Abstract
Many microorganisms are capable of producing and secreting exopolysaccharides (EPS), which have important implications in medical fields, food applications or in the replacement of petro-based chemicals. We describe an analytical platform to be automated on a liquid handling system that allows the fast and reliable analysis of the type and the amount of EPS produced by microorganisms. It enables the user to identify novel natural microbial exopolysaccharide producers and to analyze the carbohydrate fingerprint of the corresponding polymers within one day in high-throughput (HT). Using this platform, strain collections as well as libraries of strain variants that might be obtained in engineering approaches can be screened. The platform has a modular setup, which allows a separation of the protocol into two major parts. First, there is an automated screening system, which combines different polysaccharide detection modules: a semi-quantitative analysis of viscosity formation via a centrifugation step, an analysis of polymer formation via alcohol precipitation and the determination of the total carbohydrate content via a phenol-sulfuric-acid transformation. Here, it is possible to screen up to 384 strains per run. The second part provides a detailed monosaccharide analysis for all the selected EPS producers identified in the first part by combining two essential modules: the analysis of the complete monomer composition via ultra-high performance liquid chromatography coupled with ultra violet and electrospray ionization ion trap detection (UHPLC-UV-ESI-MS) and the determination of pyruvate as a polymer substituent (presence of pyruvate ketal) via enzymatic oxidation that is coupled to a color formation. All the analytical modules of this screening platform can be combined in different ways and adjusted to individual requirements. Additionally, they can all be handled manually or performed with a liquid handling system. Thereby, the screening platform enables a huge flexibility in order to identify various EPS.
Keywords: Medicine, Issue 110, Exopolysaccharide, automated, modular, screening, platform, high-throughput, different genera, carbohydrate fingerprint.
Introduction
Microbial exopolysaccharides (EPS) are a structurally highly diverse group of polymers that fulfill various biological functions. They usually are built of complex repeat units, which are distinguished by different types of monomers (sugar, sugar derivatives, sugar acids), the bonds between these monomers and their substitutions. The structural diversity of microbial polysaccharides confers rather different characteristics to the members of this molecule class, which allows their application in different fields like food1, cosmetics2,3, construction chemistry4 or water treatment5. To further extend the field of application of these bio-based and such sustainable polymers the identification of novel natural microbial polysaccharides as well as the engineering of structural variants represents promising approaches. Either way, a fast screening method is required to quickly scan a vast number of microbial strains for their polysaccharide formation, and to analyze their products. Therefore, we have recently developed a 96-well HT-screening platform for the analysis of microbial polysaccharide production from natural isolates or engineered strain variants that includes the identification of the monomeric composition6.
Applying this platform for a first screening round of ~500 natural isolates allowed us to identify only about 10 to 20% of the isolated strains as being able to produce EPS (data not shown). This means that 80-90% of the analyzed strains did not produce EPS under the conditions applied, and therefore, a further analysis of the detailed carbohydrate fingerprint was not necessary. As this highly sophisticated identification of the monomeric composition is a time consuming process, especially for data analysis, a fast pre-screening method to identify the strains positive in EPS production, would drastically increase the efficiency. Furthermore, reagents, consumables and measurement time at the UHPLC-UV-ESI-MS would be reduced. Additionally, the different analytical modules, while on one hand make the method highly reliable, are on the other hand complicating the manual handling of more than two 96-well plates in parallel, and as such restrict the full potential of the method. For these reasons, we decided to develop an automated screening platform. Therefore, we combined the modular format of the existing carbohydrate fingerprint technique with a fully automated fast detection method of the total sugar content, based on absorbance measurement.
The phenol-sulfuric-acid method still represents the method of choice for the fast determination of total carbohydrate content of bacterial and plant polysaccharides7,8. This method was first described by Dubois et al.9 and adapted for different applications and sample sizes, even for small scale in 96-well plates10,11. The phenol-sulfuric-acid method measures the total carbohydrate content by one value, summating all monomeric, oligomeric and polymeric carbohydrates of the samples.
Taking these aspects into account, the choice of a suitable cultivation medium is essential to apply this method. Complex media containing oligomeric or polymeric carbohydrate compounds like yeast extract might lead to an altered polymer content and therefore, should strictly be avoided. Furthermore, high amounts of sugars are used as C-source for the cultivation of the strains. High levels of remaining carbohydrates from the cultivation process might negatively interfere with the measurement of the EPS content.
Therefore, the use of defined and pure sugars is advised. In our experiments we used glucose for the cultivation of the cells. The remaining glucose after cultivation was reduced via a gel-filtration and determined via a glucose-assay. Finally, the glucose equivalents of the polysaccharides were calculated by subtracting the remaining glucose after the gel-filtration from the total carbohydrate content that had been detected with the phenol-sulfuric-acid method. Gel-filtration and glucose-assay in combination with the phenol-sulfuric-acid method ensure reliable results and are capable of being our first, completely automated detection system.
Two new analytical modules were included in the automated screening platform to increase the amount of information from the EPS-detection system: the precipitation and the observation of an increase in viscosity.
Many different EPS — e.g. succinoglycan, xanthan and colonic acid12 — are reported to be modified with a non-carbohydrate pyruvate ketal on sugar positions C4 and C6. Those pyruvate ketals (just as succinyl half esters and uronic acids) contribute to the polyanionic nature and therefore, to the physical properties of the polymer by interacting via divalent cation bridges13. In order to identify those particular polymers the determination of pyruvate was established as another additional analytical module. This increases the information about the polysaccharide substituents and their potential macroscopic properties.
Combining all the modules enables the identification of different EPS as well as a fast and efficient determination of EPS producers. By that approach the screening platform could be divided into two major parts (Figure 1). Within the automated screening (part I) the workflow occurs fully automated (Table 1) to quickly preselect the EPS producing strains. The carbohydrate fingerprint analysis (part II) quantitatively determines the monomer composition of the EPS produced by the preselected strains. Thereby, the data analysis was minimized in order to optimize the screening of large strain collections. This offers the possibility to analyze 384 strains in one single automated screening run and with two runs that are possible per day of 768 strains per day. Additionally, the carbohydrate fingerprint analysis gives an even more detailed overview of all the identified EPS. This enables the directed analysis and identification of only slightly differing EPS variants or completely new ones compared to the already described chemical structures of EPS.
Protocol
1. Automated Screening
Note: All liquid handling steps are done with a robotic liquid handling system. The composition of the robot worktable is presented in Figure 2. All consumables are stored in the storage carousel, unless mentioned otherwise. For all the automated screening steps a robotic manipulator (RM) moves the consumables, (deep well plate (DWP); micro titer plate (MTP); polymerase chain reaction plate (PCR-plate) and so on) between the carousel positions (CP) and the worktable positions (WTP). All the pipette steps are performed with a 96-channel-pipette-arm, except if it is mentioned otherwise. All steps are programmed and are performed automatically in 96-well format.
- Strain Cultivation (Task 1 in Figure 1) Note: Handle the inoculation of the putative EPS producing strains and the sealing of the cultivation plates under sterile conditions (laminar flow). The automated fast screening can handle four 96-well plates per run. Different strains of several genera were already tested6.
- Manually inoculate the pre-culture (1 ml EPS-media in a DWP) with a 96-pin replicator from a 96-well glycerol stock plate. Cover the plate with a breathable sealing film to avoid evaporation and to allow aeration. Incubate at 30 °C for 48 hr at 1,000 rpm on a MTP-shaker.
- Inoculate the main-culture (990 µl EPS-media in a DWP) by transferring 10 µl of the pre-culture using a 50 µl 12-channel multi-pipette and incubate under the same conditions as the pre-culture. Take a note of all the strains that do not grow.
- Preparation of the Robot Worktable and the Storage Carousel
- Provide all consumables listed in materials and equipment to the correct position in the storage carousel. Add 990 µl of double distilled water (ddH2O) in each well of the DWP for the 1:100 dilutions for the glucose-assay (carousel position 4-1 to 4-4).
- Arrange a 250 ml trough containing 150 ml of ddH2O at the worktable position (WTP) 11.
- Deposit a 250 ml trough containing 50 ml glucose-assay reagent-mix (4 ml 500 mM potassium phosphate (pH 5.7), 1.5 ml 50 mM 2.2-azinobis-(3ethylbenzthiazoline)-6-sulfonic acid, 2 ml 100 U glucose oxidase, 10 µl 1,000 U horseradish peroxidase and 42.49 ml ddH2O) at position WTP 1-2.
- Place two empty dummy F-bottom MTPs at position WTP 3-1 and 3-2.
- Remove the breathable sealing film, allocate the main-cultures in the carousel positions (CP) 1-1 to 1-4 and start the automated robotic program.
- Equilibration of the Gel-filtration Plates Note: The gel filtration plates that are used during this screening can be reused. For this, wash and centrifuge three times each with 150 µl of ddH2O at 2,000 x g for 2 min at 20 °C. Afterwards, store at 4 °C with 75 µl of 20% ethanol and cover with a silicon cap mat.
- Move the 250 ml trough (CP 5-7) with 5 mM ammonium acetate buffer (pH 5.6) to the WTP 3-3.
- Transfer the gel-filtration plates (CP 1-6, 1-7, 2-6 and 2-7) from the carousel to the WTPs 4-1 to 4-4.
- Aspirate 150 µl of ammonium acetate buffer using 200 µl tips and dispense into the center of all gel-filtration plates for equilibration.
- Transfer the gel-filtration plates into the centrifuge (2,000 x g for 2 min at 20 °C).
- Transfer the plates back to the worktable, repeat the steps 1.3.3, 1.3.4 and 1.3.3. Do not repeat the centrifugation step to avoid dehydration of the gel-filtration plates. Store the equilibrated gel-filtration plates back within the carousel.
- Cell Removal via Centrifugation (Task 1 in Figure 1) Note: In order to ensure a low background within the screening approach, analyze cell free EPS containing supernatants only and remove cells via centrifugation after cultivation.
- Transfer the main-culture DWP from the carousel (1-1 to 1-4) into the centrifuge (4,300 x g for 30 min at 20 °C).
- Prepare the worktable via the RM to process the main culture. Note: For steps 1.4.2 to 1.4.3.5 the system always handles two plates in parallel and then repeats all steps with the next two plates.
- For the precipitation before filtration move the MTP from CP 6-1 and 6-2 to WTP 4-1 and 5-1. Move the pH-indicator MTPs from CP 7-1 and 7-2 to WTP 4-2 and 5-2.
- Transfer the filtration plate together with the collector plate from the CP 2-1 and 2-2 to WTP 4-3 and 5-3. Relocate the main-culture DWP 1-1 and 1-2 from the centrifuge to WTP 4-4 and 5-4.
- Prepare the analytical modules. Note: Take 200 µl tips from the tip storage (position 2-1 to 2-4). In order to reduce consumables, use the same tip-set for step 1.4.3.1.
- Transfer 180 µl of the main-culture supernatant to the filtration plate. Aspirate 150 µl from the main-culture supernatant and dispense 50 µl into the MTP for precipitation before filtration and 100 µl to the pH-indicator plate. Note: The pH-determination is not essential; however, after adding 12.5 ml methyl red indicator (50 mg in 50 ml ethanol) in each well with a multi-step pipette it indicates high acid-producing strains. This information can be useful to avoid growth inhibition (at low pH) for further cultivations, e.g. for a second screening with higher buffer concentration. Furthermore, low pH may also induce EPS production in order to protect the cell against the acidic environment.
- Repeat step 1.4.3.1 for the second main-culture plate. Use a fresh tip-set.
- Move the filtration plates to the centrifuge and remove all other plates from the worktable back to their home positions in the carousel.
- Repeat the steps 1.4.2.1 to 1.4.3.3 for the third and fourth main-culture plate.
- Take a note of those fermentation broths, which show decreased sedimentation after centrifugation of the main culture plates, when they are transferred back to the carousel (viscosity control).
- 96-well Filtration for Complete Cell Removal (Task 2 in Figure 1) Note: To ensure cell removal from viscous fermentation broth a filtration step is included in the screening.
- Centrifuge the filtration plates at 3,000 x g for 10 min at 20 °C and bring them back to their carousel home position.
- Prepare the gel-filtration plates.
- Move the equilibrated gel-filtration plates from the carousel to the centrifuge and spin at 1,000 x g for 2 min at 20 °C.
- Transfer the gel-filtration plates from the centrifuge to WTP 4-1 to 44.
- Move the fresh gel-filtration collector plates from CP (3-1 to 3-4) to WTP (5-1 to 5-4).
- Transfer the equilibrated gel-filtration plates (WTP 4-1 to 4-4) to fresh collector plates (5-1 to 5-4).
- Clean up the worktable except positions 5-1 and 5-2 and move position 5-1 to position 4-1.
- Prepare the worktable via the RM to process the filtrates. Note: For steps 1.5.3 to 1.6.3.5 the system handles two plates in parallel and repeats all steps with the next two plates.
- Move 50 µl tips from the carousel (4-6 and 4-7) to WTP 3-3 and 3-4.
- Transfer the filtration plates from CP 3-1 to 3-2 onto the worktable (4-4 and 5-4).
- Relocate the DWP (1:100 dilution) for the detection of the glucose consumption from the CP 4-1 and 4-2 to WTP 4-2 and 5-1.
- Move the MTP for the second precipitation after filtration from CP 8-1 and 8-2 to WTP 4-3 and 5-3.
- Lift the filtration plates from the collector plates (WTP 4-4 and 5-4) and move them to WTP 3-1 and 3-2.
- Prepare the analytical modules after filtration. In order to reduce consumables, use the same tip-set for steps 1.5.4.1 and 1.5.4.3.
- Take 50 µl tips and pipette a 10 µl aliquot of the filtrate to the DWP (1:100 dilution) for the glucose-assay (glucose consumption).
- Pipette 35 µl of filtrate to the center of the equilibrated gel-filtration plate and 50 µl to the MTP that is used for precipitation.
- Repeat steps 1.5.4.1 to 1.5.4.2 for the second filtration plate.
- Move the gel-filtration plates to the centrifuge and move all other plates from the worktable back to the home positions in the carousel.
- Repeat steps 1.5.3.1 to 1.5.4.4 for the third and fourth filtration plate.
- After storage of all plates back in in the carousel take note of highly viscous supernatants, as indicated by residuals in the filter plate (high viscosity control after the filtration step). A lack of filtrate can lead to false negative results in the following analytical modules.
- Removal of the Glucose via 96-well Gel-filtration (Task 3 in Figure 1)
- Centrifuge the gel-filtration plates at 1,000 x g for 2 min at 20 °C.
- Prepare the worktable via the robotic manipulator to process the gel-filtrates.
- Provide 50 µl tips from the carousel (5-3 and 5-4) to WTP 3-3 and 3-4.
- For the determination of the remaining glucose after gel-filtration move two MTPs from CP 9-1 and 9-2 to WTP 4-1 and 5-1.
- For the phenol-sulfuric-acid method move two MTPs from CP 8-5 and 8-6 to WTP 4-2 and 5-2.
- Transfer two gel-filtration plates from the centrifuge to WTP (4-3 and 5-3).
- Lift the gel-filtration plates from the collector plate (4-3 and 5-3) and move them to WTP 3-1 and 3-2.
- Prepare the worktable via the robotic manipulator to process the gel-filtrates. Note: In order to reduce consumables, use the same tip-set for steps 1.6.3.1 and 1.6.3.2.
- To detect the remaining glucose after gel-filtration (1:10 dilution) take 50 µl tips and transfer 25 µl of ddH2O (WTP 1-1) to a fresh MTP. Aspirate 20 µl of ddH2O (WTP 1-1), 5 µl of air and 5 µl of gel-filtrate and dispense into the same plate.
- For the phenol-sulfuric-acid-method pipette 20 µl of gel-filtrate to the MTP.
- Repeat steps 1.6.3.1 and 1.6.3.2 for the second gel-filtrate plate.
- Transfer all plates from the worktable back to the home positions in their carousel.
- Repeat the steps 1.6.2.1 to 1.6.3.4 for the third and fourth gel-filtration plate.
- Glucose-assay Modules
- Prepare the worktable via the robotic manipulator to perform the glucose-assay.
- Relocate the DWP (1:100 dilution) from CP 4-1 and 4-4 to WTP 5-1 and 5-4.
- Move all MTPs from CP 9-5 and 9-8 to WTP 4-1 and 4-4.
- Take 200 µl tips and mix the dilution by aspirating and dispensing ten times the volume of 180 µl. Then pipette a 50 µl aliquot to the MTP.
- Repeat steps 1.7.1.2 and 1.7.1.3 for all four DWPs (1:100 dilution).
- Remove all DWPs (5-1 to 5-4) from the worktable.
- Prepare the worktable to start the glucose-assay.
- For the determination of the remaining glucose after gel-filtration transfer all MTPs from CP (9-1 to 9-4) to WTP (5-1 to 5-4).
- Move the glucose-assay calibration plate — containing three times 50 µl of the following glucose concentrations (90, 45, 18, 9, 4.5, 1.8, 0.9, 0.45 and 0 mg/L) — from CP 9-9 to WTP 3-3.
- Take 200 µl tips and aspirate 50 µl of glucose-assay reagent-mix from WTP 1-2, to start the first assay. Move the MTPs into the incubator (30 °C at 150 rpm for 30 min).
- Set the time schedule to start the program with 5 min delay in between the plates.
- After 30 min of incubation move the plates from the incubator to the MTP-reader and record absorbance at 418 and 480 nm.
- After the measurement move the plates to their home position in the carousel.
- Preparation of the Precipitation Note: In order to assess the EPS production perform a precipitation of the supernatant with 2-propanol before and after the first filtration step. Furthermore, evaluate the adsorption of the polymer on the filter membrane by observation of fibers and flakes in the first, but not in the second precipitation. Caution: Handle 2-propanol as a flammable liquid strictly under a fume hood.
- Remove all the precipitation plates (CP 6-1 to 6-4 and 8-1 to 8-4) from the carousel. And add manually 150 µl of 2-propanol to each well with a 12.5 ml multi-step pipette.
- Cover all MTPs with a silicone cap mat and shake at room temperature (RT) (10 min at 900 rpm). Visually observe fiber or flake formations after shaking that indicate EPS production.
- Phenol-sulfuric-acid Method Caution: Handle concentrated sulfuric-acid and phenol under a fume hood. Phenol is a corrosive and mutagenic agent.
- For the phenol-sulfuric-acid method, remove the plates (CP 8-5 to 8-8) from the carousel and place them into the liquid handling system (Position 4 to 8). Include the calibration plate with 20 µl of different glucose concentrations (10; 5; 2.5; 1; 0.5; 0.25; 0.1; 0.05; 0 g/L) in triplicates.
- Place a waste container at position 1, a 200 µl tip box at position 2 and a 250 ml trough with 110 ml phenol-sulfuric-acid (freshly prepared on ice with18.3 ml phenol 5% (w/v) in ddH2O and 91.7 ml of conc. sulfuric acid) at position 3.
- Use an 8-channel pipette with 300 µl tips and transfer 180 µl of phenol-sulfuric-acid into each row of all the plates.
- Cover all MTPs with lids, mix them by shaking (5 min at 900 rpm, RT) and incubate them for 35 min at 80 °C in an oven for color reaction. After cooling down under a fume hood, measure the extinction at 480 nm.
- Selection of EPS Positive Supernatants (Task 4 in Figure 1)
- For the evaluation of EPS production, select those strains from the automated screening that fulfill at least two of the three criteria as positives (viscosity control 2.6.1 and 2.6.2, detection of polymer 2.6.3, glucose equivalent 2.6.5). Transfer the remaining filtrates from the positive hits to a new MTP for further processing. Note: When the plates are covered with an aluminum sealing film they can be stored at -20 °C for at least one week. Within that time the determination of the carbohydrate fingerprint needs to be performed.
2. Carbohydrate Fingerprint
Note: All steps for the carbohydrate fingerprint are manually performed.
- Gel-filtration of the Positive Supernatants (Task 5 in Figure 1) Note: Gel-filtration is necessary as there is no filtrate left from the automated screening. Gel-filtration with a volume of more than 35 µl results in decreased purification efficiency.
- Prepare equilibration of the gel-filtration plates by dispensing 150 µl of ammonium acetate buffer into all wells via a 12.5 ml multi-step pipette.
- Transfer the gel-filtration plate into the centrifuge (2,000 x g for 2 min at 20 °C).
- Repeat steps 2.1.1, 2.1.2 and again 2.1.1. Centrifuge the gel-filtration plate at 1,000 x g for 2 min at 20 °C before further use.
- Pipette 35 µl of filtrate into the center of the gel-filtration plate using a 12-channel 50 µl pipette.
- Centrifuge the gel-filtration plate at 1,000 x g for 2 min at 20 °C and then lift it from the collector plate.
- Prepare the glucose-assay before hydrolysis: Perform a 1:10 dilution by adding 45 µl of ddH2O and 5 µl of gel-filtrate with a 12-channel 50 µl pipette and cover the MTPs with a silicone cap mat. Note: For the correct determination of the glucose value of the polymer, measure the glucose content before hydrolysis and subtract it from the glucose content quantified after the hydrolysis step.
- Prepare the pyruvate-assay before hydrolysis: Perform a 1:20 dilution using a 12-channel 200 µl pipette to add 95 µl of ddH2O, transfer 5 µl of gel-filtrate to each well and seal the MTP with a silicon cap mat.
- 96-well Micro Hydrolysis (Task 6 in Figure 1) Note: Heat up the incubation oven (including a sand bath) to 121 °C for at least 1.5 hr before using. A special clamping device was developed to avoid evaporation during the small scaled hydrolysis step.
- Take a new PCR-plate and transfer 20 µl of gel-filtrate with a 12-channel 50 µl pipette. Caution: Trifluoroacetic acid is a corrosive acid and toxic. Ammonium solution is corrosive. Handle both chemicals only under a fume hood.
- Add 20 µl of 4 M trifluoroacetic acid with a 1.25 ml multi-step pipette to each well. Then cover the PCR-plate with a thermoplastic elastomer (TPE) cap mat and place the PCR-plate in the special clamping device.
- Mix the solutions via inverting the clamping device ten times. Put the PCR-plate in a centrifuge and spin at 2,000 x g for 2 min to collect all the liquid on the bottom. Put the PCR-plate back to the clamping device and secure the device with screws.
- Place the secured clamping device in the pre-heated sand bath and incubate for 90 min at 121 °C.
- Remove the clamping device from the sand bath and let it cool down to RT.
- Remove the screws and spin again in a centrifuge at 2,000 x g for 2 min in order to collect all the condensate at the bottom of the wells and to prevent cross contamination during removal of the cap mat.
- Add 3.2% ammonia solution to adjust the pH to approx. 8 using a 12-channel 200 µl pipette. Cover the PCR-plate with a TPE cap mat and shake it manually using the clamping device.
- After neutralization centrifuge the PCR-plate at 2,000 x g for 2 min.
- High-throughput-1-phenyl-3-methyl-5-pyrazolone (HT-PMP)-derivatization of the Carbohydrates (Task 7 in Figure 1)
- Transfer 25 µl of the neutralized hydrolysate in a fresh PCR-plate using a 12-channel 50 µl pipette. Note: After taking the aliquot, check the neutralization of the remaining liquid in the hydrolysis plate via adding 12.5 µl phenol red indicator (0.05 g phenol red in 5 ml 20% ethanol) with a 1.25 ml multi-step pipette. All wells that do not turn into pink color (pH 8) are not correctly derivatized.
- Add 75 µl derivatization reagent-mix (125 mg PMP, 7 ml methanol, 3.06 ml ddH2O, and 438 µl 3.2% ammonium solution) and cover the plate with a TPE cap mat.
- After shaking the PCR-plate in the clamping device centrifuge the plate at 2,000 x g for 2 min to accumulate all the liquid at the bottom.
- For derivatization place the PCR-plate in a PCR-cycler at 70 °C for 100 min followed by cooling down to 20 °C.
- Transfer a 20 µl aliquot into a new MTP using a 12-channel 50 µl pipette. Then use a 12-channel 200 µl pipette and add 130 µl 19.23 mM acetic acid (0.962 ml 1 M acetic acid + 49.038 ml ddH2O) to each line. Mix directly via aspirating and dispensing (minimum six times) and transfer all the liquid to a 0.2 µm filter plate with an MTP collector plate.
- Centrifuge the plate (1,000 x g for 5 min), remove the filter plate, seal the MTP with a silicone cap mat and place the MTP into the UHPLC-UV-ESI-MS for the determination of the carbohydrate fingerprint14.
- Preparation of the Glucose-assay
- Perform a 1:10 dilution via adding 45 µl of ddH2O and 5 µl of neutralized hydrolysate using a 12-channel 50 µl pipette and cover the MTPs with a silicone cap mat. Add three times 50 µl of different glucose standards (500, 250, 100, 50, 25, 10, 5, 2.5 and 0 µM) to a new MTP used for calibration.
- Add 50 µl of glucose-assay reagent-mix (recipe step1.2.3) using a 12-channel 50 µl pipette. Then cover the plates with a silicon cap mat and incubate at 30 °C and 400 rpm for 30 min in an MTP-incubator.
- Directly after the incubation make an absorbance read in an MTP-reader at 418 and 480 nm in an MTP-reader. For the calculation of the glucose concentration perform a linear calibration as performed in step 2.6.4.
- Preparation of the Pyruvate-assay
- Perform a 1:20 dilution using a 12-channel 200 µl pipette. Add 95 µl of ddH2O and transfer 5 µl of neutralized hydrolysate to each well. Cover the MTP with a silicon cap mat.
- Add 100 µl of pyruvate-assay reagent-mix (3 ml 1 mM N-(carboxymethylamino-carbonyl)-4.4'-bis(dimethylamino)-diphenylamine sodium salt (DA-64), 300 µl 10 mM thiamine pyrophosphate, 60 µl 100 mM magnesium chloride hexahydrate, 2.4 ml 500 mM potassium phosphate puffer (pH 5.7), 30 µl 100 U pyruvate oxidase, 12 µl 1,000 U horseradish peroxidase and 24.19 ml ddH2O) using a 12-channel 200 µl pipette.
- Cover the plates with a silicon cap mat and incubate at 37 °C and 150 rpm for 30 min. Directly after the incubation measure absorbance in an MTP-reader at 727 and 540 nm.
- Evaluation of All Results of the Automated Screening and the Carbohydrate Fingerprints.
- Take note of those samples that show decreased sedimentation after the main culture plates have been centrifuged and stored back to the carousel (viscosity control step 1.4.1). Samples that do not show pellet formation are positive (criterion 1 for automated screening).
- Take note of highly viscous supernatants, which are indicated by residuals in the filter plate after the filtration step (high viscosity control step 1.5.4.6). A lack of filtrate can lead to a false negative result in the following analytical modules. Samples that maintain culture supernatants in the filter-wells are positive (also criterion 1 for automated screening).
- Visually observe fiber or flake formation after incubation. Take notes as this indicates polysaccharides. Rate no precipitation with (-); low precipitation of fibers or flakes with (+) and high precipitation with (++). Furthermore, detect adsorption of the polymer on the filter membrane via observation of fibers and flakes in the first, but not in the second precipitation (criterion 2 for automated screening).
- Perform a linear calibration for the calculation of the glucose concentration.
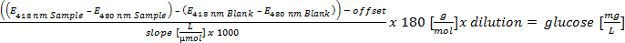 Please click here to view a larger version of this equation.
Please click here to view a larger version of this equation.
- Perform the linear calibration (absorption over concentration) between 5 to 0.1 g/L glucose. Calculation of the glucose equivalent of the hydrolyzed polymers and evaluate by following parameters: glucose equivalent: >700 mg/L = positive; 300-700 mg/L = putative positive; <300 mg/L = negative in regard of EPS formation (criterion 3 for automated screening).
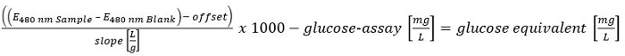 Please click here to view a larger version of this equation.
Please click here to view a larger version of this equation.
- Quantify all carbohydrates determined with the HT-PMP method. Sum up all quantities and evaluate as follows: >300 mg/L (positive); 150-300 mg/L (putative positive) and <150 mg/L (negative).
- For the calculation of the pyruvate concentration perform a linear calibration (100, 50, 25, 10, 5, 2.5, 1, 0.5 and 0 µM). To exclude the remaining pyruvate in the supernatant, correct the pyruvate content after hydrolysis via the pyruvate content before hydrolysis.
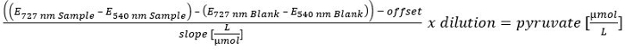 Please click here to view a larger version of this equation.
Please click here to view a larger version of this equation.
Representative Results
The validation of the phenol-sulfuric-acid method showed good results with a coefficient of determination (r²) of 0.9998 (Table 2). For the 5 g/L concentration the coefficient of variation (CV) and the accuracy showed a good performance with 1.8% and 2.2% error, but lower performance for the 0.25 g/L standard with 5.3% (CV) and 6.1% error (Bias).
The coefficients of determination of both pyruvate-assay calibration curves (with and without matrix) were >0.9999 in a calibration range of 150 µM (Table 3). The coefficients of variation (CV) for the highest and lowest calibration level were <4.6% and the accuracy showed a very good performance over the complete calibration range with less than 3.9% error. Thus, the matrix from the hydrolysis step showed no influence on the enzymatic assay, which is therefore capable to measure pyruvate before and after hydrolysis.
Table 4 shows the detailed results of three exemplary novel strains as successfully identified with the screening platform. The left part of the table displays the results of the automated screening modules concerning viscosity formation, polymer production and the glucose equivalent from the total hydrolysis which were used as evaluation parameters for detailed carbohydrate fingerprint analysis. The carbohydrate fingerprint based on calibrated sugars as well as unknown sugars, dimers and substituents are given in the right part of the table. By use of this information the monomeric composition can be calculated and compared with already known polymer structures. Furthermore, a targeted screening for interesting monomeric compositions and rare carbohydrates can be performed.
The high performance of the micro scale hydrolysis and the HT-PMP-derivatization were demonstrated in our previous work14. Furthermore, the validation of the gel-filtration and the carbohydrate fingerprint for various genera have been described in another publication6. In sum, the screening platform with its modular structure can easily be modified and adapted to individual requirements of the user. The automated screening of the platform enables an eight times higher throughput and gives reliable results. Novel analytical modules like the pyruvate-assay can be integrated and in combination with the carbohydrate fingerprint analysis they provide very detailed information about the identified EPS. Thereby, the screening platform is essential when searching for both slightly modified and completely novel EPS variants.
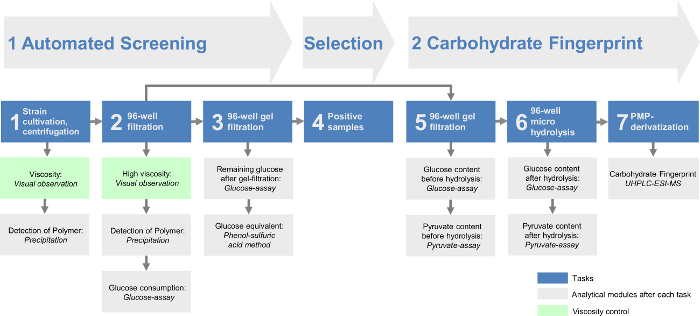 Figure 1: Overall scheme of the modular high-throughput exopolysaccharide screening platform. The automated screening includes the first three tasks. After bacteria are cultivated in 96-well plates, cells are removed by centrifugation (task 1) and a 96-well filtration (task 2). Then, the remaining monomeric sugars from the growth media are removed via a 96-well gel filtration (task 3). The EPS containing samples are evaluated in task 4. The carbohydrate fingerprint of the screening platform contains the last three tasks. The remaining filtrate of the positive hits from task 2 provides the basis for the gel-filtrate in task 5. After hydrolyzation in task 6 the carbohydrate fingerprint can be analyzed via the HT-PMP method (high-throughput 1-phenyl-3-methyl-5-pyrazolone, task 7). All tasks are followed by different analytical modules and/or a viscosity control. Please click here to view a larger version of this figure.
Figure 1: Overall scheme of the modular high-throughput exopolysaccharide screening platform. The automated screening includes the first three tasks. After bacteria are cultivated in 96-well plates, cells are removed by centrifugation (task 1) and a 96-well filtration (task 2). Then, the remaining monomeric sugars from the growth media are removed via a 96-well gel filtration (task 3). The EPS containing samples are evaluated in task 4. The carbohydrate fingerprint of the screening platform contains the last three tasks. The remaining filtrate of the positive hits from task 2 provides the basis for the gel-filtrate in task 5. After hydrolyzation in task 6 the carbohydrate fingerprint can be analyzed via the HT-PMP method (high-throughput 1-phenyl-3-methyl-5-pyrazolone, task 7). All tasks are followed by different analytical modules and/or a viscosity control. Please click here to view a larger version of this figure.
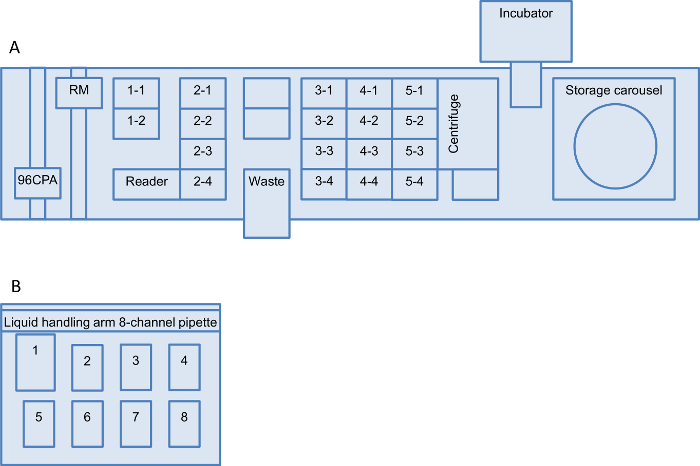 Figure 2: Robot worktable setup for the screening platform. Layouts of both liquid handling robot worktables are shown: (A) robotic liquid handling system and (B) liquid handling station. (A) The setup consists of a microplate carrier with two positions (positions 1-1 to 1-2), a carrier for disposable tips with four positions (positions 2-1 to 2-4) and three microplate carriers with four positions each (positions 3-1 to 3-4, 4-1 to 4-4 and 5-1 to 5-4). In addition, there is a storage carousel with five hotel carriers (1 to 5) each for seven deep well plates (DWP) and four hotel carriers (6-9) each for 21 micro-titer-plates. The hardware installed on the liquid handling robot is a 96-channel-pipette-arm for use with disposable tips and a robotic manipulator (RM) that moves plates/equipment between the worktable, the storage carousel, the MTP-reader, the centrifuge and the shaking-incubator. (B) The liquid handling station is equipped with a liquid handling arm and an 8-channel 300 µl pipette, a waste container at position 1, a tip adapter with 300 µl tips (position 2), a height adapter 30 with a 250 ml trough (position 3) and five height adapter 60 for MTPs (position 4 to 8). The numbering of the positions is referred to throughout this protocol. Alternative worktables can also be used if there are equivalent setups available. Please click here to view a larger version of this figure.
Figure 2: Robot worktable setup for the screening platform. Layouts of both liquid handling robot worktables are shown: (A) robotic liquid handling system and (B) liquid handling station. (A) The setup consists of a microplate carrier with two positions (positions 1-1 to 1-2), a carrier for disposable tips with four positions (positions 2-1 to 2-4) and three microplate carriers with four positions each (positions 3-1 to 3-4, 4-1 to 4-4 and 5-1 to 5-4). In addition, there is a storage carousel with five hotel carriers (1 to 5) each for seven deep well plates (DWP) and four hotel carriers (6-9) each for 21 micro-titer-plates. The hardware installed on the liquid handling robot is a 96-channel-pipette-arm for use with disposable tips and a robotic manipulator (RM) that moves plates/equipment between the worktable, the storage carousel, the MTP-reader, the centrifuge and the shaking-incubator. (B) The liquid handling station is equipped with a liquid handling arm and an 8-channel 300 µl pipette, a waste container at position 1, a tip adapter with 300 µl tips (position 2), a height adapter 30 with a 250 ml trough (position 3) and five height adapter 60 for MTPs (position 4 to 8). The numbering of the positions is referred to throughout this protocol. Alternative worktables can also be used if there are equivalent setups available. Please click here to view a larger version of this figure.
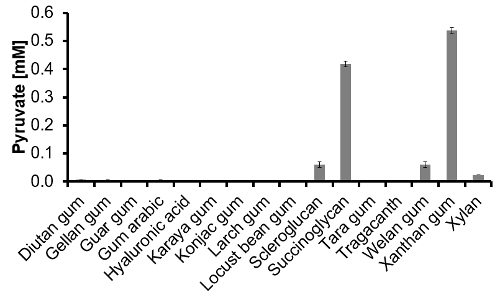 Figure 3: Pyruvate content of 16 commercially available polymers determined via pyruvate-assay. After the 1 g/L polymer solutions were hydrolyzed and neutralized, the pyruvate-assay was performed from a 1:10 dilution (n = 3). Please click here to view a larger version of this figure.
Figure 3: Pyruvate content of 16 commercially available polymers determined via pyruvate-assay. After the 1 g/L polymer solutions were hydrolyzed and neutralized, the pyruvate-assay was performed from a 1:10 dilution (n = 3). Please click here to view a larger version of this figure.
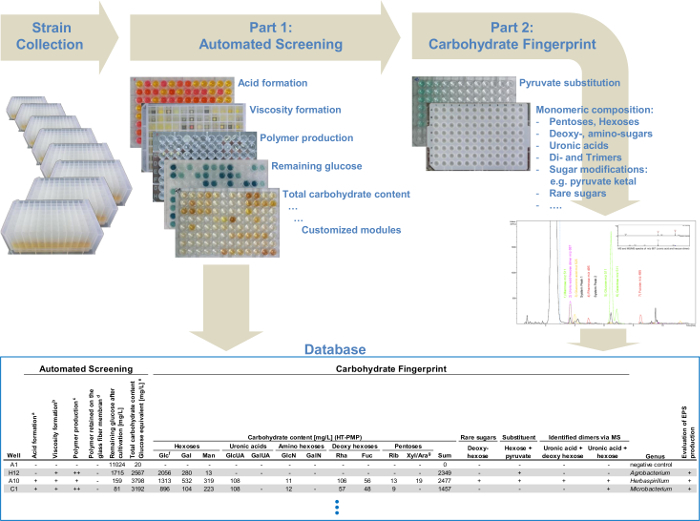 Figure 4: Flow chart of the modular high-throughput screening platform. The automated screening system, which combines different polysaccharide detection modules: analysis of viscosity formation, polymer production and determination of the total carbohydrate content. The second part provides a detailed monosaccharide analysis for all the selected EPS producers identified in the first part. All data from the automated screening and the data from the carbohydrate fingerprint via UHPLC-ESI-MS are collected in a database and enable the simple identification of structurally related variants of already known EPS or novel EPS and therefore, a targeted screening. Please click here to view a larger version of this figure.
Figure 4: Flow chart of the modular high-throughput screening platform. The automated screening system, which combines different polysaccharide detection modules: analysis of viscosity formation, polymer production and determination of the total carbohydrate content. The second part provides a detailed monosaccharide analysis for all the selected EPS producers identified in the first part. All data from the automated screening and the data from the carbohydrate fingerprint via UHPLC-ESI-MS are collected in a database and enable the simple identification of structurally related variants of already known EPS or novel EPS and therefore, a targeted screening. Please click here to view a larger version of this figure.
| Main step / Analytical module | Workflow | Observation / Description |
| Cultivation of the strains | 1 ml EPS-mediuma Pre-culture 48 hr, 30 °C, 1,000 rpma Main-culture 48 hr, 30 °C, 1,000 rpma | Production of EPS |
| Cell removal / viscosity | Centrifugation: 30 min at 4,300 x g | No pellet = increased viscosity = positive |
| Detection of Polymer: Precipitation | 50 µl supernatant + 150 µl 2-propanolb Shaking 10 min at RT and 900 rpmb | Visual: Fibers and flakes = positive precipitation of polymer |
| Cell removal / high viscosity | 180 µl supernatant of main-culture Centrifugation: 10 min at 3,000 x g 1.0 µm glass fiber membrane | No filter passing = high viscosity = positive |
| Detection of Polymer: Precipitation | 50 µl filtrate + 150 µl 2-propanolb Shaking 10 min at RT and 900 rpmb | Visual: Fibers and flakes = positive precipitation of polymer |
| Glucose consumption: Glucose-assay | Dilution 1:100: 10 µl filtrate + 990 µl ddH2O 50 µl aliquot + 50 µl reagent-mix Incubation 30 min at 30 °C 150 rpm Measurement 418-480 nm | Remaining glucose after cultivation |
| Gel-filtration | Equilibration: 3 x 150 µl NH4-acetat buffer pH 5.6 2 x 2 min at 2,000 x g 1 x 2 min at 1,000 x g Gel-filtration: 35 µl filtrate, 2 min at 1,000 x g Washing: 3 x 150 µl ddH2O, 2 min at 2,000 x g 75 µl 20% ethanol for storage | Polymer purification: Removal of salts, pyruvate, glucose and other sugar monomers from cultivation supernatant |
| Remaining glucose after gel-filtration Glucose-assay | Dilution 1:10: 25 µl ddH2O + 20 µl ddH2O and 5 µl filtrate + 50 µl reagent-mix Incubation 30 min at 30 °C, 150 rpm Measurement 418-480 nm | Subtraction of remaining glucose after gel-filtration from the phenol-sulfuric-acid method |
| Glucose equivalent: Phenol-sulfuric-acid methodc | 20 µl gel-filtrate + 180 µl phenol-sulfuric-acid (30 µl 5% (w/v) phenol in ddH2O + 150 µl conc. H2SO4 (ρ = 1.84 g/ml)) Shaking 5 min at 900 rpm Incubation 35 min at 80 °C Measurement at 480 nm | Glucose equivalent: Δ (phenol-sulfuric-acid value - remaining glucose after gel-filtration) <300 mg/L negative >300 and <700 mg/L putative positive >700 mg/L positive |
| a Handled manually under sterile conditions (laminar flow). | ||
| b Flammable liquid handled manually under a fume hood. | ||
| c Phenol-sulfuric-acid handled with Brand Liquid Handling Station (LHS) under a fume hood. |
Table 1: Complete workflow of the automated prescreening with the robotic liquid handling system and the liquid handling station. Overview of all parameters for the automated analytical modules.
| Linearity | LOD | LOQ | ||
| r²a | Slopea | Offseta | mg/L | mg/L |
| 0.9998 | 0.0007 | -0.021 | 50 | 100 |
| Standard | Meanb | Precisionb | Accuracyb | |
| mg/L | mg/L | CV% | Bias (%error) | |
| 5,000 | 5,112 | 1.8 | 2.2 | |
| 250 | 265 | 5.3 | 6.1 | |
| a Mean of eight measurements, calibration with six levels glucose from 0.1 to 5 g/L | ||||
| b Performed with a Student’s t-test (α = 0.05; n = 8). | ||||
| LOD: limit of determination, LOQ: limit of quantification, CV: coefficient of variation. |
Table 2: Validation of the phenol-sulfuric-acid method was carried out with the liquid handling station. The linearity was calculated based on a six point calibration (n = 8). Mean, precision and accuracy of two exemplarily chosen glucose concentrations are given here.
| Linearity | LOQ | |||
| r²a | Slopea | Offseta | µM | |
| without matrix | 0.99999 | 0.0223 | -0.0019 | 1 |
| 1:10 diluted matrix | 0.99999 | 0.0221 | -0.0011 | 1 |
| Standard | Meanb | Precisionb | Accuracyb | |
| µM | µM | CV% | Bias (%error) | |
| without matrix | 50 | 49.96 | 3.05 | -0.09 |
| 1 | 1.04 | 2.95 | 3.86 | |
| 1:10 diluted matrix | 50 | 49.98 | 0.44 | -0.04 |
| 1 | 1.00 | 4.58 | 0.33 | |
| a Mean of three measurements, calibration with six concentrations of pyruvate from 1 to 50 µM. | ||||
| b (n = 3) | ||||
| LOQ: limit of quantification, CV: coefficient of variation. |
Table 3: Validation of the pyruvate-assay with and without a 1:10 diluted neutralized trifluoroacetic-acid-matrix. Two six point calibrations (n = 3) with and without evaluation of matrix influences were performed. Mean, precision and accuracy of two exemplarily chosen pyruvate concentrations with and without effects of a 1:10 dilution were calculated.
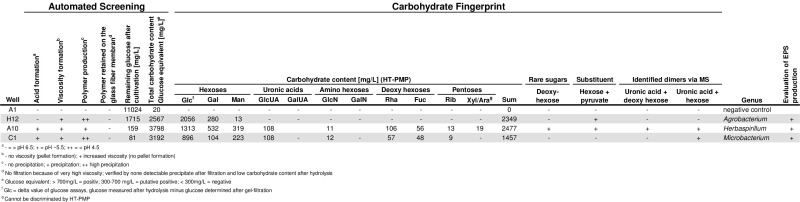 Table 4: Results of three exemplary strains screened with the platform. Data collected from the automated screening and the carbohydrate fingerprint. Please click here to download this table as a Microsoft Excel file.
Table 4: Results of three exemplary strains screened with the platform. Data collected from the automated screening and the carbohydrate fingerprint. Please click here to download this table as a Microsoft Excel file.
| Carbohydrate | Absorption max [nm] | Absorption at 480 nm mean±SD | Absorbance relative to glucose [%] | |
| Diutan gum | 470 | 0.342 | ±0.010 | 187 |
| Gellan gum | 472 | 0.334 | ±0.002 | 183 |
| Guar gum | 478 | 0.387 | ±0.017 | 212 |
| Gummi arabic | 476 | 0.393 | ±0.034 | 215 |
| Hyaluronic acid | 484 | 0.231 | ±0.011 | 126 |
| Karaya gum | 478 | 0.455 | ±0.023 | 249 |
| Konjac gum | 480 | 0.297 | ±0.009 | 163 |
| Larch gum | 480 | 0.337 | ±0.032 | 185 |
| Locust bean gum | 478 | 0.354 | ±0.033 | 194 |
| Scleroglucan | 484 | 0.168 | ±0,010 | 92 |
| Succinoglycan | 482 | 0.168 | ±0.005 | 92 |
| Tara gum | 480 | 0.318 | ±0.016 | 174 |
| Tragacanth | 478 | 0.513 | ±0.003 | 281 |
| Welan gum | 472 | 0.226 | ±0.016 | 124 |
| Xylan | 472 | 0.567 | ±0.007 | 311 |
| Xanthan gum | 482 | 0.245 | ±0.021 | 134 |
| Glucose | 484 | 0.191 | ±0.014 | 100 |
| SD: standard deviation |
Table 5: Results as obtained by the phenol-sulfuric-acid method for 16 commercially available polymers and glucose. The absorption maximum and absorption at 480 nm of 16 commercially available polymers (1 g/L) as well as glucose (1 g/L) were measured applying the phenol-sulfuric-acid method. The absorbance relative to glucose of all the polymers was calculated.
| Standard | Meana | Precisiona | Accuracya | |
| mg/L | mg/L | CV% | Bias (%error) | |
| 1:10 dilution | 450 | 460 | 1.01 | 2.14 |
| 45 | 44.7 | 1.41 | -0.70 | |
| 1:100 dilution | 4,500 | 5,026 | 1.19 | 11.6 |
| 450 | 471 | 1.16 | 4.55 | |
| b Performed with a Student’s t-test (α = 0.05; n = 8). | ||||
| CV: coefficient of variation. |
Table 6: Validation of the automated dilution for the glucose-assay. The dilution for the glucose-assay after cultivation (1:100) and after gel-filtration (1:10) were validated. Two glucose concentrations (n = 8) were diluted via the liquid handling system and evaluated. Mean, precision and accuracy were calculated.
| Theoretical glucose value | Covered with silicone cap mat | Test of evaporation (uncovered) | |||||
| Meana | Precisiona | Accuracya | Meana | Precisiona | Accuracya | Evaporation | |
| mg/L | mg/L | CV % | Bias (%error) | mg/L | CV % | Bias (%error) | %error |
| 45.0 | 45.2 | 0.69 | 0.44 | 46.0 | 0.66 | 2.05 | 1.60 |
| 18.0 | 17.7 | 0.80 | -1.68 | 18.0 | 0.72 | -0.01 | 1.69 |
| 9.0 | 8.74 | 1.20 | -2.98 | 8.92 | 0.81 | -0.95 | 2.09 |
| 4.5 | 4.50 | 1.26 | -0.04 | 4.58 | 1.57 | 1.76 | 1.80 |
| 1.8 | 1.85 | 0.74 | 2.90 | 2.01 | 2.82 | 11.6 | 8.48 |
| 0.9 | 1.03 | 1.43 | 14.1 | 1.16 | 3.52 | 28.3 | 12.4 |
| a (n = 4) | |||||||
| CV: coefficient of variation. |
Table 7: Evaluation of the evaporation effect of covered and uncovered MTP. Six different glucose standards (n = 4) were stored in the carousel for 3.5 hr at room temperature. The effect of the evaporation was evaluated by using uncovered as well as covered (silicon mat) standard samples. Mean, precision, accuracy and the evaporation in % error were calculated.
| Before gel-filtration | After gel-filtration | Remaining glucose after gel-filtration | |||
| Meana | SDa | Meana | SDa | ||
| mg/L | mg/L | mg/L | mg/L | % | |
| 1 | 8,647 | 110 | 259 | 121 | 3.00 |
| 2 | 5,108 | 56 | 116 | 37 | 2.27 |
| 3 | 2,014 | 12 | 50.8 | 14 | 2.52 |
| 4 | 1,015 | 12 | 25.1 | 8.1 | 2.47 |
| 5 | 510 | 4.9 | 12.8 | 4.3 | 2.51 |
| 6 | 223 | 8.6 | 6.6 | 1.5 | 2.94 |
| 7 | 122 | 5.6 | 4.3 | 0.9 | 3.48 |
| 8 | 75 | 6.0 | 3.1 | 0.3 | 4.18 |
| a (n=8) | |||||
| SD: standard deviation |
Table 8: Results of the gel-filtration efficiency. Eight different glucose standards were determined before and after gel-filtration to evaluate the efficiency of the gel-filtration. Mean, standard deviation and remaining glucose after gel-filtration in % were calculated.
Discussion
Polysaccharide detection with the phenol-sulfuric-acid method: Different monosaccharides show different absorption maxima and molar extinction coefficients by use of this method9. This results in different absorption maxima of polymers, which contain several sugars in different amounts. The different wavelengths of absorption maxima for 16 different commercially available polymers are given in Table 5. The polymers were dissolved (1 g/L) in ddH2O, stirred (150 rpm) overnight and measured with the phenol-sulfuric-acid-method. Diutan gum showed the lowest absorption maxima at 470 nm and scleroglucan and hyaluronic acid the highest at 484 nm. Based on these results 480 nm was chosen for this screening platform. The relative absorbance of the polymers was calculated based on the absorbance obtained with 1 g/L glucose (set as 100%). The lowest results were obtained with scleroglucan and succinoglycan, both with 92%. This was expected because scleroglucan only contains glucose and succinoglycan contains glucose and galactose in a ratio of 7:1. Both commercial polymers have different losses of drying and different ash contents, this is the reason why the theoretical value of ~110% was not reached. Xylan showed the highest relative absorbance with 311%. The reason for this is the high molar extinction coefficient achieved from xylose due to the more dominant furanose form. At a level of 0.1 g/L glucose the quantification limit was reached, as well as the detection limit at a concentration <0.05 g/L. However, the detection limit for positive strains in the screening is higher than 0.7 g/L and therefore, the assay showed a good performance. In order to get reliable results, the remaining glucose after gel-filtration was determined with a glucose-assay and this value was subtracted from the value from the phenol-sulfuric-acid method.
Automated glucose-assay dilution: The performance of the glucose determination after cultivation (dilution 1:100) was investigated. For this, 10 µl of supernatant were transferred to 990 µl of ddH2O in a deep well plate and mixed via ten times aspirating and dispensing 180 µl out of this dilution. The second critical step was the correct pipetting of only 5 µl aliquot for the 1:10 dilution from the glucose-assay after gel-filtration. In order to generate the dilution 25 µl of ddH2O were transferred with a 50 µl-tip first, afterwards 20 µl of ddH2O and 5 µl gel-filtrate were aspirated together. This ensures a better removal of the 5 µl aliquot out of the tip. Both dilution steps were verified with various glucose standards via a glucose-assay. The results for two exemplary concentrations are given in Table 6. The 1:100 dilution for the determination of the glucose content after cultivation showed high precision for both standards with a CV <1.2%. At the same time, the accuracy for the higher standard was up to 11.6 (% error). However, this is negligible as the glucose determination represents only the remaining glucose content after cultivation and therefore, is not important for the polymer detection. The 1:10 dilution for the remaining glucose after gel-filtration showed very reliable results with a CV <1.4% and an accuracy <2.1% error.
Consideration of evaporation: The screening requires 3.5 hr from the first step to the first glucose-assay. In order to find out, whether this time frame has an influence on uncapped MTP storage, 50 µl of glucose-assay calibration standards were stored with and without cover for 3.5 hr in the robot carousel. In the calibration range (45 to 4.5 mg/L) the sample concentration hardly increased. An increase — caused by evaporation — was below 2.1% and only for the two lowest concentrations (1.8 and 0.9 mg/L) it reached up to 12.4% (Table 7).
Gel-filtration: High amounts of non-metabolized glucose disturb the quantitative detection of glucose from the hydrolyzed polymer. Therefore, a gel-filtration step was required to remove the remaining glucose after cultivation. Additionally, the gel-filtration purifies the polymer containing supernatant from salts and monomeric carbohydrate compounds, others than glucose, to minimize the analytical background in the monomer analysis. At the gel-filtration step 35 µl of filtrate were placed in the center of the well. For validation of the robustness of gel-filtration in the automated system, eight calibration standards from 0.045 up to 9 g/L glucose were filtrated (n = 8). The glucose of every concentration was always reduced by more than 95% of the initial value (Table 8). In doing so, the gel-filtration showed very good results for various concentrations of glucose. Additionally, the remaining glucose after gel-filtration was also determined with a glucose-assay and subtracted from the phenol-sulfuric-acid determination to receive the correct amount of glucose equivalent for the hydrolyzed polymer.
Pyruvate-assay: First of all, it was investigated whether the neutralized and diluted (1:10) TFA-matrix from the hydrolysis step interferes with the enzymatic reaction. Therefore, the complete assay was performed twice, one time with and one time without matrix and showed reliable results. Finally, the pyruvate content of 16 commercially available polymers was successfully measured and is depicted in Figure 3. It is generally known that out of those 16 polymers only succinoglycan and xanthan naturally contain pyruvate. With our pyruvate-assay both of these polymers were correctly identified. In scleroglucan, welan gum and xylan pyruvate was also detected in significant amounts. Overall, the capability of the approach was validated and the pyruvate-assay showed a high performance. It proved to be able to detect pyruvate in different polymers after hydrolysis.
Carbohydrate fingerprint: After performing all analytical modules in the automated screening, potential EPS producers were selected for the carbohydrate fingerprint analysis. For this, several criteria were applied: 1) Positive observation of viscosity after centrifugation and/or after filtration. 2) Precipitation before and after filtration. Observed fibers and flakes were evaluated as positive. 3) Glucose equivalent value from the phenol-sulfuric-acid method. Values >700 mg/L were rated as positive and values between 300 and 700 mg/L were rated as putative EPS producers. When two or three criteria were evaluated as positive, the strains were selected for further carbohydrate fingerprint analysis. The criteria can be customized towards the individual purpose of the EPS screening (e.g. low viscosity EPS). Our approach aimed at finding efficient EPS producers. When searching for strains that only produce small amounts of EPS the evaluation limit of the glucose equivalent should be reduced.
Technical benefit and future applications: One interesting feature of this protocol is the modular character of the steps and the different analytical modules. They can be combined in different ways, adjusted to individual requirements and novel modules can easily be implemented. Furthermore, the analytical modules can be used separately, e.g. the hydrolysis module in combination with the HT-PMP-derivatization module is able to perform a monomeric composition analysis from different polymer solutions (1 g/L) in 96-well format. For laboratories without having access to a liquid handling system the complete screening can be handled manually without any changes in the pipetting scheme. However, using a liquid handling system increases the throughput to up to 768 strains (instead of 192 strains if screened manually) per day. The protocol that is described here is capable of a screening for different genera and therefore, for the screening of large strain collections to identify novel EPS producers and analyze their carbohydrate fingerprint in one approach (Figure 4). Furthermore, a targeted screening for polysaccharides containing rare sugars like fucose, uronic acids or even unknown sugars can be performed via the detailed monosaccharide analysis. Also, different sugar combinations in defined ratios can be detected. This enables the simple identification of structurally related variants of already known EPS or novel EPS.
Disclosures
We have nothing to disclose.
Acknowledgments
We sincerely thank Thomas Howe and Jörg Carsten for the programming and technical support with the liquid handling systems.
References
- Milani J, Maleki G. Ch. 2, Hydrocolloids in Food Industry. In: Valdez B, editor. Food Industrial Processes - Methods and Equipment. Vol. 2. InTech; 2012. [Google Scholar]
- Thibodeau A. Protecting the skin from environmental stresses with an exopolysaccharide formulation. Cosmet Toiletries. 2005;120(12):81–90. [Google Scholar]
- Prajapati VD, Jani GK, Zala BS, Khutliwala TA. An insight into the emerging exopolysaccharide gellan gum as a novel polymer. Carbohydr Polym. 2013;93(2):670–678. doi: 10.1016/j.carbpol.2013.01.030. [DOI] [PubMed] [Google Scholar]
- Schmidt W, Brouwers HJH, Kuehne HC, Meng B. The working mechanism of starch and diutan gum in cementitious and limestone dispersions in presence of polycarboxylate ether superplasticizers. Appl. Rheol. 2013;23(5):52903. [Google Scholar]
- Srinivasan R. Green Materials for Sustainable Water Remediation and Treatment. The Royal Society of Chemistry; 2013. Natural Polysaccharides as Treatment Agents for Wastewater; pp. 51–81. [Google Scholar]
- Rühmann B, Schmid J, Sieber V. High throughput exopolysaccharide screening platform: From strain cultivation to monosaccharide composition and carbohydrate fingerprinting in one day. Carbohydr Polym. 2015;122:212–220. doi: 10.1016/j.carbpol.2014.12.021. [DOI] [PubMed] [Google Scholar]
- Ortega-Morales BO, et al. Characterization of extracellular polymers synthesized by tropical intertidal biofilm bacteria. J Appl Microbiol. 2007;102(1):254–264. doi: 10.1111/j.1365-2672.2006.03085.x. [DOI] [PubMed] [Google Scholar]
- Xu R, Ma S, Wang Y, Liu L, Li P. Screening identification and statistic optimization of a novel exopolysaccharide producing Lactobacillus paracasei HCT. Afr. J. Microbiol. Res. 2010;4(9):783–795. [Google Scholar]
- Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956;28(3):350–356. [Google Scholar]
- Masuko T, et al. Carbohydrate analysis by a phenol-sulfuric acid method in microplate format. Anal Biochem. 2005;339(1):69–72. doi: 10.1016/j.ab.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Geater CW, Fehr WR, Wilson LA, Robyt JF. A more rapid method of total sugar analysis for soybean seed. Crop Sci. 2001;41(1):250–252. [Google Scholar]
- Sutherland IW. Ch 16, Microbial Exopolysaccharides. In: Dumitriu S, editor. Polysaccharides Structural Diversity and Functional Versatility. Marcel Dekker; 2005. pp. 431–457. [Google Scholar]
- Wingender J, Neu TR, Flemming HC. What are Bacterial Extracellular Polymeric Substances? In: Wingender J, Neu TR, Flemming HC, editors. Microbial extracellular polymeric substances: characterization, structure and function. Springer; 1999. p. 258. [Google Scholar]
- Rühmann B, Schmid J, Sieber V. Fast carbohydrate analysis via liquid chromatography coupled with ultra violet and electrospray ionization ion trap detection in 96-well format. J. Chromatogr. A. 2014;1350:44–50. doi: 10.1016/j.chroma.2014.05.014. [DOI] [PubMed] [Google Scholar]


