Abstract
Initial evaluations of large microbial libraries for potential producers of novel antimicrobial proteins require both qualitative and quantitative methods to screen for target enzymes prior to investing greater research effort and resources. The goal of this protocol is to demonstrate two complementary assays for conducting these initial evaluations. The microslide diffusion assay provides an initial or simple detection screen to enable the qualitative and rapid assessment of proteolytic activity against an array of both viable and heat-killed bacterial target substrates. As a counterpart, the increased sensitivity and reproducibility of the dye-release assay provides a quantitative platform for evaluating and comparing environmental influences affecting the hydrolytic activity of protein antimicrobials. The ability to label specific heat-killed cell culture substrates with Remazol brilliant blue R dye expands this capability to tailor the dye-release assay to characterize enzymatic activity of interest.
Keywords: Immunology, Issue 110, Microbiology, Molecular Biology, antimicrobial, lytic activity, microslide diffusion assay, dye-release assay
Introduction
Antimicrobial agents play an essential role in targeting a wide range of microorganisms such as viruses, fungi, and bacteria. The antimicrobial activities produced by various bacteria range in target specificity from broad-spectrum to species-specific, including clinically relevant bacterial pathogens 1. In nature, protein antimicrobial compounds like bacteriocins and lysins represent a microbial arsenal of tools for self-defense, replication, and nutrient acquisition 2,3. Serving as mediators of population dynamics within microbial communities, protein antimicrobials provide a competitive advantage to the producing organism over nonproducing, sensitive species 4. As recombinant enzymes and probiotics, these proteins also have an economic and societal importance as the need for new sources of pathogen control increases with each new expansion of antimicrobial resistance within the bacterial population 5.
The first evaluations of newly discovered protein antimicrobials include determining the basic physical characteristics that are inherent to the enzyme. Methods for rapid screening of potential antimicrobials allow selection of candidates with hydrolytic activities against the desired target substrates. The microslide diffusion assay and the quantitative dye-release assay allow for the use of a variety of substrates in the detection of enzymatic hydrolysis. For protein antimicrobial enzymes with activity against the bacterial cell wall, the versatility of these assays allow rapid and effective, qualitative and quantitative screening against viable bacterial cells (microslide assay only), heat-killed whole bacterial cell substrates, or purified peptidoglycan from the target bacterium. These assays have utility in antimicrobial enzyme discovery for use in fields such as alternative therapeutics, food supplements, and inhibitors of biofilm production.
The microslide diffusion assay and the dye-release assay are demonstrated methods for detection and characterization of novel protein antimicrobials. The microslide diffusion assay provides a relative (qualitative) estimate of antimicrobial activity and allows for minimal inference of enzyme concentration and activity. Using this method, activity is readily visualized as the development of a zone of lysis with the absence of activity resulting in a lack of zone formation. The rate of zone development and the zone diameter are indicative of the amount and purity of the enzyme present in the sample; however, this rate and the quality of the substrate clearing within the zone can also be affected by the presence of contaminants, the completeness of the hydrolysis reaction, and the type of substrate. Interfering contaminants can affect the rate of enzyme diffusion from the well, while incomplete hydrolysis or variation of the substrate type can result in a turbid zone of lysis 6.
The dye-release assay quantitatively assesses the amount of covalently-linked Remazol brilliant blue R dye products released into the reaction supernatant from the enzymatically hydrolyzed substrate. The method has only slight variability associated with a difference in substrate, thus allowing greater sensitivity for detection of hydrolysis as compared to the microslide diffusion assay. These properties allow the method to have utility in the characterization of biochemical properties of the enzyme and in the standardization of the assay for comparison between different antimicrobial enzymes.
In this protocol, we provide methods to perform the basic microslide diffusion assay and dye-release assay, while demonstrating the scale of detection limits and variability for each. The assays are used to perform basic evaluations of an unknown protein antimicrobial purified from the culture supernatant of an uncharacterized soil bacterial isolate. Observed activities against bacterial cell substrates within the assays allow comparison of reaction activities between a previously uncharacterized protein antimicrobial and α-chymotrypsin, a control proteolytic enzymes. These protocols provide a foundation for the application of the two techniques that can be expanded and modified to accommodate varied applications.
Protocol
1. Preparation of Bacterial and Peptidoglycan Substrates
Note: Whole cell bacterial substrates and purified peptidoglycans are used as enzyme substrates in both the microslide diffusion assay and the dye-release assay. These substrates require preparation prior to conducting the enzyme reactions. The following protocol describes their preparation.
- Whole Bacterial Cell Substrate
- Produce bacterial cell substrate by inoculating 500 ml of nutrient broth with a 2 ml overnight culture of Bacillus subtilis 168 (American Type Culture Collection; ATCC 23857). Incubate at 30 °C with shaking (125 rpm) until the culture reaches an exponential phase of growth, defined as a rapid growth phase resulting in the doubling of the bacterial culture. For the cultivation of Salmonella enterica subsp. enterica (ATCC 10708), use nutrient broth as a growth medium at 37 °C with shaking (125 rpm).
- Heat-kill each culture by autoclaving for 10 min at 121 °C under 3 atm of pressure.
- Harvest the heat-killed bacterial substrate by centrifugation for 20 min at 5,000 x g. Wash the pellet three times with Type 1 water and re-suspend in a minimal amount of water. In this study, suspend the substrates in 1,200 µl.
- Aliquot 300 µl of the bacterial cell substrates to 1.5 ml microfuge tubes and store at 20 °C.
- Purified Peptidoglycan Substrate
- Purify peptidoglycan from the target substrate bacterium 7-10 or acquire from a vendor (see Materials and Equipment Table). Purify crude peptidoglycan preparations from accessory cell wall polymers.
2. Qualitative Microslide Diffusion Assay [Modified from Lachica, et al. 11 ]
Note: The microslide diffusion assay is a qualitative method for detecting the presence of antimicrobial enzyme in a sample. As the enzyme diffuses through an agarose matrix containing substrate, a zone of clearing develops as the enzyme hydrolyzes the substrate. The following protocol describes the preparation and performance of the microslide diffusion assay for qualitatively detecting the presence of protein antimicrobials.
Determine the concentration of the antimicrobial enzyme to be evaluated using a Bicinchoninic Acid (BCA) Protein Assay Kit according to manufacturer's protocol.
Utilize phosphate-buffered saline (PBS) as an enzyme buffer; however, empirically determine the buffer appropriate for the given enzyme.
Perform a serial dilution of the antimicrobial enzyme using phosphate-buffered saline (PBS) as a reaction buffer. The serial dilution used in these microslide diffusion assays produced final assay protein masses of 0 pg, 10 pg, 100 pg, 1 ng, 10 ng, 100 ng, 1 µg, and 10 µg per reaction volume.
Dispense protein masses into individual microfuge tubes and adjust protein volumes to 20 µl using PBS in preparation for addition to microslide reaction wells.
Make a 0.5% agarose solution by dissolving 0.25 g of agarose in 50 ml of PBS. Heat the solution to a boil until the agarose is completely dissolved. Determine water lost during the melting process by weight and add back to the solution.
Add 50 µl of 10% sodium azide in water to the agarose solution and adjust the temperature of the solution to 50 °C in a water bath. The sodium azide is added to inhibit contaminating bacterial growth during incubation of the microslide diffusion assay. If viable cells are used as substrate, omit the sodium azide from the agarose solution.
Thaw the heat-killed bacterial substrate on ice.
Re-suspend the bacterial substrate in 12 ml of the agarose solution to approximately match the turbidity of a 2.0 McFarland equivalence standard (see Materials Table). Compare the turbidities of the solutions by visualizing a black line on a white background through the solutions, adding bacterial substrate to the agarose until the approximate turbidity is achieved.
Immediately pipet 3 ml of the agarose-substrate solution to each microslide (25 x 75 x 1 mm3).
After slides solidify, punch three wells in the agarose-substrate layer of each microslide using a cork borer (4.8 mm diameter). Add 20 µl of each adjusted protein serial dilution to their respective wells.
Use PBS (20 µl) or bovine serum albumin (5 µg in 20 µl PBS) in a negative control well. Use known bacteriolytic enzymes appropriate for the substrate, such as lysozyme, as a positive control.
Incubate the slide in a humidity chamber at 37 °C or the optimal activity temperature of the enzyme. The length of incubation depends upon the concentration and specific activity of the antimicrobial enzyme. A typical reaction time is overnight (approximately 16 hr).
After incubation, observe the slides under indirect light and capture images of the developing assay using a digital single-lens reflex camera with a 60 mm lens at a focal distance of approximately 30 cm. Note: Enzymatic activity of the protein antimicrobial for the given substrate is correlated with the development of a clear zone of hydrolysis, which forms around the well as the enzyme diffuses into the agarose.
To qualify the enzymatic activity, observe the sizes of zones that form around each well. High enzymatic activity qualitatively relates to a larger zone, while low enzymatic activity qualitatively relates to a smaller zone.
3. Substrate Labeling - Remazol Brilliant Blue R Dye Labeling [Modified from Zhou 12 ]
Note: In the dye-release assay, the substrate is covalently linked to Remazol brilliant blue R dye. The following protocol describes the preparation of dyed enzyme substrates.
Make a 250 mM sodium hydroxide (NaOH) solution by dissolving 1 g NaOH in 99 ml of Type I water to be used for making a 200 mM Remazol brilliant blue R dye (RBB) solution.
Make a 200 mM RBB solution by dissolving 1.25 g RBB in 98.75 ml ± 1 ml of a fresh 250 mM NaOH solution (step 3.1).
Re-suspend heat-killed bacterial cells at a concentration of 0.5 g wet weight in 30 ml of RBB solution. For purified peptidoglycan, re-suspend the peptidoglycan at a concentration of 0.3 g wet weight in 30 ml of RBB solution.
Incubate the reaction mixture in an Erlenmeyer flask on a rotating platform for 6 hr at 37 °C with gentle mixing.
Transfer the reaction mixture to a 4 °C incubator, and incubate for an additional 12 hr with gentle mixing.
After incubation, harvest the dyed substrate by centrifugation at 3,000 x g for 30 min. Decant the dye solution from the substrate pellet.
Remove non-covalently linked soluble dye from the substrates by washing the dye-labeled cells or peptidoglycan repeatedly (approximately 3-5 washes) with Type I water followed by centrifugation. With each water addition, re-suspend the pellet thoroughly. Note: When unbound, soluble dye is no longer visible in the water wash after centrifugation. The substrate should be given one additional wash. Note that the substrate will remain blue, while the supernatant of the last wash will be clear.
Store the dyed substrates suspended in a minimal amount of water at -20 °C for later use.
4. Quantitative Dye-release Assay
Note: During the dye-release assay, the hydrolysis of RBB-dyed substrate in the enzymatic reaction releases dyed products into the reaction supernatant. Colorimetric measurement of the amount of dye released indicates the amount of enzymatic activity present within the sample for a given enzyme. The quantitative method allows the comparison of different enzymes across substrates and allows for variation of environmental conditions influencing the enzymatic reaction (e.g., temperature, salinity, and pH). The following protocol describes the preparation and performance of the dye-release assay for quantitatively detecting the enzymatic activity of protein antimicrobials.
- Preparation for the Dye-release Assay
- Allow the frozen dyed substrate to return to room temperature and wash twice with assay buffer (PBS), which is empirically determined for the given enzyme.
- Calculate the volume of substrate suspension that is needed by multiplying the 200 µl reaction volume by the number of dye-release assays to be performed.
- Prepare the substrate suspension by adding dyed substrate to the volume of assay buffer, determined in 4.1.2, until an optical density (OD) of 2.0 is achieved at 595 nm using a spectrophotometer. To remain within the functional limitations of the spectrophotometer, measure the 2.0 OD595 as 1.0 for a 1:1 dilution of the concentrated solution. Use assay buffer as a blank. Note: The substrate turbidity for the reaction can be raised beyond 2.0 to match the activity levels of highly efficient enzymes.
- Suspend the protein antimicrobial to be evaluated in assay buffer at an estimated concentration of 1 mg/ml.
- Using the microplate assay of a BCA Protein Assay Kit according to manufacturer's protocol, determine the actual concentration of the protein sample to be assayed. Adjust the concentration of the stock suspension to 1 mg/ml using assay buffer.
- Using the Dye-release Assay to Determine the Optimal Incubation Conditions for a Protein Antimicrobial
- Dilute the stock protein antimicrobial suspension to 100 ng/µl. This concentration gives a final reaction mass of 1 µg of protein per volume (10 µl) added to the assay reaction.
- Determine the thermal range to be assessed for the bacteriolytic protein. The thermal range in these assays included 5 °C, 15 °C, 25 °C, 35 °C, 45 °C, 55 °C, and 65 °C.
- Perform the reaction assays in 0.5 ml microfuge tubes. For each thermal condition, add 10 µl of the stock protein to 200 µl of prepared substrate suspension.
- Incubate in a thermal cycler for 8 hr with mixing by inversion once per hour.
- Following incubation, arrest the reactions by adding 25 µl of ethanol.
- Remove undigested, insoluble substrate by centrifugation at 3,000 x g for 2 min, and transfer 150 µl of the reaction supernatant for each reaction mixture to a 96-well flat-bottom microplate, taking care not to disrupt the pellet of undigested substrate.
- Measure enzymatic activity of the protein antimicrobial for the given substrate by determining the amount of soluble RBB dye dissociated from the substrate after enzymatic hydrolysis. To quantify the enzymatic activity, measure the absorbance of the supernatant at 595 nm using a microplate spectrophotometer. Increased absorbance by the soluble dye released into the supernatant from the labeled substrate is a quantitative measurement of enzymatic activity. Note: The change in absorbance is represented by activity units (AU), where 1 AU results in a 0.01 increase in the optical density of the dye-release reaction supernatant at 595 nm. A blank reaction, incubated with all reaction components except enzyme, is used to subtract any dye that releases from the RBB-labeled substrate due to the incubation. The optimal enzymatic temperature provides the peak activity unit measurement.
- Repeat steps 4.2.1 through 4.2.7 using various incubation buffers, to determine the optimal buffer conditions for the antimicrobial enzyme. In these assays, use PBS.
- Using the Dye-release Assay to Determine the Minimal Active Concentration at the Optimal Incubation Temperature for a Protein Antimicrobial
- Perform a serial dilution of the stock antimicrobial protein suspension using assay buffer. The dilution series range will vary with the activity of the antimicrobial enzyme. The serial dilution used in these assays produced final assay protein amounts of 0 pg, 10 pg, 100 pg, 1 ng, 10 ng, 100 ng, 1 µg, and 10 µg.
- Perform each reaction assay in a 48-well flat bottom microplate. For each assay condition, add 10 µl of the respective protein suspension to 200 µl of prepared substrate suspension. Adjust any variation in volume of the protein solution added to the reaction to 10 µl using reaction buffer.
- Seal the microplate with plate sealing film to avoid volume variation due to evaporation. Incubate in a shaker incubator at the determined optimal incubation temperature for the given protein antimicrobial for 16 hr with shaking.
- Following incubation, arrest the reaction by adding 25 µl of ethanol to each well and remove undigested substrate by centrifugation at 3,000 x g for 2 min.
- Transfer 150 µl of the reaction supernatant for each reaction mixture to a 96-well flat-bottom microplate, taking care not to disrupt the pellet of the undigested substrate.
- To quantify the enzymatic activity, measure the absorbance of the supernatant at 595 nm using a microplate spectrophotometer. A blank reaction, incubated with all reaction components except enzyme, is used to subtract any dye that releases from the RBB-labeled substrate due to the incubation. Increased absorbance by the soluble dye released into the supernatant from the labeled substrate provides a quantitative measure to enzymatic activity. Note: The change in absorbance is represented by activity units (AU), where 1 AU results in a 0.01 increase in the optical density of the dye-release reaction supernatant at 595 nm.
Representative Results
The microslide diffusion assay and the quantitative dye-release assay are effective methods for screening and measuring initial investigations of new protein antimicrobials. Each assay has advantages and limitations; however, when performed in conjunction, they allow rapid initial screening and basic characterization of an antimicrobial.
The microslide diffusion assay efficiently allows for the rapid screening of microbial libraries, producing protein antimicrobials. When the enzyme concentration levels are of concern, sensitivity of detection constraints may limit the assay, requiring a greater amount of enzyme to be added to the reaction well than the dye-release assay. As illustrated in Figure 1, the qualitative properties of the assay allow the observer to compare relative enzyme amounts present within each well.
In the developing zone of lysis, illustrated in Figure 1A (25 µg of enzyme), the leading edge of the zone displays turbid or incomplete lysis of the substrate, while the zone closest to the well displays a more complete substrate lysis. This phenomenon, a product of the diminishing concentration of the diffusing enzyme at the leading edge, complicates the accurate measurement of the zone diameter. As the wells B (15 µg), C (10 µg), D (5 µg), E (1.0 µg), and F (0.1 µg) are observed in Figure 1, the diameter of the zone of complete lysis dissipates to extinction correlating to the reduced amount of enzyme. For condition F (0.1 µg), the enzyme concentration within the agarose is below the limit that will allow the diffusing enzyme to be visualized as it moves through the agarose, hydrolyzing the substrate. The microslide diffusion assay was also run using purified Bacillus subtilis peptidoglycan as substrate (Figure 2). While the zone is less defined than those observed with whole cell Salmonella enterica due to the reluctance of the peptidoglycan to evenly suspend in the agarose, the hydrolysis of the peptidoglycan by the unknown antimicrobial enzyme is apparent.
The dye-release assay is a more sensitive and versatile assay than the microslide diffusion assay, allowing a lower detection limit and variation of environmental factors affecting the enzyme reaction. In the representative assays, temperature was varied to determine the optimal temperature for the antimicrobial enzyme, determined to be 35 °C in PBS (Figure 3). This optimum is seen clearly in the reaction supernatants as increased amounts of blue color (Figure 3B) as well as represented in activity units derived from absorbance measurements at 595 nm (Figure 3A). The versatility of the dye-release assay allows the researcher to vary not only the temperatures but also the reaction buffer and buffer components to rapidly determine optimum incubation conditions for a given enzyme.
The activity level of the unknown antimicrobial enzyme (Figure 4) and the α-chymotrypsin control enzyme (Figure 5) were measured at the determined optimum incubation temperature of 35 °C in PBS against RBB-labeled Bacillus subtilis heat-killed substrate. Comparison of results from Figure 4 and Figure 5 indicates that the unknown antimicrobial enzyme has almost twice the affinity for the B. subtilis substrate. In addition, the α-chymotrypsin control did not completely digest the heat-killed B. subtilis substrate within the well (data not shown). The activity of the α-chymotrypsin control begins to plateau around 0.3 µg as compared to the continued rise in activity units across all enzyme amounts for the unknown antimicrobial enzyme (Figure 4 and Figure 5). This may indicate that the unknown enzyme has a greater sustained activity or that there are a greater number of cleavage sites available to the enzyme within the B. subtilis substrate.
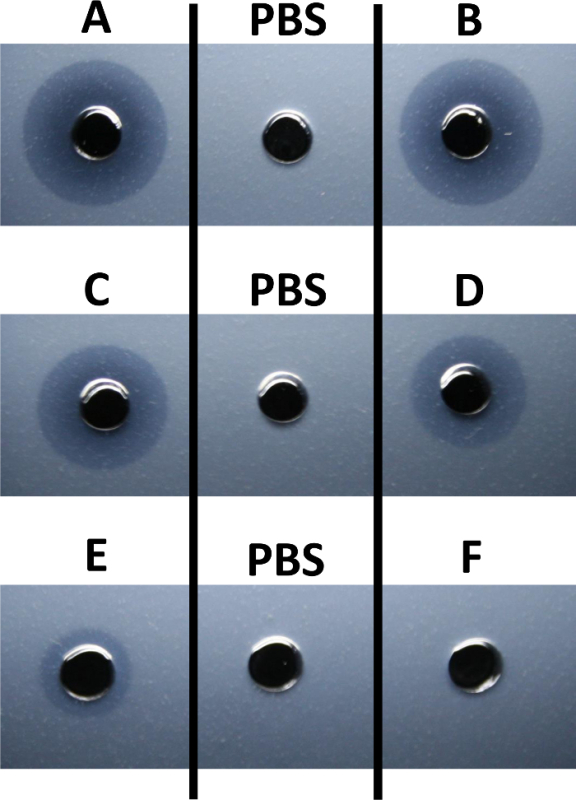 Figure 1: Enzyme Activity against Salmonella enterica Whole Cell Substrate. The microslide diffusion assay was used to qualitatively evaluate the activity of an unknown protein antimicrobial against heat-killed Salmonella enterica subsp. enterica (ATCC 10708). The protein masses of the unknown antimicrobial suspended in phosphate-buffered saline (PBS) that were added to the respective wells of the slides included 25 µg (well A), 15 µg (well B), 10 µg (well C), 5 µg (well D), 1 µg (well E), and 0.1 µg (well F). PBS alone was used for the negative controls of the assays. Zones of lysis were imaged after a 6 hr incubation. Please click here to view a larger version of this figure.
Figure 1: Enzyme Activity against Salmonella enterica Whole Cell Substrate. The microslide diffusion assay was used to qualitatively evaluate the activity of an unknown protein antimicrobial against heat-killed Salmonella enterica subsp. enterica (ATCC 10708). The protein masses of the unknown antimicrobial suspended in phosphate-buffered saline (PBS) that were added to the respective wells of the slides included 25 µg (well A), 15 µg (well B), 10 µg (well C), 5 µg (well D), 1 µg (well E), and 0.1 µg (well F). PBS alone was used for the negative controls of the assays. Zones of lysis were imaged after a 6 hr incubation. Please click here to view a larger version of this figure.
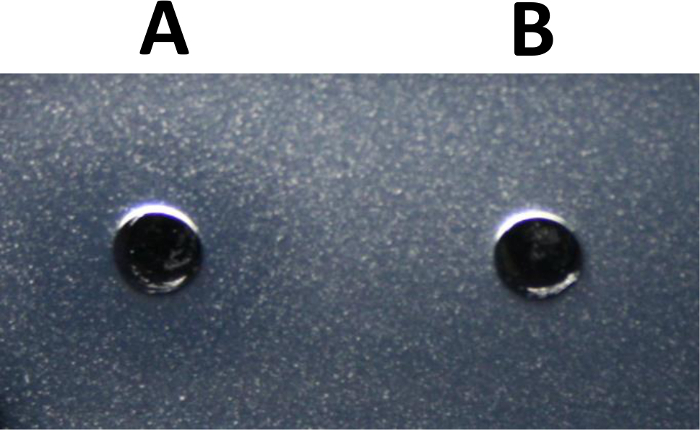 Figure 2: Enzyme Activity Against Bacillus subtilis Peptidoglycan Cell Wall. The microslide diffusion assay was used to qualitatively evaluate the activity of an unknown protein antimicrobial against peptidoglycan of Bacillus subtilis 168. Suspended in 20 µl of PBS, 10 µg of the unknown antimicrobial was added to well A of the microslides. PBS alone was used as a negative control for the assay (well B). The zone of lysis was imaged after a 6-hour incubation at 37 °C. Please click here to view a larger version of this figure.
Figure 2: Enzyme Activity Against Bacillus subtilis Peptidoglycan Cell Wall. The microslide diffusion assay was used to qualitatively evaluate the activity of an unknown protein antimicrobial against peptidoglycan of Bacillus subtilis 168. Suspended in 20 µl of PBS, 10 µg of the unknown antimicrobial was added to well A of the microslides. PBS alone was used as a negative control for the assay (well B). The zone of lysis was imaged after a 6-hour incubation at 37 °C. Please click here to view a larger version of this figure.
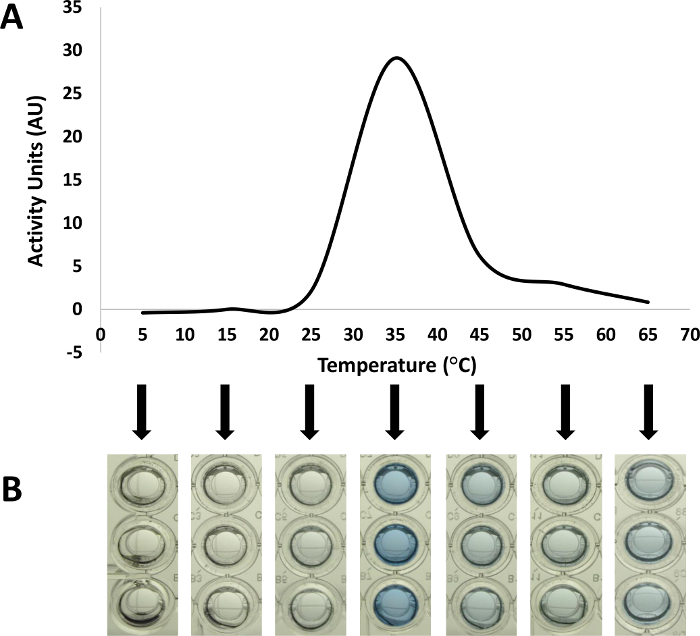 Figure 3: Optimal Reaction Temperature for a Protein Antimicrobial. The versatility of the dye-release assay allows the variation of physical and chemical parameters, such as incubation temperatures and buffers, to determine their influences on the activity of the enzyme of interest 6. As illustrated in this figure, the activity of the unknown protein antimicrobial (1 µg) was evaluated in PBS against RBB-labeled heat-killed Bacillus subtilis under a range of incubation temperatures. Displayed as activity units (AU) in (A), the absorbance of RBB-bound products released into the supernatant after hydrolysis was measured at 595 nm using a microplate spectrophotometer. At each temperature condition, reactions with no enzyme added were used to subtract the effects of any released dye that resulted from incubation at the various temperatures. The reaction supernatants corresponding to each incubation temperature were imaged prior to absorbance measurement (B). Please click here to view a larger version of this figure.
Figure 3: Optimal Reaction Temperature for a Protein Antimicrobial. The versatility of the dye-release assay allows the variation of physical and chemical parameters, such as incubation temperatures and buffers, to determine their influences on the activity of the enzyme of interest 6. As illustrated in this figure, the activity of the unknown protein antimicrobial (1 µg) was evaluated in PBS against RBB-labeled heat-killed Bacillus subtilis under a range of incubation temperatures. Displayed as activity units (AU) in (A), the absorbance of RBB-bound products released into the supernatant after hydrolysis was measured at 595 nm using a microplate spectrophotometer. At each temperature condition, reactions with no enzyme added were used to subtract the effects of any released dye that resulted from incubation at the various temperatures. The reaction supernatants corresponding to each incubation temperature were imaged prior to absorbance measurement (B). Please click here to view a larger version of this figure.
 Figure 4: Activity Range for a Protein Antimicrobial. The activity range for the unknown protein antimicrobial was measured as activity units after overnight incubations for 0, 100, 200, 300, 400, 500, 600, 700, 800, 900, and 1,000 ng, as measured by the BCA protein assay (A). For these reactions, the RBB-labeled B. subtilis substrate level was increased to give an optical density of 5.0 at 595 nm to ensure that the reaction with the highest amount of enzyme (1,000 ng) did not completely hydrolyze all available substrate within the reaction incubation period. Control reactions with no enzyme added were used to subtract the effects of any released dye caused by the incubation alone. The reaction supernatants corresponding to each enzyme amount were imaged prior to absorbance measurements (B). Please click here to view a larger version of this figure.
Figure 4: Activity Range for a Protein Antimicrobial. The activity range for the unknown protein antimicrobial was measured as activity units after overnight incubations for 0, 100, 200, 300, 400, 500, 600, 700, 800, 900, and 1,000 ng, as measured by the BCA protein assay (A). For these reactions, the RBB-labeled B. subtilis substrate level was increased to give an optical density of 5.0 at 595 nm to ensure that the reaction with the highest amount of enzyme (1,000 ng) did not completely hydrolyze all available substrate within the reaction incubation period. Control reactions with no enzyme added were used to subtract the effects of any released dye caused by the incubation alone. The reaction supernatants corresponding to each enzyme amount were imaged prior to absorbance measurements (B). Please click here to view a larger version of this figure.
 Figure 5: Activity Range for α-Chymotrypsin. The activity range for α-chymotrypsin was measured as activity units after overnight incubations for 0, 100, 200, 300, 400, 500, 600, 700, 800, 900, and 1,000 ng of enzyme (A). For these reactions, the RBB-labeled B. subtilis substrate level was increased to give an optical density of 5.0 at 595 nm to ensure that the reaction with the highest amount of enzyme (1,000 ng) did not completely hydrolyze all available substrate within the reaction incubation period. Control reactions with no enzyme added were used to subtract the effects of any released dye caused by the incubation alone. The reaction supernatants corresponding to each enzyme amount were imaged prior to absorbance measurements (B). Please click here to view a larger version of this figure.
Figure 5: Activity Range for α-Chymotrypsin. The activity range for α-chymotrypsin was measured as activity units after overnight incubations for 0, 100, 200, 300, 400, 500, 600, 700, 800, 900, and 1,000 ng of enzyme (A). For these reactions, the RBB-labeled B. subtilis substrate level was increased to give an optical density of 5.0 at 595 nm to ensure that the reaction with the highest amount of enzyme (1,000 ng) did not completely hydrolyze all available substrate within the reaction incubation period. Control reactions with no enzyme added were used to subtract the effects of any released dye caused by the incubation alone. The reaction supernatants corresponding to each enzyme amount were imaged prior to absorbance measurements (B). Please click here to view a larger version of this figure.
Discussion
The microslide diffusion assay and the dye-release assay provide advantages for detecting and characterizing previously unidentified protein antimicrobials. These rapid and sensitive methods allow high-throughput screening of organism source libraries for the identification of enzymes of interest prior to investing greater research resources. As high profile target enzymes are identified, variations of the detection assays can be used to rapidly detect the enzyme or to determine biochemical characteristics of the enzyme. For example, during a multistep protein purification scheme, the microslide diffusion assay can rapidly detect fractions containing the enzyme of interest. In addition, reaction optima for the enzyme can be determined by altering the reaction buffers and incubation temperatures in the dye-release assay.
The range of enzyme mass required for detection in the microslide diffusion assay is a function of enzyme characteristics. Detection limitations of the assay generally require the use of higher amounts of enzyme than the dye-release assay. In this study, comparison of the detection limits of the microslide diffusion assay (Figure 1) to the dye-release assay (Figure 4) illustrated that the microslide diffusion assay is a less sensitive technique than the dye-release assay. When enzyme concentration is not a concern, the microslide assay allows for rapid initial screening of libraries for the production of protein antimicrobials against target substrates with low labor and equipment demands. Although requiring more effort and equipment, the dye-release assay is more sensitive and reproducible, yielding quantitative results that allow comparison between dye-release assays of the same or different enzymes for a given substrate. The two methods are easily performed in most microbiological laboratories with no need for highly specialized instrumentation. In the representative assays, the control enzyme, α-chymotrypsin, was detected at amounts as low as 100 ng (Figure 5) using the dye-release assay, while the minimal amount detected in the microslide diffusion assay was 1 µg (data not shown).
Providing utility as a qualitative detection assay for antimicrobial enzymes, a number of variations of the diffusion assay have been reported 11,13. In reviewing these assays, the microslide diffusion assay was developed for its relatively high sensitivity as an initial screen and for its ease of reaction observance. Modified from the work of Lachica et al. 11, in which agarose overlays were used to detect the production of nucleases by strains of Staphylococcus aureus, the microslide diffusion assay operates similarly to the radial immunodiffusion method 14 as the protein diffuses from the well into the agarose containing the substrate. The radial immunodiffusion method halts the expansion of the zone of immunoprecipitation when the system reaches a complex formation equilibrium between the diffusing antigen and antibody within the agarose. In contrast, the substrate of the microslide diffusion assay is initially digested by the diffusing protein antimicrobial in a continually expanding zone of lysis surrounding the well. The increasing diameter of the zone continues until the enzyme loses activity or the substrate is exhausted. The rate of the production of the zone of lysis is quickened as the purity, and thus the specific activity, of the enzyme or the concentration of the enzyme added to the well increases.
For quantitatively assessing the enzymatic activity of protein antimicrobials against heat-killed B. subtilis, the dye-release assay was chosen. Colorimetric assays using Remazol brilliant blue R dye (RBB)-labeled substrates allow versatile and sensitive evaluation of protein antimicrobial activity. Enzymatic hydrolysis of RBB-labeled substrate results in the release of soluble blue products that can be readily measured by a spectrophotometer at 595 nm. Several variations of the RBB dye-release assay, developed to characterize other protein antimicrobial enzymes, show the flexibility of this assay for application with other substrates. For example, in determining the effects of lysostaphin bacteriolytic activity, Zhou, et al., reported a dye-release assay using RBB-dyed staphylococcal cells and staphylococcal peptidoglycan as substrates 12. In a modification of the application of this RBB dye-release assay, RBB-labeled Micrococcus luteus substrate was proposed for use as a rapid and sensitive means to diagnose and screen for diseases such as bacterial infection and the presence of a malignant carcinoma, which can be correlated to human lysozyme expression levels in blood serum 15. In addition, RBB-labeled bacterial cell substrates have also been used in a zymogram method for the detection of lysozyme 16.
We demonstrate the lowered detection limit, convenience, and reproducibility of the dye-release assay, allowing for rapid library screening for enzyme production. Modifications of the assay allow for downstream quantitative biochemical characterization assays, including effects of temperature, pH, and salinity as well as the effects of other proteolytic enzymes on the activity of antimicrobial proteins 6. In addition, the quantitative dye-release assay can be used to compare the activities of multiple enzymes for a given substrate. The RBB dye-release assay has reported utility for enzymes with activity against polysaccharides in addition to the characterization and detection of protein antimicrobial agents. Pettersson and Eriksson reported an assay for detecting endoglycanase activity using amorphous, RBB-dyed polysaccharide beads of cellulose, xylan, mannan, and chitin 17. In an expansion of these findings, a sensitive method for detection of chitinase enzymes produced by Bacillus thuringiensis Bt-107 was developed using colloidal chitin labeled with RBB 18. From these and other studies, commercially available sources of RBB-dyed substrates have emerged for use in detection of glycolytic activity, including those against keratin, amylopectin, amylose, glycogen, laminarin, d-xylan, Azo-barley glucan, and starch.
In this study, we demonstrate the utility of the dye-release assay in a microplate format, reducing the volume of RBB-labeled substrate and enzyme required to produce the colorimetric result. As illustrated in Figure 4 and Figure 5, the microplate dye-release assay provides a lower detection limit than the microslide diffusion assay for enzymatic activity detection. With regard to rapid use, conservation of labor, and ease of interpretation, the microslide diffusion assay provides utility in initial high-throughput screens for the presence of antimicrobial activity as well as the indication of antimicrobial protein presence in downstream protein isolation and purification steps. The combination of these two assays complement each other in the initial screening and ultimate characterization of novel protein antimicrobials
Disclosures
The authors declare that they have no competing financial interests. The MITRE Corporation is a not-for-profit company that operates multiple federally funded research and development centers (FFRDCs).
Acknowledgments
This work was funded by The MITRE Corporation through the MITRE Innovation Program (Project 25MSR621-CA). The authors would like to thank Mark Maybury, Ph.D., Richard Games, Ph.D., James Patton, Ellen Mac Garrigle, Ph.D., Carl Picconatto, Ph.D., and Caroline Gary for their support and review of this work.
References
- Anand TP, et al. Antimicrobial activity of marine bacteria associated with sponges from the waters off the coast of South East India. Microbiol Res. 2006;161:252–262. doi: 10.1016/j.micres.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Tagg JR, Dajani AS, Wannamaker LW. Bacteriocins of gram-positive bacteria. Bacteriol Rev. 1976;40:722–756. doi: 10.1128/br.40.3.722-756.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley MA, Gordon DM. The ecological role of bacteriocins in bacterial competition. Trends Microbiol. 1999;7:129–133. doi: 10.1016/s0966-842x(99)01459-6. [DOI] [PubMed] [Google Scholar]
- Tenover FC. Mechanisms of antimicrobial resistance in bacteria. Am J Med. 2006;119:S3–S10. doi: 10.1016/j.amjmed.2006.03.011. [DOI] [PubMed] [Google Scholar]
- Farris MH, Steinberg AD. Mitrecin A, an endolysin-like bacteriolytic enzyme from a newly isolated soil streptomycete. Lett Appl Microbiol. 2014;58:493–502. doi: 10.1111/lam.12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehart HP, Heath HE, Heath LS, Leblanc PA, Sloan GL. The Lysostaphin Endopeptidase Resistance Gene (epr) Specifies Modification of Peptidoglycan Cross Bridges in Staphylococcus simulans and Staphylococcus aureus. Appl Environ Microbiol. 1995;61:2811. doi: 10.1128/aem.61.7.2811-2811.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava KK, Siddique IH. Quantitative chemical composition of peptidoglycan of Listeria monocytogenes. Infect Immun. 1973;7:700–703. doi: 10.1128/iai.7.5.700-703.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heilmann HD. On the peptidoglycan of the cell walls of Pseudomonas aeruginosa. Eur J Biochem. 1972;31:456–463. doi: 10.1111/j.1432-1033.1972.tb02552.x. [DOI] [PubMed] [Google Scholar]
- Schumann P. In: Taxonomy of Prokaryotes. Rainey F, Oren A, editors. Vol. 38. Academic Press; 2011. [Google Scholar]
- Lachica RV, Genigeorgis C, Hoeprich PD. Metachromatic agar-diffusion methods for detecting staphylococcal nuclease activity. Appl Microbiol. 1971;21:585–587. doi: 10.1128/am.21.4.585-587.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Chen S, Recsei P. A dye release assay for determination of lysostaphin activity. Anal Biochem. 1988;171:141–144. doi: 10.1016/0003-2697(88)90134-0. [DOI] [PubMed] [Google Scholar]
- Osserman EF, Lawlor DP. Serum and urinary lysozyme (muramidase) in monocytic and monomyelocytic leukemia. J Exp Med. 1966;124:921–952. doi: 10.1084/jem.124.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini G, Carbonara AO, Heremans JF. Immunochemical quantitation of antigens by single radial immunodiffusion. Immunochemistry. 1965;2:235–254. doi: 10.1016/0019-2791(65)90004-2. [DOI] [PubMed] [Google Scholar]
- Ito Y, Yamada H, Imoto T. Colorimetric assay for lysozyme using Micrococcus luteus labeled with a blue dye, Remazol brilliant blue R, as a substrate. Chem Pharm Bull. (Tokyo) 1992;40:1523–1526. doi: 10.1248/cpb.40.1523. [DOI] [PubMed] [Google Scholar]
- Hardt M, Guo Y, Henderson G, Laine RA. Zymogram with Remazol brilliant blue-labeled Micrococcus lysodeikticus cells for the detection of lysozymes: example of a new lysozyme activity in Formosan termite defense secretions. Anal Biochem. 2003;312:73–76. doi: 10.1016/s0003-2697(02)00443-8. [DOI] [PubMed] [Google Scholar]
- Pettersson B, Eriksson KE. A standardized spectrophotometric assay of endoglycanase activities using dyed, amorphous polysaccharides. Anal Biochem. 2000;285:220–224. doi: 10.1006/abio.2000.4776. [DOI] [PubMed] [Google Scholar]
- Gomez Ramirez M, Rojas Avelizapa LI, Rojas Avelizapa NG, Cruz Camarillo R. Colloidal chitin stained with Remazol Brilliant Blue R, a useful substrate to select chitinolytic microorganisms and to evaluate chitinases. J Microbiol Methods. 2004;56:213–219. doi: 10.1016/j.mimet.2003.10.011. [DOI] [PubMed] [Google Scholar]


