Summary
Background
Partners HealthCare Personalized Medicine developed GeneInsight Clinic (GIC), a tool designed to communicate updated variant information from laboratory geneticists to treating clinicians through automated alerts, categorized by level of variant interpretation change. Objectives: The study aimed to evaluate feedback from the initial users of the GIC, including the advantages and challenges to receiving this variant information and using this technology at the point of care.
Methods
Healthcare professionals from two clinics that ordered genetic testing for cardiomyopathy and related disorders were invited to participate in one-hour semi-structured interviews and/ or a one-hour focus group. Using a Grounded Theory approach, transcript concepts were coded and organized into themes.
Results
Two genetic counselors and two physicians from two treatment clinics participated in individual interviews. Focus group participants included one genetic counselor and four physicians. Analysis resulted in 8 major themes related to structuring and communicating variant knowledge, GIC’s impact on the clinic, and suggestions for improvements. The interview analysis identified longitudinal patient care, family data, and growth in genetic testing content as potential challenges to optimization of the GIC infrastructure.
Discussion
Participants agreed that GIC implementation increased efficiency and effectiveness of the clinic through increased access to genetic variant information at the point of care.
Conclusion
Development of information technology (IT) infrastructure to aid in the organization and management of genetic variant knowledge will be critical as the genetic field moves towards whole exome and whole genome sequencing. Findings from this study could be applied to future development of IT support for genetic variant knowledge management that would serve to improve clinicians’ ability to manage and care for patients.
Keywords: Clinical informatics applications, clinical decision support, genetics, personalized medicine, laboratory software
1. Background
In the new world of personalized medicine, use of genetic testing in clinical care presents a unique and challenging situation. Although the existence of genetic variants reported through molecular diagnostic tests on germline DNA should not change over a patient’s lifetime, the clinical interpretations of these variants may be revised as new evidence emerges such as population frequency data, segregation studies, functional evidence or new case data. Such changes in variant interpretation have been documented to occur frequently and can impact care of a patient and their family members [1, 2]. The American College of Medical Genetics and Genomics (ACMG) initially suggested that “the laboratory may be in the best position to modify previously issued interpretations and should make an effort to contact physicians of previously tested patients in the event that new information changes the initial clinical interpretation of their sequence variant” [3]. However, new guidelines have modified this policy in recognition of the inability for most laboratories to support this requirement [4]. Taking on the task of modifying previously issued interpretations and contacting physicians when changes in interpretation occur would require laboratories to spend significantly more time staying up-to-date and organized around large volumes of genetic information, time that is not currently resourced or reimbursed. Furthermore, it is difficult for clinicians to keep up with the constant evolution of genetic knowledge as the scope of genetic testing grows. New information systems are needed to address these issues [5, 6].
The Laboratory for Molecular Medicine (LMM) at Partners HealthCare Personalized Medicine has developed information technology (IT) infrastructure to support, manage, and communicate clinical genetic test results. At the time of this study, this system, called GeneInsight Suite, had two application components: GeneInsight Lab (GIL), which supports the activities of geneticists in laboratories, and GeneInsight Clinic (GIC), which supports the treating clinicians and is the focus of our study [1, 7] (► Figure 1). The clinical focus of the initial sites deploying GIC was primarily cardiomyopathy and related disorders but it now supports sites testing a broad range of genetic diseases.
Fig. 1.
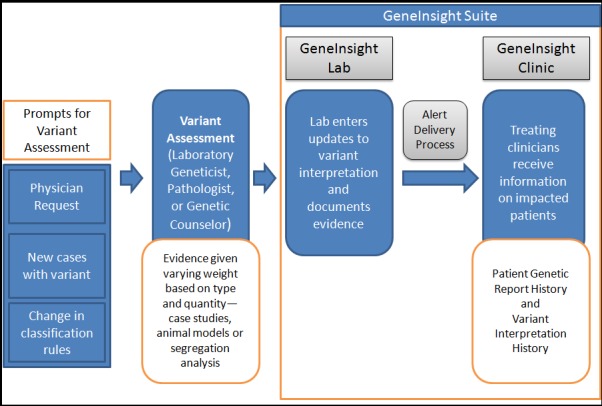
GeneInsight Lab (GIL) and GeneInsight Clinic (GIC) high level workflow
GeneInsight Lab (GIL)
The laboratory geneticists use the GIL tool as a repository for genetic testing results and variant interpretations. Because there are very few standards in the community that address terminologies for variant interpretation, the requirements for evidence, and the relative weight of each piece of evidence that would go into assigning an interpretation, the LMM developed a categorization scheme for variant interpretations that included categories benign, likely benign, pathogenic, likely pathogenic, and unknown significance (currently referred to as uncertain significance) that would be assigned based on the variant’s likelihood to be the cause of a disease. Variant interpretations may change if new evidence would change the labs determination of the variant’s effect on disease. The LMM may reassess a variant interpretation by conducting a search for new evidence on an ad-hoc basis upon physician request or through encountering the variant in a new case. Once the evidence has been vetted to support knowledge updates, the geneticists document the evidence upon which their variant classifications are based and record them in the GIL in a variant specific manner, not patient specific. The system is then able to identify all patients with that variant and append the new variant interpretation automatically to each patient record. Alerts are then generated and communicated to the clinicians through the GIC if one of their patients was affected by the change in significance. There are national databases, such as ClinVar, to which many laboratories are now regularly contributing their variant knowledge and physicians can query as needed. However, because submission of variant data is done manually, there is often a delay in representing a laboratory’s current knowledge in ClinVar. In a recent study, 19% of differences in variant classification seen in ClinVar were due to the ClinVar entries being out of sync with the lab’s internal knowledgebase [8]. Furthermore, there is currently no mechanism to alert a physician that a variant of interest has changed classification by the laboratory that issued the report.
GeneInsight Clinic (GIC)
Physicians and genetic counselors are typically responsible for reviewing genetic results and variant updates and making any decisions regarding clinical management or family testing. However, these clinicians most often do not have access to this type of information at the point of care. The GIC component has been developed to address this issue. The GIC offers four major functions: genetic test report delivery, ability to view genetic tests that have been conducted on a patient, email alerts sent to treating clinicians when new information is learned about variants previously identified in their patients, and ability to search for their patients by variant name or other demographics, all of which can be utilized at the point of care to support patient management and to provide more information to patients. The GIC uses three levels of alerts to classify changes made in variant interpretation based on the importance of the change (its impact on clinical care and testing decisions): high, medium and low, which are used in the GIC to notify clinicians.
When this study was underway in 2011, the LMM had information stored for over 10,000 unique variants in 219 genes covered by 183 tests in their GeneInsight Lab. The GeneInsight Suite was created to provide an efficient mechanism for storing, managing and transmitting genetic information between the laboratory and clinics. Building, deploying and optimizing this kind of infrastructure requires significant time and expense. Given the continued expansion of genetic information and the need for supporting IT infrastructure, it is important to identify early on what does and does not work in managing this information so as to promote best practices for communicating this information to clinicians and for integrating it into the EHR for optimal use [5, 6, 9, 10]. The research team has previously published work on the multi-aim study evaluating GIC, including a report on think-aloud usability tests conducted to assess issues with the tools’ user interface prior to its use in the clinics; and, an analysis of system audit data to understand the time it took for new genetic knowledge to be available and accessed through the GIC to support clinical care [11, 12].
2. Objectives
This manuscript reports qualitatively on the feedback from the initial users of the GeneInsight Clinic. Feedback was solicited regarding the advantages and challenges to receiving this variant information and in using this technology in practice. Interviews and a focus group with physicians and genetic counselors were used to gather feedback. The overall investigation was approved by the Partners HealthCare Human Research Committee (PHRC) and registered with ClinicalTrials.gov (ID: NCT01225978).
3. Methods
This qualitative pilot study used interviews and focus groups to gather feedback from the physicians and genetic counselors at the initial clinics using the GIC for cardiomyopathy related genetic disorders.
3.1 Sample
Staff from four clinics where GIC had been fully implemented between 2010 and 2011 and who would receive e-mails from GIC regarding changes in patients’ variant interpretations were asked to participate in the larger study evaluating the tool. These clinics sent a large number of genetic tests related to cardiomyopathy to the LMM. All of the staff members who were piloting the tool from two of the four clinics were asked to participate in this qualitative sub-study (clinic A and B). A total of 7 genetic counselors and physicians were invited. A recruitment email was sent from the principal investigator to staff from both clinics inviting them to participate in one or more activities, including an interview and focus group. Follow-up emails were sent to schedule individual interviews. Another follow-up email was sent to schedule the focus group with staff at one of the clinics.
3.2 Qualitative Data Collection
3.2.1 Interviews
During this study, the same member of our research team (PN) conducted one-hour semi-structured interviews about the GIC with participants, each of whom were asked questions from an interview guide, developed by our team, specific to their clinical role. The interview questions were drafted based on key issues the IT developers and laboratory staff were interested in assessing as well as questions the research team identified during usability tests; the questions were reviewed and finalized by the team. The interview guides used in the interviews with the genetic counselors and physicians contained questions focused on overall experience with GIC, feedback on alerting process, GIC impact on workflow and patient care, and opinions on alert categorization (Appendix A). During the interviews we referred to the definitions of significance levels assigned to variant changes in use at the time. Interviews were held in person or over the phone and were audio-recorded and transcribed.
3.2.2 Focus Group
A one-hour focus group was conducted by research team members (PN, LV) at one of the major academic medical center sites. We chose to conduct the focus group with one clinic to understand from multiple members of the same clinic how they felt about the GIC. The questions were developed using feedback from initial discussions with potential users, usability tests, and IT developers. The questions were designed to collect feedback on the communication of genetic results, the review process and patient follow-up, impact on workflow and workload, value of the tool, and future implications (Appendix B). The focus group was audio-recorded and transcribed.
3.3 Data Analysis
Four members of the research team (SN, LV, PN, SP) analyzed the transcripts from the interviews and focus group using a Grounded Theory approach and Microsoft Word was used to organize concepts to help with the qualitative analysis. Grounded theory allows for the emergence of findings grounded in data, using an open coding technique of constantly comparing each concept within a category to another concept in that category, and then categories to categories. Since the idea of GeneInsight is new and innovative, a grounded theory approach was chosen as the most appropriate approach for simultaneously collecting and analyzing data to develop a rich description of this area [13, 14].
All concepts were discussed and agreed upon by four members of the team at each level of the analysis. For the first round of analysis, the interviews and the focus group transcripts were individually analyzed and important concepts from each were identified. We used a constant comparative method whereby we discussed each concept, compared them to other concepts and analyzed them simultaneously to come to a consensus on a code for that concept [14]. As we read through each of the interview transcripts and the focus group transcripts we added to our coded concepts. We were left with one set of coded concepts based on the data from all of the transcripts and the focus group. We summarized and categorized these concepts into eight larger themes related to the participants experience with the GIC that were reviewed by the larger research group.
These themes were further combined into three broader feedback categories for presentation in this manuscript. During the entire process, a key was used to track the source interview for each comment so additional detail or clarification could be obtained if necessary.
4. Results
All seven individuals who were involved in piloting this tool at the two clinics took part in these activities. Two genetic counselors and two physicians participated in the interviews. The focus group consisted of five participants from one of the sites, including one genetic counselor and four physicians (► Table 1). Eight themes identified from participant feedback are presented in ► Table 2.
Table 1.
Participant Population Table
| Participant | Site | Role | Interview | Focus Group |
|---|---|---|---|---|
| 1 | A | Physician | X | |
| 2 | A | Physician | X | |
| 3 | A | Physician | X | X |
| 4 | A | Physician | X | |
| 5 | A | Genetic Counselor | X | X |
| 6 | B | Physician | X | |
| 7 | B | Genetic Counselor | X |
Table 2.
Major Themes from the Interviews and Focus Group
| Feedback Category | Major Theme | Example Quotes |
|---|---|---|
| I. Communicating Variant Knowledge Updates |
1. Feedback on Alert Content and Timing Alert content and timing based on the users’ perception of the alert’s priority and potential affect on clinical management is helpful. |
“It’s probably not something that’s going to change the patient management in the course of a week. So I guess making it on a scheduled weekly update would be fine. That would probably make it a little more time-efficient as opposed to happening right at that moment.” – Physician 1 |
| II. Impact of GeneInsight Clinic (GIC) in the Clinic |
2. Workflow Automated and proactive communication of information to clinicians has the potential to improve workflow. 3. Accessing and Communicating Information Having information available electronically in a central location and at the point of care can aid communication and increase efficiency in the clinic. |
“I think it has improved workflow, the knowledge that if I haven’t marked it as reviewed then I keep getting the alert in the email once a week is a reminder that I need to communicate with the patient. So I think it’s definitely helpful.” – Physician 2 “It reduced a lot of the time it was taking for communication of results back and forth within our group.” – Physician 1 |
| III. Improvement Suggestions |
4. Training Needs Introducing a novel health information technology (HIT) tool requires a discussion of expectations, best practices regarding workflow integration and training on functionality to implement and use successfully in a clinic. 5. GeneInsight Clinic (GIC) Functionality Additional functionality such as electronic health record (EHR) integration, patient follow-up tracking, and bi-directional communication between lab and clinic may be useful to consider. |
“I think giving examples [of workflows] would be useful. I imagine that not everyone has the same staffing that we have, but I think it would be helpful to know how other people do it” – Physician 2 “It would be great to connect the Electronic Health Record (EHR) and GeneInsight Clinic (GIC).” – Genetic Counselor 1 |
| IV. Broader Issues to Address |
6. Family Data Ability to access family data and having a policy in place for contacting family will be important for broader use of this tool and Whole Genome Sequencing (WGS). 7. Increase in Genetic Knowledge/ Whole Genome Sequencing The volume and complexity of information with Whole Genome Sequencing (WGS) introduces challenges in knowledge management. 8. Patient Consent & Follow-Up Longitudinal patient follow-up and defining responsibilities outside initial testing introduces challenges.(see Table IV for additional detail) |
“Finding a way to pull all that up at once, if you have a family with a variant that has changed significance, or a new alert, the ability to look at that family as a intact entity is quite important” – Physician 3 Focus Group “In the near-term future that is where we are headed. We will be testing on a genome-wide level and there is too much out there that you can’t expect a physician to know, so we will need a new system.” – Geneticist 2 “We see a fair number of people on a one time basis, and people may have other care providers. They may have just been referred in for genetic counseling, genetic testing. There is one instance where we are still trying to contact them. This is still handled by the provider, but the person has moved and we have not been able to get updated information yet.” – Genetic Counselor 2 |
4.1 Communicating Variant Knowledge Updates
The participants gave feedback on how the variant knowledge updates were communicated to them through the GIC. They shared their experiences and opinions on alert content and timing.
4.1.1 Feedback on Alert Content and Timing
Specific feedback from physicians and genetic counselors on the alert levels and content of the alerts are presented in ► Table 3 below.
Table 3.
Participant Feedback on High, Medium and Low Alert Content and Timing
| Alert Significance | Content & Alert Timing at Time of Study1 | Feedback from Participants |
|---|---|---|
| High | Content
Timing
|
Content
Timing
|
| Medium | Content Likely pathogenic to pathogenic Pathogenic to likely pathogenicTiming· Once per week |
Content
Timing
|
| Low | Content
|
Content
|
1Please note the information and associated alerting algorithms presented in this table represent those in effect at the time of the study. They have subsequently been refined to accommodate a higher degree of detail since the completion of our study.
Clinicians expressed an interest in having the tool indicate the date when the evidence was last reviewed along with the evidence for the variant knowledge update in GIC. Clinicians recommended that the tool distinguish the reason for the alert, such as an evidence change versus only a change in variant interpretation criteria that produced a change independent of any new variant evidence. Clinicians also liked having their names associated with alerts for which they were responsible, and having the summary email organized into categories so that the high alerts were prioritized at the top of the list. Clinicians felt that the variant alerts were useful and it was helpful to get the history and evidence, including references, of the variant interpretations (► Figure 2).
Fig. 2.
Screenshot of GeneInsight Clinic (GIC) variant interpretation page
4.2 Impact of GIC in the Clinic and Laboratory
Participants also provided feedback on the impact GIC had on the clinic’s workflow as well as its impact on how the clinicians accessed and communicated information.
4.2.1 Workflow
Genetic counselors reported that the GIC implementation allowed proactive, automatic communication to clinicians of changes in significance levels regarding all patients with a given variant, decreasing the frequency and urgency of clinician calls to the laboratory. Before GIC, notifications of variant knowledge classification changes were not always prompt and there was no standard for communicating these updates with clinicians [3]. In many cases, clinicians would call the LMM with little notice in advance of patient appointments to check for updates. Now genetic counselors report they will likely only contact the lab if there is a highly significant change or with questions regarding the evidence. GIC also allows original genetic test results to be sent via GIC, which clinicians found more convenient than receiving them via fax.
Clinics created different ways to integrate GIC into their practice. The GIC included a review button that was intended to help clinicians track that a patient’s record was reviewed in response to email alerts. They designed the GIC so that once a patient’s record had been opened, a user had the ability to mark the case as reviewed. Once the button was clicked, additional email alerts would be turned off. We learned from the clinicians that the processes for using the review button were dependent on the individual genetic counselor and clinician decisions about workflow. While some clinicians elected not to use the review button, many used the review button to manage their to-do list of patients.
For many participants, the weekly summary email that included all of the alerts from the week as well as unreviewed alerts served as a reminder to go into the system and check un-reviewed genetic testing reports and updated variants. In general, clinicians liked that GIC had the repeat notifications as a safety precaution when the review button had not yet been clicked. Some clinicians whose workflows included clicking the review button only once a patient had received communication liked having the weekly summary email to keep track of which patients had been contacted. Once they were in GIC, clinicians could use the “search un-reviewed items” box to find patients that still needed review.
The clinicians noted that the summary email could get very long with the genetic testing reports, variant updates and reminders, particularly for clinics that had not yet decided how to incorporate the review button into their workflow. These clinics continued to see the same alerts appear in multiple summary emails even though the alerts had been handled. Clinicians felt that marking the variant update as reviewed should not be required to indicate the information had been addressed, especially for low significance updates where time is not critical. There was also concern expressed by some clinicians that requiring use of the review button to indicate that data has been seen could raise liability issues.
4.2.2 Accessing and Communicating Information
At the time of the study, clinicians noted that although GIC’s impact on direct patient clinical care to date had been minimal, getting prompt information about variant changes had influenced decisions regarding testing of patients and families. For example, alerts indicating a variant change from likely pathogenic to unknown significance would change a recommendation from predictive testing to testing only affected family members to generate new evidence. If unaffected family members had been tested, they needed to be contacted for potential changes in care plans. Low and medium significance category changes would help to substantiate original analysis but would likely not change clinical practice.
Genetic counselors appreciated the ease with which clinicians could pull up the most updated variant knowledge available from the laboratory to discuss with patients at the point of care. They also liked using the search function to view variant interpretations when patients came for genetic test results. The ability to easily access this information made clinicians more confident that they were treating patients with the most up-to-date information available from the LMM. Additionally, having the significance category levels, as well as the ability to generate a list of items to be reviewed, helped clinicians prioritize. The counselors thought that responding to significant variant knowledge updates had the potential to increase their workload around patient contact, but changes of low significance would have minimal impact.
Clinicians noted that the tool was helpful in communicating information to patients because the reports and alerts were received quickly. In response to alerts at some clinics, clinicians would call patients with high significance changes within a week unless they had an upcoming appointment. GIC also reduced the time taken to communicate results within a clinic because it served as a central repository of information that could be accessed by multiple clinicians caring for a patient.
4.3 Improvement Suggestions
Suggestions for improvement regarding training and functionality of the system were also discussed.
4.3.1 Training Needs
Although a formal GIC training process was not provided prior to implementation (a demonstration of the system and user manuals were provided), users felt that the system was intuitive and easy to use. A few issues with the technical aspects of using the tool did arise in addition to those reported in the usability testing [11]. Clinicians recommended that at implementation it should be made clear to users that they have the ability to search additional patients using the “search un-reviewed patients” button once they have clicked into GIC from an email alert associated with another patient. Users should also be informed that unless the tool has been integrated with the electronic health record (EHR), data will not be automatically updated in the electronic medical record. Also, we learned that it is important to communicate to users that the tool, at the time, did not provide the reasons for variant knowledge change if the change was solely based on rule changes in the laboratory’s variant interpretation system. Clinicians indicated that, before implementation, it would be useful to have some discussion about workflow using the GIC and have the clinic decide who will have access, how new variant information will be handled, and how they will use the review button. Some clinicians indicated that it might be helpful to see the workflow of other practices using the tool to determine if any other processes would improve their use of the system.
4.3.2 GIC Functionality
Included in the feedback provided by participants were suggestions on changes that could be made to GIC functionality. Many participants thought that integration of the GIC tool with the EHR would be useful. They desired an integration that could ensure information from GIC is accessible within the EHR, and that could assist in tracking patients over time, manage patient-family data, and document review of patient genetic updates
Some clinicians expressed interest in being able to append GIC patient information with whether the patient was contacted and whether there were significant changes in the clinical management due to the update. They also thought that it would be useful if the tool included templates of patient/ family correspondence and educational materials to facilitate communication with patients and families.
Many participants thought that it would be advantageous for the GIC system to provide bi-directional communication between the clinic and the laboratory. The laboratory sees more patients and may have knowledge the clinic lacks, but the interpretation of this knowledge may be complicated and, therefore, could be informed by feedback from the clinic about a patient’s clinical status. A system that allows for the storage of the variant-centric information managed by the laboratory as well as the patient-centric information managed by the clinic, and communication of this information among the two was highly recommended by participants in our study.
4.4 Broader Issues and Participant Recommendations
In the course of collecting feedback on the tool, participants also brought up broader issues reflecting potential challenges with the expansion of genetic knowledge and personalized medicine that are outlined in ► Table 4 below.
Table 4.
Broader Issues and Participant Recommendations
| Broader Issue | Feedback and Recommendations from Participants on Broader Issues |
|---|---|
| Family Data Ability to access family data and having a policy in place for contacting family will be important for broader use of this tool and WGS |
|
| Increase in Genetic Knowledge/Whole Genome Sequencing The volume and complexity of information with Whole Genome Sequencing (WGS) introduces challenges in knowledge management |
|
| Patient Consent & Follow-Up Longitudinal patient follow-up and defining responsibilities outside initial testing introduces challenges. |
|
5. Discussion
In this study, seven participants (two genetic counselors and five physicians) with experience using GeneInsight Clinic took part in interviews and/or a focus group to provide feedback on the pilot release of the GIC. Clinicians found that receiving genetic variant updates automatically through GIC whenever variant knowledge was reviewed and updated by the laboratory was easier than calling the laboratory before patient visits for updates. GIC increased their access to this information, while alert categories allowed them to prioritize alerts, promoting efficiency. GIC also facilitated sharing updates with other clinicians in the clinic and with patients at the point of care. While clinicians did find benefits to implementing GIC, they also identified several areas for improvement, including making alerts less frequent with some low level alerts appearing in GIC without email notification, integrating GIC and the EHR, allowing bidirectional communication between the laboratory and the clinic, and increasing discussion of GIC functionality and potential workflow changes before GIC implementation. Broader issues in the genetic testing process surrounding whole genome sequencing, access to family data, and longitudinal patient interaction were also raised for future consideration. Since the time the study began, a few changes were made to the GIC in response to initial feedback. These included integration with the electronic health record and changes to the summary email alerts that added the reason the variant was updated and the name of the laboratory staff that approved the update. The GIC development team continues to refine the tool and respond to feedback from users.
Few studies exist in the literature that address the complexities of standardizing and communicating genetic variant information and changes in this information over time. A study by Manolio et al collected information from several groups who had implemented genomic medicine programs and underscored the value of a genetic variant knowledgebase as part of infrastructure needs [15]. Ramos et al reported the findings from a workshop convened by the National Human Genome Research Institute and the Wellcome Trust to establish requirements for collecting and sharing genetic variant information [16]. In 2015, the ACMG released new guidelines to better outline standards for defining variant interpretation categories [17]. Several data sharing efforts have also begun in this area, including the use of GeneInsight to create a share-and-share alike network called VariantWire and the Clinical Genome Resource (ClinGen) effort focused on increasing data sharing through the ClinVar database at the National Center for Biotechnology Information which aggregates information about sequence variation and its relationship to human health [18]. Our study focuses on genetic variant information in clinical care, but more specifically on the use of a genetic variant management IT system that has the ability to directly provide clinicians with genetic variant information at the point of care. GIC introduces a novel way of communicating genetic variant information to clinicians, and findings from our study can provide useful information for developing and enhancing genetic IT support for the clinic. Based on the findings from this study, the features that would be the most important to include in similar applications are (1) integration with the EHR (2) transparency regarding reason for a variant interpretation change, supporting evidence, and lab staff that approved the change (3) an alerting mechanism that prioritizes changes depending on their impact on clinical management and family testing and (4) information available at the point of care in a central electronic repository.
The integration of genetic data into the EHR ecosystem is critical to advancing genetic based precision medicine. There are many efforts underway to enhance this integration including the NIH funded eMERGE Network [19]. GIC will be used within the network to return genetic test results. In addition, GIL will be used to create a de-identified case repository for sharing genetic and phenotypic data across sites within the eMERGE network.
This study had several limitations. Firstly, study participants were self-selected. The clinics from which participants were recruited were clinics that had already agreed to implement GIC and therefore may have been biased, as these clinics must have been willing and able to integrate this tool into their practice. This evaluation was with a small number of users on a novel system and some of the findings may not be generalizable, although many of the issues raised would be important to consider in designing similar systems. Many of the themes and broader issues identified are important for clinicians and informatics professionals to think about and consider in order to realize the potential of personalized medicine. Although only a small set of genetic disorders were studied, the tool was constructed to be nonspecific. Even though the demands on the tool would be expected to be similar, the feedback we received may change once additional genetic testing and diseases are included. Because this study was conducted within a short period of time after implementation, a future study might include follow-up interviews with the clinics once they have had a chance to use the tool more in practice in order to identify additional modifications that may be useful.
6. Conclusions
The information gathered in this study regarding considerations for IT infrastructure supporting genetic variant knowledge updates indicates that there are opportunities to further optimize IT support of care. While this study collected feedback from clinicians who used GIC specifically, many of the findings can be applied more broadly to genetic variant management IT. As sequencing expands to the whole genomes, health IT infrastructure will be critical in patient care and the development of personalized medicine. Implementation of GIC showed that use of health IT infrastructure could alleviate some of the burden of managing genetic variant information. Future studies should be conducted to inform how health IT can best manage the amount of rapidly increasing genetic information and improve patient care, while taking clinician workloads, workflows, and perspectives into consideration.
Supplementary Material
Acknowledgements
The authors would like to thank the participants across the multiple clinic and laboratory sites who gave freely of their time to assist in this evaluation. This work was supported by the National Institutes of Health Challenge Grants and Partnerships as part of the American Recovery and Reinvestment Act of 2009. The project described was supported by Award Number RC1LMO10526 from the National Library of Medicine. The content is solely the responsibility of the authors and do not necessarily represent the official views of the National Library of Medicine or the National Institutes of Health.
Footnotes
Clinical Relevance
Genetics is increasingly being applied to the care of patients. However, given the complexity of the genome and continuous evolution of knowledge, IT systems, such as the GeneInsight Clinic application described here, will be necessary to realize the increasing value of genomics in medicine.
Conflicts of Interest
This research was conducted at Brigham and Women’s Hospital, which is an affiliated entity of Partners HealthCare. Partners HealthCare licensed the GeneInsight technology involved in this study in November 2012, to a company in which both Partners Healthcare and Sunquest currently have ownership interests. The Committee on Conflicts of Interest reviewed this research in light of Partners acquisition of this financial interest, and the committee, consistent with Partners policy, required that notice of Partners financial interest in the technology be provided to journals and in publications and presentations resulting from the research. No authors had personal financial interest in the GeneInsight technology beyond that associated with being employed by Partners Healthcare.
Human Subjects Protection
This study was performed in compliance with the World Medical Association Declaration of Helsinki on Ethical Principles for Medical Research Involving Human Subjects, was approved by the Partners Human Research Committee (PHRC), and registered with ClinicalTrials.gov (ID: NCT01225978).
References
- 1.Aronson SJ, Clark EH, Varugheese M, Baxter S, Babb LJ, Rehm HL. Communicating new knowledge on previously reported genetic variants. Genet Med 2012; 14: 713–719. doi:10.1038/gim.2012.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rehm HL, Berg JS, Brooks LD, Bustamante CD, Evans JP, Landrum MJ, Ledbetter DH, Maglott DR, Martin CL, Nussbaum RL, Plon SE, Ramos EM, Sherry ST, Watson MS. ClinGen — The Clinical Genome Resource. N Engl J Med 2015; 372(23):2235–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richards CS, Bale S, Bellissimo DB, Das S, Grody WW, Hegde MR, Lyon E, Ward BE. ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genetics in medicine : official journal of the American College of Medical Genetics 2008; 10(4):294–300. [DOI] [PubMed] [Google Scholar]
- 4.Rehm HL, Bale SJ, Bayrak-Toydemir P, Berg JS, Brown KK, Deignan JL, Friez MJ, Funke BH, Hegde MR, Lyon E. ACMG clinical laboratory standards for next-generation sequencing. Genet Med 2013; 15(9): 733-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffman MA, Williams MS. Electronic medical records and personalized medicine. Hum Genet 2011; 130(1): 33-39. [DOI] [PubMed] [Google Scholar]
- 6.Del Fiol G, Williams MS, Maram N, Rocha RA, Wood GM, Mitchell JA. Integrating genetic information resources with an EHR. AMIA Annu Symp Proc 2006: 904. [PMC free article] [PubMed] [Google Scholar]
- 7.Aronson SJ, Clark EH, Babb LJ, Baxter S, Farwell LM, Funke BH, Hernandez AL, Joshi VA, Lyon E, Parthum AR, Russel FJ, Varugheese M, Venman TC, Rehm HL. The GeneInsight Suite: a platform to support laboratory and provider use of DNA-based genetic testing. Human mutation 2011; 32(5): 532-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrison S, Dolinsky J, Vincent L, Johnson AK, Chao E, Azzariti D, Das S, Bale S, Rehm H. Clinical Laboratories Implement the ACMG/AMP Guidelines to Resolve Differences in Variant Interpretations Submitted to ClinVar. in ACMG Annual Clinical Genetics Meeting. 2016. Tampa, FL. [Google Scholar]
- 9.Bell GC, Crews KR, Wilkinson MR, Haidar CE, Hicks JK, Baker DK, Kornegay NM, Yang W, Cross SJ, Howard SC, Freimuth RR, Evans WE, Broeckel U, Relling MV, Hoffman JM. Development and use of active clinical decision support for preemptive pharmacogenomics. J Am Med Inform Assoc 2014; 21(e1): e93-e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kho AN, Rasmussen LV, Connolly JJ, Peissig PL, Starren J, Hakonarson H, Hayes MG. Practical challenges in integrating genomic data into the electronic health record. Genet Med 2013; 15(10): 772-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neri PM, Pollard SE, Volk LA, Newmark LP, Varugheese M, Baxter S, Aronson SJ, Rehm HL, Bates DW. Usability of a novel clinician interface for genetic results. J Biomed Inform 2012; 45(5): 950-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilcox AR, Neri PM, Volk LA, Newmark LP, Clark EH, Babb LJ, Varugheese M, Aronson SJ, Rehm HL, Bates DW. A novel clinician interface to improve clinician access to up-to-date genetic results. J Am Med Inform Assoc Feb; 21(e1): e117–21. doi: 10.1136/amiajnl-2013–001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glaser B, Straus AL. The Discovery of Grounded Theory: Strategies for Qualitative Research. Chicago, IL: Aldine Transaction; 1967. [Google Scholar]
- 14.Urquhart C, Lehmann H., Myers MD. Putting the ‘theory’ back into grounded theory: guidelines for grounded theory studies in information systems. Information Systems Journal 2010; 20(4):357–381. [Google Scholar]
- 15.Manolio TA, Chisholm RL, Ozenberger B, Roden DM, Williams MS, Wilson R, Bick D, Bottinger EP, Brilliant MH, Eng C, Frazer KA, Korf B, Ledbetter DH, Lupski JR, Marsh C, Mrazek D, Murray MF, O’Donnell PH, Rader DJ, Relling MV, Shuldiner AR, Valle D, Weinshiboum R, Green ED, Ginsburg GS. Implementing genomic medicine in the clinic: the future is here. Genet Med 2013; 15(4): 258-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramos EM, Din-Lovinescu C, Berg JS, Brooks LD, Duncanson A, Dunn M, Good P, Hubbard TJ, Jarvik GP, O’Donnell C, Sherry ST, Aronson N, Biesecker LG, Blumberg B, Calonge N, Colhoun HM, Epstein RS, Flicek P, Gordon ES, Green ED, Green RC, Hurles M, Kawamoto K, Knaus W, Ledbetter DH, Levy HP, Lyon E, Maglott D, McLeod HL, Rahman N, Randhawa G, Wicklund C, Manolio TA, Chisholm RL, Williams MS. Characterizing genetic variants for clinical action. Am J Med Genet C Semin Med Genet 2014; 166C(1):93–104.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015; 17(5): 405-424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, Maglott DR. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 2014; 42(Database issue): D980–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Electronic Medical Records and Genomics (eMERGE) Network. 2016; Available from: http://www.genome.gov/27540473. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



