Abstract
Nucleocytoplasmic protein O-GlcNAcylation is essential for embryogenesis. The dynamics of the O-GlcNAc proteome and the underlying mechanistic biology linking it to embryonic development is not understood. Harnessing the unusual properties of an O-GlcNAcase mutant that binds O-GlcNAc sites with nanomolar affinity, we uncover changes in O-GlcNAc proteins as a function of Drosophila development.
Introduction
Protein O-GlcNAcylation is a dynamic, nucleocytoplasmic post-translational modification (PTM) involving the transfer of N-acetylglucosamine (GlcNAc) to protein serine/threonine residues1. O-GlcNAc transfer is catalyzed by O-GlcNAc transferase (OGT) and its removal is catalyzed by O-GlcNAcase (OGA)1. OGT and OGA are both encoded by single genes in most animals, building an O-GlcNAc proteome of over a 1000 proteins involved in metabolic pathways, cell signalling, transcriptional and translational control, epigenetic modification, intracellular trafficking and as components of nuclear pores2. Disruption of O-GlcNAc signalling is associated with pathogenesis, including diabetes, cancer and nervous system disorders3–5. Utilising several genetic approaches, it has been demonstrated that O-GlcNAcylation is essential for embryonic development in mouse, Drosophila and zebrafish6–8, although the underpinning mechanistic biology remains to be explored. Drosophila contains single copies of both OGT (also known as super sex combs, sxc) and OGA. sxc mutants do not survive beyond the pupal stages, whereas a mutant lacking OGA protein is viable7, 9. While sxc is known to be essential for Drosophila development, changes in the O-GlcNAc proteome as a function of embryonic development have not yet been characterised.
Although the O-GlcNAc PTM was discovered three decades ago, its detection and quantification remains challenging. One of the pan-O-GlcNAc antibodies, CTD110.6, is a mouse monoclonal IgM raised against an O-GlcNAcylated peptide derived from the C-terminal domain of RNA polymerase II10. However, CTD110.6 has recently been shown to cross-react with N-glycosylated proteins11, 12, N-acetylglucosamine in the endoplasmic reticulum13 and extracellular O-GlcNAc proteins14, 15. The other commercially available O-GlcNAc antibody, RL2, shows selectivity towards glycosylated nucleoporins16. Other methods to detect nucleocytoplasmic O-GlcNAc include labeling with (UDP)-[3H]galactose using galactosyltransferase17, lectins18, chemical labelling with modified galactose and recombinant galactosyltransferase19–21 and 35S labeling using GlcNAc-specific sulfotransferases22. These techniques recognise only a subset of the O-GlcNAc proteome and/or are not compatible with more complex biological samples.
The bacterium Clostridium perfringens possesses an apparent OGA ortholog (CpOGA) of unknown function, but with a remarkable ability to strip entire O-GlcNAc proteomes of O-GlcNAc23, 24. Interestingly when a critical catalytic residue (Asp298, the catalytic acid protonating the glycosidic bond) is mutated to Asn (CpOGAD298N), catalytic activity is lost, yet substrate binding is retained23. This has been exploited to determine crystal structures of Ser-O-GlcNAc peptides in complex with CpOGA25. Here, we investigate the hypothesis that CpOGAD298N can also specifically bind to O-GlcNAcylated proteins and thus be used in a Far Western approach (Fig. 1a) to reveal changes in the O-GlcNAc proteome as a function of Drosophila embryonic development.
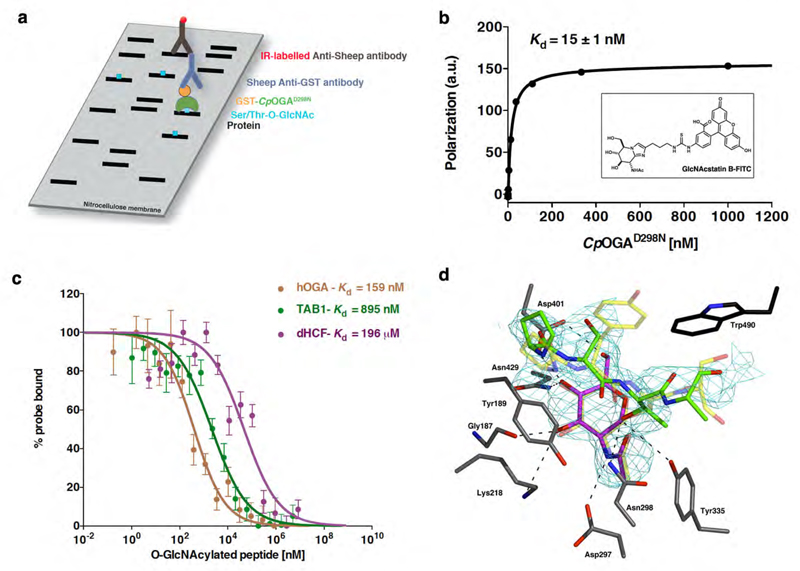
Figure 1. CpOGAD298N binds to O-GlcNAcylated peptides.
a) Cartoon outlining the Far Western approach using N-terminally GST-tagged ( ) CpOGAD298N (
) CpOGAD298N ( ). O-GlcNAc (
). O-GlcNAc ( ) on proteins (
) on proteins ( ) transferred to a nitrocellulose membrane is detected by incubating the membrane with GST-CpOGAD298N followed by anti-GST primary (
) transferred to a nitrocellulose membrane is detected by incubating the membrane with GST-CpOGAD298N followed by anti-GST primary (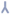 ) and IR-labelled (
) and IR-labelled ( ) secondary (
) secondary ( ) antibodies.
) antibodies.
b) Fluorescence polarization assay showing the binding of fluorescently-labelled probe (structure in the inset- see main text and methods for more details) to CpOGAD298N. Binding was measured by incubating a fixed concentration of labeled probe with varying concentrations of CpOGAD298N. Data points were fitted to a saturation binding equation using Prism (GraphPad). Experiments were performed in triplicate and error bars represent standard error of the mean.
c) Dose-response curves from fluorescence polarization assay showing the displacement from CpOGAD298N of a fixed concentration of fluorescent probe by increasing concentrations of O-GlcNAcylated peptides. Highest amount of probe bound to CpOGAD298N in the absence of competing O-GlcNAcylated peptides was arbitrarily set as 100%. Data points were fitted to a three-parameter equation for dose-dependent inhibition using Prism (GraphPad). Experiments were performed in triplicate and error bars represent standard error of the mean.
d) Structure of CpOGAD298N in complex with O-GlcNAcylated dHCF peptide (614VPSgTMSAN621). The carbons of the active site residues of CpOGAD298N are shown in grey sticks. The unbiased 2.44 Å |Fo|-|Fc|, Φcalc electron density map for the O-GlcNAcylated dHCF peptide is shown in cyan, contoured at 2.5σ. The dHCF peptide, PSgTA (as defined by the unbiased electron density), is shown as sticks with green carbons and the O-GlcNAc moiety with magenta carbons. Trp490, which is not conserved in hOGA, is shown as sticks with black carbons. Superimposed for comparison is the structure of CpOGAD298N in complex with the human TAB1 peptide, VPYgSS (PDB entry 2YDS25). The TAB1 peptide and O-GlcNAc moiety are shown in transparent sticks with yellow carbons. Hydrogen bonds are shown as dashed lines.
Results and Discussion
Since the Drosophila O-GlcNAc proteome has not previously been studied, we initially immunoprecipitated O-GlcNAc proteins from Drosophila S2 cell lysates using the anti-O-GlcNAc RL2 antibody and analysed these with ETD MS/MS. Several peptides from Drosophila host cell factor (dHCF) were found to be O-GlcNAcylated with one site conserved with the human HCF1 orthologue (Fig. S1, 614VPSgTMSAN621). Human HCF1 is one of the best-characterised OGT substrates that is heavily glycosylated and proteolytically cleaved by OGT, known to play a central role in epigenetic regulation of gene expression26, 27. We next investigated whether the dHCF Thr-O-GlcNAc peptide 614VPSgTMSAN621, containing an O-GlcNAc threonine, was able to bind CpOGAD298N. To facilitate this, we developed a fluorescence polarization assay based on a newly synthesized FITC-labelled OGA inhibitor that binds with 15 nM affinity to CpOGAD298N (Fig. 1b). In competition experiments, the dHCF O-GlcNAc peptide was shown to bind CpOGAD298N with a Kd of 19.6 µM (Fig. 1c). Encouraged by this, we also determined binding of Ser-O-GlcNAc peptides. Strikingly, 392VPYgSSA397 (derived from the O-GlcNAc site on the innate immunity signalling protein TAB128, Kd = 895 nM) and 402VAHgSGAK408 (derived from the O-GlcNAc site on human OGA (hOGA)29, Kd = 159 nM) bind with nanomolar affinity. The unglycosylated peptide controls showed no detectable binding. These data are consistent with KD values obtained from a Surface Plasmon Resonance (SPR) binding assay (Fig. S2). To explore the molecular basis of these interactions and possible differences in recognition of Ser- versus Thr-O-GlcNAc proteins, we determined the crystal structure of CpOGAD298N in complex with the dHCF-derived peptide (Fig. 1d). The structure reveals tight contacts between the GlcNAc moiety and the protein via hydrogen bonds to residues that are all conserved in hOGA, and explains why binding is lost with the D401A mutant of the enzyme (CpOGAD401A), which abrogates the interactions with the GlcNAc O4 and O6 hydroxyl groups23. Furthermore, Tyr189, conserved in hOGA, makes stacking interactions with the backbone of the O-GlcNAc peptide, explaining how this class of enzymes recognise their substrates in a sequence-independent, yet protein-dependent manner. Interestingly, compared to previously determined structures of CpOGAD298N in complex with the TAB1/hOGA derived Ser-O-GlcNAc peptides25, the apolar extra γ-methyl group of the threonine appears to be unfavourably positioned near the Tyr335 hydroxyl group, perhaps explaining the reduction in affinity compared to the Ser-O-GlcNAc peptides. Nevertheless, CpOGAD298N appears to be able to bind both Ser- and Thr-O-GlcNAc peptides, enabling its potential use as a general detector of O-GlcNAc proteins.
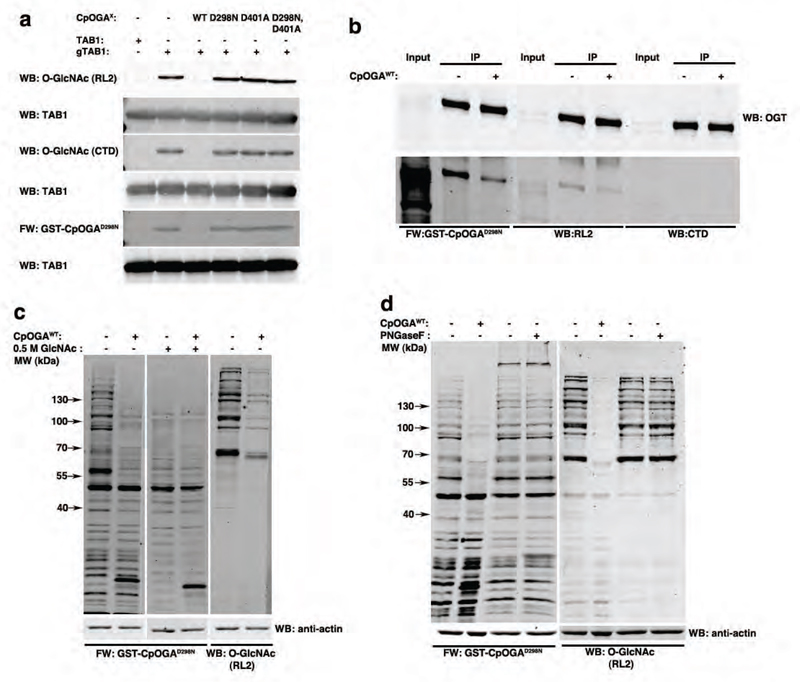
We next investigated the suitability of CpOGAD298N as a Far Western probe as outlined in Fig. 1a. Human TAB1 was O-GlcNAcylated in vitro and probed with GST-fused CpOGAD298N in a Western blot. The fusion protein was detected using sheep anti-GST antibody in combination with a Li-Cor compatible anti-sheep secondary antibody. Specific signal was detected only with GST-CpOGAD298N (Fig. 2a). However, on performing the Far Western with GST-CpOGAWT, (which hydrolyses the sugar, resulting in loss of binding), GST-CpOGAD401A or GST-CpOGAD298N,D401A (both deficient in O-GlcNAc peptide binding23) (Fig. S3a), as expected specific signal was not detected, establishing the suitability of CpOGAD298N for this purpose. When glycosylated TAB1 is pretreated with CpOGAWT, thus deglycosylating it, the specific O-GlcNAc dependent signal is no longer detectable on a Far Western blot with GST-CpOGAD298N or with the CTD110.6/RL2 O-GlcNAc antibodies (Fig. 2a). However, pre-treatment with CpOGAD298N, CpOGAD401A or CpOGAD298N,D401A did not lead to any change in the specific signal detected by GST-CpOGAD298N or with the CTD110.6/RL2 O-GlcNAc antibodies, further confirming that these mutants are indeed inactive (Fig. 2a).
Figure 2. CpOGAD298N can be utilized as a Far Western tool.
a) Immunoblots with anti-O-GlcNAc antibodies (RL2 and CTD110.6) or Far Western with GST-CpOGAD298N was performed on either naked or in vitro O-GlcNAcylated TAB1 (labels adjacent to the blots). The specificity of the signal detected was confirmed by deglycosylating O-GlcNAcylated TAB1 by pre-treatment with CpOGAWT. Pre-treatment as indicated above the blots were performed to probe the activity of all the CpOGA constructs.
b) Embryonically overexpressed DmOGT-HA was immunoprecipitated with HA antibody and immunoblotted with anti-O-GlcNAc antibodies (RL2 and CTD110.6) or subjected to Far Western with GST-CpOGAD298N. Treating the immunoprecipitate with CpOGAWT was used as a control.
c) HEK293 lysates without or with CpOGAWT pre-treatment were subjected to GST-CpOGAD298N Far Western. In addition, the GST-CpOGAD298N Far Western was performed in the presence of 0.5 M GlcNAc competition. The extreme right panel is an immunoblot of the same samples with anti-O-GlcNAc antibody, RL2.
d) HEK293 lysates without or with PNGase F/CpOGAWT pre-treatment were subjected to GST-CpOGAD298N Far Western. The right panel is an immunoblot of the same samples with anti-O-GlcNAc antibody, RL2.
Mammalian OGT is known to self-glycosylate30, and hence we investigated auto-glycosylation of Drosophila OGT (DmOGT) using CpOGAD298N. Hemagglutinin (HA) tagged DmOGTWT was embryonically overexpressed and immunoprecipitated using anti-HA antibody. O-GlcNAcylation of immunoprecipitated OGT was readily detected using the CpOGAD298N Far Western (Fig. 2b). The specificity of this reactivity was also determined by treating the immunoprecipitate with CpOGAWT, which considerably reduced the signal (Fig. 2b), thus demonstrating the utility of this Far Western approach in expanding the repertoire of detectable O-GlcNAc sites.
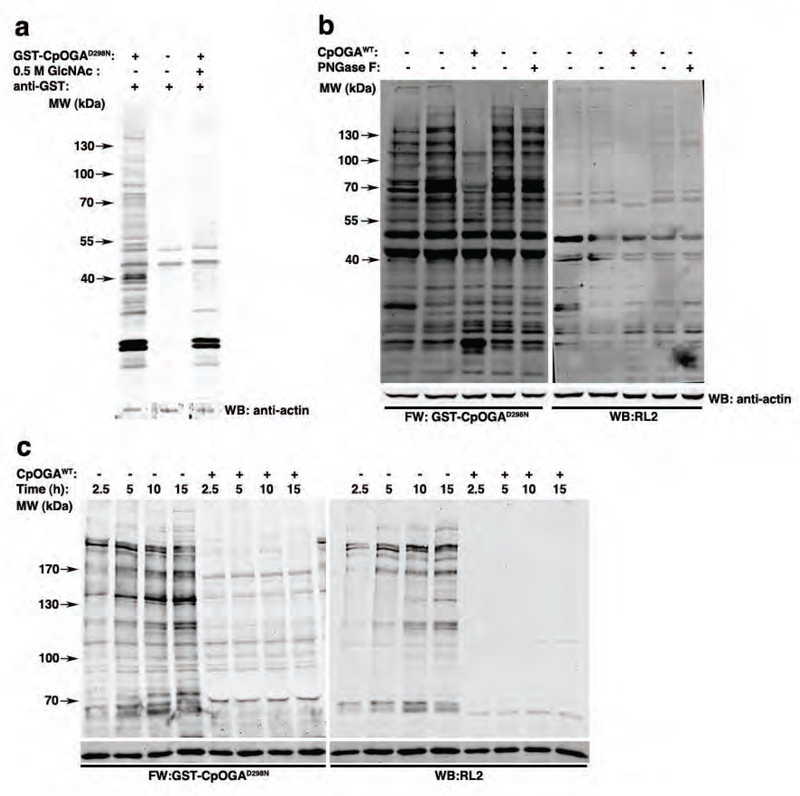
Next we explored this approach using a more complex mammalian sample, HEK293 cell lysates. Specific O-GlcNAcylated proteins were detected when the Far Western was performed using GST-CpOGAD298N, although GST-dependent reactivity is visible in the <50 kDa range (Fig. 2c). The specific signal of the >50 kDa bands diminished significantly when the lysates were pretreated with CpOGAWT in order to remove the O-GlcNAc from proteins or on competing with 0.5 M GlcNAc (Fig. 2c). Additional controls including treatment of blots with GST, anti-GST antibody or the Li-Cor anti-sheep secondary antibody alone further established the specificity of O-GlcNAc detection with GST-CpOGAD298N (Fig. S3b). The anti-O-GlcNAc antibody RL2 identified fewer bands (around 55 kDa, 100 kDa, and > 130 kDa) as compared to GST-CpOGAD298N (Fig. 2c). Since the secretory N-glycosylation pathway also utilises an N-GlcNAc(β1,4)GlcNAc core, cross-reactivity of CpOGAD298N to N-glycosylated proteins was tested. Lysates were treated with PNGase F or CpOGAWT to remove N-glycosylation or O-GlcNAcylation on proteins prior to the Far Western/Western analyses. Treatment of lysates with PNGase F distinctly reduced reactivity to Concanavalin A (ConA), a lectin routinely used to detect N-glycans (Fig. S3c), but not the GST-CpOGAD298N-dependent signal (Fig. 2d). As expected, pretreatment with CpOGAWT significantly reduced detection by CpOGAD298N, establishing its specificity for nucleocytoplasmic O-GlcNAcylation (Fig. 2d). As with the in vitro experiments, specific signal is not detected when GST-CpOGAWT, GST-CpOGAD401A or GST-CpOGAD298N,D401A are used as Far Western probes on HEK293 lysates (Fig. S3d).
Finally, we used the Far Western CpOGAD298N probe to investigate changes in the O-GlcNAc proteome in the context of Drosophila embryonic development. Specific O-GlcNAcylated proteins, spanning a large molecular weight range, were identified in 0-16 h Drosophila embryo lysates using this approach (Fig. 3a). Specificity of the detection was established by either competing the CpOGAD298N binding with 0.5 M GlcNAc or the loss of specific signal on pretreatment of the lysate with CpOGAWT (Fig. 3a,b). The reactivity of CpOGAD298N was not altered when the embryo lysates were pretreated with PNGase F, which did significantly reduce ConA binding (Fig. S3e), confirming that CpOGAD298N specificity is towards O-GlcNAc proteins and not N-glycosylated proteins in Drosophila embryo lysates.
Figure 3. The O-GlcNAc proteome across Drosophila embryogenesis is dynamic.
a) Drosophila embryonic lysates from 0-16 h collections were subjected to GST or GST-CpOGAD298N Far Western. In addition, the GST-CpOGAD298N Far Western was performed in the presence of 0.5 M GlcNAc.
b) 0-16 h Drosophila embryonic lysates without or with PNGase F/CpOGAWT pre-treatment were subjected to GST-CpOGAD298N Far Western. The right panel is an immunoblot of the same samples with anti-O-GlcNAc antibody, RL2.
c) 2.5, 5, 10 and 15 h Drosophila embryonic lysates without or with CpOGAWT pre-treatment were subjected to GST-CpOGAD298N Far Western. The right panel is an immunoblot of the same samples with anti-O-GlcNAc antibody, RL2.
To investigate changes in O-GlcNAcylation across Drosophila embryogenesis, embryos (growing at 25 °C) at the cellular blastoderm (2.5 h), germ band extension (5 h), germ band retraction (10 h) and dorsal closure (15 h) stages were collected and the Far Western experiment performed. The number of O-GlcNAc proteins was the highest at the dorsal closure stage (Fig. 3c). While there was progressive accumulation of O-GlcNAc on several proteins with developmental time, many proteins are O-GlcNAcylated at the germ band retraction stage or later (Fig. 3c). While the overall O-GlcNAc profile detected using RL2 is comparable, numerous additional proteins are detected by the Far Western approach, (for instance, around 70 kDa, 100 kDa, between 120-130 kDa and >170 kDa; Fig. 3c). These data reveal that, like protein phosphorylation, different developmental stages are associated with different O-GlcNAc proteomes offering opportunities for future identification of the proteins involved and probing the mechanistic developmental biology underlying these changes from larger-scale tissue samples.
Conclusions
The approach in this study was to investigate changes in the O-GlcNAc proteome during Drosophila embryonic development, harnessing the O-GlcNAc binding properties of CpOGAD298N. During the course of developing this tool we have established that CpOGAD298N has higher affinity to Ser-O-GlcNAc as opposed to Thr-O-GlcNAc peptides. A crystal structure of a Thr-O-GlcNAc peptide in complex with CpOGAD298N has revealed that the position of the additional Thr γ-methyl is unfavourable, possibly leading to the lower affinity of CpOGAD298N for Thr-O-GlcNAc peptides. Whether this difference in affinities of CpOGAD298N towards Ser- versus Thr-O-GlcNAc would lead to differential catalytic rates of the active enzyme or translates to physiological consequence(s) remains to be explored.
We have shown here that the O-GlcNAc proteome during Drosophila embryonic development is dynamic. Changes in the O-GlcNAc proteome during Xenopus embryogenesis have been investigated using anti-O-GlcNAc RL2 antibody31. During Xenopus embryogenesis the number of different O-GlcNAcylated proteins seems to be constant with the most obvious changes being the extent of their O-GlcNAc levels, possibly because of the restricted specificity of the RL2 antibody. However, during Drosophila embryonic development, in addition to quantitative changes in O-GlcNAc levels on particular proteins, the number of proteins O-GlcNAcylated is also dynamic. A recent report identified Polyhomeotic (Ph), a Polycomb group protein, as the major substrate for OGT during Drosophila development32. However, the work described here demonstrates that there are multiple OGT substrates during Drosophila embryogenesis. Given that sxc mutants lacking maternal OGT are late embryonic lethals, it will be feasible to identify the O-GlcNAcylated proteins during Drosophila embryogenesis and investigate the significance of their O-GlcNAc status in this model. Reduced O-GlcNAcylation of proteins (other than Ph) could possibly lead to non-lethal, subtler phenotypes that could be investigated during early embryogenesis. The impetus of this approach would be to understand the role of O-GlcNAcylation on sites conserved in mammalian orthologues using the strength of the Drosophila genetic model.
Materials and Methods
Protein production
pGEX6P1 plasmids containing N-terminally GST-tagged CpOGAWT (31-618)25 and mutants were transformed into E. coli BL21 (DE3) pLysS cells. Cells were grown overnight at 37 °C in Luria-Bertani medium containing 50 µg/ml ampicillin (LB-Amp) and used at 10 mL/ L to inoculate fresh LB-Amp. Cells were grown to an OD600 of 0.6-0.8, transferred to 18 °C and induced with 250 µM of IPTG and harvested after 16 h by centrifugation for 30 min at 3500 rpm (4 °C). Cell pellets were resuspended in 10-20 mL/L of 50 mM Tris, 250 mM NaCl at pH 7.5 (lysis buffer) supplemented with protease inhibitors (1 mM benzamidine, 0.2 mM PMSF and 5 µM leupeptin), DNAse and lysozyme prior to lysis. Cells were lysed using a continuous flow cell disrupter (Avestin, 3 passes at 20 kpsi) and the lysate was cleared by centrifugation (30 min, 48,000 g, 4 °C). Supernatants were collected and loaded on to 2 mL glutathione sepharose (GE Healthcare Life Sciences) pre-equilibrated with lysis buffer. The column was washed with 500 mL of lysis buffer. Proteins were eluted with 25 mL of lysis buffer supplemented with 50 mM reduced glutathione and dialysed into 1 x Tris-buffered saline (TBS), pH 7.5. Proteins were further purified by size exclusion chromatography using a Superdex 200, 26/60 column. TAB1 was purified as described previously33.
Synthesis of peptides and fluorescently labelled GlcNAcstatin
The glycosylated amino acid building blocks 3,4,6-triacetyl-O-GlcNAc-Fmoc-Ser-OH and 3,4,6-triacetyl-O-GlcNAc-Fmoc-Thr-OH were synthesized in house according to the published procedures. Microwave-assisted solid phase peptide synthesis was performed with a CEM Liberty automated peptide synthesizer on low-load (0.38 mmol/g) Rink amide MBHA resin 100-200 mesh (Novabiochem) using standard Fmoc chemistry protocols on a 0.05 mmol scale. After the removal of the N-terminal Fmoc group the peptidyl resin was acetylated (Ac2O, DIPEA), carbohydrate residues were deprotected (20% hydrazine, MeOH), and peptides were globally deprotected and cleaved from the resin with TFA-TIPS-water (92.5:2.5:5) cocktail for 2 h. The cleavage mixture was filtered off, the resin was washed with TFA twice and the combined filtrate was concentrated to 1/10 of the initial volume in vacuum. The oily residue was triturated with cold (0 ºC) methyl t-butyl ether, and the precipitated peptide was collected by centrifugation. The crude peptides were purified to greater than 95% purity using Waters Peptide Separation Technology 19×100 C18 column flow rate 20 mL/min, on a preparative HPLC system consisting of Gilson 331/332 pumps, Gilson UV156 detector, and Gilson 203 fraction collector controlled by Gilson Trilution LC software. The appropriate fractions were pooled and freeze dried.
Synthesis of the FITC-labelled fluorescence polarimetry probe was based on the synthetic procedure for GlcNAcstatin B34 and will be described in detail elsewhere.
Protein crystallography
CpOGAD298N was purified and crystallized as described previously25. The glycopeptide complex with dHCF (VPSgTMSAN) peptide was achieved through soaking with 10 mM glycopeptide for 0.5 h prior to cryoprotection with 20% glycerol in mother liquor. Diffraction data were collected at the Diamond Light Source (Didcot, UK) I03 (Table S1). Crystals belonged to space group P61 and contained one molecule per asymmetric unit, with 73.47% solvent content. The structure was solved by molecular replacement, using Protein Data Bank ID 2YDS25 as a search model, followed by iterative model building with COOT35 and refinement with REFMAC536 using 2% of reflections as an Rfree test set. Table S1 gives details of the data collection, processing, and refinement statistics. The model for the peptide was included when it became fully defined by the difference map upon refining the protein structure and adding hetero atoms.
Surface Plasmon Resonance
CpOGAWT (31-618) and mutants were purified as described previously25. Proteins were chemically biotinylated using the EZ-Link NHS-PEG4-Bioin kit (Thermo) according to the manufacturer’s instructions, except that a 1:1 molar ratio of biotinylation reagent to protein was used. Proteins were captured on a neutravidin surface prepared on high capacity amine sensor chip of a Mass-1 instrument (Sierra Sensors) at densities ~3,600–3,900 RU. All experiments were performed at 25 °C. Ligands were injected over captured proteins at flow rate 30 µL min−1 in running buffer (50 mM HEPES, 250 mM NaCl, 10 mM EDTA, 5 mM TCEP, 0.05% Tween20 or 25 mM Tris pH 7.5, 150 mM NaCl, 0.05% Tween20), with each compound injected in duplicates in concentration series adjusted specifically around their affinities. Association was measured for 60 s and dissociation for 120 s. All data were double referenced for blank injections of buffer and biotin-blocked Streptavidin surface. Analyser 2 (Sierra Sensors) and Scrubber 2 (BioLogic Software) were used to process and analyse the data.
Fluorescence Polarization
Experiments were performed in PerkinElmer, black, 384-well plates and millipolarization units measured using a Pherastar FS plate reader (BMG LABTECH) at excitation and emission wavelengths of 485 nm and 530 nm respectively. For determination of the equilibrium dissociation constant (Kd) of CpOGAD298N for the fluorescent probe, 5 nM probe was incubated with a range of concentrations of protein in 25 µL total reaction volume containing 1 x TBS buffer pH 7.5 and a final concentration of 1% DMSO. Reactions were allowed to stand at room temperature for 10 min and polarization was measured every 5 min for a period of 2.5 h (equilibrium was reached within 10 min). Readings were corrected for background emissions from reactions containing no CpOGAD298N and Kd was determined by fitting a non-linear regression curve with Prism (GraphPad) to readings obtained at 15 min. To avoid receptor depletion, reaction mixtures for competition binding experiments contained 5 nM fluorescent probe, 20 nM CpOGAD298N (receptor) and a range of concentrations of O-GlcNAcylated peptides in the aforementioned reaction conditions. Highest amount of fluorescent probe bound to CpOGAD298N in the absence of competing O-GlcNAcylated peptides was arbitrarily set as 100%. EC50 values were determined by fitting non-linear regression curves with Prism (GraphPad) and converted to Kd as outlined37. All experiments were performed in triplicate.
Drosophila stocks and embryo collections
w1118 wild type flies were used for all experiments. Embryos were collected on apple juice agar plates at 25 °C for 30 min and aged for an additional 2, 4.5, 10.5 or 14.5 h. To make total embryo lysates, overnight (0-16 h) embryo collections were set up. The embryos thus collected and aged were dechorionated with bleach and snap frozen in dry ice. Other lines used were UAS::OGTWT-HA and tub::GAL4/TM3. To overexpress HA-tagged OGTWT, UAS::OGTWT-HA homozygous virgins were crossed with tub::GAL4/TM3 flies and 0-16 h embryos were collected on apple juice agar plates, dechorionated with bleach and used for immunoprecipitation experiments.
Lysates and immunoprecipitation
Drosophila embryo, HEK293 and S2 cell lysates were prepared identically for Western, Far Western and immunoprecipitation experiments. The frozen embryos were homogenized in lysis buffer (LB; 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Triton-X-100, 1 μM GlcNAcstatin C, 5 mM sodium fluoride, 2 mM sodium orthovanadate, 1 mM benzamidine, 0.2 mM PMSF, 5 μM leupeptin and 1 mM DTT). Confluent HEK293 or S2 cultures were washed with 1X Phosphate Buffered Saline, pH 7.5 prior to lysis with LB. Lysates were then centrifuged at 16000 g for 10 min, supernatants were collected and protein concentrations were estimated using the 660 nm protein assay (Thermo Scientific).
For immunoprecipitation experiments, lysates of OGTWT-HA overexpressing embryos were prepared with LB and 2.5 mg protein was incubated with Protein-G dynabeads (Invitrogen) bound to 5 μg of anti-HA antibody (clone 12CA5). For immunoprecipitating O-GlcNAcylated proteins, 5 mg of S2 cell lysate was incubated with Protein-G dynabeads bound to 10 μg anti-O-GlcNAc antibody (RL2). After washing, the immunoprecipitated proteins were eluted by boiling with 1X SDS loading buffer. The proteins were then separated on 4-12% NuPAGE gels, stained with Coomassie and gel pieces at the same molecular weight range in the control IgG and RL2 immunoprecipitates were excised and processed for mass spectroscopy. Where indicated, the immunoprecipitate, after washing away the lysate was incubated with 10 μg of CpOGA for 60 min at 30 °C.
Mass spectrometry
The excised gel pieces were subjected to enzymatic digestion as previously reported with minor modifications38. Briefly, excised bands were rinsed thrice with AmBic buffer (50 mM ammonium bicarbonate in 50% methanol (HPLC grade, Merck)) followed by a reduction step with 10 mM dithiothreitol (Sigma-Aldrich). Subsequently, the gel pieces were rinsed twice with AmBic buffer and dried in a SpeedVac before alkylation with 55 mM iodoacetamide (Sigma-Aldrich) in 50 mM ammonium bicarbonate. Thereafter, the gel pieces were rinsed with AmBic buffer, dehydrated with acetonitrile (HPLC grade, Merck) and dried in a SpeedVac. The dry gel pieces were treated with Trypsin (Promega, Madison, WI; 20 ng/µl in 20 mM ammonium bicarbonate), incubating them at 37 ºC for 16 h. Peptides were extracted thrice by incubating for 20 min with 40 µl of 60% acetonitrile in 0.5% HCOOH. The resulting peptide extracts were pooled, concentrated in a SpeedVac and stored at -20 ºC until MS analysis.
Identification of the O-GlcNAcylated proteins and O-GlcNAc sites was performed by ESI-IT-ETD (ElectroSpray IonTrap Electron Transfer Dissociation) mass spectrometry coupled to a nano-LC system (Ultimate 3000 RSLC, Dionex, The Netherlands). Dried peptides were resuspended in 30 µL of 0.5% HCOOH and 10 µL of this was injected for mass spectrometric analysis. Tryptic peptides were concentrated on a trap column (2 cm x 100 µm, Dionex) at 10 µL/min and separated on a 15 cm x 75 µm Pepmap C18 reversed-phase column (Thermo Fischer Scientific). Peptides were eluted by a linear 60 min gradient of 95% A / 5% B to 10% A / 90% B (A: H2O, 0.1% HCOOH; B; 80% acetonitrile (ACN), 0.08% HCOOH) at 300 nl/min into a LTQ Velos ETD (Thermo Fisher Scientific). MS spectra were acquired in positive mode, firstly MS full scans were acquired followed by MS/MS in ETD mode. Up to ten most intense precursors were selected for ETD fragmentation with an activation time of 300 ms and non-dynamic exclusion. Proteome Discoverer v 1.4.0.288 software (Thermo Scientific) was used to process raw LCMS/MS data, applying the Mascot (version 2.4, Matrix Science, Boston, MA, USA) search engine algorithm against SwissProt database with specified taxonomy [D. melanogaster, number of sequences 5545] with the following Mascot parameters: 2+, 3+, 4+ and 5+ ions; precursor mass tolerance 100 ppm; Da; fragment tolerance 0.6 Da and up to 2 missed cleavages. The variable modifications included were: oxidation (M) (15.99 Da), dioxidation (M) (+31.98 Da) and HexNAc (ST) (+203.0794 Da). All MS/MS data and database results were manually inspected in detail to verify the automatic assignment of fragment ions using the above software. Peptides with an expectation value (Exp. Value) smaller than 0.1 are considered as a precise O-GlcNAc site assignment.
Western and Far Western analysis
10 μg of the crude lysates was subjected to SDS-PAGE and transferred onto nitrocellulose membrane before immunoblotting with 1 μg/ml RL2 (1:1000 of 1 mg/ml, Abcam), 0.6 μg/ml CTD110.6 (1:500 of 0.3 mg/ml, Cell Signalling Technologies), mouse anti-α-tubulin (1:10000, Developmental Studies Hybridoma Bank), rabbit anti-tubulin (1:2,500, Cell Signalling) and/or rabbit anti-Actin (1:5000, Sigma). For the Far Western experiments, the blots were incubated with 10 μg/ml GST-CpOGAD298N for 30 min at room temperature followed by incubation with sheep anti-GST (1:5000) antibody. For each of the experiments the respective infrared dye conjugated secondary antibodies (Li-Cor or Life Technologies, 1:10,000) were used. Signal was detected with an Odyssey® Li-Cor infrared imaging system. To ascertain the specificity of the O-GlcNAc signal, embryo lysates (made without GlcNAcstatin C) were pre-treated with 10 μg of CpOGA for 60 min at 30 °C before being processed. Control samples not treated with CpOGA were also incubated at 30 °C for 60 min. To establish that CpOGAD298N does not cross react with N-glycosylated proteins, lysates were treated with Peptide:N-glycosidase F (NEB) as per the manufacturer’s instructions. Removal of N-glycans was monitored by blotting with a specific lectin, Concanavalin-A, following a previously described protocol39.
Supplementary Material
Acknowledgements
We thank the Diamond synchrotron for beam time on beamline I03. This work was funded by a Wellcome Trust Senior Research Fellowship (WT087590MA) to D.v.A. The refined crystal structure has been deposited in the PDB (entry 4ZXL).
Footnotes
Contributions D.M. and D.v.A conceived the study. D.M. performed the cellular and Drosophila assays; N.S. performed protein expression, structural biology and FP; V.S.B. performed peptide and chemical synthesis; J.A.L., performed MS studies; A.T.F. performed molecular biology; C.S. and I.H.N performed SPR; D.M., N.S. and D.v.A. interpreted the data and wrote the manuscript.
References
- 1.Hart G, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu Rev Biochem. 2011;80:825–883. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hart GW. Three Decades of Research on O-GlcNAcylation - A Major Nutrient Sensor That Regulates Signaling, Transcription and Cellular Metabolism. Front Endocrinol (Lausanne) 2014;5:183. doi: 10.3389/fendo.2014.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vaidyanathan K, Wells L. Multiple tissue-specific roles for the O-GlcNAc post-translational modification in the induction of and complications arising from type II diabetes. The Journal of biological chemistry. 2014;289:34466–34471. doi: 10.1074/jbc.R114.591560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma Z, Vosseller K. Cancer metabolism and elevated O-GlcNAc in oncogenic signaling. The Journal of biological chemistry. 2014;289:34457–34465. doi: 10.1074/jbc.R114.577718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu Y, Shan X, Yuzwa SA, Vocadlo DJ. The emerging link between O-GlcNAc and Alzheimer disease. The Journal of biological chemistry. 2014;289:34472–34481. doi: 10.1074/jbc.R114.601351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shafi R, et al. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci U S A. 2000;97:5735–5739. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gambetta MC, Oktaba K, Muller J. Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science. 2009;325:93–96. doi: 10.1126/science.1169727. [DOI] [PubMed] [Google Scholar]
- 8.Webster DM, et al. O-GlcNAc modifications regulate cell survival and epiboly during zebrafish development. BMC Dev Biol. 2009;9:28. doi: 10.1186/1471-213X-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Radermacher PT, et al. O-GlcNAc reports ambient temperature and confers heat resistance on ectotherm development. Proc Natl Acad Sci U S A. 2014;111:5592–5597. doi: 10.1073/pnas.1322396111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Comer FI, Hart GW. Reciprocity between O-GlcNAc and O-phosphate on the carboxyl terminal domain of RNA polymerase II. Biochemistry. 2001;40:7845–7852. doi: 10.1021/bi0027480. [DOI] [PubMed] [Google Scholar]
- 11.Isono T. O-GlcNAc-specific antibody CTD110.6 cross-reacts with N-GlcNAc2-modified proteins induced under glucose deprivation. PLoS One. 6:e18959. doi: 10.1371/journal.pone.0018959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reeves RA, Lee A, Henry R, Zachara NE. Characterization of the specificity of O-GlcNAc reactive antibodies under conditions of starvation and stress. Anal Biochem. 2014;457:8–18. doi: 10.1016/j.ab.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ogawa M, Sakakibara Y, Kamemura K. Requirement of decreased O-GlcNAc glycosylation of Mef2D for its recruitment to the myogenin promoter. Biochemical and biophysical research communications. 2013;433:558–562. doi: 10.1016/j.bbrc.2013.03.033. [DOI] [PubMed] [Google Scholar]
- 14.Tashima Y, Stanley P. Antibodies that detect O-linked beta-D-N-acetylglucosamine on the extracellular domain of cell surface glycoproteins. The Journal of biological chemistry. 2014;289:11132–11142. doi: 10.1074/jbc.M113.492512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogawa M, et al. Impaired O-Linked N-Acetylglucosaminylation in the Endoplasmic Reticulum by Mutated Epidermal Growth Factor (EGF) Domain-specific O-Linked N-Acetylglucosamine Transferase Found in Adams-Oliver Syndrome. The Journal of biological chemistry. 2015;290:2137–2149. doi: 10.1074/jbc.M114.598821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snow CM, Senior A, Gerace L. Monoclonal antibodies identify a group of nuclear pore complex glycoproteins. J Cell Biol. 1987;104:1143–1156. doi: 10.1083/jcb.104.5.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torres CR, Hart GW. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J Biol Chem. 1984;259:3308–3317. [PubMed] [Google Scholar]
- 18.Roquemore EP, Chou TY, Hart GW. Detection of O-linked N-acetylglucosamine (O-GlcNAc) on cytoplasmic and nuclear proteins. Methods Enzymol. 1994;230:443–460. doi: 10.1016/0076-6879(94)30028-3. [DOI] [PubMed] [Google Scholar]
- 19.Khidekel N, et al. A chemoenzymatic approach toward the rapid and sensitive detection of O-GlcNAc posttranslational modifications. J Am Chem Soc. 2003;125:16162–16163. doi: 10.1021/ja038545r. [DOI] [PubMed] [Google Scholar]
- 20.Clark PM, et al. Direct in-gel fluorescence detection and cellular imaging of O-GlcNAc-modified proteins. J Am Chem Soc. 2008;130:11576–11577. doi: 10.1021/ja8030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ramakrishnan B, Qasba PK. Structure-based design of beta 1,4-galactosyltransferase I (beta 4Gal-T1) with equally efficient N-acetylgalactosaminyltransferase activity: point mutation broadens beta 4Gal-T1 donor specificity. The Journal of biological chemistry. 2002;277:20833–20839. doi: 10.1074/jbc.M111183200. [DOI] [PubMed] [Google Scholar]
- 22.Wu ZL, et al. Detecting O-GlcNAc using in vitro sulfation. Glycobiology. 2014;24:740–747. doi: 10.1093/glycob/cwu037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao FV, et al. Structural insights into the mechanism and inhibition of eukaryotic O-GlcNAc hydrolysis. Embo J. 2006;25:1569–1578. doi: 10.1038/sj.emboj.7601026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dorfmueller HC, et al. GlcNAcstatin: a picomolar, selective O-GlcNAcase inhibitor that modulates intracellular O-glcNAcylation levels. J Am Chem Soc. 2006;128:16484–16485. doi: 10.1021/ja066743n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schimpl M, Borodkin VS, Gray LJ, van Aalten DM. Synergy of peptide and sugar in O-GlcNAcase substrate recognition. Chem Biol. 2012;19:173–178. doi: 10.1016/j.chembiol.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zargar Z, Tyagi S. Role of host cell factor-1 in cell cycle regulation. Transcription. 2012;3:187–192. doi: 10.4161/trns.20711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Capotosti F, et al. O-GlcNAc transferase catalyzes site-specific proteolysis of HCF-1. Cell. 2011;144:376–388. doi: 10.1016/j.cell.2010.12.030. [DOI] [PubMed] [Google Scholar]
- 28.Pathak S, et al. O-GlcNAcylation of TAB1 modulates TAK1-mediated cytokine release. Embo J. 2012;31:1394–1798. doi: 10.1038/emboj.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khidekel N, et al. Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics. Nat Chem Biol. 2007;3:339–348. doi: 10.1038/nchembio881. [DOI] [PubMed] [Google Scholar]
- 30.Whelan SA, Lane MD, Hart GW. Regulation of the O-linked beta-N-acetylglucosamine transferase by insulin signaling. The Journal of biological chemistry. 2008;283:21411–21417. doi: 10.1074/jbc.M800677200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dehennaut V, et al. Survey of O-GlcNAc level variations in Xenopus laevis from oogenesis to early development. Glycoconj J. 2009;26:301–311. doi: 10.1007/s10719-008-9166-0. [DOI] [PubMed] [Google Scholar]
- 32.Gambetta MC, Muller J. O-GlcNAcylation Prevents Aggregation of the Polycomb Group Repressor Polyhomeotic. Dev Cell. 2014;31:629–639. doi: 10.1016/j.devcel.2014.10.020. [DOI] [PubMed] [Google Scholar]
- 33.Conner SH, et al. TAK1-binding protein 1 is a pseudophosphatase. Biochem J. 2006;399:427–434. doi: 10.1042/BJ20061077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dorfmueller HC, Borodkin VS, Schimpl M, van Aalten DM. GlcNAcstatins are nanomolar inhibitors of human O-GlcNAcase inducing cellular hyper-O-GlcNAcylation. Biochem J. 2009;420:221–227. doi: 10.1042/BJ20090110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta crystallographica Section D, Biological crystallography. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta crystallographica Section D, Biological crystallography. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 37.Nikolovska-Coleska Z, et al. Development and optimization of a binding assay for the XIAP BIR3 domain using fluorescence polarization. Anal Biochem. 2004;332:261–273. doi: 10.1016/j.ab.2004.05.055. [DOI] [PubMed] [Google Scholar]
- 38.Alonso J, Santaren JF. Proteomic analysis of the wing imaginal discs of Drosophila melanogaster. Proteomics. 2005;5:474–489. doi: 10.1002/pmic.200400923. [DOI] [PubMed] [Google Scholar]
- 39.Mariappa D, et al. Protein O-GlcNAcylation is required for fibroblast growth factor signaling in Drosophila. Sci Signal. 2011;4:ra89. doi: 10.1126/scisignal.2002335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.