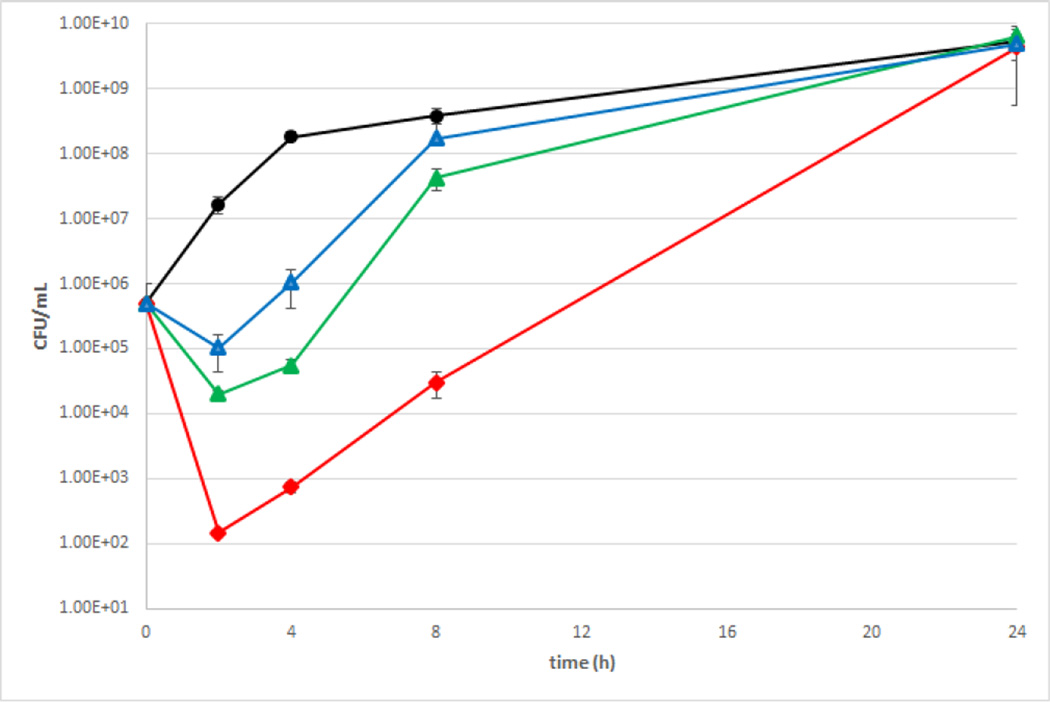
We then quantified activity by evaluating the effect of our lead compound 9e against both A. baumannii and P. aeruginosa as a function of time by constructing time kill curves. We chose to use 4106 as the representative strain for A. baumannii, as the previous study with 1 also used this strain,11 while 1133 was chosen as the representative strain for P. aeruginosa due to its high colistin resistance and sensitivity to 1 and 9e. Initially, we investigated the effect 9e had on bacterial growth in the absence of antibiotic. When A. baumannii was grown in the presence of 30 µM 9e, there was a 1.52 and 1.09 log reduction in CFUs/mL at the 2 and 4 h time points, but growth was identical to the control afterwards. This is contrast to the more pronounced effect the same concentration of 1 has on bacterial growth, where at the same time points and concentration there is a 2.25 and 3.41 log reduction in CFUs/mL respectively. Our previous study established that at 8 h the combination of 30 µM compound 1 and 2 µg/mL colistin effected a 2.4 log reduction in CFU/mL as compared to the control. With the same concentration of 9e and colistin, at the same time point, there is a 4.11 log reduction in CFU/mL, further highlighting the improved efficacy of compound 9e (Figure 4).
Figure 4.
Time kill curve for combination of 9e and colistin against A. baumannii. Black = untreated control, blue = 30 µM 9e + 0.125 µg/mL colistin, green = 30 µM 9e + 0.5 µg/mL colistin, red = 30 µM 9e + 2 µg/mL colistin.
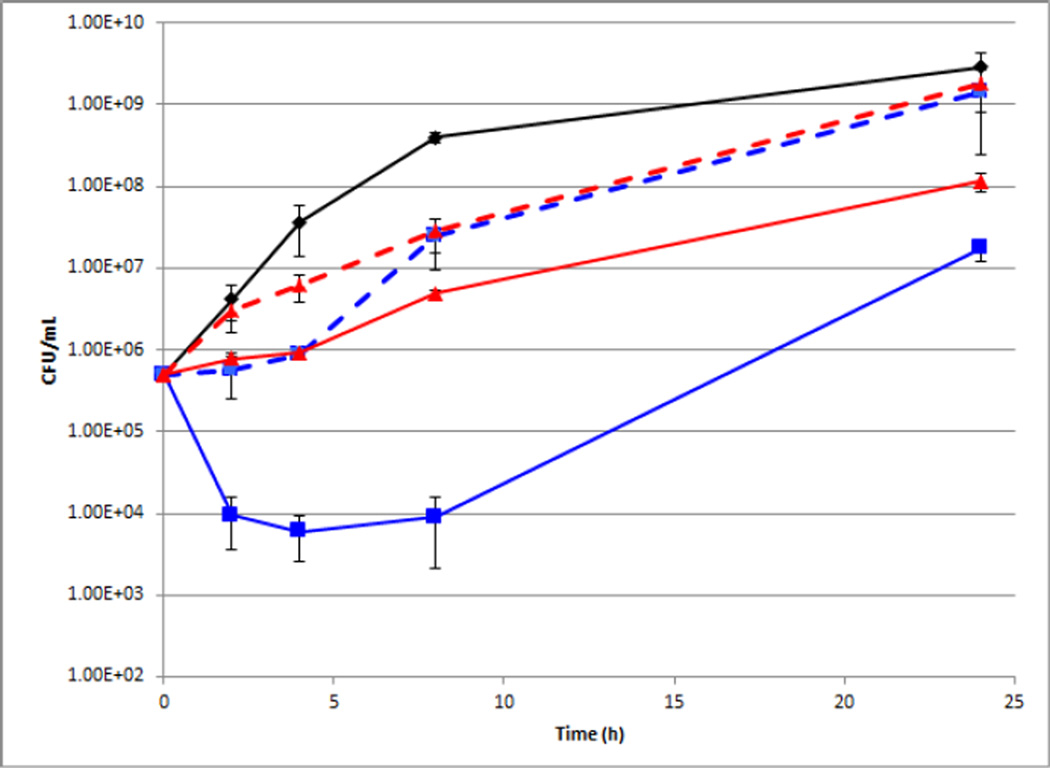
Growth of P. aeruginosa was unaffected by the presence of either compound 1 or 9e at their active concentration of 50 µM. Additionally, P. aeruginosa growth was monitored in the presence of colistin alone, revealing that at concentrations below 32 µg/mL growth was unaffected. After establishing the lack of toxicity of both compounds towards P. aeruginosa we next turned our attention towards combinations of colistin and either compound. After 4 h the combination of 50 µM compound 1 and 16 µg/mL colistin resulted in a modest 1.51 log reduction in CFU/mL as compared to the control, whereas 50 µM 9e and only 4 µg/mL colistin was able to produce the same level of reduction in CFU/mL. At the same time point, the combination of 16 µg/mL colistin and 50 µM 9e caused a 3.81 log reduction CFU/mL (Figure 5).
Figure 5.
Time kill curve for combination of 9e or 1 and colistin against P. aeruginosa. Black = untreated control, blue solid line = 50 µM 9e + 16 µg/mL colistin, red solid line = 50 µM 1 + 16 µg/mL colistin. Blue dotted line = 50 µM 9e + 4 µg/mL colistin red dashed line = 50 µM 1 + 4 µg/mL colistin..
Conclusion
Using compound 1 as inspiration we synthesized two distinct libraries and evaluated their ability to reverse colistin resistance in multiple strains of A. baumannii. From these studies, compound 9e was identified as the lead compound and was able to lower the colistin MIC up to 1024-fold, an eight-fold improvement over compound 1. Encouraged by this study, we then turned to P. aeruginosa, another bacterial pathogen for which colistin resistance has been reported. Compound 9e once again emerged as the most active compound, capable of lowering the colistin MIC up to 256-fold, a 32-fold improvement over compound 1. Time-dependent studies showed that 9e has less effect on bacterial growth than 1 at identical concentrations at early times points against A. baumannii, while also leading to an enhanced adjuvant effect thereby further decoupling intrinsic microbicidal activity from MIC suppression. Efforts to further tune the activity of 9e and evaluate adjuvant activity in vivo are currently underway.
Supplementary Material
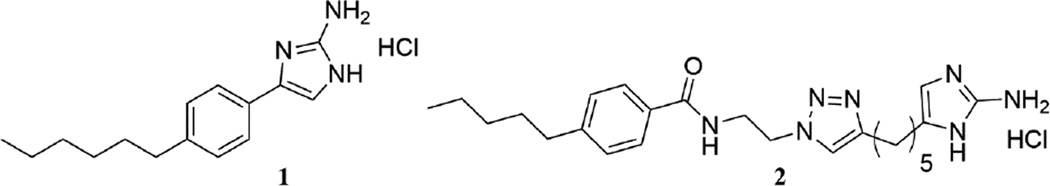
Figure 1.
Structures of compound 1 and 2
Acknowledgements
The authors would like to thank the National Institutes of Health (GM055769 to C.M, AI067653 to S.M.M) and the Cystic Fibrosis Foundation (MOSKOW13P0 to S.M.M.) for their generous support of this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.CDC. 2013 [Google Scholar]
- 2.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. Clin. Infect. Dis. 2009;48:12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 3.Coates AR, Halls G, Hu Y. Br J Pharmacol. 2011;163:184–194. doi: 10.1111/j.1476-5381.2011.01250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falagas ME, Kasiakou SK. Clin. Infect. Dis. 2005;40:1333–1341. doi: 10.1086/429323. [DOI] [PubMed] [Google Scholar]
- 5.Olaitan AO, Morand S, Rolain JM. Front Microbiol. 2014;5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beceiro A, Llobet E, Aranda J, Bengoechea JA, Doumith M, Hornsey M, Dhanji H, Chart H, Bou G, Livermore DM, Woodford N. Antimicrob Agents Chemother. 2011;55:3370–3379. doi: 10.1128/AAC.00079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gutu AD, Rodgers NS, Park J, Moskowitz SM. Antimicrob Agents Chemother. 2015;59:5377–5387. doi: 10.1128/AAC.00904-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brackett CM, Melander RJ, An IH, Krishnamurthy A, Thompson RJ, Cavanagh J, Melander C. J Med Chem. 2014;57:7450–7458. doi: 10.1021/jm501050e. [DOI] [PubMed] [Google Scholar]
- 9.Worthington RJ, Bunders CA, Reed CS, Melander C. ACS Med Chem Lett. 2012;3:357–361. doi: 10.1021/ml200290p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris TL, Worthington RJ, Melander C. Angew Chem Int Ed Engl. 2012;51:11254–11257. doi: 10.1002/anie.201206911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris TL, Worthington RJ, Hittle LE, Zurawski DV, Ernst RK, Melander C. ACS Chem Biol. 2014;9:122–127. doi: 10.1021/cb400490k. [DOI] [PubMed] [Google Scholar]
- 12.Rogers SA, Huigens RW, 3rd, Cavanagh J, Melander C. Antimicrob Agents Chemother. 2010;54:2112–2118. doi: 10.1128/AAC.01418-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furlani RE, Yeagley AA, Melander C. Eur J Med Chem. 2013;62:59–70. doi: 10.1016/j.ejmech.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 14.Su Z, Yeagley AA, Su R, Peng L, Melander C. Chem Med Chem. 2012;7:2030–2039. doi: 10.1002/cmdc.201200350. [DOI] [PubMed] [Google Scholar]
- 15.Kumar KSA, Chattopadhyay S. RSC Adv. 2015;5:19455–19464. [Google Scholar]
- 16.Gao L, Hwang GS, Ryu do H. J Am Chem Soc. 2011;133:20708–20711. doi: 10.1021/ja209270e. [DOI] [PubMed] [Google Scholar]
- 17.Su Z, Peng L, Melander C. Tetrahedron Letters. 2012;53:1204–1206. [Google Scholar]
- 18.Institute, C. a. L. S. 2012;32 [Google Scholar]
- 19.Moskowitz SM, Brannon MK, Dasgupta N, Pier M, Sgambati N, Miller AK, Selgrade SE, Miller SI, Denton M, Conway SP, Johansen HK, Hoiby N. Antimicrob Agents Chemother. 2012;56:1019–1030. doi: 10.1128/AAC.05829-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller AK, Brannon MK, Stevens L, Johansen HK, Selgrade SE, Miller SI, Hoiby N, Moskowitz SM. Antimicrob Agents Chemother. 2011;55:5761–5769. doi: 10.1128/AAC.05391-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.