Abstract
Circular peptides and proteins of great number and diversity have been discovered in bacteria, plants, and animals. Cyclotides—disulfide-knotted and head-to-tail cyclized plant peptides that exhibit various bioactivities—are by far the largest group of circular proteins. Since their first discovery over three decades ago, there has been a lot of progress in the elucidation of structural characteristics and applications of cyclotides as novel peptide drug grafting frameworks, but there is a lack of information about their native occurrence in various plant families. The “global cyclotide adventure” was initiated as a plant collection and analysis project to advance our understanding of the origin and distribution of cyclotides in flowering plants. Here, I will provide a chronological overview of the preparation of this project, including background information on plant taxonomy and morphology, summarize, and comment on the recent progress about the discovery of cyclotide-producing plants and will give an outlook on the future of cyclotide analysis and further discoveries to be made.
Keywords: bioactive peptides, Rubiaceae, Violaceae, Apocynaceae, disulfide, peptidomics
Introduction
The discovery of the first cyclotide, kalata B1, was based on its presence in a tea from the Rubiaceae species Oldenlandia affinis (R&S) DC. used in African indigenous medicine to accelerate childbirth.1 Today it is clear that many other cyclotides exist, but we lack information about their origin and distribution in plants. To understand the occurrence and evolution of cyclotides, we started the “global cyclotide adventure,” a journey dedicated to the collection and analysis of flowering plant species from all over the world for the occurrence of cyclotides.
Cyclotides are disulfide-rich plant peptides2,3 of about 30 amino acids in size and the unique structural features of a head-to-tail cyclized backbone and a knotted arrangement of three-disulfide bonds, referred to as a cyclic cystine knot (CCK) motif.2 The compact CCK motif makes cyclotides exceptionally resistant to thermal, chemical, or enzymatic degradation.3,4 In contrast to secondary plant metabolites, cyclotides are true gene products and their biosynthesis from larger precursor proteins involves enzymatic processing5,6 and protein folding events.7,8 Cyclotides exhibit various biological activities, including uterotonic,9 hemolytic,10 anti-neurotensin,11 anti-HIV,12 cytotoxic,13 antibacterial,14 anti-fouling,15 and nematocidal16,17 activities. Their natural function appears to be as plant defense molecules based on their insecticidal properties.18–20
Before the “global cyclotide adventure” only six species of the coffee family (Rubiaceae) and several plants of the violet family (Violaceae) have been known to contain cyclotides. Additionally, cyclotide-like molecules have been found in the cucurbit family (Cucurbitaceae21,22), and cyclotide-like gene sequences have been discovered in representatives of the grass family (Poaceae), including important cereal crops, such as wheat, maize, and rice.23 Today, it is evident that cyclotides are much more numerous than earlier anticipated and it has been suggested that they may surpass the well-known plant defensins in number and diversity.24,25 As is clear from other articles in this special issue, cyclotides are just one of a number of families of cyclic peptides and proteins that have been discovered in bacteria, plants, and animals26 over the last 15 years. Very recently it has been estimated that there are at least 50,000 novel cyclotides to be discovered in Rubiaceae27 and taken together with the occurrence of cyclotides in other plant families, this makes them by far the largest family of cyclic proteins. Although cyclotides appear to occur in every Violaceae species analyzed so far,24,28,29 information about their occurrence, distribution, and evolution in the Rubiaceae family and other plant families is still very limited.
In this article, I will give a chronological overview of the preparation and outcome of the global cyclotide adventure. The first part comprises an introduction to Rubiaceae taxonomy and morphology, including a field collection guide for (Bio-) Chemists. Then, I will continue with a description of the recent discovery of cyclotides in Rubiaceae and other flowering plant families, and finally I will comment on the origin and evolution of cyclotides and will give an outlook on the future discovery of cyclotides.
Field Trip Preparation—“A (Bio-) Chemists’ Guide to Botany”
Since there was not much known about the occurrence of cyclotides at the time, there was need for the systematic collection of flowering plants and their analysis for the presence or absence of cyclotides. The global cyclotide adventure27 comprised the collection of various Rubiaceae species to cover many genera and species in the family, from different types of plants (trees, shrubs, and herbs) and from different regions (climatic and geographic) and their analyses using state-of-the-art analytical chemistry instrumentation. We analyzed over 200 species of Rubiaceae from field collections, living glasshouse collections, and herbarium specimens and reported 22 potential cyclotide-producing Rubiaceae species.27 The discoveries made from the Rubiaceae screen triggered further collection, screening, and discovery of cyclotides in another flowering plant family, i.e., we analyzed >140 plant species related to Rubiaceae and Violaceae families and found 12 potential cyclotide-producing species in the dogbane family (Apocynaceae). Of the 22 Rubiaceae species shown to contain putative cyclotide masses, two species have been confirmed to contain cyclotides by direct peptide sequencing and verification of their circular peptide backbone. Cyclotide sequencing studies for the Apocynaceae species in question are currently under way. Although it is likely that all 34 species do contain cyclotides, it is important that they are still regarded as putative hits until actual sequence data, or additional evidence has been provided.
Predeparture preparation is the key to success for any botanical field trip, which includes thorough planning of field site visits (transportation, equipment, etc.) and local permit organization (field collections and plant material export). In particular, the latter can be time consuming and frustrating as many countries have a complex way of dealing with botanical field research regulations. Apart from the logistical planning, it is necessary to prepare the field trip scientifically, which includes the survey of botanical literature and local resources about the existing flora. Nevertheless, guidance from botanical experts to support the field collections and/or identification of the collected plant material is as important. Furthermore, it is useful to acquire knowledge about plant morphology and taxonomy of the families of interest. The starting point for this global adventure to discover cyclotides was the Rubiaceae family. The reasons for choosing this plant family was not only because cyclotides have been initially isolated from a Rubiaceae plant, but also due to the limited knowledge about the distribution of cyclotides in Rubiaceae, the family’s size, its global distribution, and its central placement within the order of Gentianales.
Rubiaceae: Overview, Taxonomy, and Morphology
Rubiaceae is the fourth largest angiosperm family and one of the largest and most diverse families in the plant kingdom, with approximately 13,000 species in 650 genera.30,31 Rubiaceae plants are widely distributed and have adapted to virtually every habitat, ranging from arid and desert environments to humid rainforests and from cold subarctic climates to hot tropical climatic regions.30 In temperate regions, Rubiaceae are represented by only a few herbaceous genera, but the bulk of the family is tropical and predominantly woody. Coffee, one of the most economically important plants worldwide, belongs to the Rubiaceae family (hence the synonym “coffee family”). Additionally, a wide range of medicinal, timber, ornamental, psychoactive, and aphrodisiac properties of Rubiaceae plants have been reported.27,30
Rubiaceae belong to the flowering plants or angiosperms (= Magnoliophyta). Within that division they form part of the Dicotyledons (= Magnoliopsida), a formerly recognized taxonomic group of flowering plants whose seeds typically contain two embryonic leaves (= cotyledons). Currently dicotyledonous plants are subdivided into several orders according to the angiosperm phylogeny system.32 This classification scheme characterizes flowering plants, aside of a couple of basal angiosperm lineages, into the following major groups: Monocotyledons and Eudicots (comprising the Rosids and Asterids) or the superorders Lilianeae, Rosanae, and Asteranae (according to the latest APGIII classification scheme), respectively, each of which are subdivided into several orders. Rubiaceae are placed within the order Gentianales of the Asterids, together with the families of Apocynaceae, Gelsemiaceae, Gentianaceae, and Loganiaceae.
Partly due to the large number of species and their global distribution, the taxonomical classification of Rubiaceae plants is constantly in a state of flux.30 In recent years, the division of the coffee family into either two, Cinchonoideae and Rubioideae,33 or three subfamilies, Cinchonoideae, Ixoroideae, and Rubioideae,34–38 has been suggested, based on molecular phylogenetics. This fluctuating situation of Rubiaceae classification continues at the genus and species level, especially as more phylogenetic data sets become available.
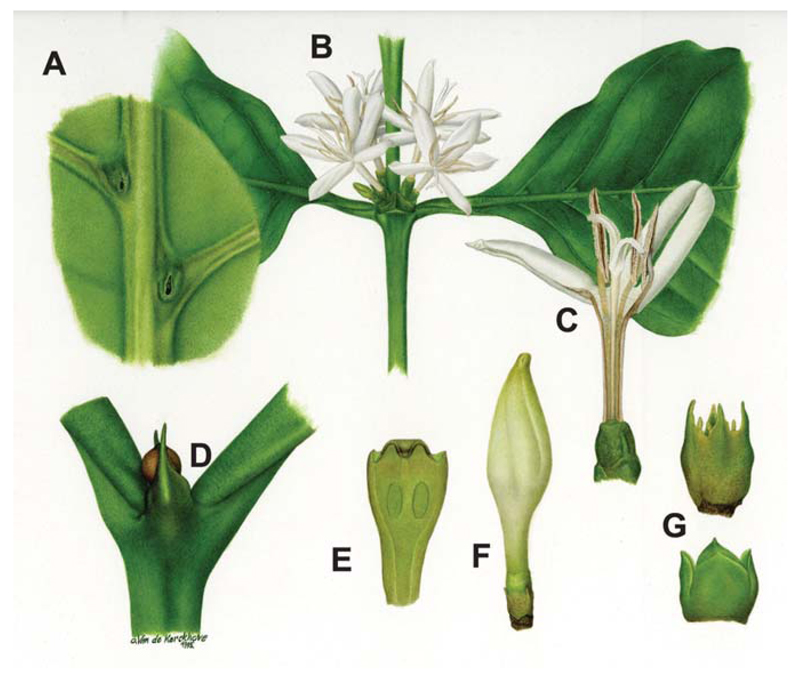
Most Rubiaceae are shrubs or small to large trees. They are easily recognized at family level by decussate and entire leaves (Table I for botanical glossary), presence of stipules, 4- or 5-merous actinomorphic flowers, and inferior ovary (Figure 1).
Table 1. Glossary of Botanical Terms.
| Botanical Term | Descriptiona |
|---|---|
| Decussate | Alternating along the stem in pairs at right angles |
| Entire leaves | Without toothing or division |
| Stipules | Appendage at the base of a petiole (leafstalk), often appearing in pairs, one on each side |
| Actinomorphic flowers | Flowers that can be divided into symmetrical halves along any diameter |
| Zygomorphic | Division only possible by one plane of symmetry |
| Inferior ovary | The flower parts grow from above the ovary |
| Bisexual | Flowers having both stamens and pistils (as opposite to unisexual) |
| Corolla | The usually shiny part of a flower that constitutes the inner whorl of the perianth |
| Inflorescences | The flowering part of a plant arranged in a special mode |
| Heterostyly | Species in which flowers are similar except that the stigmas and anthers are held at different levels relative to each other, because style length differs between plants |
| Axile placentation | Arrangement of placentas on the central axis of a septate ovary |
| Dicotyledon | Flowering plant with two cotyledons (=first leaves to appear after germination) |
| Fruit | Mature ovary of a seed plant |
| Berry | Indehiscent fleshy fruit with fleshy pericarp surrounding the seeds |
| Capsule | Dry fruit composed of more than one carpel (= simple pistil) that splits partially open at maturity to discharge seeds or spores |
| Drupe | A fleshy or pulpy fruit with the inner portion of the pericarp (= fruit wall) hard or stony |
| Schizocarp | Fruit that splits between carpels into one-seeded portions |
| Endosperm | The nutritive tissue in a seed of a flowering plant that surrounds the embryo |
Adapted and modified from the online glossary of botanical terms; available at: http://glossary.gardenweb.com.
Figure 1.
Morphological features of Rubiaceae. Typical morphological Rubiaceae features, as shown with an example of the most famous family member, namely Coffea arabica L., which include (A) underside of leaf with domatia, (B) opposite leaf pair with axillary placed flowers, (C) dissected flower showing inside of corolla tube, anthers and style, (D) tip of stem showing stipules with secretion, (E) longitudinal section of ovary, (F) floral bud, and (G) bracts (Reproduced with permission from the National Botanical Garden Belgium; Artist: O. van de Kerckhove, 1995).
Besides these characteristics, the following list of features is used to classify Rubiaceae:
Floral features: Flowers are actinomorphic (rarely zygomorphic) and bisexual (rarely unisexual); corolla connate (i.e., sympetalous); inflorescences are various; flowers often show heterostyly and their ovaries are inferior and often with axile placentation
Fruit and seed features: Dicotyledon; fruits are berries, capsules, drupe, or schizocarp; seeds are single or many and with or without endosperm
Vegetative features: Occurrence as herbs, shrubs, trees, lianas or epiphytes; leaves are simple, entire (rarely lobed), opposite, or whorled; stipules are often fused at the node and they may be leaf-like
Rubiaceae Field Guide for (Bio-) Chemists
This guide describes five characteristics of Rubiaceae, which are useful during field collections to identify plants to the family level and in a few cases to the genus level. With the help of adequate literature, it may be possible to identify plants to the species level. Please note that this guide should be used as a first “hands-on guide” only and not as a reliable tool for the correct identification of Rubiaceae plants to the species level. However, if a plant meets all the five features described below (Figures 1 and 2 for illustration), the chance of it belonging to the Rubiaceae family is very high!
Figure 2.
Illustration of cyclotide-containing Rubiaceae species. (A) Palicourea rigida Kunth is a small tree and a typical plant of the “Cerrado,” characterized by its orange inflorescences. (B) Palicourea coriacea (Cham.) K. Schum. is a shrub and another typical inhabitant of the Cerrado, characterized by its bright yellow inflorescences and very dry, but firm leaves. (C) Psychotria prunifolia (Kunth) Steyerm. is a small-sized tree and has typical five petal containing white flowers and small, pointed stipules (bi-fid). (D) Psychotria suterella Müll. Arg. is a tree with white, five-petalled flowers, typical whole, structured leaves, and blue fruits. (E) Psychotria brachyceras Müll. Arg. is a medium-sized tree. White five-petalled flowers, small, whole structured leaves, and pointed stipules are typical for this plant. (F) Geophila repens (L.) I.M. Johnst. is a ground growing herb with characteristic white, five-petalled flowers. (G) Chassalia discolour K. Schum. subsp. discolor is a medium-sized tree. Its inflorescences have a characteristic red/purple color. (H) Kadua centranthoides Hook. & Arn. growing on volcanic rock that results in the red color of the plant. Triangular stipules and actinomorphic flowers are typical Rubiaceae features. (I) Psychotria punctata Vatke with bright red fruits.
Red color: The word Rubiaceae is thought to originate etymologically from the Latin word “rubeus,” meaning red. As an example, the most famous member of the Rubiaceae family, i.e., coffee (Coffea arabica) has bright red fruits when ripe. However, the color red as a feature of Rubiaceae seems rather anecdotal, as there are many Rubiaceae plants that cannot be characterized by their red color and there are many examples of non-Rubiaceae plants which also have red-colored plant parts. In the context of a field guide to identify Rubiaceae, the color red is a useful indicator. When walking through a forest, red colored fruits, flowers, or leaves can easily be recognized and it is worth investigating that particular plant and by using secondary indicators (such as leaf and fruit morphology) to determine whether the species belongs to the Rubiaceae family or not.
Opposite leaves: Rubiaceae leaves are opposite (i.e., two leaves directly opposite—180°—attached at the same node) or in rare cases whorled (i.e., three or more leaves attached at the same node). This feature is easily recognized when seeking Rubiaceae in the field, although other plant families have opposite leaves as well.
Stipules: Stipules can often be recognized. They are an outgrowth of the lower zone of a young leaf and part of the leaf base. Sometimes they can be very small and the use of a pocket magnifying glass is necessary to recognize them.
Leaf structure: The leaves are entire or whole (i.e., they have a smooth margin) and they have abaxial veins (i.e., the veins exit the axis of the leaf in alternate fashion and bend toward the apex of the leave).
Flowers/fruits: Flowers and fruits are mostly actinomorphic, which means that the flowers and reproductive organs can be divided into symmetrical planes.
Field Collection Sites
A number of collection sites in several countries were visited. In some locations, we were able to obtain permits to collect and export plant material; however, collection was not possible in other locations.
Locations in Tanzania (Lushoto: Usambara Mountains), in Brazil (Goiás: central Brazilian plateau, “Cerrado”; Rio Grande do Sul: Atlantic rainforest, “Mata Atlântica”) and in Venezuela (Mérida: great southern American Andes mountain range) were visited and Rubiaceae species for future collections were surveyed. On Hawai’i, the field trip went to the islands of O’ahu, Kaua’i, and Hawai’i (The Big Island). With the support of local botanists and the National Tropical Botanical Garden in Kalaheo it was possible to gather many Rubiaceae plants, in particular species from the tribe Spermacoceae. Besides the field collections, a network of botanical gardens that provided plant material for cyclotide analysis from living glass house collections or Herbarium specimen was established. For example, plant material and expertise was obtained from gardens in Australia, New Zealand, the Missouri Botanical Garden and the New York Botanical Garden (both USA), but above all we have established a great collaboration with the National Botanic Garden of Belgium. A complete list of collection sites can be found in Gruber et al.27 Figure 2 illustrates the variety of collected and potential cyclotide-containing Rubiaceae species.
Distribution of Cyclotides Within the Rubiaceae
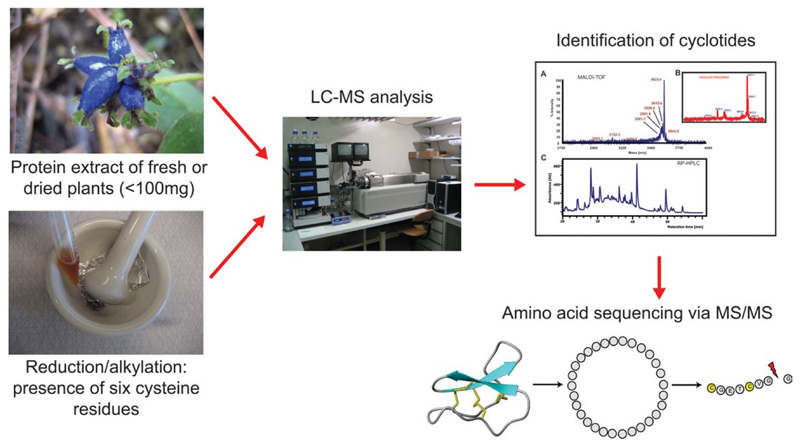
To analyze the obtained plant species, a robust and reliable screening procedure was developed to test each species for the occurrence of cyclotides (Figure 3 and Gruber et al.27). The plant peptide extracts were analyzed using HPLC and mass spectrometry (MS), before and after chemical modification of cysteine residues to characterize cyclotide properties, such as hydrophobicity, molecular weight, and number of cysteine residues. The robustness and reliability of the screening procedure was validated and exemplarily we have sequenced individual cyclotides.
Figure 3.
Cyclotide screening flowchart. Protein extracts of fresh or dried plant material were prepurified using C18 chromatography. The extracts were analyzed by LC-MS before and after reduction and alkylation of cysteine residues. This chemical modification of the extracts and all peptides in the extract was used to characterize the number of cysteine residues per peptide. Cyclotides were identified by their typical hydrophobicity, their molecular weight, and the conserved number of cysteine residues. Finally, individual cyclotides were sequenced by reduction of the disulfide bonds and enzymatic cleavage of the stable peptide backbone to make them accessible for dissociation and sequencing via tandem mass spectrometry.
By analyzing a representative number of species and by using the latest information on Rubiaceae phylogeny and taxonomy, the occurrence and distribution of cyclotides in Rubiaceae plants was characterized.27 We tested >200 Rubiaceae species belonging to the subfamilies Cinchonoideae/Ixoroideae and the subfamily Rubioideae. We identified 22 potential cyclotide-producing species, all belonging to the subfamily Rubioideae. Within the Rubioideae we analyzed plants from all three clades, namely the basal grade, the woody clade, and the herbaceous clade. The results showed that cyclotides occur in four tribes of the subfamily Rubioideae. More broadly, cyclotides occur in the basal grade, the tribe Lasiantheae, and the two main clades, namely the Psychotrieae/Palicoureeae complex and the tribe Spermacoceae sensu lato. We examined the taxonomical relationships between the positive species and compared them with other cyclotide-containing plant families. Based on the distribution of cyclotides, we estimated the number of Rubiaceae species that are likely to contain cyclotides and concluded that the number of total existing cyclotides within the plant kingdom must be far greater than earlier anticipated.
Cyclotide Discovery in Other Flowering Plant Families
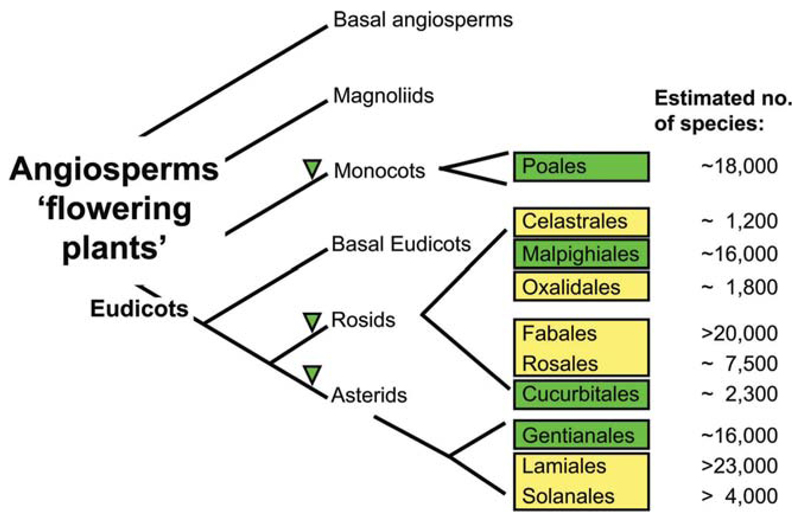
Detection of cyclotides in the basal grade led to the conclusion that cyclotide occurrence had its origin early in the evolution of the Rubioideae and hence these findings indicated that other families within the order Gentianales are likely to contain cyclotides, based on the evolution and relationships of the families within the order Gentianales.39,40 Hence, we analyzed >60 species from four plant families, Apocynaceae, Gentianaceae, Loganiaceae, and Potaliaceae and found 12 potential cyclotide containing species within the Apocynaceae s.l. (including Asclepiadaceae). Figure 4 summarizes the presently known and candidate cyclotide containing plant families.
Figure 4.
Presently known cyclotide-containing families within the flowering plants. The angiosperm phylogeny tree has been adapted and modified from the Angiosperm Phylogeny Group (Adapted and modified from Ref. 32, with permission from Academic Press). It shows the relationship of the different flowering plant classes, which are subdivided into subclasses and orders (some representative orders are shown and their number of estimated species is indicated). All cyclotide or cyclotide-like containing families are indicated in green. The family Rubiaceae belongs to the order of Gentianales (euasterids I), Violaceae to the order of Malphigiales (eurosids I), Cucurbitaceae to the order of Cucurbitales (eurosids I), and Poaceae, a monocotyledon plant, belongs to the order of Poales (commelinids). Other plant orders (e.g., Celestrales, Oxalidales, Fabales, Rosales, Lamiales, and Solanales) that are close to the respective positively-tested cyclotide containing orders are marked in yellow and could be potential candidate plant orders to continue the search for cyclotides. The number of estimated species in each order is indicated.
Evolution of Cyclotides
Cyclotides are much more abundant and diverse in the Gentianales (e.g., coffee and dogbane families)27 than earlier anticipated. Taking into account the occurrence of cyclotides in the violet family24,28,29 and cyclotide-like molecules and gene sequences in the cucurbit19,22 and grass family,23 respectively, it is evident that the number of existing and unique cyclotides may be by far greater than 50,000. Hence, the family of cyclotides could form one of the largest distinct protein families in the plant kingdom and a very large group of naturally-occurring plant compounds.
While trying to understand their number and diversity, it is tempting to speculate about the origin and evolution of cyclotides in plants. In attempting to answer this question it is important to point out the three commonly known evolutionary mechanisms of parallel occurrence of traits: (i) multiple independent gains, (ii) lateral (= horizontal) gene transfer, and (iii) descendent from a common ancestor with losses. Considering the presently known distribution of cyclotides in Rubiaceae, the most favorable explanation for their occurrence is to assume at least four independent origins within this family. This is supported by the distribution of cyclotide and cyclotide-like containing families within the other flowering plants. The four families known so far, (i) Rubiaceae, (ii) Violaceae and Cucurbitaceae, and (iii) Poaceae, belong to Asterids, Rosids and Monocots (see Figure 4), respectively. There is no direct phylogenetic link between these families, which is another indicator that cyclotides likely have originated independently more than once, and in distantly related families. Lateral gene transfer is another possibility for the evolution of cyclotides, although it is very unlikely since Rubiaceae and Violaceae have different cyclotide precursor gene structures.27 The difference in cyclotide gene structures does furthermore not favor the existence of a common cyclotide ancestor between at least these two plant families, which argues against divergent evolution. Divergent evolution would require loss of cyclotide genes from a significant portion of the Rubiaceae family, since cyclotides have been found in every Violaceae species screened but only in ~10% of Rubiaceae species and this seems unlikely. Based on the current knowledge of cyclotide occurrence and distribution throughout flowering plants and by applying a process of elimination of possible evolutionary mechanisms, it is likely that cyclotides evolved as multiple independent gains of function, i.e., by convergent evolution.
Conclusion and Outlook
In summary, during the “global cyclotide adventure” >340 flowering plant species, mainly belonging to the coffee family, were analyzed and the presence of cyclotides was confirmed in two species from Rubiaceae. An additional 20 species from Rubiaceae and 12 species from Apocynaceae contained peptides of mass and cysteine content consistent with cyclotides. Based on the distribution of cyclotides in those families, the number of possible cyclotides to be discovered in the future was estimated to be very large (>50,000) and the occurrence of cyclotides in the presently known families (Figure 4) led to the first insights into the mechanistic basis of cyclotide evolution.
However, it is too early to provide a reliable, evidence-based evolutionary mechanism of cyclotides in flowering plants. Taking together all previous cyclotide screening efforts, ~500 different species from a total of >250,000 existing flowering plant species have been analyzed to date. This makes it a mere 0.2% analyzed species and this small fraction forms the basis of the current evolutionary hypothesis for cyclotides. Hence, it has to be clear that a lot more plant species need to be tested before one can make reliable conclusions on the evolution and origin of cyclotides within the plant kingdom. A starting point for future analysis would be plant families and orders that are in close phylogenetic proximity to the already positively identified ones, e.g., Celestrales, Oxalidales, Fabales, Rosales, Lamiales, and Solanales (Figure 4). Considering the number of species in each of these orders, there is great potential for future cyclotide discoveries.
Another point to be mentioned here is the importance of the analytical procedure. Until now, the decision whether a plant contains cyclotides or not was based on MS analysis of crude and chemically treated (reduction/cysteine alkylation) extracts, followed by laborious purification, peptide sequencing using MS/MS and amino acid analysis of target cyclotides.27 Furthermore, a number of control experiments had to be performed to guarantee a negligible incidence of falsepositive and false-negative testing results. To manage cyclotide analysis for the number of plants that need to be tested for confident predictions on cyclotide evolution, more rapid screening techniques have to be developed. Recently, a state-of-the-art LC-MS/MS method was applied to the rapid screening and de novo sequencing of conotoxins (a class of peptide toxins from marine cone snails) from crude peptide extracts.41 A similar methodology could be investigated for the screening of cyclotides, although in this instance it may be more difficult to develop, since the compact structure and the circular peptide backbone of cyclotides is very difficult to be dissociated in MS/MS without prior enzymatic and/or chemical treatment. If successful, this will lead to a faster analysis per species and to increased sensitivity, which both will lead to a better and faster screening technique for future cyclotide analysis of unknown plant species. A first attempt for such a screening technique has been described in an article by Colgrave et al. in this special issue42 and together with updated tools of CyBase43 (a freely accessible database dedicated to the study of circular proteins), it is a step in the right direction for future cyclotide discovery studies.
Cyclotides are interesting targets for pharmaceutical and agrochemical applications due to their unique structural framework, range of bioactivities, and sequence diversity.44,45 Based on their predicted number, which makes them one of the largest protein families within the plant kingdom, they constitute an immense library of natural peptides46 and may therefore be accessible for drug discovery efforts.47 A more efficient and rapid analysis of the distribution and occurrence of cyclotides will not only lead to a better understanding of their diversity and evolution, but will ultimately advance their applications as pharmaceutical scaffolds and agrochemicals and the role of cyclotides as natural compounds in drug discovery.
Acknowledgments
The author thanks all persons who have contributed to study the distribution and occurrence of cyclotides in plants, in particular, David Craik and coworkers (Institute for Molecular Bioscience, Australia), Ulf Göransson and coworkers (Uppsala University, Sweden), Steven Dessein and Elmar Robbrecht (National Botanic Garden of Belgium), Piero Delprete (Institut de Recherche pour le Developpement, French Guiana), and many others who are either coauthors on earlier publications (in particular, Gruber et al.27) or have been acknowledged previously. Thanks to Christoph Dobeš (University of Vienna, Austria) and David Craik for reading the manuscript and making useful comments. The National Botanic Garden of Belgium is holder of the copyright for Figure 1 and granted permission to reprint.
Contract grant sponsors: The Swedish Institute, the Australian Research Council (ARC) and The University of Queensland and the Austrian Science Fund (FWF).
Footnotes
This article was originally published online as an accepted preprint. The “Published Online” date corresponds to the preprint version. You can request a copy of the preprint by emailing the Biopolymers editorial office at biopolymers@wiley.com
References
- 1.Gran L. Acta Pharmacol Toxicol (Copenh) 1973;33:400–408. doi: 10.1111/j.1600-0773.1973.tb01541.x. [DOI] [PubMed] [Google Scholar]
- 2.Craik DJ, Daly NL, Bond T, Waine C. J Mol Biol. 1999;294:1327–1336. doi: 10.1006/jmbi.1999.3383. [DOI] [PubMed] [Google Scholar]
- 3.Craik DJ, Daly NL, Mulvenna J, Plan MR, Trabi M. Curr Protein Pept Sci. 2004;5:297–315. doi: 10.2174/1389203043379512. [DOI] [PubMed] [Google Scholar]
- 4.Colgrave ML, Craik DJ. Biochemistry. 2004;43:5965–5975. doi: 10.1021/bi049711q. [DOI] [PubMed] [Google Scholar]
- 5.Gillon AD, Saska I, Jennings CV, Guarino RF, Craik DJ, Anderson MA. Plant J. 2008;53:505–515. doi: 10.1111/j.1365-313X.2007.03357.x. [DOI] [PubMed] [Google Scholar]
- 6.Saska I, Gillon AD, Hatsugai N, Dietzgen RG, Hara-Nishimura I, Anderson MA, Craik DJ. J Biol Chem. 2007;282:29721–29728. doi: 10.1074/jbc.M705185200. [DOI] [PubMed] [Google Scholar]
- 7.Gruber CW, Cemazar M, Clark RJ, Horibe T, Renda RF, Anderson MA, Craik DJ. J Biol Chem. 2007;282:20435–20446. doi: 10.1074/jbc.M700018200. [DOI] [PubMed] [Google Scholar]
- 8.Gruber CW, Cemazar M, Heras B, Martin JL, Craik DJ. Trends Biochem Sci. 2006;31:455–464. doi: 10.1016/j.tibs.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 9.Gran L, Sandberg F, Sletten K. J Ethnopharmacol. 2000;70:197–203. doi: 10.1016/s0378-8741(99)00175-0. [DOI] [PubMed] [Google Scholar]
- 10.Schöpke T, Hasan Agha MI, Kraft R, Otto A, Hiller K. Sci Pharm. 1993;61:145–153. [Google Scholar]
- 11.Witherup KM, Bogusky MJ, Anderson PS, Ramjit H, Ransom RW, Wood T, Sardana M. J Nat Prod. 1994;57:1619–1625. doi: 10.1021/np50114a002. [DOI] [PubMed] [Google Scholar]
- 12.Gustafson KR, McKee TC, Bokesch HR. Curr Protein Pept Sci. 2004;5:331–340. doi: 10.2174/1389203043379468. [DOI] [PubMed] [Google Scholar]
- 13.Lindholm P, Goransson U, Johansson S, Claeson P, Gulbo J, Larsson R, Bohlin L, Backlund A. Mol Cancer Ther. 2002;1:365–369. [PubMed] [Google Scholar]
- 14.Tam JP, Lu YA, Yang JL, Chiu KW. Proc Natl Acad Sci USA. 1999;96:8913–8918. doi: 10.1073/pnas.96.16.8913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Göransson U, Sjogren M, Svangard E, Claeson P, Bohlin L. J Nat Prod. 2004;67:1287–1290. doi: 10.1021/np0499719. [DOI] [PubMed] [Google Scholar]
- 16.Colgrave ML, Kotze AC, Huang YH, O’Grady J, Simonsen SM, Craik DJ. Biochemistry. 2008;47:5581–5589. doi: 10.1021/bi800223y. [DOI] [PubMed] [Google Scholar]
- 17.Colgrave ML, Kotze AC, Kopp S, McCarthy JS, Coleman GT, Craik DJ. Acta Trop. 2009;109:163–166. doi: 10.1016/j.actatropica.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 18.Barbeta BL, Marshall AT, Gillon AD, Craik DJ, Anderson MA. Proc Natl Acad Sci USA. 2008;105:1221–1225. doi: 10.1073/pnas.0710338104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gruber CW, Cemazar M, Anderson MA, Craik DJ. Toxicon. 2007;49:561–575. doi: 10.1016/j.toxicon.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 20.Jennings C, West J, Waine C, Craik D, Anderson M. Proc Natl Acad Sci USA. 2001;98:10614–10619. doi: 10.1073/pnas.191366898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiche L, Heitz A, Gelly JC, Gracy J, Chau PT, Ha PT, Hernandez JF, Le-Nguyen D. Curr Protein Pept Sci. 2004;5:341–349. doi: 10.2174/1389203043379477. [DOI] [PubMed] [Google Scholar]
- 22.Felizmenio-Quimio ME, Daly NL, Craik DJ. J Biol Chem. 2001;276:22875–22882. doi: 10.1074/jbc.M101666200. [DOI] [PubMed] [Google Scholar]
- 23.Mulvenna JP, Mylne JS, Bharathi R, Burton RA, Shirley NJ, Fincher GB, Anderson MA, Craik DJ. Plant Cell. 2006;18:2134–2144. doi: 10.1105/tpc.106.042812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simonsen SM, Sando L, Ireland DC, Colgrave ML, Bharathi R, Goransson U, Craik DJ. Plant Cell. 2005;17:3176–3189. doi: 10.1105/tpc.105.034678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trabi M, Svangard E, Herrmann A, Goransson U, Claeson P, Craik DJ, Bohlin L. J Nat Prod. 2004;67:806–810. doi: 10.1021/np034068e. [DOI] [PubMed] [Google Scholar]
- 26.Craik DJ. Science. 2006;311:1563–1564. doi: 10.1126/science.1125248. [DOI] [PubMed] [Google Scholar]
- 27.Gruber CW, Elliott AG, Ireland DC, Delprete PG, Dessein S, Goransson U, Trabi M, Wang CK, Kinghorn AB, Robbrecht E, Craik DJ. Plant Cell. 2008;20:2471–2483. doi: 10.1105/tpc.108.062331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burman R, Gruber CW, Rizzardi K, Herrmann A, Craik DJ, Gupta MP, Goransson U. Phytochemistry. 2010;71:13–20. doi: 10.1016/j.phytochem.2009.09.023. [DOI] [PubMed] [Google Scholar]
- 29.Ireland DC, Colgrave ML, Craik DJ. Biochem J. 2006;400:1–12. doi: 10.1042/BJ20060627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delprete PG. In: Flowering Plant Families of the American Tropics. Smith NP, Heald SV, Henderson A, Mori SA, Stevenson DW, editors. Princeton University Press/New York Botanical Garden Press; New York: 2004. pp. 328–333. [Google Scholar]
- 31.Govaerts R, Frodin DG, Ruhsam M, Bridson DM, Davis AP. Scripta Bot Belg. 2006;40:35. [Google Scholar]
- 32.Chase MW, Reveal JL. Bot J Linn Soc. 2009;161:122–127. [Google Scholar]
- 33.Robbrecht E, Manen JF. Syst Geogr Plants. 2006;76:85–146. [Google Scholar]
- 34.Bremer B. Opera Bot Belg. 1996;7:33–50. [Google Scholar]
- 35.Bremer B, Andreasen K, Olsson D. Ann Missouri Bot Gard. 1995;82:383–397. [Google Scholar]
- 36.Bremer B, Jansen RK. Amer J Bot. 1991;78:198–213. [Google Scholar]
- 37.Bremer B, Jansen RK, Oxelman B, Backlund M, Lantz H, Kim K-J. Syst Biol. 1999;48:413–435. doi: 10.1080/106351599260085. [DOI] [PubMed] [Google Scholar]
- 38.Rova HE, Delprete PG, Andersson L, Albert VA. Amer J Bot. 2002;89:145–159. doi: 10.3732/ajb.89.1.145. [DOI] [PubMed] [Google Scholar]
- 39.Backlund M, Oxelman B, Bremer B. Amer J Bot. 2000;87:1029–1043. [PubMed] [Google Scholar]
- 40.Struwe L, Albert VA, Bremer B. Cladistics. 1994;10:175–205. [Google Scholar]
- 41.Ueberheide BM, Fenyo D, Alewood PF, Chait BT. Proc Natl Acad Sci USA. 2009;106:6910–6915. doi: 10.1073/pnas.0900745106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Colgrave ML, Poth A, Kaas Q, Craik DJ. Biopolymers. 2010;94:592–601. doi: 10.1002/bip.21400. [DOI] [PubMed] [Google Scholar]
- 43.Kaas Q, Craik DJ. Biopolymers. 2010;94:584–591. doi: 10.1002/bip.21424. [DOI] [PubMed] [Google Scholar]
- 44.Craik DJ, Cemazar M, Daly NL. Curr Opin Drug Discov Devel. 2007;10:176–184. [PubMed] [Google Scholar]
- 45.Craik DJ, Clark RJ, Daly NL. Expert Opin Investig Drugs. 2007;16:595–604. doi: 10.1517/13543784.16.5.595. [DOI] [PubMed] [Google Scholar]
- 46.Wang CK, Kaas Q, Chiche L, Craik DJ. Nucleic Acids Res. 2008;36:D206–D210. doi: 10.1093/nar/gkm953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gruber CW, Muttenthaler M, Freissmuth M. Curr Pharm Des. 2010 Aug 5; doi: 10.2174/138161210793292474. [BSP/CPD/E-Pub/00142] [DOI] [PMC free article] [PubMed] [Google Scholar]