Abstract
Imaging of hyperpolarized substrates by magnetic resonance shows great clinical promise for assessment of critical biochemical processes in real time. Due to fundamental constraints imposed by the hyperpolarized state, exotic imaging and reconstruction techniques are commonly used. A practical system for characterization of dynamic, multi-spectral imaging methods is critically needed. Such a system must reproducibly recapitulate the relevant chemical dynamics of normal and pathological tissues. The most widely utilized substrate to date is hyperpolarized [1-13C]-pyruvate for assessment of cancer metabolism. We describe an enzyme-based phantom system that mediates the conversion of pyruvate to lactate. The reaction is initiated by injection of the hyperpolarized agent into multiple chambers within the phantom, each of which contains varying concentrations of reagents that control the reaction rate. Multiple compartments are necessary to ensure that imaging sequences faithfully capture the spatial and metabolic heterogeneity of tissue. This system will aid the development and validation of advanced imaging strategies by providing chemical dynamics that are not available from conventional phantoms, as well as control and reproducibility that is not possible in vivo.
Keywords: Medicine, Issue 110, Hyperpolarized, Pyruvate, Carbon 13, Magnetic Resonance Imaging, Phantom, Sequence Development, Quality Assurance
Introduction
The clinical impact of hyperpolarized magnetic resonance imaging (MRI) of 13C-labeled compounds is critically dependent on its ability to measure chemical conversion rates through real time magnetic resonance spectroscopy and spectroscopic imaging1-5. During sequence development and verification, dynamic chemical conversion is generally achieved through in vivo or in vitro models6-9 that offer limited control and reproducibility. For robust testing and quality assurance, a more controlled system that preserves the chemical conversion endemic to this measurement would be preferred. We outline a method to achieve this conversion in a reproducible manner using a dynamic single enzyme phantom.
Most studies with hyperpolarized 13C agents focus on imaging hyperpolarized substrates in a functioning biological environment. This is the obvious choice if the goal is to study biological processes or determine potential for impact on clinical care. However, if characterization of some measurement system or data processing algorithm is desired, biological models have numerous drawbacks such as inherent spatial and temporal variability10. However, conventional static phantoms lack the chemical conversion that drives the primary clinical interest in MRI of hyperpolarized substrates, and cannot be used to characterize detection of conversion rates or other dynamic parameters11. Using a single enzyme system we can provide controllable and reproducible chemical conversion, enabling rigorous examination of dynamic imaging strategies.
This system is directed to investigators who are developing imaging strategies for hyperpolarized substrates and wish to characterize performance for comparison against alternate approaches. If static measurements are the desired endpoint then static 13C-labled metabolite phantoms will suffice11. On the other end if more complex biological characterization is critical to the method (delivery, cellular density, etc.) then actual biological models will be needed12-14. This system is ideal for assessment of imaging strategies that aim to provide a quantitative measure of apparent chemical conversion rates.
Protocol
NOTE: (Phantom Design) Two 3 ml chambers were machined out of Ultem and fitted with PEEK tubing (1.5875 mm OD and 0.762 mm ID) for injection and exhaust. The Chambers were placed in a 50 ml centrifuge tube filled with water (Figure 1). To avoid signal voids created by bubbles, the chambers and the lines were pre filled with deionized water (dH2O).
1. Solution Preparation
Prepare 1 L buffer solution (81.3 mM Tris pH 7.6, 203.3 mM NaCl). Weigh out 11.38 g Trizma preset crystals pH 7.6 and 11.88 g NaCl and dissolve in 1 L of dH2O.
Prepare 50 mM NADH solution. Weigh out 17.8 mg of β-nicotinamide adenine dinucleotide (NADH), reduced dipotassium salt and dissolve in 280 µl of the buffer solution that was prepared in step one.
Prepare 250 U/ml Enzyme solution. Weigh out 78.75 activity units of lactate dehydrogenase (LDH) and dissolve in 315 µl buffer from step one.
Prepare pyruvic acid mix. Weigh out 21.4 mg Ox063 trityl radical and dissolve in 1.26 g (~1 ml) [1-13C] pyruvic acid.
Prepare dissolution media (40 mM Tris pH 7.6, 40 mM NaOH, 0.27 mM EDTA and 50 mM NaCl). Weigh out 5.96 g of Trizma preset crystals pH 7.6, 1.6 g of NaOH, 0.1 g ethylenediaminetetraacetic acid disodium salt dehydrate (EDTA) and 2.9 g NaCl and dissolve in 1 L dH2O.
Prepare 1:10 gadoteridol solution (50 mM). Mix 1 µl of gadoteridol in 9 µl of dH2O. Preparation of 8 M [13C] urea. Weigh out 1.465 5 [13C] urea and dissolve in 3 ml dH2O.
2. Preparation of Hyperpolarized Pyruvate
In a sample cup for a Dynamic Nuclear Polarization (DNP) system, pipette 0.3 µl of the gadoteridol solution and 13 mg (~10 µl) of the pyruvic acid solution.
Briefly stir this mixture in the sample cup with the pipette tip.
- Insert Sample into the DNP system.
- Ensure the door to the DNP system is closed. Begin the sample insertion process by clicking the insert sample button on the DNP system console. On the sample wizard select normal sample and press next.
- Keeping the sample cup vertical gently place the insertion rod over the top of the sample cup. When prompted, open the DNP system and insert the cup into the variable temperature insert (VTI) using the insertion rod.
- Pull on the plunger at the end of the sample insertion rod to release the sample in the variable temperature index. Remove the sample insertion rod from the system and click the next button on the DNP system console.
- Initiate the polarization.
- Click the start polarization button on the DNP system console. In the RINMR software type .HYPERSENSENMR to launch polarization monitoring software. Set build up configuration to 1 and press enter. Then click solid build up.
- Set the location and name of the save file. Select the profile for 13-C in the drop down tab on the DNP system console and click next. Check the box to sample during the buildup, set sample time to 300 sec and click finish.
Measure out 3.85 g (~4 ml) of the dissolution media either by volume with a 5 ml syringe or by weight using a scale.
3. Preparation of the Enzyme Phantom
Fill a microcentrifuge tube with ~3 ml [13C] urea solution and place it in the 50 ml centrifuge tube. Fill the 50 ml centrifuge tube with dH2O.
Pre-fill the two enzyme chambers and the lines with dH2O by injecting ~3 ml dH2O into the injection lines of the phantom, taking care to flush any bubbles formed.
Place the phantom in the center of the magnet with easy access to the injection lines. Ensure that there is some container to catch the liquid that will vent out to the exhaust line.
Prepare high activity enzyme mixture (17.14 mM NADH, 44.57 U/ml LDH). Mix together 240 µl NADH solution, 125 µl LDH solution and 335 µl buffer and keep in a 3 ml syringe that can be attached to the injection line. NOTE: Once combined with the 500 µl of 40 mM pyruvate from the DNP system, the final phantom volume will be 1.2 ml with concentrations of 16.7 mM Pyruvate, 10 mM NADH, and 26 U/L LDH in a tris buffered solution with pH ~ 7.5.
Prepare low activity enzyme mixture (17.14 mM NADH 26.79 U/ml LDH) Mix together 240 µl NADH solution, 75 µl LDH solution and 385 µl buffer and keep in a separate 3 ml syringe that can be attached to the injection line. NOTE: Once combined with the 500 µl of 40 mM pyruvate from the DNP system the final phantom volume will be 1.2 ml with concentrations of 16.7 mM Pyruvate, 10 mM NADH, and 15.625 U/L LDH in a tris buffered solution with pH ~ 7.5.
4. Run Any Quality Assurance (QA) and Positioning Scans
- Initial positioning.
- Load a new FLASH positioning scan in operation mode [1H] TX/RX Volume mode. Change set up dimensions to 2: Spectrometer Control Tool -> Edit GS -> Setup Dimensions -> 2. Press GSP on the Spectrometer Control and move phantom until centered in the magnet. Press STOP then Press GOP on the Spectrometer Control.
- Pilot Scan.
- Load a new TriPiolt positioning scan in operation mode [1H] TX/RX Volume mode. Position Slice: Scan Control -> Slice Tool- > move slices (hold M key click and drag; select slice to move with the slice package slider).
- Wobble 1H Coil: Spectrometer Control Tool - > Acq -> Wobble. Tune and match 1H coil behind the magnet and press STOP. While holding the shift key press the traffic light on the scan control window.
5. Radial Echo Planar Spectral Imaging Scan Setup
Load a new radial echo planar spectroscopic imaging (radEPSI) scan in operation mode [13C] TX/RX Volume mode. Position Slice: Slice Tool on the Scan control and move slices (hold M key click and drag; select slice to move with the slice package slider).
Wobble 13C Coil by clicking Spectrometer Control Tool - > Acq -> Wobble. Set the receiver gain to 1,000-2,000 on the Spectrometer.
Perform the final system check. Depending on the sequence, observe carbon 13 signal from the urea chamber in a scout protocol. NOTE: This ensures that the system is set up properly before beginning the irreversible dissolution process.
6. Run Dissolution
When the pyruvate has attained >90% polarization (~ 1 hr), the solutions and phantom are ready, and the scan is configured click the run dissolution button on the DNP system console.
When prompted move the dissolution stick into its operating position and inject dissolution media. Close the DNP system and click the finished button on the DNP system console. Move dissolution stick back to resting position when prompted then click finish.
When the DNP system delivers the hyperpolarized pyruvate (~ 2 min after heating starts) withdraw 500 µl of the pyruvate solution into each the high and low enzyme concentration solution syringes. Slowly (~10 sec) inject each syringe into an injection line. NOTE: Scanning could have been initiated prior to injection or anytime up to 3 min post injection depending on the scan protocol used.
7. Image Processing
NOTE: This phantom was designed for use with many imaging strategies. See Figure 2, as an example of how the rad-EPSI images were processed using Matlab.
Load raw data from the fid file. Reshape the data to match the number of projections, read out points, echoes and account for data stored as real and imaginary pairs. Separate out the even and odd echo points.
Fourier transform either the even or odd echoes along the echo dimensions. Visually identify the frequency bands for pyruvate and lactate. For simplicity the absolute value of the spectrum was used.
Separate each metabolite band and Fourier transform along the frequency encode direction to yield isolated sinograms for each metabolite. Inverse radon transform the separate sinograms to produce image of either lactate or pyruvate.
Representative Results
Slice-selective 2D images were acquired using a snapshot radEPSI sequence. Metabolite images were reconstructed using filtered back projection. The metabolite images were well aligned with proton images, as seen in Figure 2. In this system hyperpolarized lactate signal can only be generated from the enzymatic conversion of hyperpolarized pyruvate. In Figure 4, the bottom chamber, with higher LDH concentration, has a stronger lactate and weaker pyruvate signal as compared to the top chamber. Using the relative metabolite signals as an estimation of enzyme concentration the lactate to pyruvate ratio is 1.47 times higher in the lower chamber. The actual enzyme concentration ratio between the lower and top compartments was 1.66 times which is in rough agreement with the signal ratios (Figure 4).
Time course images were generated using an IDEAL sequence (Figure 3). Strong agreement between the metabolite images and the proton reference was again observed. Note that no urea phantom was present for this study. In the high enzyme chamber at right, a very strong initial lactate signal is observed. The reaction was almost completely catalyzed by the time it was delivered into the chamber (9 sec delivery, 12 sec peak lactate). The reaction progressed more slowly in the low enzyme concentration chamber (left). It is also notable that less pyruvate signal was observed in the high enzyme chamber as more of the signal was converted into lactate.
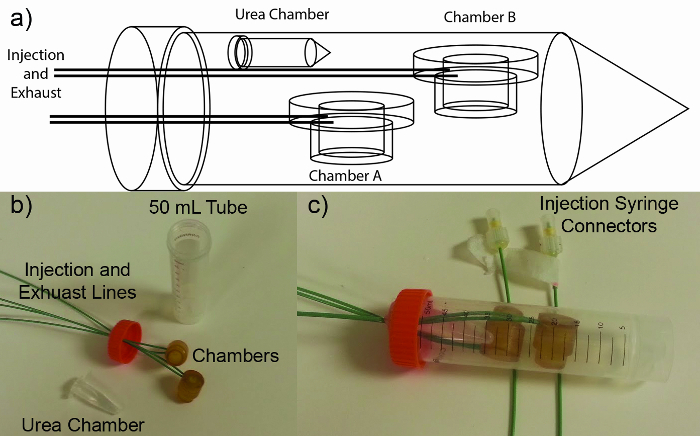 Figure 1. Phantom Schematic: (A) Diagram showing the chamber design. Chambers A and B fitted for injections and exhaust lines. The urea chamber is sealed as it does not need to be injected during acquisition. (B) Photo of disassembled phantom showing the chambers, injection lines and 50 ml tube. (C) The assembled phantom system with the injection lines wrapped around so the syringe connectors at the end of the lines are in the frame. Please click here to view a larger version of this figure.
Figure 1. Phantom Schematic: (A) Diagram showing the chamber design. Chambers A and B fitted for injections and exhaust lines. The urea chamber is sealed as it does not need to be injected during acquisition. (B) Photo of disassembled phantom showing the chambers, injection lines and 50 ml tube. (C) The assembled phantom system with the injection lines wrapped around so the syringe connectors at the end of the lines are in the frame. Please click here to view a larger version of this figure.
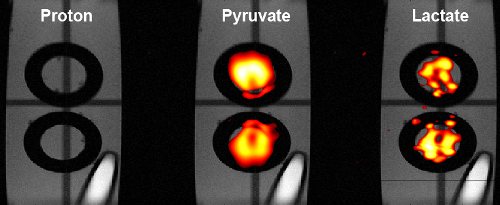 Figure 2. Metabolite images using rad-EPSI. Coronal images generated from two separate spectral bands, pyruvate (center) and lactate (right). The top chamber has noticeably more pyruvate signal and less lactate signal than the bottom chamber. A proton image (left) was acquired after imaging hyperpolarized agents. Please click here to view a larger version of this figure.
Figure 2. Metabolite images using rad-EPSI. Coronal images generated from two separate spectral bands, pyruvate (center) and lactate (right). The top chamber has noticeably more pyruvate signal and less lactate signal than the bottom chamber. A proton image (left) was acquired after imaging hyperpolarized agents. Please click here to view a larger version of this figure.
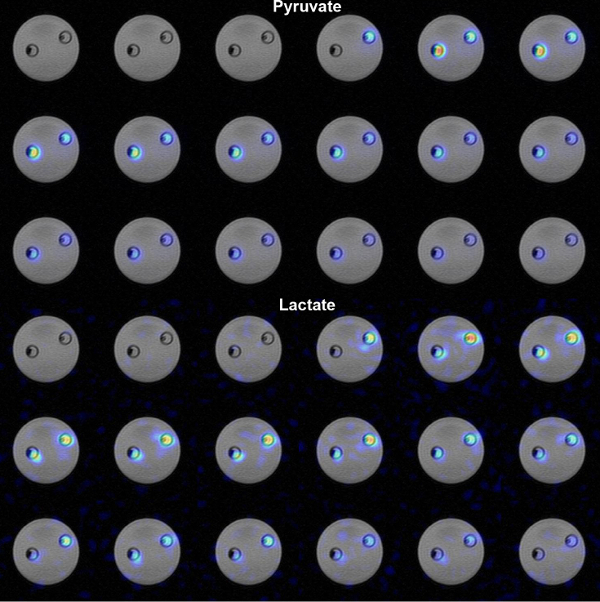 Figure 3. Metabolite time course images using an IDEAL sequence. Axialimages in this series were acquired in 3-sec intervals. Pyruvate (top) and lactate (bottom) are displayed over the grayscale proton image. Strong lactate and weaker pyruvate signals are observed in the right chamber that contains the higher enzyme concentration. The left chamber, with lower enzyme concentration, showed less lactate and stronger pyruvate signal. The chambers are void of hyperpolarized signal for the first 9 sec because the scan was initiated prior to injection. Please click here to view a larger version of this figure.
Figure 3. Metabolite time course images using an IDEAL sequence. Axialimages in this series were acquired in 3-sec intervals. Pyruvate (top) and lactate (bottom) are displayed over the grayscale proton image. Strong lactate and weaker pyruvate signals are observed in the right chamber that contains the higher enzyme concentration. The left chamber, with lower enzyme concentration, showed less lactate and stronger pyruvate signal. The chambers are void of hyperpolarized signal for the first 9 sec because the scan was initiated prior to injection. Please click here to view a larger version of this figure.
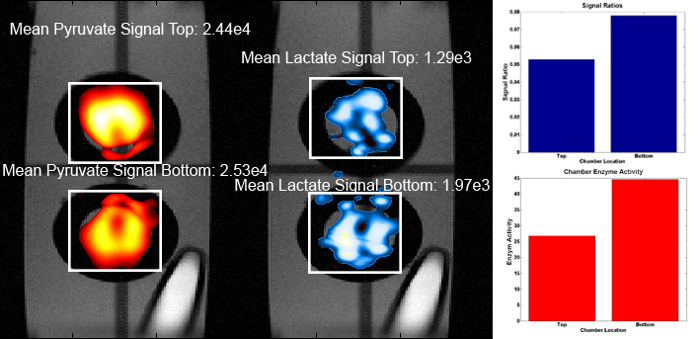 Figure 4. Processed Images: The average signal magnitudes for each chamber shown over the combined metabolite images (left). The difference in lactate to pyruvate ratios for the top and bottom chambers qualitatively match the difference in enzyme activities used, shown as bar graphs (right).
Figure 4. Processed Images: The average signal magnitudes for each chamber shown over the combined metabolite images (left). The difference in lactate to pyruvate ratios for the top and bottom chambers qualitatively match the difference in enzyme activities used, shown as bar graphs (right).
Discussion
Real time imaging of hyperpolarized metabolites has many unique challenges for sequence design, validation, and quality control. The ability to resolve spatiotemporal and spectral heterogeneity offers substantial clinical potential but precludes QA and validation methods associated with conventional MRI. Complex imaging sequences or reconstruction algorithms can have subtle dependencies that render them difficult to characterize or validate outside of the imaging experiment. Biological heterogeneity and other practical concerns limit the use of in vivo and in vitro models to characterize or validate sequences, hardware or data processing algorithms.
With multiple spatial compartments and dynamic chemical evolution, it is possible to recapitulate the critical features associated with imaging hyperpolarized substrates in vivo but in a controlled manner. Regions of high and low enzyme activity provide reproducible dynamic contrast for assessment of qualitative and quantitative imaging biomarkers. Spatial variations ensure geometric distortions, misalignments, and other common sources of error are accounted for properly.
While more controllable than living systems the reaction rates catalyzed by LDH are still very sensitive experimental conditions and are a major source of uncertainty. Critically, LDH is sensitive to temperature. If subsequent measurements are to be compared great care must be taken that the room and liquid temperatures remain constant across all measurements. Additionally the high concentrations of pyruvate associated with hyperpolarized studies forces the use of high concentrations of the coenzyme NADH and the enzyme LDH. These high concentrations generally do not allow the assumptions of little to no enzyme inhibition or a particular substrate being in excess concentration. If rigorous characterization of the chemical kinetics was desired likely difference concentrations of reagents would be needed to recover these assumptions. However, this formulation will result in the production of hyperpolarized lactate at variable rates in each compartment which should be sufficient for most imaging validation.
If little or no lactate signal is observed a first place to check would be the activity of the enzyme, which may need to be replaced. Additionally variation in delivery into the phantom compartments causes uncertainty in the amount of hyperpolarized signal. To reduce these sources of variability it is critical that the amount of hyperpolarized pyruvate added to each chamber is measured accurately and it is added smoothly. The inherent signal decay of hyperpolarized pyruvate complicates this task by placing strict time limits on the injection. Future work will focus on implementing an automated injection mechanism that will allow the process of hyperpolarized pyruvate introduction to the enzyme mixture and subsequent injection into the phantom to be machine controlled. This system will remove the human error in measuring the amount of pyruvate added to each chamber as well as increase the speed of delivery to the scanner. Notably the hyperpolarized pyruvate was withdrawn into the syringes containing the enzyme mixture before the entire volume was injected into the chambers. This was done to allow the injection process to aid in mixing the pyruvate with the enzyme mixture. A limitation of this method is that the reaction of pyruvate to lactate is initiated before the mixture is in the scanner and therefore the early part of the reaction is not measured. Future refinement of the chamber design will aim to allow the chambers to be pre filled with the enzyme mixture and only the hyperpolarized pyruvate injected while still ensuring sufficient mixing of the chamber contents as pyruvate arrives.
Compared to static phantoms this system has some limitations. The dependence on some dynamic chemical reaction limits these phantoms to single use. After injection the lines and chambers will need to be cleaned and fresh reagent solutions will need to be used, or thawed from aliquots. While the single enzyme phantom captures some of dynamic chemical behavior it does not faithfully recapitulate an actual biologic system. In living systems pyruvate is involved in multiple complex biochemical pathways and the simple conversion of pyruvate to lactate in the presence of co-enzyme with a single enzymatic isoform is a substantial simplification. Additionally, if vascular or some other form of delivery as well as any type of cellular uptake are critical to the measurement this system will likely be an insufficient model. The single enzyme phantom is a compromise that focuses on some chemical reaction, it is not as repeatable or practical as a static phantom but is still more repeatable that living systems. The reproducibility of this system is characterized by variation in repeated measurement of 15 to 20 percent as compared to animal studies which have between 25 to 40 percent intra group variability10. Additionally it does not model every aspect of living cells or tissue but maintains the critical dynamic behavior that is lost when using a static phantom.
This phantom system was designed to operate with a range of imaging techniques. With such a flexible platform the QA scans and hyperpolarized acquisitions performed above are interchangeable with the sequence or protocols used by another institution. To illustrate this we performed a similar study on a GE 3T scanner using a custom IDEAL sequence with similar results.
Hyperpolarized MRI of [1-13C]-pyruvate shows great promise due to its ability to probe in vivo cancer metabolism in real time. Interestingly, the sensitivity of this technique poses a great challenge when developing or validating measurement techniques. Using a single enzyme phantom it is possible to recreate the spatial and temporal dynamics that are critical to measurement of hyperpolarized agents by MRI in a practical, controllable and repeatable manner necessary to be useful for development and verification.
Disclosures
Publication of this video-article is supported by Bruker corporation.
Acknowledgments
This work was supported in part by the Cancer Prevention and Research Institute of Texas (RP140021-P5), a Julia Jones Matthews Cancer Research Scholar CPRIT research training award (RP140106, CMW), and the National Institutes of Health (P30-CA016672).
References
- Merritt ME, et al. Hyperpolarized 13C allows a direct measure of flux through a single enzyme-catalyzed step by NMR. Proceedings of the National Academy of Sciences of the United States of America 104. 2007;104:19773–19777. doi: 10.1073/pnas.0706235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues TB, et al. Magnetic resonance imaging of tumor glycolysis using hyperpolarized 13C-labeled glucose. Nature medicine. 2014;20:93–97. doi: 10.1038/nm.3416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day SE, et al. Detecting tumor response to treatment using hyperpolarized 13C magnetic resonance imaging and spectroscopy. Nature medicine. 2007;13:1382–1387. doi: 10.1038/nm1650. [DOI] [PubMed] [Google Scholar]
- Keshari KR, et al. Hyperpolarized 13C dehydroascorbate as an endogenous redox sensor for in vivo metabolic imaging. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18606–18611. doi: 10.1073/pnas.1106920108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher FA, et al. Magnetic resonance imaging of pH in vivo using hyperpolarized 13C-labelled bicarbonate. Nature. 2008;453:940–943. doi: 10.1038/nature07017. [DOI] [PubMed] [Google Scholar]
- Larson PE, et al. Investigation of tumor hyperpolarized [1-13C]-pyruvate dynamics using time-resolved multiband RF excitation echo-planar MRSI. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2010;63:582–591. doi: 10.1002/mrm.22264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham CH, Dominguez Viqueira W, Hurd RE, Chen AP. Frequency correction method for improved spatial correlation of hyperpolarized 13C metabolites and anatomy. NMR in biomedicine. 2014;27:212–218. doi: 10.1002/nbm.3055. [DOI] [PubMed] [Google Scholar]
- Larson PE, et al. Fast dynamic 3D MR spectroscopic imaging with compressed sensing and multiband excitation pulses for hyperpolarized 13C studies. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2011;65:610–619. doi: 10.1002/mrm.22650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer D, et al. Application of subsecond spiral chemical shift imaging to real-time multislice metabolic imaging of the rat in vivo after injection of hyperpolarized 13C1-pyruvate. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2009;62:557–564. doi: 10.1002/mrm.22041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CM, et al. A Catalyzing Phantom for Reproducible Dynamic Conversion of Hyperpolarized [1-C-13]-Pyruvate. PloS one. 2013;8:e71274. doi: 10.1371/journal.pone.0071274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin YS, Mayer D, Yen YF, Hurd RE, Spielman DM. Optimization of fast spiral chemical shift imaging using least squares reconstruction: application for hyperpolarized (13)C metabolic imaging. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2007;58:245–252. doi: 10.1002/mrm.21327. [DOI] [PubMed] [Google Scholar]
- von Morze C, et al. Simultaneous multiagent hyperpolarized (13)C perfusion imaging. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2014;72:1599–1609. doi: 10.1002/mrm.25071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogaard LV, Schilling F, Janich MA, Menzel MI, Ardenkjaer-Larsen JH. In vivo measurement of apparent diffusion coefficients of hyperpolarized (1)(3)C-labeled metabolites. NMR in biomedicine. 2014;27:561–569. doi: 10.1002/nbm.3093. [DOI] [PubMed] [Google Scholar]
- Patrick PS, et al. Detection of transgene expression using hyperpolarized 13C urea and diffusion-weighted magnetic resonance spectroscopy. Magnetic resonance in medicine : official journal of the Society of Magnetic Resonance in Medicine / Society of Magnetic Resonance in Medicine. 2015;73:1401–1406. doi: 10.1002/mrm.25254. [DOI] [PubMed] [Google Scholar]


