Summary
The protein kinase C (PKC) family of serine-threonine kinases plays a central role in T lymphocyte activation. Here, we identify NR2F6, a nuclear zinc-finger orphan receptor, as a critical PKC substrate and essential regulator of CD4+ T cell activation responses. NR2F6 potently antagonized the ability of T helper 0 (Th0) and Th17 CD4+ T cells to induce expression of key cytokine genes such as interleukin-2 (IL-2) and IL-17. Mechanistically, NR2F6 directly interfered with the DNA binding of nuclear factor of activated T cells (NF-AT):activator protein 1 (AP-1) but not nuclear factor κB (NF-κB) and, subsequently, transcriptional activity of the NF-AT-dependent IL-17A cytokine promoter. Consistent with our model, Nr2f6-deficient mice had hyperreactive lymphocytes, developed a late-onset immunopathology, and were hypersusceptible to Th17-dependent experimental autoimmune encephalomyelitis. Our study establishes NR2F6 as a transcriptional repressor of IL-17 expression in Th17-differentiated CD4+ T cells in vitro and in vivo.
Introduction
Immune responses are exquisitely controlled, requiring multiple finely tuned activation and inactivation signals. Engagement of the T cell receptor (TCR) and coreceptors triggers immediate activation of the critical transcription factors nuclear factor κB (NF-κB), nuclear factor of activated T cells (NF-AT), and activator protein 1 (AP-1), whose activation results in, e.g., interleukin-2 (IL-2) expression and, ultimately, in T cell activation and differentiation into effector-memory subsets. Protein kinase C (PKC) isotypes are serine-threonine kinases that play a key role in these cellular signaling pathways in lymphocytes (Baier, 2003; Spitaler and Cantrell, 2004; Tan and Parker, 2003). PKCθ has been shown to be critical in cytokine responses in vitro (Altman et al., 2004; Pfeifhofer et al., 2003; Sun et al., 2000) and T cell immune responses in vivo including T helper 17 (Th17)-mediated autoimmunity (Anderson et al., 2006; Chaudhary and Kasaian, 2006; Tan et al., 2006). Additionally, studies have shown that also the classical PKC isotypes, PKCα and PKCβ, are critical for T cell activation processes (Pfeifhofer et al., 2006, Volkov et al., 2001). PKC-mediated signals induce NF-AT:AP-1 transactivation in T cells (Isakov and Altman, 2002; Tan et al., 2006); nevertheless, direct effectors of PKC, as well as the biochemical basis by which PKC isotypes mediate this, have remained enigmatic. The rise of intracellular Ca2+ triggered by antigen binding to the TCR leads to the activation of the phosphatase activity of calcineurin followed by dephosphorylation of NF-AT and, finally, to nuclear import of NF-AT. In effector CD3+ T cells, this Ca2+-calcineurin-NF-AT pathway crosstalks with the Ras-MAPK-AP-1 signaling pathway, and NF-AT forms complexes with the transcription-factor family of AP-1 proteins in order to bind with high affinity to DNA (Rao et al., 1997; Macian et al., 2001).
Here we describe a mechanism by which NF-AT activities induced by antigen-receptor stimulation are regulated in CD4+ T cells. This involves the nuclear orphan receptor NR2F6 (nuclear receptor subfamily 2, group F, member 6; previously known as “Ear2” [Nuclear Receptors Nomenclature Committee, 1999; Miyajima et al., 1988; Giguere, 1999]) as a player in T cell attenuation. A defining feature of the three mammalian NR2F subfamily members, NR2F1, NR2F2, and NR2F6, is their role in the regulation of organogenesis, neurogenesis, and cellular differentiation during embryonic development (Park et al., 2003; Takamoto et al., 2005; You et al., 2005; Zhang and Dufau, 2004). NR2F6 as transcription factor binds to a TGACCT direct-repeat motif and has been established in controlling facets of central nervous system (CNS) (Liu et al., 2003; Warnecke et al., 2005); however, a function for NR2F6 in the immune system has yet to be demonstrated. Mechanistically, lymphocyte-expressed NR2F6 acts as a critical regulatory factor during T lymphocyte activation, potently antagonizing antigen-receptor-induced cytokine responses in vitro and in vivo. DNA binding of NR2F6 is under the direct control of TCR-induced PKC signaling, the latter known to positively adjust activation thresholds in CD3+ T cells (Baier, 2003; Tan and Parker, 2003).
Physiologically, NR2F6 appeared of particular importance in the effector-memory CD4+ T cell lineage—known as Th17 cells—that produces IL-17. Th17 cells are critically involved in the pathophysiology of tissue inflammation and autoimmunity (Bettelli et al., 2007; Harrington et al., 2005, 2006; McKenzie et al., 2006; Ivanov et al., 2007; Jankovic and Trinchieri, 2007; Weaver et al., 2007; Sundrud and Rao, 2008). Th17 differentiation is directed by two lineage-specific nuclear orphan receptors, RORα (NR1F1) and RORγ (NR1F3), that positively regulate IL-17 transcription (Yang et al., 2008). Opposite to the established roles of RORα and RORγ isotypes, here we found that NR2F6 acted as a nuclear repressor of IL-17 expression and suppressed Th17 cell functions. Therefore, NR2F6 represents a nuclear orphan receptor family member that acts as a transcriptional suppressor of the Th17 CD4+ T cell subset.
Results
Ser-83 on Recombinant NR2F6 Is a PKC Substrate Site
We employed a PKC-phosphorylation-site prediction approach to identify PKC substrates (Fujii et al., 2004) and identified Ser-83 on nuclear orphan receptor NR2F6 (nuclear receptor subfamily 2, group F, member 6; previously known as “Ear2” [Nuclear Receptors Nomenclature Committee, 1999]; domain schematic in (Figure S1A available online) as a candidate PKC substrate site in silico. NR2F6 has been established in controlling facets of CNS development (Warnecke et al., 2005). Endogenous expression of Nr2f6 mRNA has been reported to be high in the embryonic brain and in the developing liver (Miyajima et al., 1988; Warnecke et al., 2005). We detected Nr2f6 expression in the thymus, spleen, lymph node, and bone marrow (Figures 1A and 1B), as well as in CD3+ T and CD19+ B lymphocytes (not shown), indicating a potential function for NR2F6 in the immune system. Of note, a decrease of Nr2f6 mRNA expression was associated with T cell activation, suggesting a silencing effect on Nr2f6 gene transcription by the TCR-mediated signaling pathway (Figure S1B).
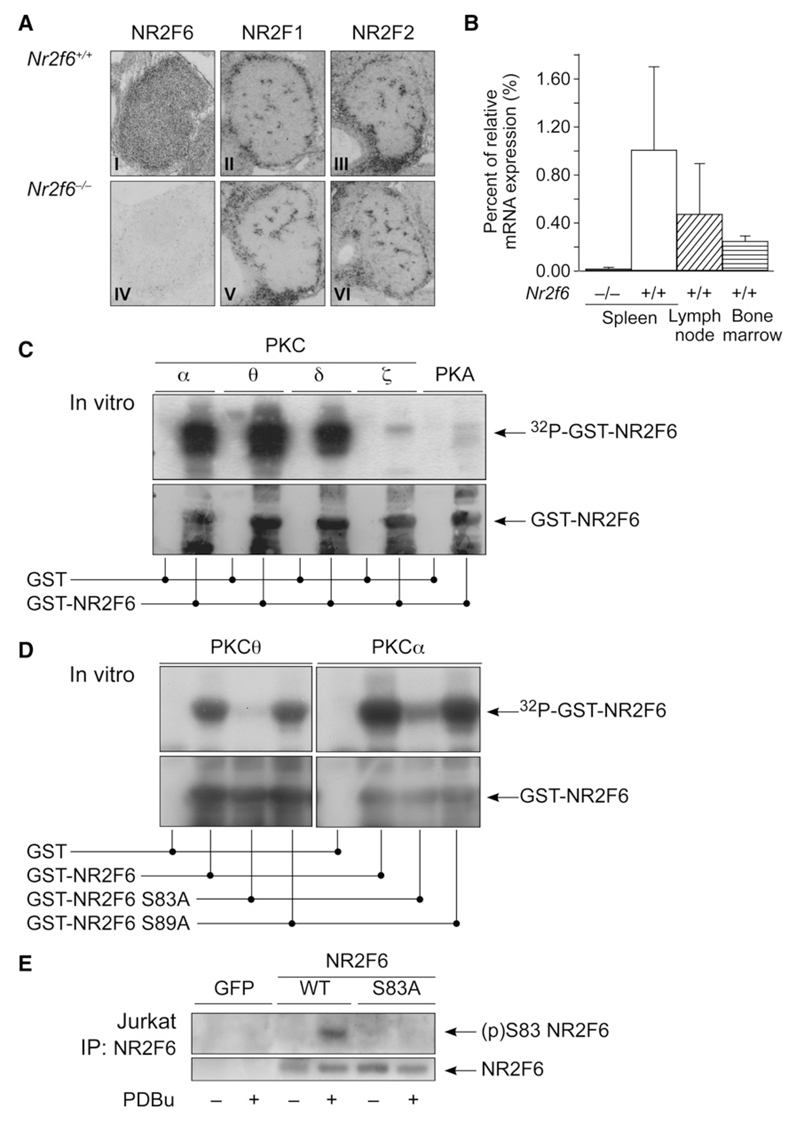
Figure 1. (p)Ser-83 on NR2F6 Is a PKC Phosphorylation Site.
(A) Nr2f6 in situ hybridization was positive in thymus sections of wild-type (I) but not Nr2f6−/− (IV) E14.5 embryonic thymus sections. Neither the expression pattern nor the intensity of the other COUP-TF family members NR2F1 (II, V) and NR2F2 (II, VI) was altered in the Nr2f6−/− thymus.
(B) qRT-PCR revealed Nr2f6 expression in spleen, lymph node, and bone marrow in the wild-type mouse. Nr2f6−/− samples served as specificity negative controls, data were normalized to GAPDH, and expression in the spleen was arbitrarily taken as 1. Mean of three independent experiments is shown; error bars represent standard error.
(C and D) Kinase assays of full-length GST-NR2F6 (wild-type or S83A or S89A mutant) incubated with recombinant PKC family members (α, θ, δ, and ζ) or protein kinase A (PKA) as control. Specific phosphorylation of NR2F6 by the PKC isotypes α, θ, δ, and, to a much lower extent, PKC ζ and PKA could be observed. This specific PKC-mediated phosphorylation was mostly lost after mutation of the S83 site (but not Ser-89) into a S83A site. The anti-GST immunoblot (lower panel) confirmed equal loading.
(E) Jurkat cells were transfected with GFP, NR2F6 wild-type, or S83A mutant, and NR2F6 was immunoprecipitated from resting cells (–) or cells stimulated for 20 min with 50 nM phorbol ester (PDBu) (+) and immunoblotted with the anti-(p)Ser-83 NR2F6-specific pAb.
We confirmed the in silico prediction by biochemical analysis of NR2F6 and PKC. PKCα, δ, and θ, as well as, to a much lower extent, PKCζ, PKA, and PKB, were able to phosphorylate recombinant NR2F6 in vitro (Figure 1C and data not shown). Mutation of Ser-83 (but not Ser-89) to alanine strongly reduced PKC-mediated NR2F6 phosphorylation, confirming Ser-83 as the major PKC phosphorylation site in NR2F6 (Figure 1D). Consistent with its identification as a PKC substrate, transfected NR2F6 bound to endogenous PKCα and PKCθ (but not PKB) in pulldown assays from T cell lysates (Figure S2A). The presence of this phosphosite on NR2F6 was confirmed with a phosphospecific (p)Ser-83 antiserum that reacted with wild-type NR2F6 but not with the S83A mutant in PDBu-stimulated Jurkat T cells (Figure 1E). Electrophoretic-mobility band-shift assays (EMSAs) revealed that in nuclear extracts from unstimulated cells transiently expressing NR2F6, wild-type NR2F6 bound to its established TGACCT direct-repeat DNA motif (Figure 2A and Figure S2B). Notably, wild-type NR2F6 DNA binding decreased in CD3 plus CD28-activated cells. The phosphomimetic replacement of Ser-83 with glutamic acid (S83E) completely abrogated DNA binding of NR2F6 in nuclear extracts in both resting and stimulated cells (Figures 2A and 2B), suggesting that the DNA-binding capacity of NR2F6 is antagonized by a (p)Ser-83 switch on NR2F6.
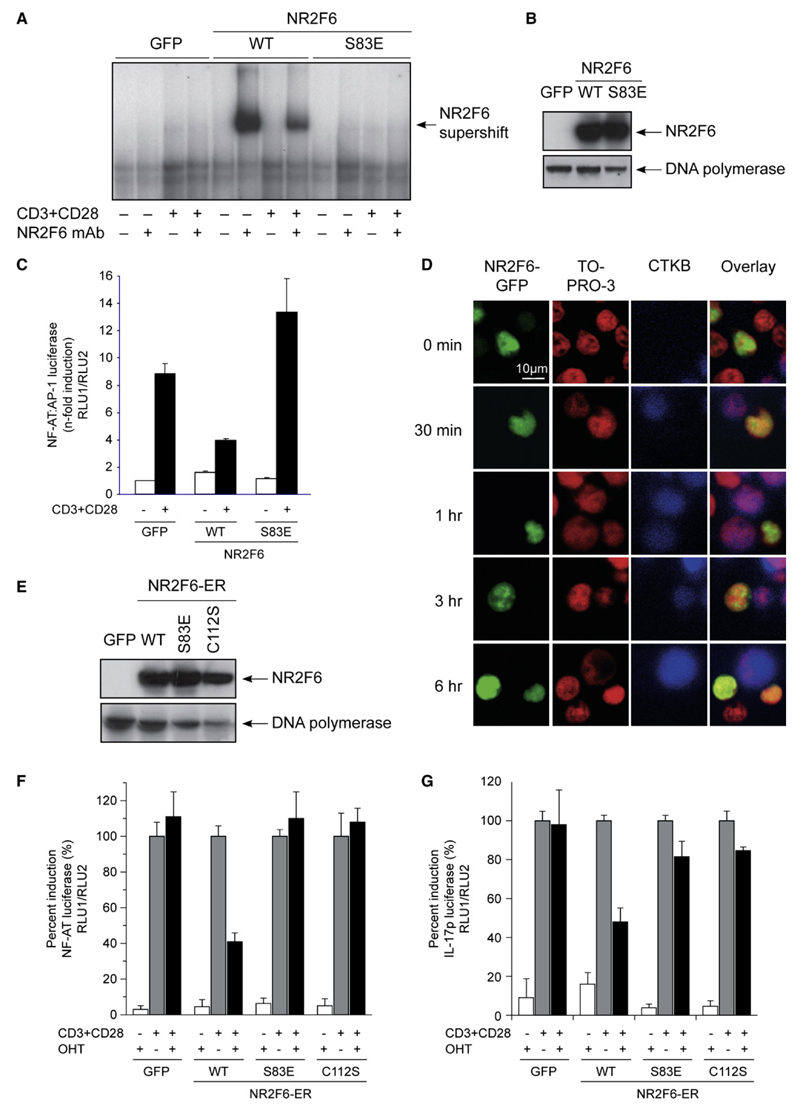
Figure 2. (p)Ser-83 on NR2F6 Regulates NR2F6 DNA Binding in Jurkat T Cells.
(A) NR2F6 Ser-83 phosphorylation directly influenced DNA binding in nuclear extracts as analyzed by EMSA. Jurkat T cells were transfected with NR2F6 wild-type, S83E mutant, or GFP control and were left unstimulated (–) or were CD3 plus CD28 stimulated (+). Supershift analysis was performed with a mAb for NR2F6. In contrast to the NR2F6 wild-type, the S83E mutant did not bind DNA.
(B) Equal expression of NR2F6 wild-type and S83E mutant proteins in the nuclear fractions was confirmed by immunoblotting (DNA polymerase served as loading control). NR2F6 wild-type and mutant protein expression remained unaltered also during CD3 plus CD28 stimulation (not shown).
(C) Transfected wild-type NR2F6 interfered with CD3 plus CD28-induced transcriptional activation of the NF-AT:AP-1 promoter luciferase reporter; this repressor activity was abolished by the S83E mutation in NR2F6.
(D) Jurkat T cells transfected with NR2F6-GFP (green) were used to form conjugates with SEE-pulsed B cells stained with the cell tracker blue (CTKB). Cells were then fixed and nuclei were stained with TOPRO-3 (red). Conjugates were analyzed for NR2F6 localization at different time points. Similar results were obtained with an RGS-His6-tagged NR2F6 and an anti-RGSHis6 mAb staining (not shown).
(E–G) Cotransfection experiments in Jurkat T cells showed that recombinant NR2F6-ER wild-type but not DNA-binding-defective mutants, S83E and C112S, induced repression of CD3 plus CD28-induced NF-AT-dependent reporter luciferase gene transcription. CD3 plus CD28-induced IL-17-dependent promoter luciferase reporter was similarly repressed by recombinant NR2F6-ER wild-type in transfected Jurkat T cells. Reporter induction rates were normalized for the transfected cells, and CD3 plus CD28-induced reporter activity without OHT treatment was arbitrarily set as 100%. Mean of at least two independent experiments analyzed in triplicates is shown. Error bars represent standard error. Equal expression of NR2F6 wild-type and S83E and C112S mutant proteins in the nuclear fractions was confirmed by immunoblotting (DNA polymerase served as loading control).
We extended the NR2F6 DNA-binding results to examine a role of NR2F6 as transcriptional regulator of the PKC-dependent NF-AT:AP-1 signaling pathway established to cooperatively regulate IL-2 target-gene expression in T cells (Macian et al., 2001). Transfection of wild-type NR2F6 in Jurkat cells resulted in a repression of the NF-AT:AP-1-dependent promoter luciferase reporter expression, containing the composite NF-AT:AP-1 DNA-binding element derived from the IL-2 minimal promoter region (Figure 2C). The S83E mutation in NR2F6 abolished its repressor activity on the NF-AT:AP-1 reporter, confirming the observation that the phosphorylation of Ser-83 abrogates DNA-binding capacity and, subsequently, transcriptional repressor function of NR2F6 (Figure 2C). Subcellular location analysis by confocal microscopy also defined NR2F6 as nuclear protein in both unstimulated and TCR-stimulated Jurkat T cells (Figure 2D).
To establish whether NR2F6 is directly involved in the regulation of NF-AT, we monitored the transcriptional activity of a cis-acting NF-AT-promoter reporter construct (containing multiple copies of only the NF-AT enhancer element) in a conditionally activated NR2F6 estrogen-receptor (ER) fusion mutant (NR2F6-ER; see Figure S3 for illustration) expressing Jurkat T cells. Exposure to 4-hydroxytamoxifen (OHT), the selective agonist of ERmut-LBD within the recombinant NR2F6-ER, selectively induced transrepression of the TCR-activation-induced NF-AT reporter gene transcription in NR2F6-ER-expressing Jurkat T cells (Figures 2E and 2F). NF-AT reporter activity was not affected by expression of NR2F6-ER without OHT treatment, nor by OHT treatment in control transfected cells (Figure 2F and data not shown). Similarly, and consistent with published studies that defined NF-AT as cis-acting transactivator for IL-17A gene transcription (Liu et al. [2004] and, for review, Chen et al. [2007]), NR2F6-ER also repressed TCR-induced IL-17A promoter-reporter activity (Figure 2G). This indicates that NF-AT is one critical transcription factor downstream of NR2F6 in the regulation of IL-17A promoter transcription. Again, phosphorylation status of Ser-83 affected NR2F6-ER cellular function, because the S83E mutation on NR2F6-ER completely rescued reporter transcription to control levels. Similarly, the C112S zinc-finger mutant of NR2F6, established to be defective in DNA binding (Liu et al., 2003 and data not shown), lost its transcriptional repressor activity, indicating that NR2F6-mediated transcriptional repression is strictly dependent on its DNA-binding ability (Figures 2E–2G). These findings validate NR2F6 as a critical negative modulator of both NF-AT DNA binding and NF-AT-dependent IL-17A promoter transcriptional responses in Jurkat T cells.
T and B Cell Development Is Normal in NR2F6-Deficient Mice
Nr2f6−/− mice are viable and fertile (Warnecke et al., 2005) and show normal thymocyte development (Table 1A). Nr2f6−/− thymocytes demonstrated normal susceptibility to TCR-ligation-mediated apoptosis both in vitro (not shown) and in vivo (Figure S4), suggesting normal sensitivity to negative selection signals of immature thymocytes. B lymphocyte development in the bone marrow of Nr2f6−/− mice was not different from wildtype controls, resulting in normal mature B cell subsets (Figure S5 and Table 1B). Consistent with this observation, FACS analysis of spleen and lymph nodes of 6–10-week-old Nr2f6−/− mice revealed no gross differences in the distribution of CD3-, CD4-, CD8-, and CD19-positive cells. In the spleen, surface expression of CD3, CD4, CD8, CD44, CD62L, ICOS, TCR, Vβ8, CD19, B220, IgM, IgD, CD43, CD5, CD21, CD23, Ger1, Mac1, and Thy1, as well as ratios of T and B cell populations, was comparable to that of Nr2f6+/+ mice (Table 1B and data not shown), suggesting that NR2F6 is dispensable for normal lymphocyte development.
Table 1. T Cell Development and T and B Lymphocyte Subsets in Young and Old Nr2f6+/+ and Nr2f6−/− Mice.
| A. Thymus | ||||
|---|---|---|---|---|
| Nr2f6+/+ | Nr2f6−/− | p Values | n | |
| Total (x 106) | 241.6 ± 9.0 | 206.6 ± 20.1 | 0.156 | 6 |
| CD4+CD8- | 20.3 ± 4.1 | 17.5 ± 4.4 | 0.343 | 6 |
| CD4−CD8+ | 7.1 ± 0.6 | 3.8 ± 0.8 | 0.010 | 6 |
| CD4+CD8+ | 200.4 ± 8.3 | 175.9 ± 16.3 | 0.223 | 6 |
| B. Spleen (6–10-Week-Old Mice) | ||||
| Nr2f6+/+ | Nr2f6−/− | p Values | n | |
| Total (x 106) | 92.6 ± 10.6 | 79.0 ± 8.2 | 0.7166 | 10 |
| T Cells | ||||
| CD4+ | 16.2 ± 1.1 | 18.8 ± 1.4 | 0.6777 | 8 |
| CD44lo+CD62Lhi | 9.6 ± 1.5 | 12.7 ± 1.7 | 0.5441 | 8 |
| CD44hi+CD62Llo | 7.4 ± 2.4 | 4.9 ± 0.6 | 0.7817 | 8 |
| CD8+ | 11.3 ± 0.8 | 8.6 ± 0.7 | 0.1421 | 8 |
| CD44lo+CD62Lhi | 7.4 ± 0.8 | 6.4 ± 0.8 | 0.1478 | 8 |
| CD44hi+CD62Llo | 3.2 ± 1.3 | 1.6 ± 0.4 | 0.6351 | 8 |
| B Cells | ||||
| CD19+ | 29.1 ± 4.1 | 35.0 ± 4.4 | 0.4783 | 8 |
| IgM+IgD | 34.1 ± 3.7 | 41.9 ± 5.0 | 0.3082 | 8 |
| T1 | 2.8 ± 0. 4 | 2.7 ± 0.4 | 0.9605 | 8 |
| MZ | 3.4 ± 1.0 | 6.1 ± 1.8 | 0.2638 | 8 |
| C. Spleen (1-Yr-Old Mice) | ||||
| Nr2f6+/+ | Nr2f6−/− | p Values | n | |
| Total (x 106) | 64.8 ± 6.2 | 131 ± 26 | 0.0065 | 11 |
| T Cells | ||||
| CD3+ | 26.6 ± 7.1 | 45.3 ± 11 | 0.0331 | 11 |
| CD4+ | 13.6 ± 2.7 | 30.1 ± 6.2 | 0.0039 | 11 |
| CD44lo+CD62Lhi | 7.1 ± 0.4 | 11.6 ± 0.9 | 0.0677 | 5 |
| CD44hi+CD62Llo | 4.8 ± 0.6 | 13.0 ± 5.5 | 0.3486 | 5 |
| CD8+ | 8.7 ± 1.3 | 8.9 ± 2.7 | 0.6769 | 11 |
| CD44lo+CD62Lhi | 3.9 ± 0.6 | 3.6 ± 0.4 | 0.7918 | 5 |
| CD44hi+CD62Llo | 6.3 ± 0.9 | 8.0 ± 2.0 | 0.5778 | 5 |
| B Cells | ||||
| CD19+ | 32.6 ± 3.5 | 62.4 ± 15.5 | 0.1250 | 11 |
| IgM+IgD | 17.5 ± 2.1 | 35.4 ± 9.3 | 0.0460 | 11 |
| T1 | 3.6 ± 0.8 | 6.5 ± 1.5 | 0.5591 | 7 |
| T2 | 1.5 ± 0.2 | 2.5 ± 0.3 | 0.0087 | 7 |
| MZ | 2.8 ± 0.5 | 5.6 ± 1.5 | 0.1210 | 7 |
Nr2f6−/− Mice Have a Hyperreactive Immune Phenotype
The critical function of NR2F6 in lymphocytes was revealed by a late-onset autoimmune pathology. Of all 1-yr-old Nr2f6−/− mice investigated, 55% displayed enlarged spleens (Figure 3A and Figure S6) with increased numbers of T (p = 0.033) and B (p = 0.046) lymphocytes (Figures 3B–3D). Consistently, untreated Nr2f6−/− mice revealed high titers of IgG1 (Figure 3E and Figure S7) and autoantibodies against nuclear antigens (Figures 3F–3H) were detected in sera from these animals. Nr2f6−/− CD4+ T (p = 0.025) and B (p = 0.036) cells were less sensitive to apoptosis induced by antigen-receptor ligation in vitro (Figure S8). The hyperplasia of CD4+ T and B cells in 1-yr-old Nr2f6−/− mice confirmed this observation in vivo (Table 1C).
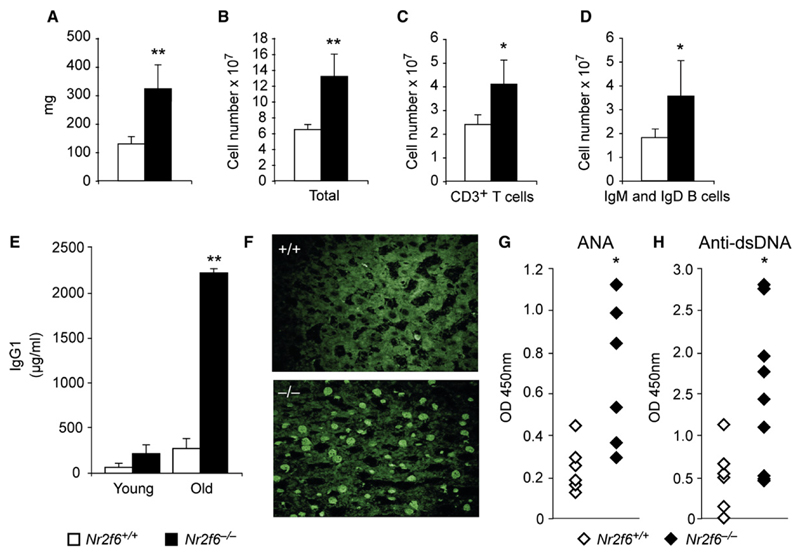
Figure 3. Nr2f6−/− Mice Develop a Late-Onset Immunopathology.
(A–D) Twelve-month-old Nr2f6−/− mice displayed enlarged spleens with increased lymphocyte numbers in Nr2f6−/− mice. (A) Splenic weight, (B) total cellularity, and (C) T cell (CD3+) and (D) mature B cell numbers (IgM+ and IgD+) are shown (Nr2f6+/+, n = 11; Nr2f6−/−, n = 11; unpaired t test; (A) p = 0.009, (B) p = 0.006, (C) p = 0.033, (D) p = 0.046; means are shown with error bars).
(E) Serum immunoglobulin levels of IgG1 young (6–10 weeks) and old (>12 months) Nr2f6−/− mice were determined via ELISA. Twelve-month-old Nr2f6−/− mice show significantly elevated IgG1 plasma titers (unpaired t test, Nr2f6+/+, n = 8; Nr2f6−/−, n = 8; p = 0.007).
(F–H) Aged Nr2f6−/− mice generate autoantibodies against nuclear antigens (ANA; unpaired t test, Nr2f6+/+, n = 9; Nr2f6−\−, n = 9; p = 0.023) and double-stranded (ds) DNA (Nr2f6+/+ n = 9; Nr2f6−/− n = 9; p = 0.037) as determined by staining of rat liver sections with mouse serum (F) and ELISA (G and H). Means are shown; error bars represent standard error.
To determine the functional capacity of peripheral T and B cells in the absence or presence of NR2F6, we analyzed the activation thresholds. Nr2f6−/− T cells were hyperreactive to stimulation, and the CD4+ T subset (unlike the CD8+ T cells) showed an enhanced proliferation response upon CD3 plus CD28 stimulation (Figures S9A and S9B). Stimulated Nr2f6−/− CD4+ T cells produced more IL-2 (p = 0.0007) than wild-type T cells (Figure 4A), whereas IFN-γ secretion in Nr2f6−/− CD8+ T cells was not elevated much more than wild-type (Figure S9C). Nr2f6−/− B cells displayed an enhanced proliferative response upon IgM plus IL-4 stimulation (Figure S10). We used siRNA and overexpression analysis to independently confirm the suppressor function of NR2F6 in CD4+ T cells. The IL-2 secretion response in Nr2f6 siRNA-transfected cells was higher (Figure 4B), whereas NR2F6-overexpressing cells showed diminished IL-2 activation responses (Figure S11A) when compared to control transfected cells. For delivery controls, both the increase in Nr2f6 mRNA expression (5-fold) in plasmid-transfected cells (see Figure S11B) and the reduction in Nr2f6 mRNA expression (>95%) in siRNA-transfected (Figure S12) was measured.
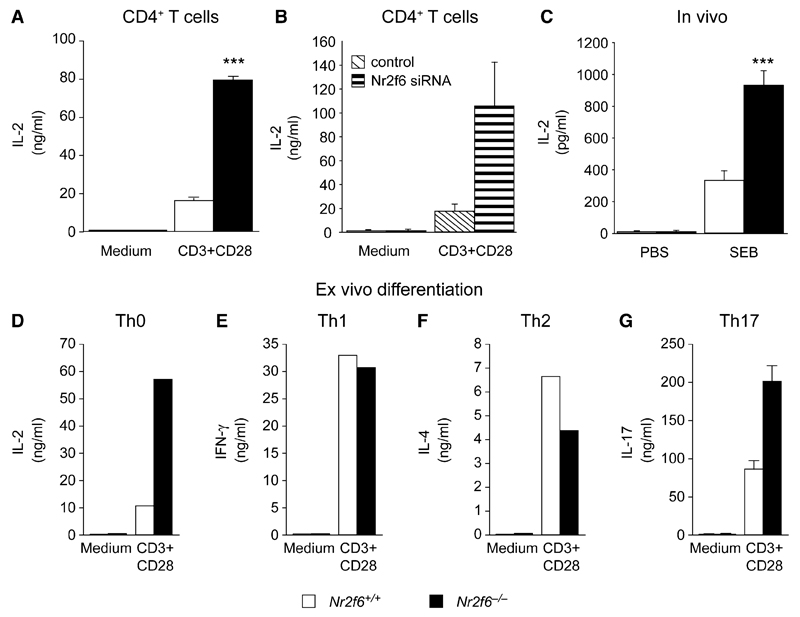
Figure 4. Nr2f6−/− T Cells Hyperrespond to Antigen-Receptor Stimulation.
(A) CD3 plus CD28-induced IL-2 cytokine secretion responses of Nr2f6-deficient T cells (black bars) were significantly higher than wild-type controls (white bars). Data shown are the mean of three independent experiments performed in duplicate (split-plot ANOVA, p = 0.0007).
(B) Similarly, siRNA-mediated Nr2f6 knockdown in CD4+ T cells resulted in enhanced IL-2 cytokine secretion upon CD3 plus CD28 stimulation, compared to siRNA nontargeting controls (n = 2).
(C) IL-2 cytokine concentrations in plasma taken 2 hr after injection with SEB i.p. (10 μg/kg) were significantly higher in Nr2f6−/− mice (unpaired t test; Nr2f6+/+, n = 10; Nr2f6−/−, n = 10; p = 0.0004).
(D–G) Naive CD4+ T cells were differentiated under neutral Th0 (D), (E) Th1, (F) Th2, and (G) Th17 conditions (conditions are defined in the Experimental Procedures), and relevant cytokines were measured from the supernatant after 4–5 days. One of two independent experiments with consistent results is shown for Th0, Th1, and Th2. IL-17 cytokine secretion in Nr2f6-deficient Th17 cells (black bars) was significantly higher than wild-type controls (white bars) (split-plot ANOVA Nr2f6+/+, n = 5; Nr2f6−/−, n = 5; p = 0.004162) Error bars represent standard error.
Consistent with an in vivo role of NR2F6 in signaling attenuation, Nr2f6 deficiency resulted in a profound augmentation of IL-2 in the plasma 2 hr after intraperitoneal (i.p.) injection of superantigen staphylococcal enterotoxin B (SEB), which selectively activated the TCR-Vβ8-positive T cells (Figure 4C). These elevated activation responses were not due to an upregulation of either CD3 or TCR-Vβ8 receptor expression on the surface of T cells (Figures S13A and S13B). Ex vivo differentiation experiments of naive CD4+ T cells thereby defined the selective role of NR2F6 in Th0 and Th17 (but not Th1 and Th2) cells (Figures 4D–4G and Figure S14A and S14B).
NR2F6 Represses NF-AT:AP-1 DNA Binding in CD4+ T Cells
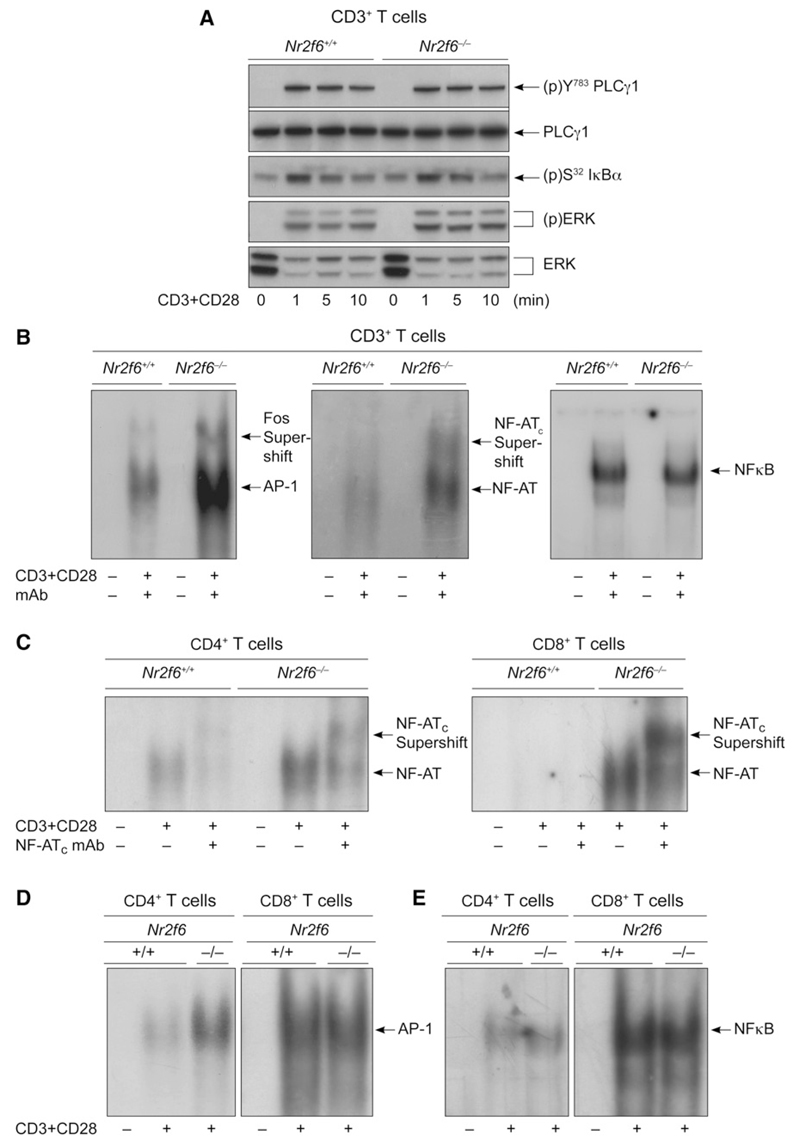
Mechanistically, no differences in the extent or kinetics of membrane-proximal phosphorylation events, such as (p)Tyr-783 on PLCγ1, (p)Ser-32 on I-κBα, or the activation loops on (p)ERK1 and (p)ERK2, were observed upon CD3 plus CD28 stimulation of Nr2f6 wild-type versus Nr2f6-deficient CD3+ T cells (Figure 5A). EMSA analysis of nuclear extracts with sequences containing critical enhancer elements demonstrated amplified NF-AT:AP-1 DNA binding in CD3+ T cells derived from Nr2f6−/− mice (Figure 5B). Similarly, activated Nr2f6−/− CD4+ T cells showed a strong augmentation in DNA binding of the NF-AT:AP-1 complex in comparison to wild-type controls (Figures 5C and 5D). However, AP-1-binding ability was the same in both wild-type and Nr2f6−/− CD8+ T cell subsets (Figure 5D). Unlike NF-AT:AP-1, NF-κB DNA binding was not affected by the absence of NR2F6 in either T cell subset (Figures 5B and 5E). These data physiologically confirm the observation that NR2F6 selectively functions as a negative regulator of both NF-AT:AP-1 transactivation responses in CD4+ T cells.
Figure 5. NR2F6 Acts as Repressor of NF-AT:AP-1 DNA-Binding Capability.
(A) CD3+ T cells (5 × 106 per lane) from Nr2f6+/+ and Nr2f6−/− mice were stimulated with CD3 plus CD28 for the indicated time periods, and the phosphorylation status of proteins was detected by immunoblotting, as indicated. One representative experiment of two is shown.
(B–E) Analysis of DNA-binding activity in nuclear extracts of Nr2f6−/− CD4+ and Nr2f6−/− CD8+ T cells showed that NF-AT DNA binding is higher in both CD4+ and CD8+ T cells, whereas AP-1 DNA binding is higher only in CD4+ T cells. No change was detected in NF-κB binding. Supershift analysis was performed with antibodies against c-fos, NF-ATc, and p50, as indicated. One representative experiment out of four is shown.
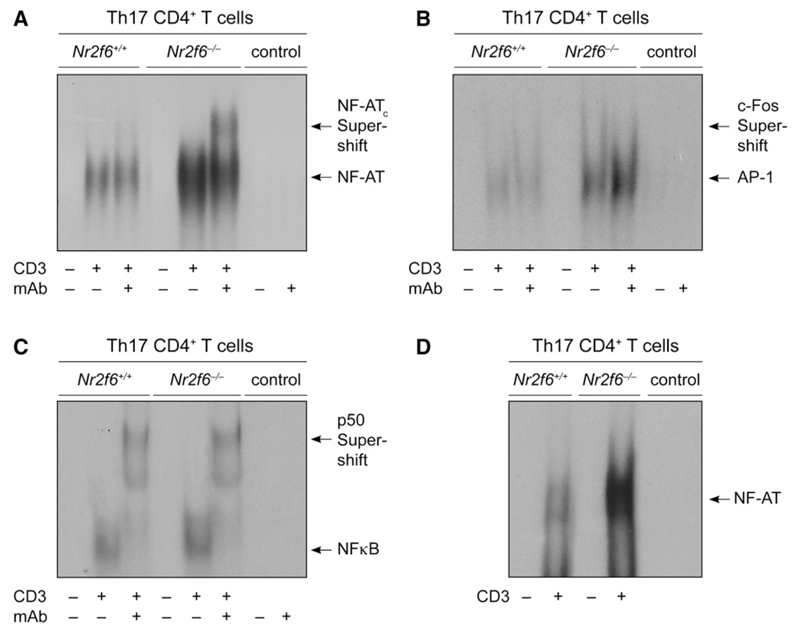
EMSA analysis of ex vivo-differentiated Th17 effector-memory cells established NR2F6 as a Th17 cell-intrinsic repressor of the DNA-binding capabilities of NF-AT:AP-1 (Figures 6A–6C). Consistent with the proposed model of NR2F6 as repressor of NF-AT-dependent IL-17 expression, NR2F6 directly interfered with NF-AT DNA binding to an IL-17-promoter-derived enhancer sequence in Th17 cells (Figure 6D). In contrast to DNA binding and transcriptional-activity regulation of NF-AT, NR2F6 deficiency had neither an effect on activation-induced upregulation (not shown) nor on nuclear translocation of NF-AT in activated primary CD4+ as well as NR2F6-ER-overexpressing Jurkat T cells (Figures S15A and S15B). Taken together, these results substantiate the biological significance of NR2F6 in Th17 cells and provide mechanistic data of NR2F6 as a nuclear repressor of NF-AT DNA binding and NF-AT-dependent transactivation in the context of the IL-17A promoter. This newly discovered repression pathway of the PKC substrate NR2F6 in CD4+ T cells thereby extends the mechanism of action of NF-AT regulation.
Figure 6. NR2F6 Suppresses NF-AT:AP-1 DNA Binding Specifically in CD4+ Th17 Effector-Memory T Cells.
(A–C) EMSA analysis of nuclear extracts prepared from Th17-differentiated and αCD3-antibody-re-stimulated cells (IL-23, TGF-β, IL-6, αIL-4, αIFN-γ). NF-AT:AP-1 DNA binding was higher in Nr2f6-deficient extract when the NF-AT:AP-1 derived from the minimal IL-2 promoter was used, whereas NF-κB remained unchanged.
(D) To distinguish between NF-AT:AP-1 and NF-AT-only binding, we used the NF-AT-specific probe #3 derived from the IL17A minimal promoter region (Liu et al., 2004); this again revealed a higher NF-AT binding in the Nr2f6-deficient nuclear Th17 cell extracts when compared to the wild-type control. Supershift analysis was performed with antibodies against c-fos, NF-ATc, and p50 as indicated. Controls are the radiolabeled probe with or without the supershifting Ab. One representative experiment out of two is shown.
NR2F6-Deficient Mice Are Hypersusceptible to Antigen-Induced Autoimmunity
To gain more insight into NR2F6 immune function in vivo, we immunized 6–10-week-old female mice with the myelin component MOG35-55 to induce experimental autoimmune encephalomyelitis (EAE), a multiple sclerosis-like autoimmune disease. The numbers and ratios of T and B cell populations, including FOXP3+ Treg and Th17 cell lineages, in EAE-diseased mice did not differ between the genotypes (Figure S16). Thus the initial commitment to differentiation of Th17 cells appears mostly independent of NR2F6, both in vitro and in vivo.
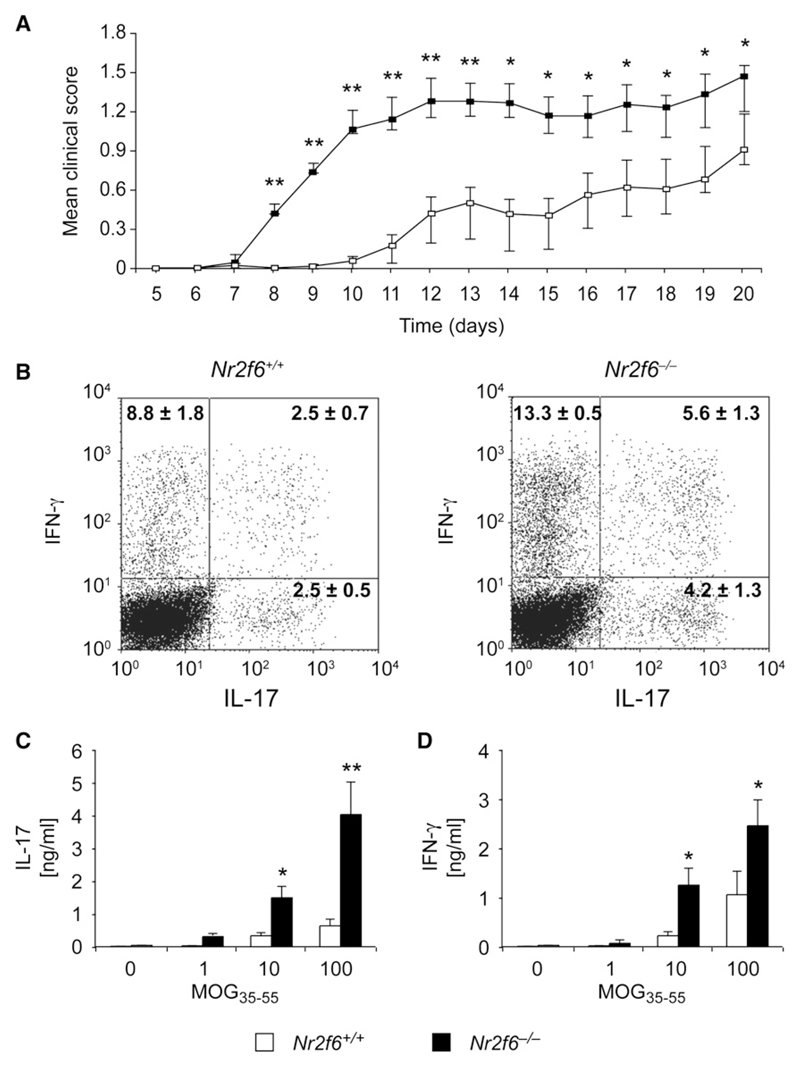
Nevertheless, Nr2f6−/− mice demonstrated both a faster onset and an overall higher clinical score than wild-type mice when progressive paralysis was scored from tail to head (Figure 7A). Accelerated disease in the Nr2f6−/− mice was associated with higher numbers of CNS-infiltrating IL-17-IFN-γ double-positive CD4+ effector T cells (p = 0.049) (Figure 7B). Furthermore, an increase of IL-17 (p = 0.00019) and IFN-γ (p = 0.0277) cytokine response in MOG35-55 antigen-dependent recall assays ex vivo (Figures 7C and 7D) could be observed, indicating that Th17 cell functions were hyperreactive in Nr2f6−/− mice. Th17 cell-intrinsic defects in Nr2f6−/− mice were already independently confirmed by the elevated IL-17 cytokine expression of ex vivo-differentiated Nr2f6−/− Th17 cells, shown also by single-cell flow cytometry and qRT-PCR analysis (Figure 4G, Figure S14, and data not shown).
Figure 7. Nr2f6−/− Th17 Effector Cells Are Hyperreactive Ex Vivo and in Experimental Autoimmune Encephalomyelitis.
(A) EAE disease course in age-matched female mice (Nr2f6+/+, n = 20; Nr2f6−/−, n = 20) is significantly higher in the Nr2f6−/− mice. Mean disease scores were calculated by comparison of the mean values of wild-type and Nr2f6−/− scores at the onset (day 7–day 13) and at disease progression (day 14–day 20) via a Welch two-sample t test. A significant difference was found in both periods (day 7–day 13, p = 0.0004; day 14–day 20, p = 0.025), although the difference was more profound during the disease onset.
(B) Cyokine production by CD4+ T cells isolated from CNS mononuclear cells 14 days after disease induction. Cells were stimulated for 4 hr with PDBu plus ion-omycin in the presence of Golgi stop and analyzed for IFN-γ and IL-17 expression. A representative profile and mean ± SD of cytokine staining are shown. A significant increase of IL-17-IFN-γ double-positive cells (Nr2f6+/+, n = 7; Nr2f6−/−, n = 7; p = 0.0494) could be observed.
(C and D) Production of (C) IL-17 and (D) IFN-γ in recall assays of splenocytes from MOG35-55-immunized mice was significantly enhanced in Nr2f6−/− T cells (split-plot ANOVA (C); Nr2f6+/+, n = 14; Nr2f6−/−, n = 14; p = 0.00019; (D) p = 0.0277).
Discussion
IL-17 production of Th17 cells has been linked to the “decision making” that regulates the balance between immunological tolerance versus autoimmunity (Bettelli et al., 2007; Harrington et al., 2005, 2006; McKenzie et al., 2006; Ivanov et al., 2007; Jankovic and Trinchieri, 2007; Weaver et al., 2007; Sundrud and Rao, 2008). The IL-17A promoter has been shown to contain two cis-acting NF-AT sites (Liu et al. [2004]; see for review Chen et al. [2007]). Consistently, pharmacological inhibition studies employing the calcineurin inhibitor CsA abrogated MOG35-55-stimulated IL-17 expression in antigen-dependent recall assays ex vivo (data not shown), confirming the observation that IL-17 gene regulation strictly depends on the Ca2+-calcineurin-NF-AT pathway. Complementary data, employing both NR2F6 overexpression and Nr2f6-deficient T cells, thus demonstrated that NR2F6 is a nuclear attenuator that directly interferes with DNA binding of NF-AT and, subsequently, transcriptional activity of the NF-AT-dependent IL-17 expression. The antagonistic crosstalk of NF-AT and NR2F6 in Th17 cells provided a plausible explanation for the hypersusceptibility of Nr2f6−/− mice to EAE induction. Nevertheless, at the moment, one cannot exclude the possibility that Th17 cell-intrinsic NR2F6-mediated transcriptional suppression of other transcription factors (both dependent and independent of NF-AT repression) also contributes to the observed phenotype of Nr2f6−/− mice.
NR2F6 negatively interfered with transcriptional cytokine responses and acted as a barrier against autoimmunity. Consistent with this observation, several other nuclear receptors (NRs), predominately the steroid receptors but also PPARs, LXR, RXR, and RAR, are documented to repress the ability of e.g., NF-κB and/or AP-1 to transcribe its target genes (Moore et al., 2006). The nuclear orphan receptor superfamily also includes several prominent molecular regulators of adaptive immunity (Winoto and Littman, 2002). Nur77 has been reported to abrogate NF-κB activation and acts as regulator of TCR-mediated clonal deletion of immature thymocytes (Harant and Lindley, 2004; Lin et al., 2004; Sohn et al., 2007). RORγ-t, a specific splice variant of the orphan nuclear receptor RORγ, is critical in the differentiation program of lymphoid tissues, lymph nodes, Peyer’s patches, lymphoid tissue inducer (LTi) cells, and the proinflammatory Th17 subset (Eberl and Littman, 2004; Eberl et al., 2004; Ivanov et al., 2006). Similarly, RORα has been established to be essential in Th17 differentiation and positively direct Th17 development in vitro and in vivo (Yang et al., 2008). Strikingly, RORα and RORγ double-deficient mice were completely protected against EAE disease induction (Yang et al., 2008). Opposite to RORα and RORγ, however, NR2F6 acted as suppressor in Th17 cells and Nr2f6-deficient mice and demonstrated both increased progression and severity of EAE scores.
In the present study, we described an autoimmune-augmented phenotype of Nr2f6-deficient mice unique to those previously described in other nuclear orphan receptor knockouts. The transcription factor NR2F6 appeared to be essential for the attenuation of a robust Th17 function; exaggerated production of the IL-17 cytokine in Nr2f6-deficient mice indicated a previously unknown biochemical mechanism for fine tuning the activation threshold of Th17-dependent autoimmune pathologies. NR2F6 may be a critical player in the mechanism that relays signals between induction and/or maintenance of peripheral immunological tolerance and autoimmunity.
Experimental Procedures
Nr2f6−/− Mice
These mice were described previously (Warnecke et al., 2005) and were kept under specific pathogen-free (SPF) conditions. All experiments comply with the current laws of Austria.
Protein Kinase Assay
Recombinant E. coli-expressed GST-NR2F6 fusion proteins were purified with glutathion-sepharose (Amersham). PKC isotype-dependent phosphorylation was measured by incorporation of 32Pi from γ32P ATP and incubation of 200 ng of purified recombinant GST-NR2F6 in kinase assay buffer (40 mM Tris [pH 7.5], 40 mM MgCl2, 0.2 mM DTT, 0.0002% Triton X-100, 0.3 mg ml−1 BSA) containing 1 μM ATP, 2 μCi [32P-ATP], 1 μM PDBu, and 160 μM phosphatidylserine. After 20 min at 30°C, the reaction was stopped by addition of stop solution (10 mM ATP, 5 mM EGTA [pH 7.5], 0.1% Triton X-100). Incorporated radioactivity was measured by SDS-PAGE and X-ray autoradiography.
Immunoblot Analysis
T cells were stimulated with solid-phase hamster anti-CD3 (clone 145–2C11) and hamster anti-CD28 (clone 37.51; BD Biosciences) at 37°C for various time periods. Cells were lysed in ice-cold lysis buffer (5 mM NaP2P, 5 mM NaF, 5 mM EDTA, 50 mM NaCl, 50 mM Tris [pH 7.3], 2% Nonidet P-40, and 50 μg ml−1 each aprotinin and leupeptin) and centrifuged at 15,000 × g for 15 min at 4°C. Protein lysates were subjected to immunoblotting with antibodies against (p)Y-783 PLCγ1 (Cell Signaling); PLCγ1, Fyn and, DNA polymerase (Santa Cruz Biotechnology); (p)S-32 I-κBα (Cell signaling); (p)ERK, ERK, and PKCθ (Cell Signaling); PKCα (UBI); and NR2F6 (Perseus Proteomics). The (p)S-83 NR2F6 polyclonal antiserum was raised in rabbits against the 12 amino acid NH2-SFFKR-(p)S-IRRNL–COOH phosphopeptide.
RNA Transcript Analysis by qRT-PCR
RNA was prepared from defined cells and tissues with either Trizol (Invitrogen) or the MagAttract direct mRNA M48 kit (Quiagen). First-strand cDNA synthesis was performed with oligo(dT) primers (Promega) with the Quiagen Omniscript RT kit according to the instructions of the supplier. Expression analysis for Nr2f6 was performed via real-time PCR done in duplicates on an ABI PRIM 7000 Sequence Detection System (Applied Biosystems) with TaqMan gene expression assays for Nr2f6 (Mm01340321-m1); expression was normalized to GAPDH.
siRNA Transfection
CD4+ T cells were negatively selected by magnetic cell sorting (Miltenyi Biotec). T cell populations were typically 95% CD4+, as determined by staining and flow cytometry. Cells were resuspended in solution from Nucleofector mouse T cell kit and program X-01 following the Amaxa guidelines for cell-line transfection. In brief, up to 1 × 107 cells mixed with 1.5 μM of synthetic ON-TARGETplus siRNA predisgened by Dharmacon (#045088) or ON-TARGETplus siCONTROL nontargeting pool were nucleofected with the Amaxa Nucleofector apparatus and immediately transferred into 37°C prewarmed culture medium and cultured for a minimum of 24 hr before experimental analysis. Plasmid transfections of CD4+ T cells were performed identically, employing 6–10 μg of plasmid DNA and cultured for 24 hr before experimental analysis.
Analysis of Proliferation Responses
Antibody- and alloantigen-induced proliferation was described (Pfeifhofer et al., 2003, 2006).
Th0, Th1, Th2, and Th17 In Vitro Differentiation
For T cell differentiation, naive CD4+ cells were isolated via the CD4+ CD62L+ T Cell Isolation Kit II (Miltenyi Biotec). Polarization of T cells into Th0, Th1, Th2, or Th17 cells was performed by solid-phase anti-CD3 (5 μg/ml) and soluble anti-CD28 (1 μg/ml) in the absence (for neutral differentiation) or presence of polarizing cytokines (Th0: IL-2 [30 ng/ml]; Th1: m IL-12 [10 n g/ml], αIL-4 [5 μg/ml]; Th2: IL-4 [10 ng/ml], α IL-12 [5 μg/ml] and αIFN-γ [5 μg/ml]; Th17: IL-23 [10 ng/ml], TGF-β [5 ng/ml], IL-6 [20 ng/ml], αIL-4 [2 μg/ml], αIFN-γ [2 μg/ml]) as described (Yang et al., 2008). Supernatant was collected on day 4 or 5 and analyzed via BioPlex multianalyte technology (BioRad). For FACS analysis, cells were washed and restimulated with phorbol 12-myristate 13-acetate (PMA) and ionomycin in the presence of Golgi-plug for 5 hr, after which IL-17-produc-ing cells were analyzed via intracellular staining.
Analysis of Cytokine Production
IL-2 in the plasma was determined 2 hr after 1 μg of staphylococcus enterotoxin B (SEB) injection i.p. into Nr2f6−/− or Nr2f6+/+ controls. IL-2, IL-4, IL-17, and IFN-γ cytokine amounts from culture supernatant of the relevant CD3+, CD8+ naive, or differentiated CD4+ T cells were measured by BioPlex multianalyte technology (BioRad).
Induction of EAE
MOG35-55 peptide was synthesized by NeoSystems, France. Female 8–12-week-old mice were immunized in the hind flank (200 μl) by one injection of 200 μg MOG35-55 peptide in CFA, supplemented with 5 μg ml−1 Mycobacterium tuberculosis H37 Ra (Difco Laboratories) emulsified 1:1 in PBS (200 μl). In addition, 200 ng of pertussis toxin (Sigma Aldrich) dissolved in 200 μl PBS was injected 24 and 72 h later intravenously (i.v.) (tail vein). Mice were monitored daily for clinical signs of EAE and graded on a scale of increasing severity from 0 to 4 by two independent investigators (Schmidt et al., 2005).
Preparation of CNS Mononuclear Cells
We followed the protocol of Korn et al. (2007) to purify mononuclear cells from the CNS; in brief, mice were perfused through the left cardiac ventricle. The forebrain and the cerebellum were dissected and flushed with PBS. CNS tissue was digested with collagenase D (2.5 mg/ml, Roche Diagnostics) and DNaseI (1 mg/ml, Sigma) at 37°C for 45 min. Mononuclear cells were isolated by passing the tissue through a cell strainer (70 μm), followed by a percoll gradient (70%-30%) centrifugation. Mononuclear cells were removed from the interphase, washed two times, and resuspended in RPMI complete medium.
Antigen Recall Assay
On day 21, splenocyte suspensions were isolated from MOG35-55 peptide-immunized Nr2f6−/− or Nr2f6+/+ mice along with PBS-treated control mice. Splenocytes from individual mice, depleted of RBC with lysing buffer (R&D), were plated in duplicates (5 × 105/well) in 200 μl proliferation medium (RPMI supplemented with 10% FCS, 2 mM L-glutamine, and 50 U ml−1 penicillin/streptomycin) containing 0, 1, 10, or 100 μg ml−1 MOG35-55 peptide and cultured at 37°C in 5% C02. After 60 hr, cytokine production after MOG35-55 peptide restimulation was measured in the cell-culture supernatants. Cytokines amounts were analyzed with BioPlex multianalyte technology (BioRad).
Gel Mobility-Shift Assays
Nuclear proteins from naive (Pfeifhofer et al., 2003, 2006) and Th17 cells (restimulated overnight in the presence of 5 μg/ml anti-CD3) were prepared and used in electrophoretic mobility-shift assays (EMSAs). The following oligonu-cleotides were used as probes and competitors: NF-κB, 5′-GCCATGGGGG GATCCCCGAAGTCC-3′; AP-1, 5′-CGCTTGATGACTCAGCCGGAA-3′; IL-2p NF-AT, 5′-GCCCAAAGAGGAAAATTTGTTTCATACAG-3′ (Nushift, Active Motif); IL-17p NF-AT oligo 3 (Liu et al., 2004), 5′-CATTGGGGGCGGAAATTTTAAC CAAA-3′; NR2F6 EMSAs were performed with the probe set for the COUP-TF family member NR2F1 (Panomics). This contains the same binding sequence as for NR2F6 (5′-GTGTCAAAGGTCGTGTCAAAGGTC-3′).
NR2F6 Localization by Confocal Microscopy
Jurkat E6-1 cells transfected with NR2F6-GFP construct (containing a COOH-terminal GFP tag) were stimulated with SEE presented by Raji B cells. Raji cells were labeled with cell tracker blue (CTKB) (Molecular Probes) and incubated in the presence or absence of 10 ng/mL SEE at 37°C for 90 min. Jurkat E6.1 cells (106) were then mixed with an equal number of Raji B cells and incubated at 37°C for 30 min, 1 hr, 3 hr, or 6 hr. Jurkat-Raji conjugates were transferred on polylysine-coated slides and then fixed with 4% paraformaldehyde and 4% sucrose in PBS. Nuclei were stained with TOPRO-3 (Molecular Probe). Immunofluorescence was analyzed with Zeiss LSM 510 confocal laser scanning microscope and Zeiss LSM software v3.2.
Reporter Gene Assays
Jurkat-TAg cells (a kind gift from G.R. Crabtree, Stanford University, CA) have been transiently transfected with circular plasmid DNA by electroporation (BTX-T820 ElectroSquarePorator, ITC, Biotech, Heidelberg, Germany), with predetermined optimal conditions (1 × 107 cells in 200 μl RPMI medium at 450 V/cm and 5 pulses of 99 ms), yielding ~40% transfection efficiency. NF-AT:AP-1 reporter gene expression was measured in transient cotransfection assays using 10 μg pSRα-CD28, 15 μg of the GFP or Nr2f6 expression vectors, and 15 μg of the promoter firefly luciferase reporter (RLU1). The latter were a NF-AT:AP-1 composite site reporter (Macian et al., 2001), a NF-AT reporter construct containing three tandem copies of the NF-AT minimal consensus sequence, and the proximal IL-17A promoter reporter (Liu et al., 2004). For normalization, 0.3 μg of the renilla luciferase reporter vector pTK-Renilla-Luc (Promega, Medison, WI) (RLU2) has been used. After 21 hr, cells were stimulated with solid-phase CD3 and CD28 agonistic antibodies for 16 hr or left un-stimulated, as indicated, and the firefly luciferase (RLU1) and Renilla luciferase (RLU2) activity in the cell lysates was measured with the dual luciferase detection kit (Promega, Madison, WI) and the βJet-Luminometer (WALLAC, Turku, Finland).
Statistical Analysis
Statistical analysis was performed with the statistical package R as described by Dalgaard (2002) and Everitt and Rabe-Hesketh (2001).
Supplemental Data
Supplemental data include Supplemental Experimental Procedures and sixteen figures and can be found with this article online at http://www.immunity.com/cgi/content/full/29/2/205/DC1/.
Acknowledgments
We are grateful to H. Dietrich, N. Krumböck, C. Mayerl, M. Hermann, and G. Böck (all from Innsbruck) for animal care, expert technical assistance, and FACS analysis. This work was supported by a grant from the FWF Austrian Science Fund (SFB-021 and P19505-B05) and the Foundation pour la Recherche Médicale (S.K.).
References
- Altman A, Kaminski S, Busuttil V, Droin N, Hu J, Hipskind RA, Villalba M. Positive feedback regulation of PLCγ1/Ca2+ signaling by PKCθ in restimulated T cells via a Tec kinase-dependent pathway. Eur J Immunol. 2004;34:2001–2011. doi: 10.1002/eji.200324625. [DOI] [PubMed] [Google Scholar]
- Anderson K, Fitzgerald M, Dupont M, Wang T, Paz N, Healy A, Xu Y, Ocain T, Schopf L, Jaffee B, et al. Mice deficient in PKCθ demonstrate impaired in vivo T cell activation and protection from T cell-mediated inflammatory diseases. Autoimmunity. 2006;39:469–478. doi: 10.1080/08916930600907954. [DOI] [PubMed] [Google Scholar]
- Baier G. The PKC gene module: Molecular biosystematics to resolve its T cell functions. Immunol Rev. 2003;192:64–79. doi: 10.1034/j.1600-065x.2003.00018.x. [DOI] [PubMed] [Google Scholar]
- Bettelli E, Oukka M, Kuchroo VK. T(H)-17 cells in the circle of immunity and autoimmunity. Nat Immunol. 2007;8:345–350. doi: 10.1038/ni0407-345. [DOI] [PubMed] [Google Scholar]
- Chaudhary D, Kasaian M. PKCθ: A potential therapeutic target for T-cell-mediated diseases. Curr Opin Investig Drugs. 2006;7:432–437. [PubMed] [Google Scholar]
- Chen Z, Laurence A, O’Shea JJ. Signal pathways and transcriptional regulation in the control of Th17 differentiation. Semin Immunol. 2007;19:400–408. doi: 10.1016/j.smim.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgaard P. Introductory Statistics with R. New York: Springer; 2002. [Google Scholar]
- Eberl G, Littman DR. Thymic origin of intestinal αβ T cells revealed by fate mapping of RORγt+ cells. Science. 2004;305:248–251. doi: 10.1126/science.1096472. [DOI] [PubMed] [Google Scholar]
- Eberl G, Marmon S, Sunshine MJ, Rennert PD, Choi Y, Littman DR. An essential function for the nuclear receptor RORγ-t in the generation of fetal lymphoid tissue inducer cells. Nat Immunol. 2004;5:64–73. doi: 10.1038/ni1022. [DOI] [PubMed] [Google Scholar]
- Everitt B, Rabe-Hesketh S. Analyzing Medical Data Using S-Plus. New York: Springer; 2001. [Google Scholar]
- Fujii K, Zhu G, Liu Y, Hallam J, Chen L, Herrero J, Shaw S. Kinase peptide specificity: Improved determination and relevance to protein phosphorylation. Proc Natl Acad Sci USA. 2004;101:13744–13749. doi: 10.1073/pnas.0401881101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giguere V. Orphan nuclear receptors: From gene to function. Endocr Rev. 1999;20:689–725. doi: 10.1210/edrv.20.5.0378. [DOI] [PubMed] [Google Scholar]
- Harant H, Lindley IJ. Negative cross-talk between the human orphan nuclear receptor Nur77/NAK-1/TR3 and nuclear factor-κB. Nucleic Acids Res. 2004;32:5280–5290. doi: 10.1093/nar/gkh856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington LE, Hatton RD, Mangan PR, Turner H, Murphy TL, Murphy KM, Weaver CT. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6:1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- Harrington LE, Mangan PR, Weaver CT. Expanding the effector CD4 T-cell repertoire: The Th17 lineage. Curr Opin Immunol. 2006;18:349–356. doi: 10.1016/j.coi.2006.03.017. [DOI] [PubMed] [Google Scholar]
- Isakov N, Altman A. Protein kinase Cθ in T cell activation. Annu Rev Immunol. 2002;20:761–794. doi: 10.1146/annurev.immunol.20.100301.064807. [DOI] [PubMed] [Google Scholar]
- Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, Littman DR. The orphan nuclear receptor RORγt directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Ivanov II, Zhou L, Littman DR. Transcriptional regulation of Th17 cell differentiation. Semin Immunol. 2007;19:409–417. doi: 10.1016/j.smim.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankovic D, Trinchieri G. IL-10 or not IL-10: That is the question. Nat Immunol. 2007;8:1281–1283. doi: 10.1038/ni1207-1281. [DOI] [PubMed] [Google Scholar]
- Korn T, Reddy J, Gao W, Bettelli E, Awasthi A, Petersen TR, Backstrom BT, Sobel RA, Wucherpfennig KW, Strom TB, et al. Myelin-specific regulatory T cells accumulate in the CNS but fail to control autoimmune inflammation. Nat Med. 2007;13:423–431. doi: 10.1038/nm1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B, Kolluri SK, Lin F, Liu W, Han YH, Cao X, Dawson MI, Reed JC, Zhang XK. Conversion of Bcl-2 from protector to killer by interaction with nuclear orphan receptor Nur77/TR3. Cell. 2004;116:527–540. doi: 10.1016/s0092-8674(04)00162-x. [DOI] [PubMed] [Google Scholar]
- Liu X, Huang X, Sigmund CD. Identification of a nuclear orphan receptor (Ear2) as a negative regulator of renin gene transcription. Circ Res. 2003;92:1033–1040. doi: 10.1161/01.RES.0000071355.82009.43. [DOI] [PubMed] [Google Scholar]
- Liu XK, Lin X, Gaffen SL. Crucial role for nuclear factor of activated T cells in T cell receptor-mediated regulation of human interleukin-17. J Biol Chem. 2004;279:52762–52771. doi: 10.1074/jbc.M405764200. [DOI] [PubMed] [Google Scholar]
- Macian F, Lopez-Rodriguez C, Rao A. Partners in transcription: NFAT and AP-1. Oncogene. 2001;20:2476–2489. doi: 10.1038/sj.onc.1204386. [DOI] [PubMed] [Google Scholar]
- McKenzie BS, Kastelein RA, Cua DJ. Understanding the IL-23-IL-17 immune pathway. Trends Immunol. 2006;27:17–23. doi: 10.1016/j.it.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Miyajima N, Kadowaki Y, Fukushige S, Shimizu S, Semba K, Yamana-shi Y, Matsubara K, Toyoshima K, Yamamoto T. Identification of two novel members of erbA superfamily by molecular cloning: The gene products of the two are highly related to each other. Nucleic Acids Res. 1988;16:11057–11074. doi: 10.1093/nar/16.23.11057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JT, Collins JL, Pearce KH. The nuclear receptor super-family and drug discovery. ChemMedChem. 2006;1:504–523. doi: 10.1002/cmdc.200600006. [DOI] [PubMed] [Google Scholar]
- Nuclear Receptors Nomenclature Committee. A unified nomenclature system for the nuclear receptor superfamily. Cell. 1999;97:161–163. doi: 10.1016/s0092-8674(00)80726-6. [DOI] [PubMed] [Google Scholar]
- Park JI, Tsai SY, Tsai MJ. Mechanism of chicken ovalbumin upstream promoter-transcription factor (COUP-TF) actions. Keio J Med. 2003;52:174–181. doi: 10.2302/kjm.52.174. [DOI] [PubMed] [Google Scholar]
- Pfeifhofer C, Gruber T, Letschka T, Thuille N, Lutz-Nicoladoni C, Hermann-Kleiter N, Braun U, Leitges M, Baier G. Defective IgG2a/2b class switching in PKCα−/− mice. J Immunol. 2006;176:6004–6011. doi: 10.4049/jimmunol.176.10.6004. [DOI] [PubMed] [Google Scholar]
- Pfeifhofer C, Kofler K, Gruber T, Tabrizi NG, Lutz C, Maly K, Leitges M, Baier G. Protein kinase C θ affects Ca2+ mobilization and NFAT activation in primary mouse T cells. J Exp Med. 2003;197:1525–1535. doi: 10.1084/jem.20020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: Regulation and function. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- Schmidt CS, Zhao J, Chain J, Gitter B, Sandusky G, Chintalacharuvu S, Glasebrook A, Na S. Resistance to myelin oligodendrocyte glycoprotein-induced experimental autoimmune encephalomyelitis by death receptor 6-deficient mice. J Immunol. 2005;175:2286–2292. doi: 10.4049/jimmunol.175.4.2286. [DOI] [PubMed] [Google Scholar]
- Sohn SJ, Thompson J, Winoto A. Apoptosis during negative selection of autoreactive thymocytes. Curr Opin Immunol. 2007;19:510–515. doi: 10.1016/j.coi.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Spitaler M, Cantrell DA. Protein kinase C and beyond. Nat Immunol. 2004;5:785–790. doi: 10.1038/ni1097. [DOI] [PubMed] [Google Scholar]
- Sun Z, Arendt CW, Ellmeier W, Schaeffer EM, Sunshine MJ, Gandhi L, Annes J, Petrzilka D, Kupfer A, Schwartzberg PL, et al. PKCθ is required for TCR-induced NF-κB activation in mature but not immature T lymphocytes. Nature. 2000;404:402–407. doi: 10.1038/35006090. [DOI] [PubMed] [Google Scholar]
- Sundrud MS, Rao A. Regulation of T helper 17 differentiation by orphan nuclear receptors: It’s not just RORγt anymore. Immunity. 2008;28:5–7. doi: 10.1016/j.immuni.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Takamoto N, You LR, Moses K, Chiang C, Zimmer WE, Schwartz RJ, DeMayo FJ, Tsai MJ, Tsai SY. COUP-TFII is essential for radial and anteroposterior patterning of the stomach. Development. 2005;132:2179–2189. doi: 10.1242/dev.01808. [DOI] [PubMed] [Google Scholar]
- Tan SL, Parker PJ. Emerging and diverse roles of protein kinase C in immune cell signalling. Biochem J. 2003;376:545–552. doi: 10.1042/BJ20031406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan SL, Zhao J, Bi C, Chen XC, Hepburn DL, Wang J, Sedgwick JD, Chintal SR, Na S. Resistance to experimental autoimmune encephalomyelitis and impaired IL-17 production in PKCθ-deficient mice. J Immunol. 2006;176:2872–2879. doi: 10.4049/jimmunol.176.5.2872. [DOI] [PubMed] [Google Scholar]
- Volkov Y, Long A, McGrath S, Ni Eidhin D, Kelleher D. Crucial importance of PKCßI in LFA-1-mediated locomotion of activated T cells. Nat Immunol. 2001;2:508–514. doi: 10.1038/88700. [DOI] [PubMed] [Google Scholar]
- Warnecke M, Oster H, Revelli JP, Alvarez-Bolado G, Eichele G. Abnormal development of the locus coeruleus in Ear2(Nr2f6)-deficient mice impairs the functionality of the forebrain clock and affects nociception. Genes Dev. 2005;19:614–625. doi: 10.1101/gad.317905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver CT, Hatton RD, Mangan PR, Harrington LE. IL-17 family cytokines and the expanding diversity of T cell lineages. Annu Rev Immunol. 2007;25:821–852. doi: 10.1146/annurev.immunol.25.022106.141557. [DOI] [PubMed] [Google Scholar]
- Winoto A, Littman DR. Nuclear hormone receptors in T lymphocytes. Cell. 2002;109(Suppl):S57–S66. doi: 10.1016/s0092-8674(02)00710-9. [DOI] [PubMed] [Google Scholar]
- Yang XO, Pappu BP, Nurieva R, Akimzhanov A, Kang HS, Chung Y, Ma L, Shah B, Panopoulos AD, Schluns KS, et al. T helper 17 lineage differentiation is programmed by orphan nuclear receptors RORα and RORγ. Immunity. 2008;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You LR, Lin FJ, Lee CT, DeMayo FJ, Tsai MJ, Tsai SY. Suppression of Notch signaling by COUP-TFII transcription factor regulates vein identity. Nature. 2005;435:98–104. doi: 10.1038/nature03511. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Dufau ML. Gene silencing by nuclear orphan receptors. Vitam Horm. 2004;68:1–48. doi: 10.1016/S0083-6729(04)68001-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.