Abstract
Nonalcoholic fatty liver disease (NAFLD) is caused by hepatic steatosis, which can progress to nonalcoholic steatohepatitis, fibrosis/cirrhosis, and hepatocellular carcinoma in the absence of excessive alcohol consumption. Nonalcoholic fatty liver disease will become the number one cause of liver disease worldwide by 2020. Nonalcoholic fatty liver disease is correlated albeit imperfectly with obesity and other metabolic diseases such as diabetes, hyperlipidemia, and cardiovascular disease, but exactly how having one of these diseases contributes to the development of other metabolic diseases is only now being elucidated. Development of NAFLD and related metabolic diseases is genetically influenced in the population, and recent genome-wide association studies (GWASs) have discovered genetic variants that associate with these diseases. These GWAS-associated variants cannot only help us to identify individuals at high risk of developing NAFLD, but also to better understand its pathophysiology so that we can develop more effective treatments for this disease and related metabolic diseases in the future.
Keywords: nonalcoholic liver disease, nonalcoholic steatohepatitis, gene, genetics, polymorphism
With the rise in obesity, we are seeing an increase in nonalcoholic fatty liver disease (NAFLD) which is reaching epidemic proportions. Nonalcoholic fatty liver disease is expected to become the number one cause of liver disease worldwide by 2020.1 Nonalcoholic fatty liver disease is correlated with the presence of other metabolic diseases including obesity, diabetes, dyslipidemia, hypertension, and cardiovascular disease, but imperfectly so.2,3 The extent to which these correlated metabolic diseases contribute to the development of NAFLD or to which NAFLD contributes to the development of these correlated metabolic diseases is currently being investigated. Like obesity, NAFLD is very common in the United States, with an overall prevalence of approximately 30%.3–5 Nonalcoholic fatty liver disease varies in prevalence across ancestries, however.4–6 In the United States, the prevalence of NAFLD in individuals of Hispanic ancestry is higher (34–58%) than in individuals of European ancestry (28–45%) than in individuals of African ancestry (19–35%).4–6 The majority of this difference in the prevalence of disease across ancestries is due to genetic differences in the predisposition to develop this disease.7,8
Nonalcoholic fatty liver disease is a spectrum of disease that includes steatosis (fat deposition in hepatocytes), nonalcoholic steatohepatitis (NASH; inflammation around the hepatic fat), and fibrosis/cirrhosis (scarring of the liver) in individuals that do not drink significant amounts of alcohol.9 In some individuals, NAFLD can predispose to the development of hepatocellular carcinoma (HCC).10 Steatosis is the hallmark of the disease, but can be absent in advanced stages of the disease. As a consequence, many causes of cryptogenic cirrhosis may actually be due to NAFLD that was not previously recognized as NAFLD.
To develop NAFLD there must be an imbalance in hepatic lipid homeostasis between fatty acid inputs and outputs. Inputs include fatty acids from dietary sources, adipose breakdown, de novo lipogenesis, and diet, while outputs include fatty acid oxidation and fatty acid export in the form of very low-density lipoprotein (VLDL; reviewed in 11). Some studies have shown that lipotoxicity and mitochondrial dysfunction may contribute to hepatocyte toxicity and death.11 Furthermore, there is evidence that activation of Kupffer cells and stellate cells can initiate the inflammatory and fibrotic processes, respectively, that lead to progressive liver disease in the form of NASH and fibrosis. Although it was thought in the past that hepatic steatosis was “benign,” more evidence suggests that hepatic steatosis can cause hepatocyte toxicity and trigger development of advanced liver disease. Indeed, HCC can develop in patients who do not have cirrhosis12 and some cases of HCC develop in the presence of just “simple steatosis,”13,14 suggesting that even hepatic steatosis may merit intervention to prevent development of subsequent liver disease.
NAFLD Measurement
Nonalcoholic fatty liver disease can be measured using various modalities. It can be measured using liver histology, which is the gold standard for the diagnosis of the disease. With histology, the complete range of the disease can be measured including steatosis, inflammation, and fibrosis/cirrhosis. Because liver biopsies carry a risk of complications, they are not usually performed on normal individuals unless there is an indication. Indications for biopsy may include many risk factors for development of advanced liver disease, including advanced age and concomitant metabolic disease. Thus, studies based on using histology may not accurately reflect population-based NAFLD. Hepatic steatosis can also be measured using noninvasive imaging, but these imaging modalities are not very sensitive for detection of NASH or early fibrosis. Magnetic resonance spectroscopy (MRS) is the most sensitive and specific for measuring triglycerides in liver, but requires specialized expertise to implement and is not widely used in clinical practice.15 Magnetic resonance imaging (MRI), computed tomography (CT), or ultrasound imaging are affordable, widely available, and confer minimal risk to the patient; thus, they are most often used in clinical practice and in the population for evaluating for the presence of hepatic steatosis.15 The sensitivity and specificity of MRI is greater for quantifying hepatic steatosis than CT, which is greater than ultrasound.15 Finally, elevations in serum ala-nine aminotransferase (ALT) can be seen with NAFLD, but this biomarker is not sensitive or specific for NAFLD16 (e.g., it can fail to be elevated in people with NAFLD or be elevated without NAFLD as a consequence of many other liver diseases) . Therefore, they require further follow-up to confirm that the elevations are due to NAFLD and not some other liver disease.
Heritability Estimates for NAFLD
Population-based NAFLD has been found to be heritable, or genetic, in various twin and family studies. In individuals of Hispanic ancestry, the heritability of population-based NAFLD has been found to be 31 to 38%.17,18 In the first study, 44 children (probands) and their parents were characterized for the presence of hepatic steatosis using MRI and histology; the heritability of NAFLD was found to be 38%.17 In the second, 843 individuals from the Insulin Resistance Atherosclerosis Study (IRAS) family study were measured for hepatic steatosis using CT-measured liver attenuation and heritability determined to be 31%.18 In 6,629 individuals from three European ancestry family-based studies (Family Heart Study, The Old Order Amish, and The Framingham Heart Study) liver steatosis was measured using computed tomography liver attenuation and heritability was calculated to be 26 to 27%.19 Finally, in individuals of African ancestry from the Insulin Resistance and Atherosclerosis Study, Family Heart Study, and Genetic Epidemiology of Arteriopathy (GENOA) Study the heritability was found to be 22 to 34%.7 Thus in all ancestries examined there is a substantial heritable component to the disease, ranging from 22 to 38%. Because there is statistical variation in these heritability measures, it cannot be determined that the heritability is more in one ancestry than the other. A recent study of 60 pairs of twins revealed that the heritability of hepatic steatosis and fibrosis using MRI proton-based fat fraction was 52% and 50%, respectively.20 The slightly higher estimate of steatosis heritability in this study compared with the family-based studies noted above is likely due to an overestimate of heritability in twin studies. In particular, heritability estimates based on phenotypic resemblance that may be due to unmeasured environmental variables which are more likely to be shared by monozygotic twins (because they often do live in the same environment and may be treated more similarly) than more distantly related individuals often inflate the heritability calculated for traits in twin studies.21
Candidate Gene Studies
Many studies have tested genetic variants in or near candidate genes for association with NAFLD measures.22 These include associations with single nucleotide polymorphisms (SNPs) in or near IRS1, ENPP1, SOD2, p21, USF1, KLF6, INFL4, IL28B, and APOC3.22–29 These associations have not been reproduced in completely independent studies to date; thus, their validity remains to be determined. Some associations have, however, already turned out to be false-positives. In 2010 Peterson et al reported an association with variants near APOC3,29 but reports by others since then have not substantiated this association.30,31 Similarly, the association of variants in or near IL28B with NAFLD has been challenged.32 In this latter case, authors of the original study suggest that the IL28B allele may have a larger effect in individuals that do not have many other NAFLD predisposing risk factors as the reason for discrepancy between studies,33 but this remains to be substantiated.
GWASs for Nonalcoholic Fatty Liver Disease
To identify specific genetic elements that associate with NAFLD in an unbiased way, several genome-wide association studies (GWASs) of NAFLD have been performed (►Table 1). Romeo et al used a custom chip of over 9,000 nonsynonymous variants across the genome to genotype individuals of European, Hispanic, and African American ancestries who had liver fat measured by proton MRS.8 They found that one variant in the gene PNPLA3, the G-allele at rs738409, encoding an I148M missense mutation, increased hepatic fat levels across European American, African American, and Hispanic American ancestries. The frequency of this variant was highest in individuals of Hispanic ancestry, followed by those of European and African American ancestries, respectively,8 a finding consistent with the known prevalence of NAFLD in these ethnic groups.5 Since the discovery of this variant, other groups have replicated the association of this variant with hepatic steatosis as well as other measures of NAFLD. Specifically, associations have been noted with elevated serum ALT levels,34–38 imaging-based steatosis,4,19,39 or with histologic NAFLD including steatosis, NASH, and fibrosis/cirrhosis (►Table 2).19,40–44 A recent meta-analysis of up to 2,937 individuals with NAFLD measured by histology revealed that individuals with GG at rs738409 in PNPLA3 had 73% higher lipid fat content, 3.24-fold greater risk of higher necroinflammatory scores, 3.2-fold greater risk of developing fibrosis, and 3.44 higher odds of developing NASH compared with CC individuals.45 More than 70% of the difference in prevalence between individuals of diverse ancestries is likely due to genetic influence—and most of that is due to variation in the prevalence of fatty liver promoting the PNPLA3 allele.7,8
Table 1.
Genome-wide association studies (GWASs) of nonalcoholic fatty liver disease (NAFLD)
| Study | Primary phenotype | Ancestry | Genotyping array | Discovery panel size (Cases/controls) | Replication panel size (Cases/controls) | Loci/genes | Scientific impact |
|---|---|---|---|---|---|---|---|
| Yuan et al (2008)34 | Clinical biochemistry (ALT levels) | European | Affymetrix 500/Illumina HumanHap 550/Illumina HumanHap 300/Perlegen Sciences custom array |
7,715 | 4,704 | PNPLA3-SAMM50 genomic locus, CPN1-ERLIN1-CHUK genomic locus | First GWAS identifying multiple loci/genes influencing plasma levels of liver enzymes |
| Romeo et al (2008)8 | MRS measured hepatic steatosis | Hispanic, African American, European | Perlegen Sciences custom array (12k) | 2,111 individuals with MRS measured hepatic steatosis | none | PNPLA3 | First exome analysis of hepatic steatosis. Frequency differences of PNPLA3 effect allele across ancestries account for much of the differences in NAFLD population prevalance |
| Chalasani et al (2010)51 | Hepatic histology | European American | Illumina CNV370 | 236 non-Hispanic white women | none | none genome-wide significant | First GWAS of NAFLD histology phenotypes |
| Speliotes et al (2011)19 | CT measured hepatic steatosis | European American | Affymetrix 550/Illumina CNV370/Illumina 1 million SNP chip/Affymetrix 500K |
7,126 with CT measured hepatic steatosis | 592/1405 histology | PNPLA3, NCAN, PPP1R3B, CCKR, LYPLAL1 | Largest GWAS of hepatic steatosis. Multiple loci/genes identified and characterized, including pleiotropies that suggest disease subtypes |
| Chambers et al (2011)35 | Clinical serum chemistry phenotypes (ALT levels) | European | Affymetrix, Illumina, and Perlegen Sciences arrays | 61,089 | none | PNPLA3, TRIB1, CPN1, loci near HSD17B13 and MAPK10 | Largest GWAS identifying multiple loci/genes influencing plasma levels of liver enzymes |
| Kozlitina et al (2014)48 | MRS measured hepatic steatosis | Hispanic, African American, European | HumanExome BeadChip, Illumina | 2,736 | none | PNPLA3, TM6SF2 | First exome chip association study of liver fat content. Identified TM6SF2 as a probable causal gene for NAFLD in chr19p13.11 locus |
Abbreviations: ALT, alanine aminotransferase; MRS, magnetic resonance spectroscopy; CT, computed tomography; SNP, single nucleotide polymorphism.
Table 2.
Association with nonalcoholic fatty liver disease (NAFLD) and related metabolic traits
| Gene SNP (effect allele, EAF) | CAD/MI (OR 95% CI)46 | Fasting glucose (effect SE) mmol/L19 | HOMA-IR (effect SE)19 | LDL-C (effect SE) mg/dL47 | TG (effect SE) mg/dL47 | TC (effect se) mg/dL47 | ALT (effect 95% CI) IU/L35 | ALP (effect 95% CI) IU/L35 | GGT (effect 95% CI) IU/L35 | Fatty liver (effect SE)a,19 | NASH/fibrosis (OR 95% CI)19 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PNPLA3 rs738409 (G, 0.23) | NA | NS | NS | NS | NS | NA | 6.0 (5.0–7.0) | NS | NS | 0.26 (0.02) | 3.26 (2.11–7.21) |
| TM6SF2 rs58542926 (T, 0.07)b | 0.87 (0.79–0.95) | NA | NA | –3.11 (0.38) | –7.83 (0.82) | –6.6 (1.2)46 | NA | – c,48 | NS | 0.24 (0.03)d | 1.88 (1.41–2.5) |
| GCKR rs780094 (T, 0.39) | NS | –0.03 (0.004) | –0.02 (0.004) | NS | 8.76 (0.40) | 1.91 (0.19) | NS | NS | 3.2 (2.4–4.0) | 0.06 (0.02) | 1.45 (1.19–1.86) |
| PPP1 R3B rs4240624 (A, 0.92) | NS | –0.03(0.006) | NS | 2.22 (0.29) | NS | 3.14 (0.32) | NS | 2.7 (1.1–4.4) | NS | 0.29 (0.03) | NS |
| LYPLAL1 rs12137855 (C, 0.79) | NA | NS | NS | NS | NS | NS | NS | NS | NS | 0.08 (0.02) | 1.37 (1.17–1.57) |
| TRIB1 rs2954021 (A, 0.52)e | NA | NA | NA | –1.84 (0.17) | –5.64 (0.39) | –2.30 (0.19) | 1.6 (0.6–2.6) | 1.4 (0.5–2.3) | NS | – f | NA |
Abbreviations: EAF, effect allele frequency in Europeans; CAD/MI, coronary artery disease/myocardial infarction; effect SE, effect per effect allele with standard error of mean in units given; mmol/L, millimole per liter; HOMA-IR, homeostatic model assessment for insulin resistance; LDL-C, low density lipoprotein cholesterol; TG, triglycerides; TC, total cholesterol; ALT, alanine aminotransferase; ALP, alkaline phosphatase; GGT, gamma-glutamyl transferase; NASH, nonalcoholic steatohepatitis; NS, not significant; NA, not available, source of values in table are from the references in the headers except when cited separately.
Effect measured as the increase in inverse normalized fatty liver by computed tomography for per effect allele (from19).
TC, LDL-C, TG results are for rs10401969 (LD r2= 0.98 with rs58542926 in EUR) (from47).
Direction of effect on serum alkaline phosphatase is decreased for the T allele of rs58542926 with a p-value = 4.3 × 10–7 (from48).
Data for SNP rs2228603 (LD r2= 0.80 with rs58542926 in EUR) (from19).
TG data for rs2954029 (LD r2= 0.81 with rs2954021 in EUR). TC and LDL-C data for rs2954022 (LD r2=0.84 with rs2954021 in EUR) (both from47).
Effect given as the odds of having >5% fat by histology for each effect allele of rs2954021 and is 1.53 (95% CI 1.25–1.88) (from52).
Two different GWASs of liver function tests identified variants at four loci that associated with elevated ALT levels. These are in or near the PNPLA3 (rs2281135, rs738409)–SAMM50 (rs2143571) genomic locus,34,35 the CPN1–ERLIN1–CHUK gene cluster (rs10883437, rs11597390, rs11591741, rs11597086),34,35 TRIB1 (rs2954021),34,35 and near HSD17B13/MAPK10 (rs6834314).35 The SNPs in the genomic region of PNPLA3–SAMM50 are correlated with each other, suggesting that they may represent the same genetic signal (e.g., are in moderate to high linkage disequilibrium [LD] with each other with a r2 [correlation]= 0.34 between rs738409 and rs2143571, r2 = 0.63 between rs2281135 and rs2143571 and r2 = 0.86 between rs2281135 and rs738409). Follow-up studies by independent groups found that variants in PNPLA3-rs738409 (see last paragraph) and near TRIB1 (rs2954021) reproducibly affect the development of NAFLD.49 Specifically, variants (rs2954021) near TRIB1 have been found to associate with histologic NAFLD in a Japanese population.49 Further, a SNP (rs6982502) in an enhancer near TRIB1 was significantly (p = 9.39 × 10−7) associated with ultrasonographically diagnosed NAFLD in a population of 5,570 individuals.50 These two SNPs that are located in or near TRIB1 are highly correlated with each other with an LD r2 = 0.94.
Another GWAS on NAFLD focused on 236 non-Hispanic white women that were genotyped with the Illumina CNV370 platform and assessed for various histologic parameters related to NAFLD.51 After correcting for multiple hypothesis testing, however, none of the SNPs managed to pass the genome-wide significance threshold of a p value less than 5 × 10−8.
One of the largest GWASs was a meta-analysis across four groups, all of European ancestry, that were genotyped either on the Affymetrix or Illumina platforms, imputed to the 2.8 million SNPs in HapMap.19 These SNPs were tested for associations with hepatic steatosis measured by CT in each of the four groups separately and combined by meta-analysis for a total of 7,176 assayed individuals19 controlling for age, gender, and principal components. Top associating variants from this meta-analysis were taken forward for assessment of effects on 592 cases of histology proven NASH/ fibrosis genotypically matched to 1405 controls, all of European ancestry.19 Variants in or near the genes PNPLA3, LYPLAL1, PPP1R3B, NCAN/TM6SF2, and GCKR were found to be associated with hepatic steatosis.19 Variants that increased hepatic steatosis at all loci except PPP1R3B were also found to be associated with NASH/fibrosis (►Table 2).19 The associations of variants at PNPLA3 have been confirmed in subsequent studies as noted above. The association of variants at GCKR and NCAN/TM6SF2 with NAFLD/NASH/fibrosis have also been confirmed by independent subsequent studies.48,49,52
The most significant variants at PNPLA3 and GCKR either were themselves missense variants or in high-linkage disequilibrium with missense variants in those genes.19 Indeed, fine mapping of these loci across ancestries suggests that the PNPLA3(I148M) and GCKR(P446L) are the variants most likely to be causally related to development of NAFLD.7 Variants at NCAN/TM6SF2 have been fine mapped to likely a missense variant in TM6SF2(E167K) as possibly the causal variant in promoting NAFLD in a recent study of exonic variants (variants in the coding parts of genes). These were assayed using the Illumina Human Exome chip for association with NAFLD measured using MRS in the Dallas Heart Study.48 This variant can account for the association signal with NAFLD seen at this locus, which in the literature also goes by the name NCAN, CILP2, and the 19p13.11 locus.48 The TM6SF2 variant rs58542926 that causes a nucleotide change of C to T, is a nonsynonymous change causing a glutamate to lysine amino acid substitution at residue 167 (E167K); the glutamate is highly conserved across mammals.48 The putative causal variant is suggested to cause a decreased function in TM6SF2 to promote NAFLD.48 The NAFLD-associated variants at LYPLAL1 or PPP1R3B fall in noncoding regions near these genes. The best associating NALFD promoting variant at PPP1R3B increases PPP1R3B expression, suggesting that this could be a functional expression quantitative trait locus (eQTL) variant.19
Pleiotropies
Effects on Related Metabolic Traits and Diseases
Some but not all NAFLD-associated variants affect the risk of developing related metabolic diseases (►Table 2). The G allele at rs738409 in PNPLA3 has not been found to associate with measures of obesity, impaired fasting glucose, diabetes, low-serum high-density lipoprotein cholesterol (HDL-C), high-serum triglycerides, and hypertension.7,34 One study did find some associations of this allele with decreased total cholesterol levels in a study of > 100,000 individuals,53 but this remains to be replicated. The fatty liver promoting allele at TM6SF2/NCAN associates not only with decreased serum low-density lipoprotein cholesterol (LDL-C), serum triglycerides, and increased serum HDL-C levels, but also with decreased risk of coronary artery disease.14,54 The fatty liver increasing allele at GCKR associates with increased serum LDL-C, increased serum triglycerides, serum gamma-glutamyl transferase, and decreased fasting glucose and homeostatic model assessment insulin resistance levels.14,35 The fatty liver increasing variant near PPP1R3B exhibited similar behavior to GCKR in associating significantly with increased serum LDL-C, increased serum HDL-C, and decreased fasting glucose,14 but also associated with increased alkaline phosphatase.35 The serum ALT level increasing allele at TRIB1 (rs2954021) significantly associates with increased serum levels of total cholesterol, serum LDL-C, serum triglycerides, and serum alkaline phosphatase, but decreased levels of serum HDL-C.35,53 Variants near TRIB1 have also been associated with adiponectin.55
The finding that some, but not all genetic variants that predispose to development of NAFLD predispose to related metabolic disease, suggests that genetics may help explain in part the imperfect correlation of these traits with each other. That is, some of the correlation seen between developing metabolic disease may be due to having shared genetic elements that predispose to more than one metabolic abnormality. The finding that not all genetic variants that predispose the NAFLD equally affect related metabolic disease suggests, however, that depending on the sets of genetic variants individuals carry, they may have different risks of developing NAFLD and concomitant related metabolic diseases. The exciting thing is that now with this new genetic information we may in the future be able to make more precise predictions on who will not only develop NAFLD but also related metabolic diseases. Because some genetic changes result in patterns of effects across traits that are similar to each other, but different from those of other variants, we may in the future also be able to use these genetic changes to subclassify individuals into disease types. These pleiotropy data are also interesting and useful from a therapeutic standpoint because they suggest that interfering with the function of some of these genes may lead to pleiotropic effects, whether desired or undesired, if targeted for medical intervention. For example, if one were to decrease the function of TM6SF2 globally to decrease serum lipid levels to prevent heart attacks, there is ample data for these studies to suggest that this would cause hepatic steatosis and advanced liver disease. Thus, this approach to reducing cardiovascular disease risk may not be optimal. The other striking finding from these genetic data is that most genes, when targeted, will have pleiotropic effects (i.e., side effects). Thus, a better strategy suggested from the genetic work is to make weak agonists and antagonists (which is what nature has done in creating these variants of small effect) that when used together, may maximally affect the desired phenotype, but have low if not canceling effects on related traits and thus can maximally affect the targeted phenotype while minimizing side effects.
Effects in Other Liver Diseases
Alcoholic liver disease (ALD), more than any other liver disease, has a similar spectrum of histopathologic features as NAFLD. This includes the development of alcoholic steatosis, alcoholic hepatitis, alcoholic fibrosis, alcoholic cirrhosis, and a predisposition to the development of HCC.64,65 The inciting event is the development of alcoholic fatty liver, which is a pathologic condition that shares many features with NAFLD.66 To try to distinguish ALD from NAFLD, cutoffs of drinking > 21 drinks a week for men and > 14 for women have been suggested as a guideline for determining what constitutes significant alcohol consumption.67 Nevertheless, given the high prevalence of metabolic disease in the population and the high prevalence of alcohol consumption in the population, it is likely that in clinical practice the two diseases often coexist, even though by formal definition they are mutually exclusive. Indeed, there is evidence to suggest that ALD can be exacerbated by the presence of obesity and that some of the pathophysiology of the two conditions may overlap.66,68,69
To date, the I148M at PNPLA3 has been most extensively studied for effects on ALD. A recent meta-analysis shows that this allele does affect development of ALD.56 More than 4,000 cases of ALD and 4,000 controls without ALD across 10 studies45,49,50,56,67–72 were analyzed.56 The PNPLA3 G-allele at rs738409 has been found to increase the risk of alcoholic liver injury with an odds ratio (OR) of 1.45 (95% confidence interval [CI] 1.24–1.69) and increase the risk for development of alcoholic cirrhosis 2.09 (95% CI 1.79–2.44).56 There was not a clear association of the I148M PNPLA3 allele with the development of alcoholic fatty liver, however.56 This could be because the allele may not have an effect in alcoholic fatty liver, but could also be due to this analysis being underpowered (with only 239 individuals being assessed) or to fatty liver being so prominent in ALD that the genetic change was not of large enough magnitude to be statistically significant across groups. A recent GWAS study has also found that variants at PNPLA3 and TM6SF2 affect development of alcoholic cirrhosis at genome-wide significance levels.145
Infection with both hepatitis B and C virus can lead to hepatitis, fibrosis, and cirrhosis and can predispose to the development of HCC (reviewed in 70,71). Association of hepatic steatosis with hepatitis C infection is well documented and most pronounced for hepatitis C genotype 3, which induces hepatic steatosis more than the other subtypes.72 Hepatic steatosis has been found to promote liver disease progression from hepatitis C, but not from hepatitis B (reviewed in 73). Hepatic steatosis has been found to exacerbate liver disease progression from nongenotype 3 hepatitis C.57 Obesity and insulin resistance seems to also exacerbate hepatitis C disease progression in most studies examined (reviewed in 73). Steatosis is not a prominent feature of hepatitis B infection,74 but concomitant presence of metabolic syndrome with hepatitis B infection does lead to exacerbated liver disease development.75 For hepatitis C, it has also been shown that hepatic steatosis can affect the ability of patients to attain sustained virologic response in nondiabetic, but not diabetic patients.76 Given the exacerbation of hepatitis C- and hepatitis B-caused liver disease by metabolic disease, genetic variants that affect NAFLD may have effects on liver disease in patients with hepatitis B and C.
The first study to assess NAFLD-associated genetic variants for effects in hepatitis C was of 819 patients from Italy with chronic hepatitis C.57 They found that the rs738409 GG genotype was associated with steatosis independently of age, sex, body mass index (BMI), diabetes, alcohol intake, and viral genotype (OR = 1.90, 95% CI 1.4–2.7; p < 0.001).57 The association with rs738409 genotype was confirmed for severe steatosis, was independent of serum liver function test levels for ALT and gamma-glutamyl transferase (GGT), and was observed in all viral genotypes but genotype 3.57 There was a trend toward the same pattern of effect also in genotype 3 hepatitis C, which was their smallest subgroup, and thus may have been underpowered to see a statistical significance. The rs738409 GG genotype was also associated with fibrosis stage and cirrhosis (OR = 1.47, 95% CI 1.2–1.9; p = 0.002) and with worse treatment response in terms of attaining sustained virologic response (n = 470; OR = 0.63, 95% CI 0.4–0.8; p = 0.006). Around the same time, another large study from Belgium, Germany, and France (n = 537) of mostly genotype 1 hepatitis C patients showed that after adjustment for age, sex, BMI, alcohol consumption, and diabetes, rs738409 mutant G allele homozygote carriers remained at higher risk for steatosis (OR = 2.55, 95% CI 1.08–6.03; p = 0.034), fibrosis (OR = 3.13, 95% CI 1.50–6.51; p = 0.002), and fibrosis progression (OR = 2.64, 95% CI 1.22–5.67; p = 0.013).77 They found that rs738409 was not independently associated with treatment failure (OR = 1.07, 95% CI 0.46–2.49; p = 0.875) and did not influence clinical or biologic variables.77 More recent studies have confirmed that the PNPLA3 I148M variant is a risk factor for the development of severe steatosis or fibrosis progression in chronic hepatitis C in individuals of European ancestry.78,79 Similarly, in 276 Japanese patients with chronic hepatitis C, the GG genotype for rs738409 was independently associated with the presence of steatosis (OR = 2.58, 95% CI 1.37–4.84; p = 0.003), severe necroinflammatory activity (OR = 2.16, 95% CI 1.12–4.16; p = 0.02), and advanced fibrosis (OR = 2.10, 95% CI 1.07–4.11; p = 0.03), after adjustment for age, sex, BMI, and diabetes.80 The effect of the I148M allele on sustained viro-logic response, however, is controversial. Valenti et al57 noted that the G allele at rs738409 was significantly associated with a lower sustained virologic response (SVR) rate in patients with genotype 1 and 4 hepatitis C virus (HCV) infection.57 In a subsequent study, they noted that the I148M variant associated with SVR in patients with genotype 1 and 4 HCV and bridging fibrosis, but not in genotypes 1 and 4 HCV without bridging fibrosis or in those with genotypes 2 and 3 HCV.81 Trepo et al,77 as well as various other groups,53,82 have not found an association of the I148M allele and SVR, however. Whether these differences are due to power or to uncorrected confounders remains to be determined.
The E167K variant at TM6SF2 has also been associated with histologic liver disease in chronic hepatitis C. One study of 148 consecutive patients from southern Italy with biopsy proven anti-HCV/HCV-RNA-positive chronic hepatitis, mostly of genotype 1 and 2, naive for antiviral therapy, were genotyped for TM6SF2 E167K.54 The liver steatosis score was higher in the 18 patients with TM6SF2 E167K variant than in the 130 homozygotes for TM6SF2 167E allele.54 There was no difference in necroinflammatory or fibrosis scores found between the two groups.54 In another study of 815 Italian patients mostly of genotype 1 and 2 with replication in 645 Swiss/German patients, the authors found that TM6SF2 E167K was associated with histologic severity of steatosis, necroinflammation, and fibrosis.83 After adjustment for steatosis severity, the E167K variant associated with cirrhosis (OR = 2.22, 95% CI 1.20–4.03; p = 0.010). The association with clinically significant fibrosis (F2-F4) was replicated in 645 Swiss/German patients (OR = 1.81, 95% CI 1.12–3.02; p = 0.016). A third recent study of 694 chronic hepatitis C patients from Italy, however, did not see an association with steatosis severity after adjustment for age, gender, BMI, and homoeostasis model assessment score with TM6SF2 rs58542926 (OR = 1.48, 95% CI 0.82–2.69; p = 0.19) or with fibrosis (OR = 0.75, 95% CI 0.34–1.63; p = 0.47).78 It is not clear whether the differences in effects between studies are due to differences in power, to differences in the population, or to the presence of unaccounted confounders.
The PNPLA3 I148M allele has also been associated with hepatic steatosis in 235 hepatitis B patients (OR = 1.62, 95% CI 1.00–7.00) after correction for BMI, age, and diabetes/impaired fasting glucose.58 A similar result has been reported in a separate study of 99 individuals with hepatitis B in which the I148M homozygotes had an OR = 13.9 (95% CI 2.2–86.9; p = 0.005) for the presence of severe steatosis compared with individuals lacking this allele.55 This allele has not been associated with fibrosis or cirrhosis in multiple studies.55,59,60
Effects on Graft Outcomes after Liver Transplantation
Liver transplantation provides a unique situation in which the effects of genetic variants within and outside the liver can be evaluated (►Table 3). After transplantation the liver genotype may not match the recipient's genotype and thus forms a genetic mosaic. In one study of 101 hepatitis C-infected individuals who underwent transplantation, the time to Ishak stage 3 fibrosis or HCV-related mortality/graft loss was analyzed using Cox regression modeling adjusting for HCV-Donor Risk Index, warm ischemic time, pretransplant Model for Endstage Liver Disease (MELD), and viral load in 620 days of follow-up after transplantation.63 The rs738409 donor GC or GG variants had 2.53 times the risk of developing fibrosis (95% CI 1.25–5.02; p = 0.008) compared with CC variants.63 In the alternative endpoint: stage 3 fibrosis or all-cause mortality/graft loss, the effect of donor genotype was attenuated but remained significant at 1.98 (95% CI 1.11–3.53).63 This result is interesting as it suggests that PNPLA3's effect is to influence liver disease development and progression. The authors did not observe an association between the PNPLA3 I148M variant, neither in donors nor recipients, with posttransplant hepatic steatosis, however.63 Another larger study, however, did not see an association of the I148M PNPLA3 allele with development of advanced fibrosis in 176 individual with hepatitis C that underwent liver transplantation.84 Also, unlike Dunn et al, Finkenstedt et al found that the PNPLA3 I148M variant in recipients, that is, recipients who carried rs738409-GG, had a higher risk of graft steatosis than recipients who carried rs738409-CC, independent of recipient age or weight gain after liver transplantation.85 This latter result is interesting because it suggests that PNPLA3 may have effects outside of the liver that may be important for development of graft steatosis. Further work will be needed to determine whether these observations are reproducible; if so they may become important to help physicians establish a fibrosis progression management plan after transplantation.
Table 3.
Association of PNPLA3 NAFLD-associated variants with other liver diseases
| Gene SNP (effect allele, EAF) |
Alcoholic steatosis OR (95% CI) |
Alcoholic liver injury OR (95% CI) |
Alcoholic cirrhosis OR (95% CI) |
Hepatitis C steatosis OR (95% CI) |
Hepatitis C cirrhosis OR (95% CI) |
Hepatitis C SVR OR (95% CI) |
Hepatitis B steatosis OR (95% CI) |
Hepatitis B cirrhosis OR (95% CI) |
NAFLD HCC OR (95% CI) |
Alcoholic HCC OR (95% CI) |
Hepatitis C HCC OR (95% CI) |
Liver fibrosis after transplant |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PNPLA3 rs738409 (G, 0.23) | NS56 | 1.45 (1.24–1.69)56 | 2.09 (1.79–2.44)56 | 1.90 (1.4–2.7)57 | 1.47 (1.2–1.9)57 | 0.63 (0.4–0.8)57 | 1.62 (1.00–7.00)58 | NS55,59,60 | 2.26 (1.23–4.14)61 | 2.20 (1.80–2.67)62 | 1.55 (1.03–2.34)62 | 2.53 (1.25–5.02)63 |
Abbreviations: CI, 95% confidence interval; EAF, effect allele frequency; HCC, hepatocellular carcinoma; NAFLD, nonalcoholic fatty liver disease; NS, not significant; OR, odds ratio; SNP, single nucleotide polymorphism; SVR, sustained virologic response.
Effects on Hepatocellular Carcinoma
Hepatocellular carcinoma, like obesity and NAFLD, is rising in prevalence and has now become the number one cause of obesity-related deaths in middle-aged men (reviewed in10) (►Table 3). Hepatocellular carcinoma can develop from many liver insults, although in the United States major causes are hepatitis C, ALD, NAFLD, and hepatitis B.65 The development of HCC was previously thought to require liver cirrhosis as a precursor, but more recent work suggests otherwise. In particular, 10 to 75% of HCC can develop in patients who do not have cirrhosis.12 It has indeed been reported that HCC can develop in patients with simple steatosis, or more specifically, patients who do not have any signs of inflammation or fibrosis.13,14 Because steatosis is a clear risk factor for the development of HCC, it is not surprising that variants that affect hepatic steatosis have been tested for effects on development of HCC.
In obese individuals from the Swedish Obese Subjects Cohort, the G allele at rs738409 in PNPLA3 increased risk of developing HCC with a hazard ratio = 5.9 (95% CI 1.5–23.8) in individuals that did not have bariatric surgery to treat their obesity.86 Further, in individuals with HCC from NAFLD compared with 275 controls with histologically characterized NAFLD, each G allele at rs738409 in multivariate analysis adjusted for age, gender, diabetes, BMI, and presence of cirrhosis, conferred an additive risk for HCC (OR = 2.26, 95% CI 1.23–4.14; p = 0.0082), with GG homozygotes exhibiting a fivefold (95% CI 1.47–17.29; p = 0.01) increased risk over CC homozygotes.61 When compared with the UK general population (1958 British Birth Cohort, n = 1476), the risk-effect was more pronounced (GC vs. CC: unadjusted OR = 2.52, 95% CI 1.55–4.10; p = 0.0002; GG vs. CC: OR = 12.19, 95% CI 6.89–21.58; p < 0.0001).61
Multiple groups reported associations of I148M with HCC in the setting of alcoholic- and hepatitis C-caused liver disease as well.57,59,77,87–90 A meta-analysis of 1,374 individuals with ALD and 945 individuals with chronic hepatitis C found that each I148M allele incurred a higher risk of developing HCC from ALD (OR = 2.20, 95% CI 1.80–2.67) than from chronic hepatitis C (OR = 1.55, 95% CI 1.03–2.34) although both were statistically significant.62 In another meta-analysis of 357 individuals the odds of developing HCC in alcoholic cirrhotics was 2.87 (95% CI 1.61–5.10) for GC versus CC individuals and 12.41 (95% CI 6.99–22.03) for GG versus CC individuals,56 which is similar to what was calculated in the study by Trepo et al.62 This suggests that the pathophysiology of ALD, which is known to have significant steatosis, may in conjunction with a problem in lipid handling incurred by the PNPLA3 mutation, lead to more advance liver pathology than simply having the I148M genetic variant along with concomitant liver disease from hepatitis C.
Improved Understanding of the Biology of Common NAFLD
Following the GWASs for NAFLD, studies into the function of several genes implicated by these GWASs has given us new insights into the pathophysiology of NAFLD (►Fig. 1, ►Table 4).
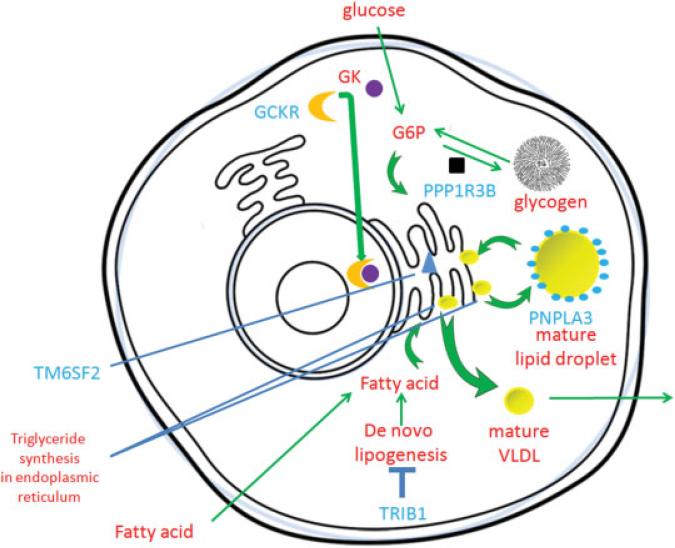
Fig. 1.
Cellular localization of some genes/gene products associated with nonalcoholic fatty liver disease (NAFLD) from genome-wide association studies (GWASs). Genes implicated by GWASs in blue. Glucokinase regulator protein (GCKR) binds to and inactivates glucokinase (GK) by bringing it into the nucleus, thus decreasing phosphorylation of glucose to glucose 6-phosphate, which is a precursor for both glycogen and triglyceride synthesis. Loss of GCKR thus might promote triglyceride synthesis and hepatic steatosis. Protein phosphatase 1 (PPP1R3B) promotes glycogen synthesis and inhibits glycogen breakdown to phosphorylated glucose which can be used to make triglycerides and promote hepatic steatosis; patatin-like phospholipase domain-containing protein 3 (PNPLA3) is present on lipid droplets, but its exact biochemical function is debated; the NAFLD promoting I148M mutation, however, results in increased triglyceride storage in lipid droplets and hepatic steatosis. Transmembrane protein 6 (TM6SF2) is in the perinuclear endoplasmic reticulum- interference with its function results in impaired very low-density lipoprotein cholesterol (VLDL) excretion, accumulation of triglycerides in the cell, and hepatic steatosis. Tribbles 1 homolog (TRIB1) inhibits de novo lipogenesis so its loss thus promotes fatty acid production, triglyceride (TG) synthesis, and hepatic steatosis. G6P, glucose 6-phosphate.
Table 4.
Properties of proteins encoded by genes associated with NAFLD
| Gene | Protein | Function | Cellular location | Catalytic activity | Pathway/biologic function |
Chromosome location |
Tissue |
|---|---|---|---|---|---|---|---|
| PNPLA3 | Patatin-like phospholipase domain-containing protein 3 | Multifunctional enzyme which has both triacylglycerol lipase and acylglycerol O-acyltransferase activities91 | Lipid droplets92 | Triglyceride lipase activity and acylglycerol O-acyltransferase91 Triacylglycerol + H2O ⇌ diacylglycerol + a carboxylate acyl-CoA + 2-acylglycerol ⇌ CoA + diacylglycerol | Triacylglycerol degradation and in glycero-lipid metabolism | chr22:44,319,619–44,343,448 | Liver, gallbladder, kidney, exocrine pancreas, seminal vesicles, intestine, and salivary gland93 |
| LYPLAL1 | Lysophospholipase-like protein 1 | Has depalmitoylating activity. Does not exhibit phospholipase or triacylglycerol lipase activity94 | Cytoplasm94 | Lysophospholipase activity Palmitoyl-protein + H2O ⇌ palmitate + protein94 | Negative regulation of Golgi to plasma membrane protein transport | chr1:219,347,192–219,386,207 | Liver95 Bone marrow96 Placenta97 |
| PPP1R3B | Protein phosphatase 1 regulatory subunit 3B | Acts as a glycogen-targeting subunit for phosphatase PP198 | Glycogen granule99 | Suppresses the rate at which PP1 dephosphorylates (inactivates) glycogen Phosphorylase and enhances the rate at which it activates glycogen synthase and therefore limits glycogen breakdown100 | Carbohydrate metabolism, glycogen metabolism | chr8:8,993,764–9,008,220 | Highly expressed in the liver, and at lower levels in skeletal muscle101 |
| TM6SF2 | Transmembrane 6 superfamily member 2 | Regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content6 | Endoplasmic reticulum (ER) and the ER-Golgi intermediate compartment102 | Regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content102 | Promotes very low-density lipoprotein export43,103 | chr19:19,375,174–19,384,074 | Liver and intestine11 |
| GCKR | Glucokinase regulatory protein | Inhibits glucokinase (GK) by forming an inactive complex with this enzyme. The affinity of GCKR for GK is modulated by fructose metabolites104 | Cytoplasm, nucleus105 | Binding of fructose 6-phosphate and fructose 1-phosphate104 | Carbohydrate metabolism | chr2:27,719,706–27,746,550 | Liver and pancreas106 |
| TRIB1 | Tribbles homolog 1, G-protein-coupled receptor-induced gene 2 protein | Interacts with mitogen-activated protein kinases (MAPKs) and regulates activation of MAPKs. May not display kinase activity107 | Cytoplasm, nucleus105 | Protein kinase inhibitor107 | Inhibits lipogenesis40,88 | chr8:126,442,563–126,450,644 | Expressed in most human tissues including liver with the highest levels in skeletal muscle, thyroid gland, pancreas, peripheral blood leukocytes, and bone marrow107 |
PNPLA3
PNPLA3 stands for patatin-like phospholipase domain containing 3, but has also been called adiponutrin (ADPN) and acylglycerol O-acyltransferase or calcium-independent phospholipase A2-epsilon (iPLA2-epsilon).108 The exact biochemical mechanism by which PNPLA3 acts to cause NAFLD is not yet clear. PNPLA3 is found on the surface of lipid droplets in liver.92 There are data to suggest that wild-type PNPLA3 can break down triglycerides and thus act as a triglyceride hydrolase,92 but others have found that it has lysophosphatidic acid acyltransferase activity also.109 Numerous studies suggest that altering PNPLA3 function can affect lipid trafficking in hepatocytes. Overexpression of the PNPLA3 (I148M) mutant allele in human hepatocyte culture results in lipid accumulation, but overexpression of the wild type of allele does not,92 suggesting a possible gain of function (new function compared with what it does normally) phenotype caused by the I148M variation. Consistent with this result, mice knocked out for PNPLA3 do not develop hepatic steatosis,110,111 whereas mice carrying a PNPLA3 (I148M) knock in mutation and fed a high sucrose diet develop NAFLD confirming that this allele can cause fatty liver disease.103 Human PNPLA3 (I148M) and the mouse equivalent Pnpla347A/A both produce proteins that are catalytically inactive for TG hydrolysis, but as noted above, a knockout of the gene alone is not sufficient to cause steatosis. The presence of the catalytically inactive protein is required to develop the fatty liver phenotype, implying that its presence may interfere with normal lipid metabolism by binding to and increasing the size of lipid droplets and/or blocking lipolysis.103 PNPLA3 mRNA levels are increased by carbohydrate feeding and this increase is mediated by SREBP-1c.112 PNPLA3 degradation is also reduced with the addition of fatty acids to hepatocytes without affecting mRNA levels.112
TRIB1
TRIB1 stands for tribbles pseudokinase 1, but has also been called phosphoprotein regulated by mitogenic pathways, g protein coupled receptor-induced protein, tribbles homolog 1, tribbles-like protein 1, and tribbles homolog 1.113 TRIB1 has been found to affect hepatic lipogenesis and VLDL export via interacting proteins to mediate its NAFLD-promoting effects.50,114,115 Overexpression of Trib1 in mouse livers decreased serum lipids, whereas knockdown increased liver triglyceride content and glycogen as well as serum\plasma glucose, TG, and cholesterol levels.40,88 Knockdown of Trib1 in two separate studies increased the levels of multiple genes involved in lipogenesis, repressed multiple genes involved in fatty acid oxidation, and also increased secretion of VLDL from liver although the exact levels of these genes varied between the two studies.40,88 Pull-down and mammalian two-hybrid analyses revealed novel molecular interaction between TRIB1 and a hepatic lipogenic master regulator, MLXIPL (also known as carbohydrate response element binding protein [ChREBP]).50 MLXIPL binds to MLX to form a heterodimeric complex that binds and activates, in a glucose-dependent manner, carbohydrate response element (ChoRE) motifs in the promoter of lipogenic enzymes. Presumably some of the effects of knocking down Trib1 are due to activation of MLXIPL. TRIB1 was also found to interact with Sin3A-associated protein (SAP18), which when knocked down increased decreased microsomal triglyceride transfer protein (MTTP) levels and led to increasing hepatic lipid levels while decreasing serum lipid levels.87 Because MTTP transfers lipids onto APOB as part of VLDL formation, presumably this results in decreased VLDL export from liver to cause the increased hepatic steatosis while simultaneously causing reduced serum levels of lipids.50 Because variants near TRIB1 that associate with NAFLD increase serum levels of triglycerides, it remains to be seen whether any of the effects of this variant is via SAP18. TRIB1 is also known to be upregulated during inflammatory events such as chronic inflammation of atherosclerotic arteries or chronic antibody-mediated rejection of transplanted organs.116 Recently Akira et al, working with Trib1 knockout mice, demonstrated that mice lacking Trib1 in hematopoietic cells exhibited severe lipodystrophy due to increased lipolysis; while in a high-fat diet, these mice exhibited hypertriglyceridemia, insulin resistance, together with increased proinflammatory cytokine production.117 They suggested that Trib1 is critical for adipose tissue maintenance and suppression of metabolic disorders by controlling the differentiation of tissue-resident anti-inflammatory-like macrophages. Thus, we cannot rule out the possibility that decreased TRIB1 function outside of liver contributes to the development of insulin resistance and higher levels of circulating triglycerides that can secondarily lead to the development of hepatic steatosis.
GCKR
GCKR stands for glucokinase regulator, but has also been called hexokinase 4 regulator and glucokinase regulatory protein.118 GCKR is a regulator of the cellular location of glucokinase (GK). When low levels of glucose are present, GCKR binds to and keeps GK localized to the nucleus where it is not active.119 High glucose concentrations lead to the release of GK from GCKR, allowing GK to be released into the cytoplasm where it can mediate phosphorylation of glucose to glucose 6-phosphate.120 Glucose 6-phosphate can be used either in glycogen synthesis or converted via the pentose phosphate shunt to the precursors required for de novo lipogenesis.121 Variants in GCKR may promote NAFLD by resulting in glucose to triglyceride shifts; this model is being tested in cell biologic systems presently. Indeed, the GCKR P446L variant that associates with NAFLD results in increased activity of glucokinase which phosphorylates glucose122 and in this way121 may promotes glucose to triglyceride shifts.
TM6SF2
TM6SF2 stands for transmembrane 6 superfamily 2 and is also known as KIAA1926.123 TM6SF2 is a membrane protein with 7 to 10 predicted transmembrane domains whose function is not known.48 Kozlitina et al showed that abundance of the mutant form of TM6SF2, (E167K) is lower than the wild-type TM6SF2 protein due to mis-folding and degradation, consistent with this being a loss of function allele.48 They showed that knockdown of mouse Tm6sf2 resulted in increased liver lipid accumulation and decreased serum TG and LDL. They also found that knockdown of mouse Tm6sf2 resulted in decreased VLDL secretion. In a separate study employing confocal microscopy, TM6SF2 was localized to the endoplasmic reticulum (ER) and the ER-Golgi compartment.102 By siRNA knockdown studies, it was found that TM6SF2 is associated with the secretion of triglyceride-rich lipoproteins and inhibition of TM6SF2 leads to increased triglyceride and lipid droplet accumulation.102 From these data, it appears that TM6SF2 may play a role in VLDL excretion.102
PPP1R3B
PPP1R3B stands for protein phosphatase 1, regulatory subunit 3B and is also known as PP1 subunit R4, hepatic glycogen-targeting protein phosphatase 1 regulatory subunit GL, protein phosphatase 1 regulatory subunit 4 (PPP1R4), and hepatic glycogen-targeting subunit, G(L).124 PPP1R3B regulates protein phosphatase-1 (PP-1) catalytic subunit (PPP1CA) and increases PP-1 dephosphorylation of glycogen synthase and phosphorylase kinase.98,100 Glycogen synthase is activated by dephosphorylation and phosphorylase kinase is inactivated by dephosphorylation.125 Thus PPP1R3B promotes glycogen synthesis and inhibits glycogen breakdown to glucose 1-phosphate, which can be converted to glucose 6-phosphate by phosphoglucomutase.126 As discussed in the section on GCKR, glucose 6-phosphate can be converted to precursors of de novo lipid synthesis via either the TCA cycle or the pentose phosphate shunt121
LYPLAL1
LYPLAL1 stands for lysophospholipase-1.127 As has been seen with some genes linked to NAFLD, how LYPLAL1 promotes the development of NAFLD is not fully understood. The LYPLAL1 gene encodes a protein with sequence similarity to APT1, a member of the lysophospholipase family of proteins that have acyl protein thioesterases activity.128 The most well-characterized member of the family is APT1, which has been shown to depalmitoylate proteins such as Ras and other Gα signaling proteins.129 Unlike other lipid modifications of proteins, palmitoylation, also known as S-acylation, like phosphorylation, is a covalent protein modification that can be enzymatically reversed. The cycle of palmitoylation and depalmitoylation of proteins such as N-Ras and HRas has been demonstrated to change the cellular localization of the protein, from the plasma membrane to the Golgi apparatus.129 Although there were suggestions that LYPLAL1 might function as a TG lipase,130 more recent biochemical and X-ray crystallographic studies indicate that it functions as a lysophospholipase.128 Studies of insulin regulated protein palmitoylation have identified several candidates for palmitoylation, but not a linkage to steatosis.131
Clinical Applications
The effect sizes of PNPLA3 and three of the four new NAFLD-associated variants have some of the largest ORs to date for affecting NASH/fibrosis per allele (1.37–3.26)19 (►Table 2), making them common variants with relatively strong effect sizes. These also increase the risk of developing HCC, again with ORs that range from 1.45–2.53 (►Table 3). These variants not only promote the development of advanced liver disease in patients with NAFLD, but also in patients with other liver diseases. Thus, it may not be too long before we may have specific recommendations by genotype for care of patients that carry these genetic variants. Already, these variants alone or in combination have been incorporated into clinical algorithms where they do have significant effects on predicting advanced liver disease.
One of the limits of using already diagnosed metabolic disease (obesity, dyslipidemia, hypertension, and dysglycemia) to predict prevalent NAFLD is that disease is already present (i.e., the individual has developed metabolic disease that is being used to predict more metabolic disease). Ideally, we should inform people of their risk of developing disease while they are still disease free, and make recommendations to prevent or minimize future disease. Here, genetics can be quite useful, as genetic variation can be determined at birth and predates all disease development. One group has determined that the rs738409 variant in PNPLA3 is able to statistically predict prevalent NAFLD above and beyond traditional risk factors; however, in the model alone it did not increase the c statistic very much compared with associated traditional metabolic risk factors.132 However, this study had a high proportion of individuals with metabolic disease; thus, it may not be as generalizable to more population-based samples and may underestimate the true power of genetic studies. Another study used a combination of variants in or near PNPLA3, TM6SF2, GCKR, LYPLAL1, and PPP1R3B and found that this genetic risk score improved prediction of NAFLD in individuals of Mexican ancestry with morbid obesity.133 The combined genetic risk score was significantly associated with higher hepatic triglyceride and total cholesterol content (p = 1.0 × 10−4 and 0.048, respectively), steatosis stage (p = 0.029), and higher ALT levels (p = 0.002). Subjects with a genetic risk score ≥ 6 showed a significantly increased risk of NASH (OR = 2.55, p = 0.045) compared with those with a score ≤ 5. However, the genetic risk score did not predict NASH status, as area under the receiver operating curve was 0.56 (p = 0.219). Overall then, these markers are able to provide information above and beyond traditional risk factors, but alone are not (yet) predictive of developing NAFLD.
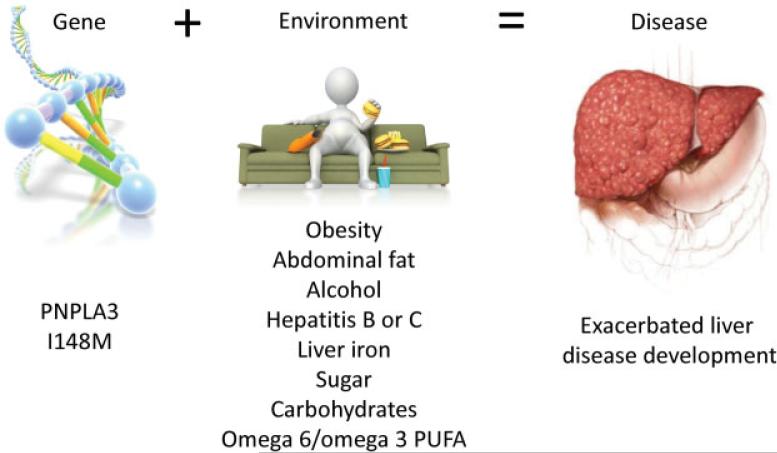
A key part to using genetics to predict and more importantly prevent disease is understanding what factors in the environment these genes interact with to cause disease. In particular, identifying environmental factors that interact with the genetic variant to not only increase risk but increase it in an exponential (gene–environment interaction effect) versus additive manner are especially important to find, as these can substantially affect outcomes. Toward that end, PNPLA3 I148M has been found to interact with obesity,38 abdominal fat,110,134,135 excessive alcohol consumption,136 chronic hepatitis B and C,57,58 or liver iron overload137 to trigger progressive liver disease (►Fig. 2). The magnitude of increase in liver enzymes in I148M carriers is correlated to high dietary carbohydrate, sugar,138–140 and increased omega6/omega3 polyunsaturated fatty acid ratio141,142 intake (►Fig. 2). In the future, incorporating not only one's genetic, but also environmental exposures such as this may help us to not only predict development of NAFLD better than we can presently but in addition to make tailored recommendations on how to mitigate this increased risk.
Fig. 2.
Factors that interact with PNPLA3 I148M to exacerbate liver disease development.22 PUFA, polyunsaturated fatty acid.
The above findings suggest that by avoiding obesity and alcohol and other disease-promoting triggers we may be able to prevent liver disease in those that carry the I148M allele at PNPLA3. This is the premise behind the precision medicine initiatives that are currently underway, where, after knowing a person's genetic risk we can make more directed recommendations to prevent disease (i.e., PNPLA3 I148M carriers may have more benefit from losing weight than noncarriers143). Prospective studies should be done soon to evaluate the effects of various recommendations on outcomes especially for alleles that confer substantial increased risk of developing advanced liver disease such as PNPLA3. At some loci, however, such as TM6SF2, the non-steatosis predisposing allele predisposes to development of elevated serum lipids and cardiovascular disease,46 suggesting that interventions that increase TM6SF2 function may not be completely benign. In particular, augmenting or decreasing TM6SF2 function may predispose to dyslipidemia/cardiovascular disease or to NAFLD/advanced liver disease/HCC, respectively, so the patient's overall risk of developing these diseases would have to be assessed using not only this allele, but also many others before advising patients on the best course of action.144
Acknowledgments
The concept, design, and drafting of the original manuscript was done by EKS; revision of the manuscript was undertaken by BK, BH, and EKS.
BK, BH, and EKS were supported by the Doris Duke Medical Foundation; NIH grant R01DK106621–01; the University of Michigan Internal Medicine Department, Division of Gastroenterology; the University of Michigan Biological Sciences Scholars Program; and The Central Society for Clinical Research.
Abbreviations
- ADPN
adiponutrin
- ALD
alcoholic liver disease
- ALT
alanine aminotransferase
- BMI
body mass index
- ChoRE
carbohydrate response element
- ChREBP
carbohydrate response element binding protein
- CI
confidence interval
- CT
computed tomography
- eQTL
expression quantitative trait locus
- ER
endoplasmic reticulum
- GCKR
glucokinase regulator
- GENOA
Genetic Epidemiology of Arteriopathy
- GK
glucokinase
- GWASs
genome-wide association studies
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- HDL-C
high-density lipoprotein cholesterol
- LD
linkage disequilibrium
- LDL-C
low-density lipoprotein cholesterol
- LYPLAL1
lysophospholipase-1
- MELD
Model for Endstage Liver Disease
- MRI
magnetic resonance imaging
- MRS
magnetic resonance spectroscopy
- MTTP
microsomal triglyceride transfer protein
- NAFLD
nonalcoholic fatty liver disease
- NASH
nonalcoholic steatohepatitis
- OR
odds ratio
- PNPLA3
patatin-like phospholipase domain containing 3
- PPP1R3B
protein phosphatase 1
- SNPs
single nucleotide polymorphisms
- SVR
sustained virologic response
- TM6SF2
transmembrane 6 superfamily 2
- TRIB1
tribbles pseudokinase 1
- VLDL
very low-density lipoprotein
References
- 1.Bellentani S, Marino M. Epidemiology and natural history of non-alcoholic fatty liver disease (NAFLD). Ann Hepatol. 2009;8(Suppl 1):S4–S8. [PubMed] [Google Scholar]
- 2.Speliotes EK, Massaro JM, Hoffmann U, et al. Fatty liver is associated with dyslipidemia and dysglycemia independent of visceral fat: the Framingham Heart Study. Hepatology. 2010;51(6):1979–1987. doi: 10.1002/hep.23593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mellinger JL, Pencina KM, Massaro JM, et al. Hepatic steatosis and cardiovascular disease outcomes: An analysis of the Framingham Heart Study. J Hepatol. 2015;63(2):470–476. doi: 10.1016/j.jhep.2015.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hernaez R, McLean J, Lazo M, et al. Genetics of Obesity-Related Liver Disease (GOLD) Consortium. Association between variants in or near PNPLA3, GCKR, and PPP1R3B with ultrasound-defined steatosis based on data from the third National Health and Nutrition Examination Survey. Clin Gastroenterol Hepatol. 2013;11(9):1183–1190. e2. doi: 10.1016/j.cgh.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Browning JD, Szczepaniak LS, Dobbins R, et al. Prevalence of hepatic steatosis in an urban population in the United States: impact of ethnicity. Hepatology. 2004;40(6):1387–1395. doi: 10.1002/hep.20466. [DOI] [PubMed] [Google Scholar]
- 6.Williams CD, Stengel J, Asike MI, et al. Prevalence of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis among a largely middle-aged population utilizing ultrasound and liver biopsy: a prospective study. Gastroenterology. 2011;140(1):124–131. doi: 10.1053/j.gastro.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 7.Palmer ND, Musani SK, Yerges-Armstrong LM, et al. Characterization of European ancestry nonalcoholic fatty liver disease-associated variants in individuals of African and Hispanic descent. Hepatology. 2013;58(3):966–975. doi: 10.1002/hep.26440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Romeo S, Kozlitina J, Xing C, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2008;40(12):1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neuschwander-Tetri BA, Caldwell SH. Nonalcoholic steatohepatitis: summary of an AASLD Single Topic Conference. Hepatology. 2003;37(5):1202–1219. doi: 10.1053/jhep.2003.50193. [DOI] [PubMed] [Google Scholar]
- 10.Michelotti GA, Machado MV, Diehl AM. NAFLD, NASH and liver cancer. Nat Rev Gastroenterol Hepatol. 2013;10(11):656–665. doi: 10.1038/nrgastro.2013.183. [DOI] [PubMed] [Google Scholar]
- 11.Duwaerts CC, Maher JJ. Mechanisms of liver injury in non-alcoholic steatohepatitis. Curr Hematol Rep. 2014;13(2):119–129. doi: 10.1007/s11901-014-0224-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ertle J, Dechene A, Sowa JP, Penndorf V, Herzer K, Kaiser J, Schlaak JF, Gerken G, Syn WK, Canbay A. Non-alcoholic fatty liver disease progresses to hepatocellular carcinoma in the absence of apparent cirrhosis. Int J Cancer. 2011;128(10):2436–2443. doi: 10.1002/ijc.25797. [DOI] [PubMed] [Google Scholar]
- 13.Paradis V, Zalinski S, Chelbi E, et al. Hepatocellular carcinomas in patients with metabolic syndrome often develop without signifi-cant liver fibrosis: a pathological analysis. Hepatology. 2009;49(3):851–859. doi: 10.1002/hep.22734. [DOI] [PubMed] [Google Scholar]
- 14.Guzman G, Brunt EM, Petrovic LM, Chejfec G, Layden TJ, Cotler SJ. Does nonalcoholic fatty liver disease predispose patients to hepatocellular carcinoma in the absence of cirrhosis? Arch Pathol Lab Med. 2008;132(11):1761–1766. doi: 10.5858/132.11.1761. [DOI] [PubMed] [Google Scholar]
- 15.Lee SS, Park SH, Kim HJ, et al. Non-invasive assessment of hepatic steatosis: prospective comparison of the accuracy of imaging examinations. J Hepatol. 2010;52(4):579–585. doi: 10.1016/j.jhep.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 16.Kunde SS, Lazenby AJ, Clements RH, Abrams GA. Spectrum of NAFLD and diagnostic implications of the proposed new normal range for serum ALT in obese women. Hepatology. 2005;42(3):650–656. doi: 10.1002/hep.20818. [DOI] [PubMed] [Google Scholar]
- 17.Schwimmer JB, Celedon MA, Lavine JE, et al. Heritability of nonalcoholic fatty liver disease. Gastroenterology. 2009;136(5):1585–1592. doi: 10.1053/j.gastro.2009.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wagenknecht LE, Scherzinger AL, Stamm ER, et al. Correlates and heritability of nonalcoholic fatty liver disease in a minority cohort. Obesity (Silver Spring) 2009;17(6):1240–1246. doi: 10.1038/oby.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Speliotes EK, Yerges-Armstrong LM, Wu J, et al. NASH CRN; GIANT Consortium; MAGIC Investigators; GOLD Consortium. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet. 2011;7(3):e1001324. doi: 10.1371/journal.pgen.1001324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loomba R, Schork N, Chen CH, et al. Genetics of NAFLD in Twins Consortium. Heritability of hepatic fibrosis and steatosis based on a prospective twin study. Gastroenterology. 2015 doi: 10.1053/j.gastro.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Visscher PM, Hill WG, Wray NR. Heritability in the genomics era—concepts and misconceptions. Nat Rev Genet. 2008;9(4):255–266. doi: 10.1038/nrg2322. [DOI] [PubMed] [Google Scholar]
- 22.Dongiovanni P, Valenti L, Rametta R, et al. Genetic variants regulating insulin receptor signalling are associated with the severity of liver damage in patients with non-alcoholic fatty liver disease. Gut. 2010;59(2):267–273. doi: 10.1136/gut.2009.190801. [DOI] [PubMed] [Google Scholar]
- 23.Petta S, Valenti L, Marchesini G, et al. PNPLA3 GG genotype and carotid atherosclerosis in patients with non-alcoholic fatty liver disease. PLoS ONE. 2013;8(9):e74089. doi: 10.1371/journal.pone.0074089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Al-Serri A, Anstee QM, Valenti L, et al. The SOD2 C47T polymorphism influences NAFLD fibrosis severity: evidence from case-control and intra-familial allele association studies. J Hepatol. 2012;56(2):448–454. doi: 10.1016/j.jhep.2011.05.029. [DOI] [PubMed] [Google Scholar]
- 25.Aravinthan A, Mells G, Allison M, et al. Gene polymorphisms of cellular senescence marker p21 and disease progression in non-alcohol-related fatty liver disease. Cell Cycle. 2014;13(9):1489–1494. doi: 10.4161/cc.28471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Wang BF, Tong J, Chang B, Wang BY. USF-1 genetic polymorphisms confer a high risk of nonalcoholic fatty liver disease in Chinese population. Int J Clin Exp Med. 2015;8(2):2545–2553. [PMC free article] [PubMed] [Google Scholar]
- 27.Miele L, Beale G, Patman G, et al. The Kruppel-like factor 6 genotype is associated with fibrosis in nonalcoholic fatty liver disease. Gastroenterology. 2008;135(1):282–291. e1. doi: 10.1053/j.gastro.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eslam M, Hashem AM, Leung R, et al. International Hepatitis C Genetics Consortium (IHCGC). Interferon-λ rs12979860 geno-type and liver fibrosis in viral and non-viral chronic liver disease. Nat Commun. 2015;6:6422. doi: 10.1038/ncomms7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petersen KF, Dufour S, Hariri A, et al. Apolipoprotein C3 gene variants in nonalcoholic fatty liver disease. N Engl J Med. 2010;362(12):1082–1089. doi: 10.1056/NEJMoa0907295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozlitina J, Boerwinkle E, Cohen JC, Hobbs HH. Dissociation between APOC3 variants, hepatic triglyceride content and insulin resistance. Hepatology. 2011;53(2):467–474. doi: 10.1002/hep.24072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crosby J, Peloso GM, Auer PL, et al. TG and HDL Working Group of the Exome Sequencing Project, National Heart, Lung, and Blood Institute. Loss-of-function mutations in APOC3, triglycerides, and coronary disease. N Engl J Med. 2014;371(1):22–31. doi: 10.1056/NEJMoa1307095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garrett ME, Abdelmalek MF, Ashley-Koch A, et al. IL28B rs12979860 is not associated with histologic features of NAFLD in a cohort of Caucasian North American patients. J Hepatol. 2013;58(2):402–403. doi: 10.1016/j.jhep.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Petta S, Craxi A. Reply to: “IL28B rs12979860 is not associated with histologic features of NAFLD in a cohort of Caucasian North American patients”. J Hepatol. 2013;58(2):403–404. doi: 10.1016/j.jhep.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 34.Yuan X, Waterworth D, Perry JR, et al. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am J Hum Genet. 2008;83(4):520–528. doi: 10.1016/j.ajhg.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chambers JC, Zhang W, Sehmi J, et al. Alcohol Genome-wide Association (AlcGen) Consortium; Diabetes Genetics Replication and Meta-analyses (DIAGRAM+) Study; Genetic Investigation of Anthropometric Traits (GIANT) Consortium; Global Lipids Genetics Consortium; Genetics of Liver Disease (GOLD) Consortium; International Consortium for Blood Pressure (ICBP-GWAS); Meta-analyses of Glucose and Insulin-Related Traits Consortium (MAGIC). Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet. 2011;43(11):1131–1138. doi: 10.1038/ng.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kantartzis K, Peter A, Machicao F, et al. Dissociation between fatty liver and insulin resistance in humans carrying a variant of the patatin-like phospholipase 3 gene. Diabetes. 2009;58(11):2616–2623. doi: 10.2337/db09-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romeo S, Sentinelli F, Cambuli VM, et al. The 148M allele of the PNPLA3 gene is associated with indices of liver damage early in life. J Hepatol. 2010;53(2):335–338. doi: 10.1016/j.jhep.2010.02.034. [DOI] [PubMed] [Google Scholar]
- 38.Romeo S, Sentinelli F, Dash S, et al. Morbid obesity exposes the association between PNPLA3 I148M (rs738409) and indices of hepatic injury in individuals of European descent. Int J Obes. 2010;34(1):190–194. doi: 10.1038/ijo.2009.216. [DOI] [PubMed] [Google Scholar]
- 39.Kollerits B, Coassin S, Kiechl S, et al. A common variant in the adiponutrin gene influences liver enzyme values. J Med Genet. 2010;47(2):116–119. doi: 10.1136/jmg.2009.066597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Speliotes EK, Butler JL, Palmer CD, Voight BF, Hirschhorn JN. GIANT Consortium; MIGen Consortium; NASH CRN. PNPLA3 variants specifically confer increased risk for histologic nonalcoholic fatty liver disease but not metabolic disease. Hepatology. 2010;52(3):904–912. doi: 10.1002/hep.23768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rotman Y, Koh C, Zmuda JM, Kleiner DE, Liang TJ. NASH CRN. The association of genetic variability in patatin-like phospholipase domain-containing protein 3 (PNPLA3) with histological severity of nonalcoholic fatty liver disease. Hepatology. 2010;52(3):894–903. doi: 10.1002/hep.23759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valenti L, Alisi A, Galmozzi E, et al. I148M patatin-like phospholipase domain-containing 3 gene variant and severity of pediatric nonalcoholic fatty liver disease. Hepatology. 2010;52(4):1274–1280. doi: 10.1002/hep.23823. [DOI] [PubMed] [Google Scholar]
- 43.Valenti L, Al-Serri A, Daly AK, et al. Homozygosity for the patatin-like phospholipase-3/adiponutrin I148M polymorphism influences liver fibrosis in patients with nonalcoholic fatty liver disease. Hepatology. 2010;51(4):1209–1217. doi: 10.1002/hep.23622. [DOI] [PubMed] [Google Scholar]
- 44.Sookoian S, Castaño GO, Burgueño AL, Gianotti TF, Rosselli MS, Pirola CJ. A nonsynonymous gene variant in the adiponutrin gene is associated with nonalcoholic fatty liver disease severity. J Lipid Res. 2009;50(10):2111–2116. doi: 10.1194/jlr.P900013-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sookoian S, Pirola CJ. Meta-analysis of the influence of I148M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of non-alcoholic fatty liver disease. Hepatology. 2011;53(6):1883–1894. doi: 10.1002/hep.24283. [DOI] [PubMed] [Google Scholar]
- 46.Holmen OL, Zhang H, Fan Y, et al. Systematic evaluation of coding variation identifies a candidate causal variant in TM6SF2 influencing total cholesterol and myocardial infarction risk. Nat Genet. 2014;46(4):345–351. doi: 10.1038/ng.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466(7307):707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kozlitina J, Smagris E, Stender S, et al. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat Genet. 2014;46(4):352–356. doi: 10.1038/ng.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kitamoto A, Kitamoto T, Nakamura T, et al. Association of polymorphisms in GCKR and TRIB1 with nonalcoholic fatty liver disease and metabolic syndrome traits. Endocr J. 2014;61(7):683–689. doi: 10.1507/endocrj.ej14-0052. [DOI] [PubMed] [Google Scholar]
- 50.Ishizuka Y, Nakayama K, Ogawa A, et al. Jichi Medical University Promotion Team of Large-Scale Human Genome Bank for All over Japan. TRIB1 downregulates hepatic lipogenesis and glycogenesis via multiple molecular interactions. J Mol Endocrinol. 2014;52(2):145–158. doi: 10.1530/JME-13-0243. [DOI] [PubMed] [Google Scholar]
- 51.Chalasani N, Guo X, Loomba R, et al. Nonalcoholic Steatohepatitis Clinical Research Network. Genome-wide association study identifies variants associated with histologic features of nonalcoholic fatty liver disease. Gastroenterology. 2010;139(5):1567–1576. 1576.e1–1576.e6. doi: 10.1053/j.gastro.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gorden A, Yang R, Yerges-Armstrong LM, et al. GOLD Consortium. Genetic variation at NCAN locus is associated with inflammation and fibrosis in non-alcoholic fatty liver disease in morbid obesity. Hum Hered. 2013;75(1):34–43. doi: 10.1159/000346195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moritou Y, Ikeda F, Iwasaki Y, et al. Impact of comorbid hepatic steatosis on treatment of chronic hepatitis C in Japanese patients and the relationship with genetic polymorphism of IL28B, PNPLA3 and LDL receptor. Acta Med Okayama. 2014;68(1):17–22. doi: 10.18926/AMO/52139. [DOI] [PubMed] [Google Scholar]
- 54.Coppola N, Rosa Z, Cirillo G, et al. TM6SF2 E167K variant is associated with severe steatosis in chronic hepatitis C, regardless of PNPLA3 polymorphism. Liver Int. 2015;35(8):1959–1963. doi: 10.1111/liv.12781. [DOI] [PubMed] [Google Scholar]
- 55.Zampino R, Coppola N, Cirillo G, et al. Patatin-like phospholipase domain-containing 3 I148M variant is associated with liver steatosis and fat distribution in chronic hepatitis B. Dig Dis Sci. 2015;60(10):3005–3010. doi: 10.1007/s10620-015-3716-7. [DOI] [PubMed] [Google Scholar]
- 56.Salameh H, Raff E, Erwin A, et al. PNPLA3 gene polymorphism is associated with predisposition to and severity of alcoholic liver disease. Am J Gastroenterol. 2015;110(6):846–856. doi: 10.1038/ajg.2015.137. [DOI] [PubMed] [Google Scholar]
- 57.Valenti L, Rumi M, Galmozzi E, et al. Patatin-like phospholipase domain-containing 3 I148M polymorphism, steatosis, and liver damage in chronic hepatitis C. Hepatology. 2011;53(3):791–799. doi: 10.1002/hep.24123. [DOI] [PubMed] [Google Scholar]
- 58.Viganò M, Valenti L, Lampertico P, et al. Patatin-like phospholipase domain-containing 3 I148M affects liver steatosis in patients with chronic hepatitis B. Hepatology. 2013;58(4):1245–1252. doi: 10.1002/hep.26445. [DOI] [PubMed] [Google Scholar]
- 59.Falleti E, Fabris C, Cmet S, et al. PNPLA3 rs738409C/G polymorphism in cirrhosis: relationship with the aetiology of liver disease and hepatocellular carcinoma occurrence. Liver Int. 2011;31(8):1137–1143. doi: 10.1111/j.1478-3231.2011.02534.x. [DOI] [PubMed] [Google Scholar]
- 60.Tong J, Guo J, Hu J, Hou S, Zhang Y, Li Q. Correlation between patatin-like phospholipase domain-containing protein 3 gene polymorphisms and liver cirrhosis in a Chinese Han population with chronic hepatitis B. Hepat Mon. 2014;14(8):e18943. doi: 10.5812/hepatmon.18943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu YL, Patman GL, Leathart JB, et al. Carriage of the PNPLA3 rs738409 C >G polymorphism confers an increased risk of non-alcoholic fatty liver disease associated hepatocellular carcinoma. J Hepatol. 2014;61(1):75–81. doi: 10.1016/j.jhep.2014.02.030. [DOI] [PubMed] [Google Scholar]
- 62.Trépo E, Nahon P, Bontempi G, et al. Association between the PNPLA3 (rs738409 C>G) variant and hepatocellular carcinoma: Evidence from a meta-analysis of individual participant data. Hepatology. 2014;59(6):2170–2177. doi: 10.1002/hep.26767. [DOI] [PubMed] [Google Scholar]
- 63.Dunn W, O'Neil M, Zhao J, et al. Donor PNPLA3 rs738409 geno-type affects fibrosis progression in liver transplantation for hepatitis C. Hepatology. 2014;59(2):453–460. doi: 10.1002/hep.26758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.O'Shea RS, Dasarathy S, McCullough AJ. Practice Guideline Committee of the American Association for the Study of Liver Diseases; Practice Parameters Committee of the American College of Gastroenterology. Alcoholic liver disease. Hepatology. 2010;51(1):307–328. doi: 10.1002/hep.23258. [DOI] [PubMed] [Google Scholar]
- 65.Younossi ZM, Otgonsuren M, Henry L, et al. Association of non-alcoholic fatty liver disease (NAFLD) with hepatocellular carcinoma (HCC) in the United States from 2004-2009. Hepatology. 2015 doi: 10.1002/hep.28123. [DOI] [PubMed] [Google Scholar]
- 66.Purohit V, Russo D, Coates PM. Role of fatty liver, dietary fatty acid supplements, and obesity in the progression of alcoholic liver disease: introduction and summary of the symposium. Alcohol. 2004;34(1):3–8. doi: 10.1016/j.alcohol.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 67.Chalasani N, Younossi Z, Lavine JE, et al. The diagnosis and management of non-alcoholic fatty liver disease: practice Guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Hepatology. 2012;55(6):2005–2023. doi: 10.1002/hep.25762. [DOI] [PubMed] [Google Scholar]
- 68.Naveau S, Giraud V, Borotto E, Aubert A, Capron F, Chaput JC. Excess weight risk factor for alcoholic liver disease. Hepatology. 1997;25(1):108–111. doi: 10.1002/hep.510250120. [DOI] [PubMed] [Google Scholar]
- 69.Raynard B, Balian A, Fallik D, et al. Risk factors of fibrosis in alcohol-induced liver disease. Hepatology. 2002;35(3):635–638. doi: 10.1053/jhep.2002.31782. [DOI] [PubMed] [Google Scholar]
- 70.Rosen HR. Clinical practice. Chronic hepatitis C infection. N Engl J Med. 2011;364(25):2429–2438. doi: 10.1056/NEJMcp1006613. [DOI] [PubMed] [Google Scholar]
- 71.Lee SH, Chung YH, Kim JA, et al. Histological characteristics predisposing to development of hepatocellular carcinoma in patients with chronic hepatitis B. J Clin Pathol. 2011;64(7):599–604. doi: 10.1136/jclinpath-2011-200036. [DOI] [PubMed] [Google Scholar]
- 72.Lonardo A, Adinolfi LE, Restivo L, et al. Pathogenesis and significance of hepatitis C virus steatosis: an update on survival strategy of a successful pathogen. World J Gastroenterol. 2014;20(23):7089–7103. doi: 10.3748/wjg.v20.i23.7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dyal HK, Aguilar M, Bhuket T, et al. Concurrent obesity, diabetes, and steatosis increase risk of advanced fibrosis among HCV patients: a systematic review. Dig Dis Sci. 2015;60(9):2813–2824. doi: 10.1007/s10620-015-3760-3. [DOI] [PubMed] [Google Scholar]
- 74.Persico M, Iolascon A. Steatosis as a co-factor in chronic liver diseases. World J Gastroenterol. 2010;16(10):1171–1176. doi: 10.3748/wjg.v16.i10.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang CC, Tseng TC, Kao JH. Hepatitis B virus infection and metabolic syndrome: fact or fiction? J Gastroenterol Hepatol. 2015;30(1):14–20. doi: 10.1111/jgh.12700. [DOI] [PubMed] [Google Scholar]
- 76.Lok AS, Everhart JE, Chung RT, et al. HALT-C Trial Group. Hepatic steatosis in hepatitis C: comparison of diabetic and nondiabetic patients in the hepatitis C antiviral long-term treatment against cirrhosis trial. Clin Gastroenterol Hepatol. 2007;5(2):245–254. doi: 10.1016/j.cgh.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 77.Trépo E, Pradat P, Potthoff A, et al. Impact of patatin-like phospholipase-3 (rs738409 C>G) polymorphism on fibrosis progression and steatosis in chronic hepatitis C. Hepatology. 2011;54(1):60–69. doi: 10.1002/hep.24350. [DOI] [PubMed] [Google Scholar]
- 78.Petta S, Maida M, Grimaudo S, et al. TM6SF2 rs58542926 is not associated with steatosis and fibrosis in large cohort of patients with genotype 1 chronic hepatitis C. Liver Int. 2015 doi: 10.1111/liv.12918. [DOI] [PubMed] [Google Scholar]
- 79.Rüeger S, Bochud PY, Dufour JF, et al. Impact of common risk factors of fibrosis progression in chronic hepatitis C. Gut. 2015;64(10):1605–1615. doi: 10.1136/gutjnl-2014-306997. [DOI] [PubMed] [Google Scholar]
- 80.Yasui K, Kawaguchi T, Shima T, et al. Effect of PNPLA3 rs738409 variant (I148 M) on hepatic steatosis, necroinflammation, and fibrosis in Japanese patients with chronic hepatitis C. J Gastroenterol. 2015;50(8):887–893. doi: 10.1007/s00535-014-1018-z. [DOI] [PubMed] [Google Scholar]
- 81.Valenti L, Aghemo A, Stättermayer AF, et al. Implications of PNPLA3 polymorphism in chronic hepatitis C patients receiving peginterferon plus ribavirin. Aliment Pharmacol Ther. 2012;35(12):1434–1442. doi: 10.1111/j.1365-2036.2012.05109.x. [DOI] [PubMed] [Google Scholar]
- 82.Clark PJ, Thompson AJ, Zhu Q, et al. The association of genetic variants with hepatic steatosis in patients with genotype 1 chronic hepatitis C infection. Dig Dis Sci. 2012;57(8):2213–2221. doi: 10.1007/s10620-012-2171-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Milano M, Aghemo A, Mancina RM, et al. Transmembrane 6 superfamily member 2 gene E167K variant impacts on steatosis and liver damage in chronic hepatitis C patients. Hepatology. 2015;62(1):111–117. doi: 10.1002/hep.27811. [DOI] [PubMed] [Google Scholar]
- 84.do O NT, Eurich D, Trautwein C, Neuhaus P, Neumann UP, Wasmuth HE. The common I148 M variant of PNPLA3 does not predict fibrosis progression after liver transplantation for hepatitis C. Hepatology. 2011;54(4):1483–1484. doi: 10.1002/hep.24535. [DOI] [PubMed] [Google Scholar]
- 85.Finkenstedt A, Auer C, Glodny B, et al. Patatin-like phospholipase domain-containing protein 3 rs738409-G in recipients of liver transplants is a risk factor for graft steatosis. Clin Gastroenterol Hepatol. 2013;11(12):1667–1672. doi: 10.1016/j.cgh.2013.06.025. [DOI] [PubMed] [Google Scholar]
- 86.Burza MA, Pirazzi C, Maglio C, et al. PNPLA3 I148M (rs738409) genetic variant is associated with hepatocellular carcinoma in obese individuals. Dig Liver Dis. 2012;44(12):1037–1041. doi: 10.1016/j.dld.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 87.Corradini SG, Burza MA, Molinaro A, Romeo S. Patatin-like phospholipase domain containing 3 sequence variant and hepatocellular carcinoma. Hepatology. 2011;53(5):1776. doi: 10.1002/hep.24244. author reply 1777. [DOI] [PubMed] [Google Scholar]
- 88.Guyot E, Sutton A, Rufat P, et al. PNPLA3 rs738409, hepatocellular carcinoma occurrence and risk model prediction in patients with cirrhosis. J Hepatol. 2013;58(2):312–318. doi: 10.1016/j.jhep.2012.09.036. [DOI] [PubMed] [Google Scholar]
- 89.Hamza H, Cao J, Li X, Zhao S. In vivo study of hepatitis B vaccine effects on inflammation and metabolism gene expression. Mol Biol Rep. 2012;39(3):3225–3233. doi: 10.1007/s11033-011-1090-x. [DOI] [PubMed] [Google Scholar]
- 90.Nischalke HD, Berger C, Luda C, et al. The PNPLA3 rs738409 148M/M genotype is a risk factor for liver cancer in alcoholic cirrhosis but shows no or weak association in hepatitis C cirrhosis. PLoS ONE. 2011;6(11):e27087. doi: 10.1371/journal.pone.0027087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jenkins CM, Mancuso DJ, Yan W, Sims HF, Gibson B, Gross RW. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J Biol Chem. 2004;279(47):48968–48975. doi: 10.1074/jbc.M407841200. [DOI] [PubMed] [Google Scholar]
- 92.He S, McPhaul C, Li JZ, et al. A sequence variation (I148M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. J Biol Chem. 2010;285(9):6706–6715. doi: 10.1074/jbc.M109.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. [August 30, 2015];[Anonymous] Human Protein Atlas. Available at: http://www.proteinatlas.org/ENSG00000100344-PNPLA3/tissue.
- 94.Tian L, McClafferty H, Knaus HG, Ruth P, Shipston MJ. Distinct acyl protein transferases and thioesterases control surface expression of calcium-activated potassium channels. J Biol Chem. 2012;287(18):14718–14725. doi: 10.1074/jbc.M111.335547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bian Y, Song C, Cheng K, et al. An enzyme assisted RP-RPLC approach for in-depth analysis of human liver phosphoproteome. J Proteomics. 2014;96:253–262. doi: 10.1016/j.jprot.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 96.Gerhard DS, Wagner L, Feingold EA, et al. MGC Project Team. The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC). Genome Res. 2004;14(10B):2121–2127. doi: 10.1101/gr.2596504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ota T, Suzuki Y, Nishikawa T, et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet. 2004;36(1):40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- 98.Doherty MJ, Moorhead G, Morrice N, Cohen P, Cohen PT. Amino acid sequence and expression of the hepatic glycogen-binding (GL)-subunit of protein phosphatase-1. FEBS Lett. 1995;375(3):294–298. doi: 10.1016/0014-5793(95)01184-g. [DOI] [PubMed] [Google Scholar]
- 99.Browne GJ, Delibegovic M, Keppens S, Stalmans W, Cohen PT. The level of the glycogen targetting regulatory subunit R5 of protein phosphatase 1 is decreased in the livers of insulin-dependent diabetic rats and starved rats. Biochem J. 2001;360(Pt 2):449–459. doi: 10.1042/0264-6021:3600449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gasa R, Jensen PB, Berman HK, Brady MJ, DePaoli-Roach AA, Newgard CB. Distinctive regulatory and metabolic properties of glycogen-targeting subunits of protein phosphatase-1 (PTG, GL, GM/RGl) expressed in hepatocytes. J Biol Chem. 2000;275(34):26396–26403. doi: 10.1074/jbc.M002427200. [DOI] [PubMed] [Google Scholar]
- 101.Munro S, Cuthbertson DJ, Cunningham J, Sales M, Cohen PT. Human skeletal muscle expresses a glycogen-targeting subunit of PP1 that is identical to the insulin-sensitive glycogen-targeting subunit G(L) of liver. Diabetes. 2002;51(3):591–598. doi: 10.2337/diabetes.51.3.591. [DOI] [PubMed] [Google Scholar]
- 102.Mahdessian H, Taxiarchis A, Popov S, et al. TM6SF2 is a regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content. Proc Natl Acad Sci U S A. 2014;111(24):8913–8918. doi: 10.1073/pnas.1323785111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Smagris E, BasuRay S, Li J, et al. Pnpla3I148M knockin mice accumulate PNPLA3 on lipid droplets and develop hepatic steatosis. Hepatology. 2015;61(1):108–118. doi: 10.1002/hep.27242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Choi JM, Seo MH, Kyeong HH, Kim E, Kim HS. Molecular basis for the role of glucokinase regulatory protein as the allosteric switch for glucokinase. Proc Natl Acad Sci U S A. 2013;110(25):10171–10176. doi: 10.1073/pnas.1300457110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jin L, Guo T, Li Z, et al. Role of glucokinase in the subcellular localization of glucokinase regulatory protein. Int J Mol Sci. 2015;16(4):7377–7393. doi: 10.3390/ijms16047377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hayward BE, Dunlop N, Intody S, et al. Organization of the human glucokinase regulator gene GCKR. Genomics. 1998;49(1):137–142. doi: 10.1006/geno.1997.5195. [DOI] [PubMed] [Google Scholar]
- 107.Kiss-Toth E, Bagstaff SM, Sung HY, et al. Human tribbles, a protein family controlling mitogen-activated protein kinase cascades. J Biol Chem. 2004;279(41):42703–42708. doi: 10.1074/jbc.M407732200. [DOI] [PubMed] [Google Scholar]
- 108.GeneCards [09/15/15];PNPLA3 gene (protein coding) Available at: http://www.genecards.org/cgi-bin/carddisp.pl?gene=PNPLA3.
- 109.Kumari M, Schoiswohl G, Chitraju C, et al. Adiponutrin functions as a nutritionally regulated lysophosphatidic acid acyltransferase. Cell Metab. 2012;15(5):691–702. doi: 10.1016/j.cmet.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen W, Chang B, Li L, Chan L. Patatin-like phospholipase domain-containing 3/adiponutrin deficiency in mice is not associated with fatty liver disease. Hepatology. 2010;52(3):1134–1142. doi: 10.1002/hep.23812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Basantani MK, Sitnick MT, Cai L, et al. Pnpla3/Adiponutrin deficiency in mice does not contribute to fatty liver disease or metabolic syndrome. J Lipid Res. 2011;52(2):318–329. doi: 10.1194/jlr.M011205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Huang Y, He S, Li JZ, et al. A feed-forward loop amplifies nutritional regulation of PNPLA3. Proc Natl Acad Sci U S A. 2010;107(17):7892–7897. doi: 10.1073/pnas.1003585107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.GeneCards [09/15/15];TRIB1 gene (protein coding) Available at: http://www.genecards.org/cgi-bin/carddisp.pl?gene=TRIB1&keywords=TRIB1.
- 114.Makishima S, Boonvisut S, Ishizuka Y, Watanabe K, Nakayama K, Iwamoto S. Sin3A-associated protein, 18 kDa, a novel binding partner of TRIB1, regulates MTTP expression. J Lipid Res. 2015;56(6):1145–1152. doi: 10.1194/jlr.M057802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Burkhardt R, Toh SA, Lagor WR, et al. Trib1 is a lipid- and myocardial infarction-associated gene that regulates hepatic lipogenesis and VLDL production in mice. J Clin Invest. 2010;120(12):4410–4414. doi: 10.1172/JCI44213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dugast E, Kiss-Toth E, Soulillou JP, Brouard S, Ashton-Chess J. The Tribbles-1 protein in humans: roles and functions in health and disease. Curr Mol Med. 2013;13(1):80–85. [PubMed] [Google Scholar]
- 117.Akira S, Misawa T, Satoh T, Saitoh T. Macrophages control innate inflammation. Diabetes Obes Metab. 2013;15(Suppl 3):10–18. doi: 10.1111/dom.12151. [DOI] [PubMed] [Google Scholar]
- 118.GeneCards [09/15/15];GCKR gene (protein coding) Available at: http://www.genecards.org/cgi-bin/carddisp.pl?gene=GCKR&keywords=gckr.
- 119.Shiota C, Coffey J, Grimsby J, Grippo JF, Magnuson MA. Nuclear import of hepatic glucokinase depends upon glucokinase regulatory protein, whereas export is due to a nuclear export signal sequence in glucokinase. J Biol Chem. 1999;274(52):37125–37130. doi: 10.1074/jbc.274.52.37125. [DOI] [PubMed] [Google Scholar]
- 120.Jetton TL, Shiota M, Knobel SM, Piston DW, Cherrington AD, Magnuson MA. Substrate-induced nuclear export and peripheral compartmentalization of hepatic glucokinase correlates with glycogen deposition. Int J Exp Diabetes Res. 2001;2(3):173–186. doi: 10.1155/EDR.2001.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bechmann LP, Hannivoort RA, Gerken G, Hotamisligil GS, Trauner M, Canbay A. The interaction of hepatic lipid and glucose metabolism in liver diseases. J Hepatol. 2012;56(4):952–964. doi: 10.1016/j.jhep.2011.08.025. [DOI] [PubMed] [Google Scholar]
- 122.Beer NL, Tribble ND, McCulloch LJ, et al. The P446L variant in GCKR associated with fasting plasma glucose and triglyceride levels exerts its effect through increased glucokinase activity in liver. Hum Mol Genet. 2009;18(21):4081–4088. doi: 10.1093/hmg/ddp357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.GeneCards [09/15/15];TM6SF2 gene (protein coding) Available at: http://www.genecards.org/cgi-bin/carddisp.pl?gene=TM6SF2&keywords=TM6SF2.
- 124.GeneCards [09/15/15];PPP1R3B gene (Protein Coding) Available at: http://www.genecards.org/cgi-bin/carddisp.pl?gene=PPP1R3B&keywords=pPP1R3B.
- 125.Johnson LN. Glycogen phosphorylase: control by phosphorylation and allosteric effectors. FASEB J. 1992;6(6):2274–2282. doi: 10.1096/fasebj.6.6.1544539. [DOI] [PubMed] [Google Scholar]
- 126.Weber G, Cantero A. Behavior of enzymes involved in glucose-6-phosphate utilization during 6-day fasting; study of hepatic glucose-6-phosphatase, phosphohexoseisomerase and phosphoglucomutase. Exp Cell Res. 1958;14(3):596–607. doi: 10.1016/0014-4827(58)90164-2. [DOI] [PubMed] [Google Scholar]
- 127.GeneCards [09/15/15];LYPLAL1 gene (Protein Coding) Available at: http://www.genecards.org/cgi-bin/carddisp.pl?gene=LYPLAL1&keywords=LYPLAL1.
- 128.Bürger M, Zimmermann TJ, Kondoh Y, et al. Crystal structure of the predicted phospholipase LYPLAL1 reveals unexpected functional plasticity despite close relationship to acyl protein thioesterases. J Lipid Res. 2012;53(1):43–50. doi: 10.1194/jlr.M019851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Rocks O, Peyker A, Kahms M, et al. An acylation cycle regulates localization and activity of palmitoylated Ras isoforms. Science. 2005;307(5716):1746–1752. doi: 10.1126/science.1105654. [DOI] [PubMed] [Google Scholar]
- 130.Steinberg GR, Kemp BE, Watt MJ. Adipocyte triglyceride lipase expression in human obesity. Am J Physiol Endocrinol Metab. 2007;293(4):E958–E964. doi: 10.1152/ajpendo.00235.2007. [DOI] [PubMed] [Google Scholar]
- 131.Wei X, Song H, Semenkovich CF. Insulin-regulated protein palmitoylation impacts endothelial cell function. Arterioscler Thromb Vasc Biol. 2014;34(2):346–354. doi: 10.1161/ATVBAHA.113.302848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kotronen A, Peltonen M, Hakkarainen A, et al. Prediction of non-alcoholic fatty liver disease and liver fat using metabolic and genetic factors. Gastroenterology. 2009;137(3):865–872. doi: 10.1053/j.gastro.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 133.León-Mimila P, Vega-Badillo J, Gutiérrez-Vidal R, et al. A genetic risk score is associated with hepatic triglyceride content and non-alcoholic steatohepatitis in Mexicans with morbid obesity. Exp Mol Pathol. 2015;98(2):178–183. doi: 10.1016/j.yexmp.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 134.Giudice EM, Grandone A, Cirillo G, et al. The association of PNPLA3 variants with liver enzymes in childhood obesity is driven by the interaction with abdominal fat. PLoS ONE. 2011;6(11):e27933. doi: 10.1371/journal.pone.0027933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Graff M, North KE, Franceschini N, et al. PNPLA3 gene-by-visceral adipose tissue volume interaction and the pathogenesis of fatty liver disease: the NHLBI family heart study. Int J Obes. 2013;37(3):432–438. doi: 10.1038/ijo.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tian C, Stokowski RP, Kershenobich D, Ballinger DG, Hinds DA. Variant in PNPLA3 is associated with alcoholic liver disease. Nat Genet. 2010;42(1):21–23. doi: 10.1038/ng.488. [DOI] [PubMed] [Google Scholar]
- 137.Valenti L, Maggioni P, Piperno A, et al. Patatin-like phospholipase domain containing-3 gene I148M polymorphism, steatosis, and liver damage in hereditary hemochromatosis. World J Gastroenterol. 2012;18(22):2813–2820. doi: 10.3748/wjg.v18.i22.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Davis JN, Lê KA, Walker RW, et al. Increased hepatic fat in overweight Hispanic youth influenced by interaction between genetic variation in PNPLA3 and high dietary carbohydrate and sugar consumption. Am J Clin Nutr. 2010;92(6):1522–1527. doi: 10.3945/ajcn.2010.30185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Nobili V, Liccardo D, Bedogni G, et al. Influence of dietary pattern, physical activity, and I148M PNPLA3 on steatosis severity in at-risk adolescents. Genes Nutr. 2014;9(3):392. doi: 10.1007/s12263-014-0392-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sevastianova K, Santos A, Kotronen A, et al. Effect of short-term carbohydrate overfeeding and long-term weight loss on liver fat in overweight humans. Am J Clin Nutr. 2012;96(4):727–734. doi: 10.3945/ajcn.112.038695. [DOI] [PubMed] [Google Scholar]
- 141.Santoro N, Savoye M, Kim G, et al. Hepatic fat accumulation is modulated by the interaction between the rs738409 variant in the PNPLA3 gene and the dietary omega6/omega3 PUFA intake. PLoS ONE. 2012;7(5):e37827. doi: 10.1371/journal.pone.0037827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Nobili V, Bedogni G, Donati B, Alisi A, Valenti L. The I148M variant of PNPLA3 reduces the response to docosahexaenoic acid in children with non-alcoholic fatty liver disease. J Med Food. 2013;16(10):957–960. doi: 10.1089/jmf.2013.0043. [DOI] [PubMed] [Google Scholar]
- 143.Sevastianova K, Kotronen A, Gastaldelli A, et al. Genetic variation in PNPLA3 (adiponutrin) confers sensitivity to weight loss-induced decrease in liver fat in humans. Am J Clin Nutr. 2011;94(1):104–111. doi: 10.3945/ajcn.111.012369. [DOI] [PubMed] [Google Scholar]
- 144.Kahali B, Liu YL, Daly AK, Day CP, Anstee QM, Speliotes EK. TM6SF2: catch-22 in the fight against nonalcoholic fatty liver disease and cardiovascular disease? Gastroenterology. 2015;148(4):679–684. doi: 10.1053/j.gastro.2015.01.038. [DOI] [PubMed] [Google Scholar]
- 145.Buch S, Stickel F, Trépo E, et al. A genome-wide association study confirms PNPLA3 and identifies TM6SF2 and MBOAT7 as risk loci for alcohol-related cirrhosis. [October 19, 2015];Nature Genet. 2015 doi: 10.1038/ng.3417. Available at http://www.nature.com/ng/journal/vaop/ncurrent/full/ng.3417.html. [DOI] [PubMed]