Abstract
We have recently developed a simple, reusable and coupled whole-cell biocatalytic system with the capability of cofactor regeneration and biocatalyst immobilization for improved production yield and sustained synthesis. Described herewith is the experimental procedure for the development of such a system consisting of two E. coli strains that express functionally complementary enzymes. Together, these two enzymes can function co-operatively to mediate the regeneration of expensive cofactors for improving the product yield of the bioreaction. In addition, the method of synthesizing an immobilized form of the coupled biocatalytic system by encapsulation of whole cells in calcium alginate beads is reported. As an example, we present the improved biosynthesis of L-xylulose from L-arabinitol by coupling E. coli cells expressing the enzymes L-arabinitol dehydrogenase or NADH oxidase. Under optimal conditions and using an initial concentration of 150 mM L-arabinitol, the maximal L-xylulose yield reached 96%, which is higher than those reported in the literature. The immobilized form of the coupled whole-cell biocatalysts demonstrated good operational stability, maintaining 65% of the yield obtained in the first cycle after 7 cycles of successive re-use, while the free cell system almost completely lost the catalytic activity. Therefore, the methods reported here provides two strategies that could help improve the industrial production of L-xylulose, as well as other value-added compounds requiring the use of cofactors in general.
Keywords: Chemistry, Issue 110, whole-cell biocatalyst, cell-free extract, cofactor regeneration, enzyme-coupled system, immobilization, L-xylulose, multi-protein synergy
Introduction
Reductive whole-cell biotransformation using microorganisms has become a widespread method for the chemo-enzymatic synthesis of commercially and therapeutically important biomolecules1-3. It presents several advantages over the use of isolated enzymes, especially the elimination of cost-intensive downstream purification processes and the demonstration of an extended lifetime4-7. For biocatalytic pathways where cofactors are required for product formation, whole-cell systems have the potential to provide in situ cofactor regeneration via the addition of inexpensive electron-donating co-substrates5,8,9. However, this capacity is diminished for reactions that require a stoichiometric concentration of rare or expensive co-substrates10-13. Together with poor reusability of whole cells, this impedes the establishment of a scalable and continuous production system. Strategic modifications of whole-cell systems for these cofactor-dependent biotransformations are required to overcome the aforementioned limitations. Specifically, the combination of whole-cell biocatalysts that work cooperatively have been shown to significantly enhance the productivity and stability of the harbored enzymes14. These factors, which are often critical for enabling large-scale production of commercially viable products, can be optimized further by co-immobilizing biocatalytic microbes15. We have recently developed a simple and reusable whole-cell biocatalytic system that allows both cofactor regeneration and biocatalyst immobilization for the L-xylulose production16. In this study, this system was utilized as an example to illustrate the experimental procedures of applying these two strategies for improved biotransformation production yield and biocatalyst reusability.
L-xylulose belongs to a class of biologically useful molecules named rare sugars. Rare sugars are unique monosaccharides or sugar derivatives that occur very rarely in nature, but play crucial roles as recognition elements in bioactive molecules17,18. They have a variety of applications ranging from sweeteners, functional foods to potential therapeutics19. L-xylulose can be used as a potential inhibitor of multiple α-glucosidases, and may also be used as an indicator of hepatitis or liver cirrhosis17,20. High efficiency conversion of xylitol to L-xylulose in whole-cell systems has been reported previously in Pantoea ananatis21,22, Alcaligenes sp. 701B23, Bacillus pallidus Y2524,25 and Escherichia coli26. In E. coli, however, it was achieved only using low (<67 mM) xylitol concentrations26 due to potential inhibitory effects of an initial xylitol concentration higher than 100 mM on xylitol-4-dehydrogenase activity21,26. The thermodynamic equilibrium between xylulose and xylitol has been shown to strongly favor the formation of xylitol25,27. Additionally, xylulose yield is limited by the amount of expensive cofactors that have to be supplied in the absence of an in situ cofactor regeneration system. Together, these factors limit the potential for scaling into sustainable systems for L-xylulose biosynthesis.
To overcome these limitations and improve the L-xylulose biotransformation yield, the strategy of cofactor regeneration was employed first by establishing a coupled whole-cell biocatalytic system. Specifically, L-Arabinitol 4-dehydrogenase (EC 1.1.1.12) from Hypocrea jecorina (HjLAD), an enzyme in the L-arabinose catabolic pathway of fungi, was selected to catalyze the conversion of L-arabinitol into L-xylulose28,29. Like many biosynthetic enzymes, a major limitation of HjLAD is that it requires a stoichiometric amount of the expensive nicotinamide adenine dinucleotide cofactor (NAD+, the oxidized form of NADH) to carry out this conversion. NADH oxidase found in Streptococcus pyogenes (SpNox) has been shown to display high cofactor-regeneration activity30,31. Taking advantage of this attribute of SpNox, E. coli cells expressing HjLAD for the production of L-xylulose were coupled with E. coli cells expressing SpNox for the regeneration of NAD+ to boost the L-xylulose production depicted by the coupled reaction shown in Figure 1A. Under optimal conditions and using an initial concentration of 150 mM L-arabinitol, the maximal L-xylulose yield reached 96%, making this system much more efficient than those reported in literature.
The strategy of whole-cell immobilization was employed next to further enhance the reusability of the coupled biocatalytic system. Commonly used methods for whole-cell immobilization include adsorption/covalent linking to solid matrices, cross-linking/entrapment and encapsulation in polymeric networks32. Among these approaches, the most suitable method for cell immobilization is encapsulation in calcium alginate beads. Their mild gelation properties, inert aqueous matrix and high porosity help preserve the physiological properties and functionality of the encapsulated biologicals33. Therefore, the coupled biocatalyst system containing both E. coli cells harboring HjLAD or SpNox was immobilized in calcium alginate beads to enable multiple cycles of L-xylulose production (Figure 2).The immobilized biocatalyst system demonstrated good operational stability, maintaining 65% of the conversion yield of the first cycle after 7 cycles of successive re-use, while the free cell system almost completely lost its catalytic activity.
Protocol
1. Whole-cell Biocatalysts Preparation
NOTE: The recombinant E. coli cells harboring pET28a-SpNox31 or pET28a-HjLAD28 are hereafter referred to as E. coliSpNox and E. coliHjLAD, respectively.
Inoculate a single colony of E. coliHjLAD in 3 ml of Luria-Bertani (LB) medium supplemented with kanamycin (50 µg/ml) and incubate in an incubator shaker O/N at 37 oC, 250 rpm.
Dilute the culture by 1:100 in 200 ml of fresh LB containing 50 µg/ml kanamycin and incubate at 37 oC, 250 rpm until the OD600 reaches ~0.6.
- Induce HjLAD protein expression by adding 0.1 mM isopropyl β-D-thiogalactopyranoside (IPTG) to the culture medium and incubate at 16 oC, 180 rpm for 16 hr.
- Alternatively, perform induction at 25 oC, 200 rpm for 6 hr if the L-xylulose biosynthesis (Step 2 below) is performed the same day.
Harvest the induced E. coliHjLAD cells by centrifugation at 3,200 x g for 20 min at 4 oC. Discard the supernatant and proceed to Step 2 to process the cell pellet.
In parallel, perform steps 1.1 - 1.4 for E. coliSpNox.
2. Biosynthesis of L-xylulose by Coupling E. coliHjLAD and E. coliSpNox for Cofactor Regeneration
Resuspend the cell pellets of E. coliHjLAD and E. coliSpNox separately in 50 mM Tris-HCl buffer (pH 8.0) at a cell density of 5.0 g dry cell weight (gDCW)/L. Note: A correlation between gDCW and the optical density measured at 600 nm (OD600) can be established to facilitate the experiment. The formula used in this protocol is 1 gDCW/L=0.722*OD600 - 0.0965, which can vary among different spectrometers.
Mix 600 µl of 5.0 gDCW/L E. coliHjLAD, 600 µl of 5.0 gDCW/L E. coliSpNox, 100 µl of 20 mM NAD+, and 150 µl of 2 M L-arabinitol in a 14 ml round-bottom tube and bring the reaction volume to 2 ml by adding 550 µl of 50 mM Tris-HCl (pH 8.0). Note: The ratio of the two whole-cell biocatalysts amount can be optimized to improve the biosynthesis of the product. For the described system, a ratio of E. coliSpNox:E. coliHjLAD = 1:1 was found to be optimal for L-xylulose biosynthesis (Figure 1B).
Incubate the reaction mixture at 30 oC, 200 rpm for 8 hr.
Collect the supernatant after centrifugation at 4 oC, 3,200 x g for 10 min and proceed to quantify the L-xylulose production as described in Step 3 below.
3. Colorimetric Assay for L-xylulose Quantification
Aspirate 100 µl of the reaction supernatant collected from step 2.4 into a 1.5 ml tube.
Add 50 µl of 1.5% cysteine, 900 µl of 70% sulfuric acid, and 50 µl of 0.1 % carbazole dissolved in ethanol and mix gently by inverting the tube 3 times.
Incubate the reaction mixture at 37 oC, 200 rpm for 20 min.
- Measure the optical absorbance of the reaction mixture at 560 nm (A560) using a spectrophotometer.
- Dilute the reaction mixture if the A560 reading is above 1.
4. Immobilization of Recombinant Whole-cell Catalysts in Calcium Alginate Beads
Dissolve 4 g sodium alginate in 100 ml of distilled water. Prepare alginate solution by adding sodium alginate to water to avoid the formation of clumps. Heat the mixture if needed.
Add 600 µl of 5.0 gDCW/L E. coliHjLAD and 600 µl of 5.0 gDCW/L E. coliSpNox in 1.2 ml 4% alginate prepared in step 4.1 and mix the cells and alginate by gentle pipetting to avoid bubble formation.
Aspirate the alginate/cell suspension into a syringe using a needle and add the mixture drop-wise into a 0.3 M calcium chloride (CaCl2) solution in a 100 ml beaker with continuous stirring. Note: The volume of CaCl2 solution used for alginate bead formation should be enough for the alginate droplets to be completely submerged. Additionally, the distance between the syringe needle and the surface of the calcium chloride solution must be maintained within an optimal range to ensure formation of uniformly spherical beads. The optimum distance range can be determined experimentally and depends on the inner diameter of the needle. As an estimate, a distance of ~15 ± 5 cm was found to be optimal for a 0.6 cm (inner diameter) syringe needle.
Leave the beads in the CaCl2 solution for 2 - 3 hr at RT without stirring to allow crosslinking and gel beads formation.
Decant the CaCl2 solution without disturbing the beads by carefully pouring the CaCl2 solution into a 50 ml conical tube, and transfer the remaining beads in CaCl2 solution into another 50 ml conical tube.
- Wash the beads with 10 ml of 50 mM Tris-HCl (pH 8.0) buffer three times to remove excessive CaCl2 and un-encapsulated cells. NOTE:At any given step, do not centrifuge the beads as doing so will rupture them. To separate the beads from solution, allow the suspension to stand undisturbed for 3 - 5 min. The beads will settle at the bottom and the wash buffer can be decanted into another container.
- Do not discard the used wash buffer. Pool the used Tris-HCl wash buffer (30 ml) with the used CaCl2 solution collected from step 4.5.
Pellet the un-immobilized E. coli cells by centrifugation of the pooled CaCl2 and Tris-HCl solution collected from step 4.6 at 3,200 x g for 20 min. To determine the immobilization efficiency, calculate the density of the pelleted un-encapsulated cells in gDCW/L as described in step 2.1.
Transfer the washed beads from step 4.6 into a tube. Follow steps 2.2 - 3.4 to evaluate the L-xylulose biosynthesis using all of the washed beads in place of cell pellets.
5. Stability Assay of Immobilized Biocatalysts for L-xylulose Production
Collect the beads from step 4.8 and wash twice with 10 ml of 50 mM Tris-HCl (pH 8.0) buffer without centrifugation (as described in Step 4.6).
Use all of the washed beads to perform the reaction as described in steps 2.2 - 3.4.
Repeat steps 5.1 - 5.2 for desired number of production cycles and measure the amount of L-xylulose produced in the reaction supernatant in each cycle.
Representative Results
To enable cofactor regeneration, L-xylulose synthesis was carried out in a coupled whole-cell biocatalytic system containing E. coliHjLAD and E. coliSpNox cells. Following the optimization of various parameters, the reusability of this system was improved by immobilizing it in calcium alginate beads (Figure 2).
L-xylulose Production with Cofactor Regeneration by Coupling E. coliHjLAD and E. coliSpNox cells. To enhance the bioconversion of L-arabinitol into L-xylulose, the E. coliHjLAD and E. coliSpNox cells were coupled to enable cofactor regeneration. To account for the differences in the protein expression and cofactor availability in E. coliSpNox and E. coliHjLAD cells, a comparison study of the L-xylulose yield at various E. coliSpNox-to-E. coliHjLAD ratios was performed. As shown in Figure 1B, the yield increased as the E. coliSpNox-to-E. coliHjLAD ratio increased from 0.13:1 to 1:1. The yields obtained for ratios 1:1 and 1.33:1 were equivalent and maximal for this coupled system. All further experiments were performed with the E. coliSpNox-to-E. coliHjLAD ratio of 1:1. Starting with an initial concentration of 150 mM L-arabinitol, the use of E. coliHjLAD biocatalyst alone produced 118 mM L-xylulose after 8 hr (saturation point for L-xylulose concentration), representing a yield of 79% (Figure 1C). In comparison, the use of the E. coliHjLAD and E. coliSpNox cells at a 1:1 ratio produced 144 mM L-xylulose, improving the yield to 96%.
Optimization of Whole-cell Biocatalyst Immobilization Immobilization of biocatalysts in calcium alginate beads offers many advantages, such as improved viability, as a result of the mild gel environment that protects the cells from the physical stresses in a bioreactor. The properties of these alginate beads is largely determined by formulation and processing parameters34. To optimize the production yield of L-xylulose by an immobilized form of the coupled whole-cell biocatalyst described above, two main parameters, namely sodium alginate concentration and CaCl2 concentration, were evaluated. These are the main parameters that determine the integrity and rigidity of the calcium alginate beads, which in turn affect the biocatalyst immobilization efficiency and molecular diffusion.
As shown in Figure 3A, the biocatalyst immobilization efficiency demonstrated an increasing trend with increasing sodium alginate concentration in the range of 1 - 3% (w/v). When the sodium alginate concentration used in the immobilization matrix was less than 1%, the resulting beads were fragile and easily broken due to low mechanical stability, releasing most of the cells into the surrounding CaCl2 solution (data not shown). Upon increasing the sodium alginate concentration to greater than 2%, the resulting beads hardened excessively leading to problems in molecular diffusion35 (Figure 4). Varying the CaCl2 concentration yielded a similar trend in the biocatalyst immobilization efficiency. For a fixed concentration of sodium alginate (2% w/v) and in the range of 0.1 - 0.4 M CaCl2, lower CaCl2 concentrations resulted in decreased rigidity of the beads and increased leakage of cells into the surrounding solution. This is due to the limited availability of Ca2+ ions for cross-linking the guluronate blocks on the alginate polymer chains36 (Figure 3B). However, increasing the CaCl2 concentration from 0.3 M to 0.4 M improved the biocatalyst immobilization efficiency only marginally. Due to the high immobilization efficiency (>90%), CaCl2 concentrations beyond 0.4 M were not evaluated. When CaCl2 concentrations of less than 0.1 M were used, the sodium alginate droplet containing the biocatalyst fell apart in the CaCl2 solution and no spherical beads were formed (data not shown).
To maximize the biocatalyst immobilization efficiency without significantly slowing down the molecular diffusion in the calcium alginate beads, the sodium alginate and CaCl2 concentration were optimized next for improved L-xylulose production. Using the same concentration range as for immobilization efficiency (1 - 3% w/v sodium alginate and 0.1 - 0.4 M CaCl2), an optimal sodium alginate concentration of 2% was observed to achieve maximal yield of L-xylulose production (Figure 4A). Beyond this optimal concentration, the yield showed a decreasing trend. This was expected, owing to the issues in diffusion associated with the excessive rigidity of the calcium alginate beads. Similarly, the optimal CaCl2 concentration was found to be 0.3 M (Figure 4B). Combined with the data shown in Figure 3, the optimal sodium alginate concentration (2%) and CaCl2 concentration (0.3 M) that allowed the formation of beads with suitable rigidity and permeability was established, enabling minimal cell leakage and maximal L-xylulose production yield.
It should be noted that, another factor that affects the overall catalytic efficiency of the immobilized biocatalyst is the weight of the cell culture encapsulated. The catalytic efficiency increases with increasing cell inoculum up to an optimal inoculum weight, beyond which it shows a decreasing trend37. A possible explanation for this decrease is cellular overcrowding, which hinders molecular diffusion throughout the calcium alginate beads and decreases the catalytic efficiency of the system. Using the optimized immobilization condition above, the highest L-xylulose production was obtained with a cell loading of 3.75 (gDCW)/L16 (data not shown).
Reusability of the Immobilized and Free Whole-cell Biocatalysts for L-xylulose Production. Using the established optimized condition of 1:1 E. coliSpNox to E. coliHjLAD ratio, 2% (w/v) sodium alginate, and 0.3 M CaCl2, the immobilized coupled whole-cell biocatalyst output a maximal L-xylulose yield of 64%, compared to a much higher yield of 96% for the free coupled whole-cell system (cycle 1 of Figure 5). Note that the yield of immobilized E. coliHjLAD cell alone was only 32.5% (data not shown), lower than that of the immobilized coupled system as well as the free single biocatalyst system (79%, Figure 1C). This reduction was expected as immobilization is known to decrease the activity of biocatalysts, possibly due to substrate diffusion limitations, but was compensated by the advantage of improved reusability of a biocatalyst upon immobilization. This effect on the stability of the biocatalyst is demonstrated by the decreasing trend in L-xylulose yield observed on subjecting the free and immobilized systems repeatedly to a batch reaction process, shown in Figure 5. The yield for the free system showed a much steeper decline than that of the immobilized system, indicating a more rapid loss of enzyme activity in the free system. As a result, both systems had a comparable L-xylulose yield for production cycle 2. At the end of 7 cycles of successive re-use, the encapsulated biocatalysts maintained their stability and retained 65% of the original yield while the free whole-cell system retained less than 10% of its ability to produce L-xylulose. This demonstrates that the immobilized biocatalysts could be reused effectively for the production of L-xylulose and that the benefit of improved reusability and stability brought on by immobilization far outweighs the reduction in activity of the biocatalyst, conferring a significant advantage to the production process over a free whole-cell system.
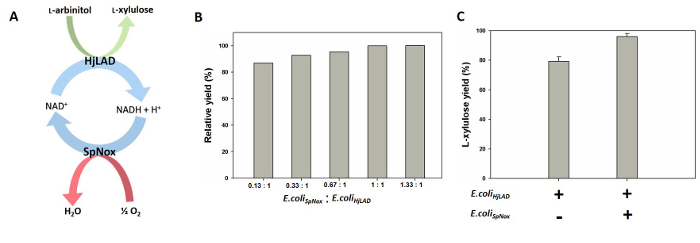 Figure 1. Coupled Whole-cell System for the Biocatalytic Production of L-xylulose. (A) Schematic illustrating the cofactor regeneration reaction occurring in a coupled whole-cell system comprising E. coli cells harboring HjLAD or SpNox, respectively. (B) Variation in the yield of L-xylulose with different E. coliSpNox to E. coliHjLAD ratios. (C) L-xylulose yield from a whole-cell biocatalyst comprised of only E. coliHjLAD compared to that of a coupled whole-cell system comprised of E. coliHjLAD and E. coliSpNox cells capable of cofactor regeneration. The plots shown represent average and standard deviation of three independent experiments. Please click here to view a larger version of this figure.
Figure 1. Coupled Whole-cell System for the Biocatalytic Production of L-xylulose. (A) Schematic illustrating the cofactor regeneration reaction occurring in a coupled whole-cell system comprising E. coli cells harboring HjLAD or SpNox, respectively. (B) Variation in the yield of L-xylulose with different E. coliSpNox to E. coliHjLAD ratios. (C) L-xylulose yield from a whole-cell biocatalyst comprised of only E. coliHjLAD compared to that of a coupled whole-cell system comprised of E. coliHjLAD and E. coliSpNox cells capable of cofactor regeneration. The plots shown represent average and standard deviation of three independent experiments. Please click here to view a larger version of this figure.
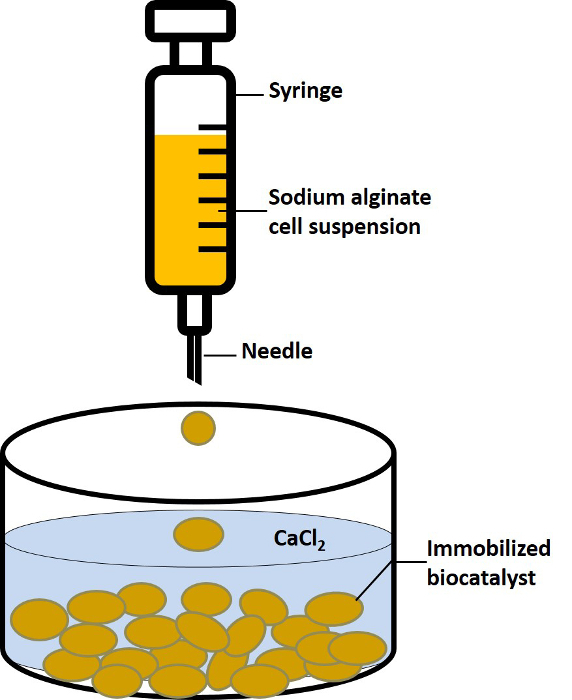 Figure 2. Immobilization of the Whole-cell Biocatalysts via Encapsulation in Alginate Beads. Schematic representing the formation of uniformly sized Ca2+ alginate beads by dropwise addition of the sodium alginate-cell suspension to a solution of calcium chloride. Please click here to view a larger version of this figure.
Figure 2. Immobilization of the Whole-cell Biocatalysts via Encapsulation in Alginate Beads. Schematic representing the formation of uniformly sized Ca2+ alginate beads by dropwise addition of the sodium alginate-cell suspension to a solution of calcium chloride. Please click here to view a larger version of this figure.
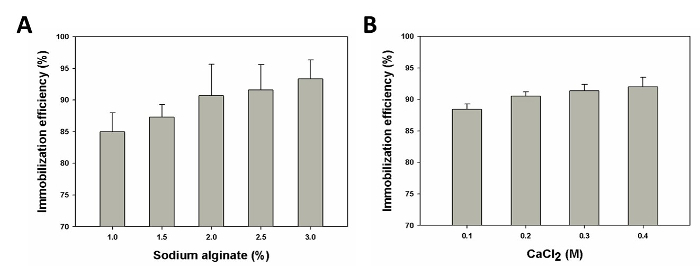 Figure 3. Parameters Influencing the Efficiency of Immobilization of the Whole-cell Biocatalyst in Alginate Beads. Immobilization efficiency as a function of (A) sodium alginate (%w/v) (B) CaCl2 (M) concentration. The plots shown represent the average and standard deviation of three independent experiments. Please click here to view a larger version of this figure.
Figure 3. Parameters Influencing the Efficiency of Immobilization of the Whole-cell Biocatalyst in Alginate Beads. Immobilization efficiency as a function of (A) sodium alginate (%w/v) (B) CaCl2 (M) concentration. The plots shown represent the average and standard deviation of three independent experiments. Please click here to view a larger version of this figure.
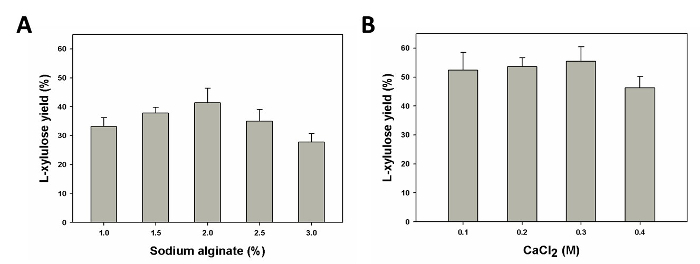 Figure 4. Evaluation of the Yield of L-xylulose from the Immobilized Coupled Whole-cell Biocatalyst as a function of (A) sodium alginate (% w/v) (B) CaCl2 (M) concentration. The plots shown represent the average and standard deviation of three independent experiments. Please click here to view a larger version of this figure.
Figure 4. Evaluation of the Yield of L-xylulose from the Immobilized Coupled Whole-cell Biocatalyst as a function of (A) sodium alginate (% w/v) (B) CaCl2 (M) concentration. The plots shown represent the average and standard deviation of three independent experiments. Please click here to view a larger version of this figure.
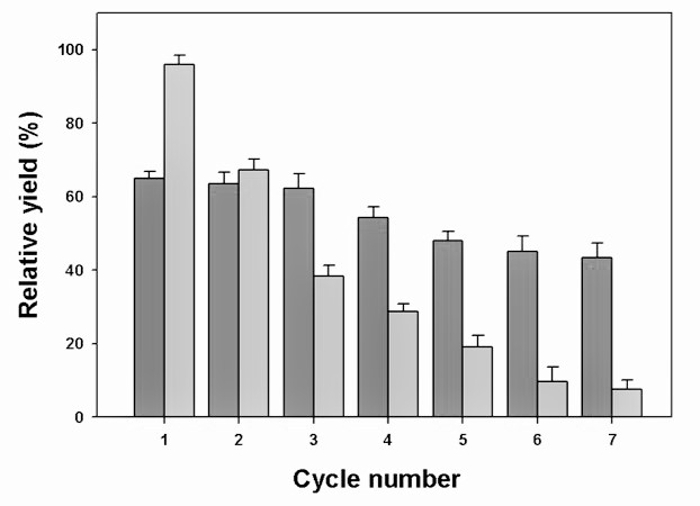 Figure 5. Assessment of the Biocatalyst Reusability. The yield of L-xylulose production for 7 successive cycles of reuse of immobilized and free coupled whole-cell biocatalyst is shown in dark and light gray, respectively. The plot shown represents the average and standard deviation of three independent experiments. Please click here to view a larger version of this figure.
Figure 5. Assessment of the Biocatalyst Reusability. The yield of L-xylulose production for 7 successive cycles of reuse of immobilized and free coupled whole-cell biocatalyst is shown in dark and light gray, respectively. The plot shown represents the average and standard deviation of three independent experiments. Please click here to view a larger version of this figure.
Discussion
Recent technological advancements have enabled a surge in the commercialization of recombinant biotherapeutics, resulting in a gradual rise in their market value in the biotechnology industry. One such advancement is the advent of metabolic engineering in recombinant microorganisms, which has shown a great promise in establishing scalable industrial systems38. As with most processes, the successful commercialization of recombinant biomolecules produced by genetically engineered microbes is highly dependent on the production yield of the system39. This has led to the rapid development of "brute force" genetics39, where directed evolution of biocatalytic enzymes has been implemented in order to improve biocatalyst activity3. However, for certain metabolic pathways, such as redox pathways, mere biocatalyst improvement is not sufficient to influence economization of production owing to the significant effect of other essential reaction requirements, such as expensive cofactors. Scale-up of such a system, even with improved biocatalysts, will only serve to increase production costs. Thus, additional strategic methods are required for the commercialization of bioreactions involving stoichiometric ratios of cofactors. In addition, the recyclability of a biocatalyst is also necessary for improving the economic feasibility of a bioprocess. To overcome the limiting effect of rapid cofactor consumption and poor reusability of whole-cell biocatalysts, we have previously developed an immobilized, coupled whole-cell biocatalytic system for cofactor regeneration and stable re-use16. Described in the present study are the experimental procedures required to couple and co-immobilize two whole-cell biocatalysts for improved biotransformation yield and reusability, along with the representative results.
A critical factor to be considered for coupling the whole-cell biocatalysts is the maintenance of a precise control over the conditions established for protein induction and biocatalytic product formation to ensure reproducibility of the system. Variations in parameters like induction OD600, induction time, and IPTG concentration should be avoided to minimize batch-to-batch variation. Additionally, re-evaluation of the cell-to-cell ratio is required for different biotransformations. Note that the E. coliHjLAD:E. coliSpNox ratio of 1:1 reported in this study was determined by experimental optimization rather than the theoretical stoichiometric ratio. For immobilization of the recombinant whole-cell biocatalysts in calcium alginate beads, there are several critical steps to ensure uniformity with regard to the bead rigidity and distribution of cells. Firstly, the alginate solution must be prepared by slowly adding sodium alginate into water, and not vice versa to prevent clumping that could affect the local alginate concentration and thus the extent of cross-linking. Secondly, gentle pipetting should be employed when handling the cell-alginate suspension to avoid the formation of air bubbles, which inflict shear stress on the encapsulated cells, impede efficient mass transfer and can even cause the beads to float. In case air bubbles are introduced into the suspension, allow the cell-alginate suspension to stand undisturbed for 20 - 30 min to let the bubbles escape. Thirdly, when making the beads, the cell-alginate suspension must be added slowly and carefully in a drop-wise manner into a sufficient volume of calcium chloride solution for uniform size and morphology. Beads submerged incompletely in calcium chloride will have an irregular shape and poor rigidity that can contribute to cell leakage. Lastly, to avoid variability in the reaction volume from residual wash buffer, the beads in Step 4.8 can be lightly blotted with filter paper (do not air dry).
Common strategies for cofactor regeneration include co-culture of cells harboring functionally complementary enzymes and co-expression of these enzymes in the same cell. The described system employs a different strategy of coupling separate E. coliSpNox and E. coliHjLAD cell cultures, offering a better control in maintaining a desired ratio between the two enzymes. Additionally, cells expressing single proteins are subjected to less stress than those co-expressing multiple proteins, which could lead to reduced protein misfolding and increased protein expression40,41. Thus, direct whole-cell coupling presents the ability to mediate multi-enzyme catalytic processes without significantly compromising product yield. However, established strategies such as careful selection of bacterial promoters with varying strength42,43 and improvements in ribosome binding sites44 can be employed to mitigate the limited control over the ratio of target enzymes in co-culture or co-expression systems. Among the many immobilization techniques, encapsulation presents multiple advantages for immobilizing whole-cell biocatalysts over alternative methods such as cross-linking and adsorption, which are more suited for purified enzymes. Encapsulation of cells can also be performed in a variety of biomaterials like agar, chitosan, gums, carrageenan, gelatin and alginate45 for improving the reusability of the biocatalysts. However, calcium alginate encapsulation has been shown to be the most effective approach with respect to immobilization efficiency and biomolecule production46,47.
One major limitation of utilizing bacterial whole-cell biocatalysts is the minimal ability for post-translational modification of heterologous proteins in wild-type E. coli48. This restricts the successful use of eukaryotic biocatalysts within this system. An alternative to overcome this limitation could be the use of organisms such as yeast that are better equipped with eukaryotic post translational machinery. Another limitation of the described system is related to the use of immobilization for improved reusability. Immobilization is a common cost-effective strategy employed to counteract the poor stability of biocatalysts and improve their turnover rate by simplifying the bioprocess49-51. The main parameters that affect the catalytic performance of encapsulated whole-cell biocatalysts include sodium alginate concentration, CaCl2 concentration, cell loading and bead size. It is important to recognize that these parameters are not independent from each other. As such, a comprehensive analysis of all factors within a single study is very time-consuming, thus it is necessary to select the most crucial parameters for optimization. Evaluated and presented here are the effects of two parameters, sodium alginate and CaCl2 concentration. Although not discussed in detail here, we have previously performed the optimization of cell loading, comparing the dependence of yield on the weight of cells immobilized16. Reports in literature indicate that variation of bead size could also modulate the production yield by increasing enzyme encapsulation, improving activity via increased surface area of the immobilized system or by changing the diffusion of substrates and products due to altered transport properties. Depending on the biocatalytic reactions under consideration, selection of alternate critical factors may be required, which could lead to the generation of a different set of optimal conditions. A detailed study for establishment of optimal conditions for additional parameters provides scope for the further improvement in performance of the coupled whole-cell system.
In summary, we report the experimental implementation of a coupled whole-cell biocatalytic system immobilized in calcium alginate beads, illustrating the improved production of L-xylulose. By expressing different recombinant enzymes that facilitate other cofactor dependent biocatalytic pathways, high yields of many biologically relevant molecules can be obtained using the protocol described here. In addition, this system can be expanded to accommodate more than two recombinant biocatalysts for one-pot multi-enzyme biosynthesis of products52 and applications other than biocatalytic production, such as microbial biosensors for biochemical oxygen demand53. Taking advantage of the synergistic feature of this system, encapsulation of symbiotic microbial consortia can be used for a myriad of applications such as bioremediation54-56, whole-cell bioprocessing for biofuels57, plant growth promotion by soil bioaugmentation58 and biomining for metal recovery59. Taken together, the strategies of cofactor regeneration, multi-cell coupling and biocatalyst immobilization described herein can be used as a promising platform for improved production of important biomolecules pursued by others in the field.
Disclosures
The authors declare no competing financial interests. The paper aims at reporting detailed methodology to generate a coupled whole-cell biocatalytic system immobilized in alginate beads. Scientific novelties have been reported in a previous study16.
Acknowledgments
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (NRF-2013R1A1A2012159 and NRF-2013R1A1A2007561), Konkuk University, and the Department of Chemical Engineering and MCubed Program at the University of Michigan.
References
- Carballeira JD, et al. Microbial cells as catalysts for stereoselective red-ox reactions. Biotech Adv. 2009;27(6):686–714. doi: 10.1016/j.biotechadv.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Sun B, Kantzow C, Bresch S, Castiglione K, Weuster-Botz D. Multi-enzymatic one-pot reduction of dehydrocholic acid to 12-keto-ursodeoxycholic acid with whole-cell biocatalysts. Biotechnol. Bioeng. 2013;110(1):68–77. doi: 10.1002/bit.24606. [DOI] [PubMed] [Google Scholar]
- Smith MR, Khera E, Wen F. Engineering novel and improved biocatalysts by cell surface display. Ind. Eng. Chem. Res. 2015;54(16):4021–4032. doi: 10.1021/ie504071f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen F, Sun J, Zhao H. Yeast surface display of trifunctional minicellulosomes for simultaneous saccharification and fermentation of cellulose to ethanol. Appl. Environ. Microbiol. 2009;76(4):1251–1260. doi: 10.1128/AEM.01687-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siedler S, Bringer S, Bott M. Increased NADPH availability in Escherichia coli: improvement of the product per glucose ratio in reductive whole-cell biotransformation. Appl. Microbiol. Biotechnol. 2011;92(5):929–937. doi: 10.1007/s00253-011-3374-4. [DOI] [PubMed] [Google Scholar]
- Kim CS, Seo JH, Kang DG, Cha HJ. Engineered whole-cell biocatalyst-based detoxification and detection of neurotoxic organophosphate compounds. Biotech Adv. 2014;32(3):652–662. doi: 10.1016/j.biotechadv.2014.04.010. [DOI] [PubMed] [Google Scholar]
- Tufvesson P, Lima-ramos J, Nordblad M, Woodley JM. Guidelines and cost analysis for catalyst production in biocatalytic processes. Org. Process Res. Dev. 2011;15(1):266–274. [Google Scholar]
- Mouri T, Michizoe J, Ichinose H, Kamiya N, Goto M. A recombinant Escherichia coli whole cell biocatalyst harboring a cytochrome P450cam monooxygenase system coupled with enzymatic cofactor regeneration. Appl Microbiol Bioechnol. 2006;72(3):514–520. doi: 10.1007/s00253-005-0289-y. [DOI] [PubMed] [Google Scholar]
- Xiao Z, et al. A novel whole-cell biocatalyst with NAD+ regeneration for production of chiral chemicals. PloS one. 2010;5(1):e8860. doi: 10.1371/journal.pone.0008860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, van der Donk WA. Regeneration of cofactors for use in biocatalysis. Curr. Opin. Biotechnol. 2003;14(6):583–589. doi: 10.1016/j.copbio.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Thomas SM, Dicosimo R, Nagarajan V. Biocatalysis: applications and potentials for the chemical industry. Trends Biotechnol. 2002;20(6):238–242. doi: 10.1016/s0167-7799(02)01935-2. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li L, Ma C, Gao C, Tao F, Xu P. Engineering of cofactor regeneration enhances (2S,3S)-2,3-butanediol production from diacetyl. Sci Rep. 2013;3:2643. doi: 10.1038/srep02643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uppada V, Bhaduri S, Noronha SB. Cofactor regeneration - an important aspect of biocatalysis. Curr. Sci. India. 2014;106(7):946–957. [Google Scholar]
- Fu N, Peiris P, Markham J, Bavor J. A novel co-culture process with Zymomonas mobilis and Pichia stipitis for efficient ethanol production on glucose/xylose mixtures. Enzyme Microb. Technol. 2009;45(3):210–217. [Google Scholar]
- Chen H-Y, Guan Y-X, Yao S-J. A novel two-species whole-cell immobilization system composed of marine-derived fungi and its application in wastewater treatment. J. Chem. Technol. Biotechnol. 2014;89(11):1733–1740. [Google Scholar]
- Gao H, et al. Repeated production of L-xylulose by an immobilized whole-cell biocatalyst harboring L-arabinitol dehydrogenase coupled with an NAD+ regeneration system. Biochem. Eng. J. 2015;96:23–28. [Google Scholar]
- Beerens K, Desmet T, Soetaert W. Enzymes for the biocatalytic production of rare sugars. J Ind Microbiol Biotechnol. 2012;39(6):823–834. doi: 10.1007/s10295-012-1089-x. [DOI] [PubMed] [Google Scholar]
- Poonperm W, Takata G, Izumori K. Polyol conversion specificity of Bacillus pallidus. Biosci., Biotechnol., Biochem. 2008;72(1):231–235. doi: 10.1271/bbb.70475. [DOI] [PubMed] [Google Scholar]
- Leviiz G, Zehner L, Saunders J, Beedle J. Sugar substitutes: their energy values, bulk characteristics and potential health benefits. Am. J. Clin. Nutr. 1995;62(5):1161S–1168S. doi: 10.1093/ajcn/62.5.1161S. [DOI] [PubMed] [Google Scholar]
- Zhang Y-W, Tiwari MK, Jeya M, Lee J-K. Covalent immobilization of recombinant Rhizobium etli CFN42 xylitol dehydrogenase onto modified silica nanoparticles. Appl Microbiol Bioechnol. 2011;90(2):499–507. doi: 10.1007/s00253-011-3094-9. [DOI] [PubMed] [Google Scholar]
- Aarnikunnas JS, Pihlajaniemi A, Palva A, Leisola M, Nyyssölä A. Cloning and expression of a Xylitol-4-Dehydrogenase gene from Pantoea ananatis. Appl. Environ. Microbiol. 2006;72(1):368–377. doi: 10.1128/AEM.72.1.368-377.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doten RC, Mortlock RP. Production of D- and L-Xylulose by mutants of Klebsiella pneumoniae and Erwinia uredovora. Appl. Environ. Microbiol. 1985;49(1):158–162. doi: 10.1128/aem.49.1.158-162.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AR, Tokunaga H, Yoshida K, Izumori K. Conversion of Xylitol to L-Xylulose by Alcaligenes sp. 701B-cells. J. Ferment. Bioeng. 1991;72(6):488–490. [Google Scholar]
- Poonperm W, Takata G, Morimoto K, Granström TB, Izumori K. Production of L-xylulose from xylitol by a newly isolated strain of Bacillus pallidus Y25 and characterization of its relevant enzyme xylitol dehydrogenase. Enzyme Microb. Technol. 2007;40(5):1206–1212. [Google Scholar]
- Takata G, Poonperm W, Morimoto K, Izumori K. Cloning and overexpression of the xylitol dehydrogenase gene from Bacillus pallidus and its application to L-xylulose production. Biosci., Biotechnol., Biochem. 2010;74(9):1807–1813. doi: 10.1271/bbb.100144. [DOI] [PubMed] [Google Scholar]
- Usvalampi A, Kiviharju K, Leisola M, Nyyssölä A. Factors affecting the production of L-xylulose by resting cells of recombinant Escherichia coli. J. Ind. Microbiol. Biotechnol. 2009;36(10):1323–1330. doi: 10.1007/s10295-009-0616-x. [DOI] [PubMed] [Google Scholar]
- Rizzi M, Harwart K, Bui-Thananh N-A, Dellweg H. A kinetic study of the NAD+-Xylitol-Dehydrogenase from the yeast Pichia stipitis. J. Ferment. Bioeng. 1989;67(1):25–30. [Google Scholar]
- Pail M, et al. The metabolic role and evolution of L-arabinitol 4-dehydrogenase of Hypocrea jecorina. Eur. J. Biochem. 2004;271(10):1864–1872. doi: 10.1111/j.1432-1033.2004.04088.x. [DOI] [PubMed] [Google Scholar]
- Tiwari MK, et al. pH-rate profiles of L-arabinitol 4-dehydrogenase from Hypocrea jecorina and its application in L-xylulose production. Bioorg. Med. Chem. Lett. 2014;24(1):173–176. doi: 10.1016/j.bmcl.2013.11.047. [DOI] [PubMed] [Google Scholar]
- Gao H, et al. Role of surface residue 184 in the catalytic activity of NADH oxidase from Streptococcus pyogenes. Appl. Microbiol. Biotechnol. 2014;98(16):7081–7088. doi: 10.1007/s00253-014-5666-y. [DOI] [PubMed] [Google Scholar]
- Gao H, Tiwari MK, Kang YC, Lee J-K. Characterization of H2O-forming NADH oxidase from Streptococcus pyogenes and its application in L-rare sugar production. Bioorg. Med. Chem. Lett. 2012;22(5):1931–1935. doi: 10.1016/j.bmcl.2012.01.049. [DOI] [PubMed] [Google Scholar]
- Roy I, Gupta MN. Hydrolysis of starch by a mixture of glucoamylase and pullulanase entrapped individually in calcium alginate beads. Enzyme Microb. Technol. 2004;34(1):26–32. [Google Scholar]
- Gombotz WR, Wee SF. Protein release from alginate matrices. Adv Drug Deliv Rev. 1998;31(3):267–285. doi: 10.1016/s0169-409x(97)00124-5. [DOI] [PubMed] [Google Scholar]
- Smrdel P, Bogataj M, Mrhar A. The influence of selected parameters on the size and shape of alginate beads prepared by ionotropic Gelation. Sci Pharm. 2008;76(1):77–89. [Google Scholar]
- Idris A, Wahidin S. Effect of sodium alginate concentration, bead diameter, initial pH and temperature on lactic acid production from pineapple waste using immobilized Lactobacillus delbrueckii. Process Biochem. 2006;41(5):1117–1123. [Google Scholar]
- Lee KY, Mooney DJ. Alginate: properties and biomedical applications. Prog. Polym. Sci. 2012;37(1):106–126. doi: 10.1016/j.progpolymsci.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X-H, Wang X-T, et al. Immobilization of Acetobacter sp. CCTCC M209061 for efficient asymmetric reduction of ketones and biocatalyst recycling. Microb. Cell Fact. 2012;11(1):119–131. doi: 10.1186/1475-2859-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav VG, et al. The future of metabolic engineering and synthetic biology: Towards a systematic practice. Metab. Eng. 2012;14(3):233–241. doi: 10.1016/j.ymben.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demain AL. From natural products discovery to commercialization: a success story. J Ind Microbiol Biotechnol. 2006;33(7):486–495. doi: 10.1007/s10295-005-0076-x. [DOI] [PubMed] [Google Scholar]
- Baneyx F, Mujacic M. Recombinant protein folding and misfolding in Escherichia coli. Nature Biotechnol. 2004;22(11):1399–1408. doi: 10.1038/nbt1029. [DOI] [PubMed] [Google Scholar]
- De Marco A, Deuerling E, Mogk A, Tomoyasu T, Bukau B. Chaperone-based procedure to increase yields of soluble recombinant proteins produced in E. coli. BMC Biotechnol. 2007;7:32–40. doi: 10.1186/1472-6750-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terpe K. Overview of bacterial expression systems for heterologous protein production: from molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 2006;72(2):211–222. doi: 10.1007/s00253-006-0465-8. [DOI] [PubMed] [Google Scholar]
- Alper H, Fischer C, Neviogt E, Stephanopoulos G. Tuning genetic control through promoter engineering. PNAS. 2006;103(8):12678–12683. doi: 10.1073/pnas.0504604102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salis HM, Mirsky EA, Voigt CA. Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol. 2009;27(10):946–950. doi: 10.1038/nbt.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riaz QU, Masud T. Recent trends and applications of encapsulating materials for probiotic stability. Crit Rev Food Sci Nutr. 2013;53(3):231–244. doi: 10.1080/10408398.2010.524953. [DOI] [PubMed] [Google Scholar]
- Hossain GS, Li J, et al. One-step biosynthesis of α-keto-γ-methylthiobutyric acid from L-methionine by an Escherichia coli whole-cell biocatalyst expressing an engineered L-amino acid deaminase from Proteus vulgaris. PloS one. 2014;9(12):e114291. doi: 10.1371/journal.pone.0114291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan B, Pal A, Jain V. Improved enzyme catalytic characteristics upon glutaraldehyde cross-linking of alginate entrapped xylanase Isolated from Aspergillus flavus MTCC 9390. Enzyme Res. 2015. [DOI] [PMC free article] [PubMed]
- Chen R. Bacterial expression systems for recombinant protein production: E. coli and beyond. Biotech Adv. 2012;30(5):1102–1107. doi: 10.1016/j.biotechadv.2011.09.013. [DOI] [PubMed] [Google Scholar]
- Truppo MD, Hughes G. Development of an improved immobilized CAL-B for the enzymatic resolution of a key intermediate to odanacatib. Org. Process Res. Dev. 2011;15(5):1033–1035. [Google Scholar]
- Twala BV, Sewell BT, Jordaan J. Immobilisation and characterisation of biocatalytic co-factor recycling enzymes, glucose dehydrogenase and NADH oxidase, on aldehyde functional ReSyn polymer microspheres. Enzyme Microb. Technol. 2012;50(6-7):331–336. doi: 10.1016/j.enzmictec.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Wang X-T, et al. Biocatalytic anti-Prelog stereoselective reduction of ethyl acetoacetate catalyzed by whole cells of Acetobacter sp. CCTCC M209061. J. Biotechnol. 2013;163(3):292–300. doi: 10.1016/j.jbiotec.2012.10.023. [DOI] [PubMed] [Google Scholar]
- Rios-Solis L, et al. Modelling and optimisation of the one-pot, multi-enzymatic synthesis of chiral amino-alcohols based on microscale kinetic parameter determination. Chem. Eng. Sci. 2015;122:360–372. [Google Scholar]
- Wang B, Barahona M, Buck M. A modular cell-based biosensor using engineered genetic logic circuits to detect and integrate multiple environmental signals. Biosens Bioelectron. 2013;40(1):368–376. doi: 10.1016/j.bios.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saratale RG, Saratale GD, Kalyani DC, Chang JS, Govindwar SP. Enhanced decolorization and biodegradation of textile azo dye Scarlet R by using developed microbial consortium-GR. Bioresour. Technol. 2009;100(9):2493–2500. doi: 10.1016/j.biortech.2008.12.013. [DOI] [PubMed] [Google Scholar]
- Malik A. Metal bioremediation through growing cells. Environ. Int. 2004;30(2):261–278. doi: 10.1016/j.envint.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Bazot S, Lebeau T. Effect of immobilization of a bacterial consortium on diuron dissipation and community dynamics. Bioresour. Technol. 2009;100(18):4257–4261. doi: 10.1016/j.biortech.2009.03.067. [DOI] [PubMed] [Google Scholar]
- Brethauer S, Studer MH. Consolidated bioprocessing of lignocellulose by a microbial consortium. Energy Environ Sci. 2014;7(4):1446. [Google Scholar]
- Malusá E, Sas-Paszt L, Ciesielska J. Technologies for beneficial microorganisms inocula used as biofertilizers. Sci World J. 2012;2012(491206) doi: 10.1100/2012/491206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brune KD, Bayer TS. Engineering microbial consortia to enhance biomining and bioremediation. Front Microbiol. 2012;3(203) doi: 10.3389/fmicb.2012.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]


