Abstract
Please cite this paper as: De Vleeschauwer et al. (2011) Cross‐protection between antigenically distinct H1N1 swine influenza viruses from Europe and North America. Influenza and Other Respiratory Viruses 5(2), 115–122.
Background An avian‐like H1N1 swine influenza virus (SIV) is enzootic in swine populations of Western Europe. The virus is antigenically distinct from H1N1 SIVs in North America that have a classical swine virus‐lineage H1 hemagglutinin, as does the pandemic (H1N1) 2009 virus. However, the significance of this antigenic difference for cross‐protection among pigs remains unknown.
Objectives We examined protection against infection with a North American triple reassortant H1N1 SIV [A/swine/Iowa/H04YS2/04 (sw/IA/04)] in pigs infected with a European avian‐like SIV [A/swine/Belgium/1/98 (sw/B/98)] 4 weeks earlier. We also examined the genetic relationships and serologic cross‐reactivity between both SIVs and with a pandemic (H1N1) 2009 virus [A/California/04/09 (Calif/09)].
Results After intranasal inoculation with sw/IA/04, all previously uninfected control pigs showed nasal virus excretion, high virus titers in the entire respiratory tract at 4 days post‐challenge (DPCh) and macroscopic lung lesions. Most pigs previously infected with sw/B/98 tested negative for sw/IA/04 in nasal swabs and respiratory tissues, and none had lung lesions. At challenge, these pigs had low levels of cross‐reactive virus neutralizing and neuraminidase inhibiting (NI) antibodies to sw/IA/04, but no hemagglutination‐inhibiting antibodies. They showed similar antibody profiles when tested against Calif/09, but NI antibody titers were higher against Calif/09 than sw/IA/04, reflecting the higher genetic homology of the sw/B/98 neuraminidase with Calif/09.
Conclusions Our data indicate that immunity induced by infection with European avian‐like H1N1 SIV affords protection for pigs against North American H1N1 SIVs with a classical H1, and they suggest cross‐protection against the pandemic (H1N1) 2009 virus.
Keywords: Avian‐like, cross‐protection, H1N1 swine influenza virus, pandemic (H1N1) 2009, triple reassortant
Introduction
Influenza viruses of H1N1, H3N2 and H1N2 subtypes are enzootic in swine populations worldwide, but they show different genetic and antigenic constellations in different parts of the world. 1 The complex epidemiology of swine influenza viruses (SIVs) is well illustrated by the nature of H1N1 SIVs in North America versus Europe. In North America, viruses of the ‘classical swine’ H1N1 lineage were the predominant SIVs until 1998. These viruses are descendants of the first SIV isolated in 1930 and are related to the 1918 human pandemic H1N1 virus. Since 1998, reassortant H3N2 SIVs with genes of classical swine, avian and human influenza virus origin have become established in the swine population in North America. These viruses further reassorted with co‐circulating classical H1N1 SIVs leading to the current triple reassortant H1N1 SIVs that contain a classical swine‐lineage H1 hemagglutinin (HA). 2 In Europe, the prevailing H1N1 SIV is of entirely avian origin and therefore designated ‘avian‐like’ H1N1. It was introduced from wild ducks into the pig population in 1979 and has become the dominant H1N1 European SIV strain. 3 , 4 Surveillance studies show high seroprevalence rates for avian‐like H1N1 SIVs in swine‐dense regions of Western Europe. In Belgium, for example, 81% of sows and 42% of fattening pigs tested positive for antibodies. 5 , 6 In Asia, multiple H1N1 lineages appear to circulate, including avian‐like, classical swine‐lineage and triple reassortant H1N1 SIVs. 7 , 8 Reassortment between the avian‐like and triple reassortant SIV lineages has been occasionally reported in Thailand and China. 8 , 9 , 10 From May 2009 to May 2010, the pandemic (H1N1) 2009 influenza virus has been reported in swine in 22 countries in five continents. 11 This pandemic virus is as a reassortant of at least two circulating SIVs. Six gene segments, including the classical H1 HA, originate from North American triple reassortant SIVs, while the genes encoding the neuraminidase (NA) and matrix (M) proteins are closely related to those in European and/or Asian avian‐like SIVs. 12 The emergence of the pandemic (H1N1) 2009 influenza virus further complicates the SIV epidemiology.
The HAs of avian‐like and classical swine‐lineage H1N1 SIVs are serologically distinct when compared by antigenic analyses with monoclonal antibodies. 13 , 14 Sequence alignment of the HA1 regions of the HA gene of these viruses likewise revealed several amino acid substitutions at putative antigenic sites. 14 The significance of these differences for cross‐protection remains unknown, and there are no published cross‐protection studies with H1N1 SIVs of different lineages in pigs. In this study, we aimed to examine a) to what extent immunity to a European avian‐like H1N1 SIV may protect pigs from infection with a North American triple reassortant H1N1 SIV and b) the antigenic and genetic relationships between both viruses. In an attempt to extrapolate our findings to protection against the pandemic (H1N1) 2009 virus, a prototype pandemic virus was also included in the genetic and antigenic analyses.
Materials and methods
Viruses and sequence analysis
A/swine/Belgium/1/98 (sw/B/98) is representative of prevailing avian‐like H1N1 SIVs in Western Europe (GenBank accession numbers ACN 67524‐28). 15 A/swine/Iowa/H04YS2/2004 (sw/IA/04) is a triple reassortant H1N1 SIV belonging to the North American ‘H1ß (rH1N1‐like)’ SIV cluster (GenBank accession numbers GQ452235‐42). 16 A/California/04/09 (Calif/09) is a prototype human pandemic (H1N1) 2009 virus (GenBank accession numbers ACP 41102‐11). 12
The HA, NA, matrix (M1) and nucleoprotein (NP) of the 3 viruses were compared at the nucleotide and protein level by BLAST software (NCBI). Amino acid (aa) differences at putative antigenic sites of the HA, as defined by Brownlee and Fodor, 17 were identified by alignment (CLC Sequence Viewer 6) with the PR8 H1 reference strain. In total, 327 aa residues (H1 open reading frame numbering) were examined with special attention to 50 aa located at putative antigenic sites. Amino acid differences in the NA were identified by multiple alignment. In total, 469 NA aa residues were examined of which 195 were located in antigenic domains as described by Maurer‐Stroh and co‐workers. 18
Experimental design
Twenty‐five 6‐week‐old pigs from a conventional herd with a high health status and free of influenza A virus were used. The pigs were also serologically negative for porcine reproductive and respiratory syndrome virus and for porcine circovirus type 2. Pigs were randomly assigned to three groups. Two groups were inoculated initially with sw/B/98 or sw/IA/04 and challenged 4 weeks later with sw/IA/04 (sw/B/98‐sw/IA/04, n = 8 and sw/IA/04‐sw/IA/04, n = 8). The third group served as a challenge control group and was challenged with sw/IA/04 along with the previously sw/B/98 or sw/IA/04 inoculated pigs (sw/IA/04‐control, n = 9). All inoculations were performed intranasally at 7·0 log10 egg infective doses 50% (EID50). Inoculations were performed on unanesthetized animals. Pigs were held in a vertical position with the neck stretched. The inoculum was gradually instilled into the middle nasal cavity by insertion of a 15‐mm plastic cannula attached to a syringe. All pigs were monitored daily for clinical signs from 5 days before until 7 days after initial inoculation (DPI), and from 5 days before until 7 days after challenge (DPCh), or until euthanasia. A daily clinical score was recorded for each pig as follows. A score of 1 each was given for the presence of fever (rectal temperature ≥40·0°C), depression, tachypnea (respiratory rate ≥45 per minute), dyspnea and forced abdominal respiration, resulting in a minimum clinical score of 0 and a maximum score of 5 per pig. To obtain the group clinical score per day, the individual scores of each day were added and divided by the maximum score possible (i.e. 40 for sw/B/98‐sw/IA/04 and sw/IA/04‐sw/IA/04, and 45 for sw/IA/04‐control). To determine the extent of virus excretion after the initial inoculations and after the challenge with sw/IA/04, nasal swabs were collected daily from all pigs from 0 until 7 DPI, and from 0 until 7 DPCh, or until euthanasia. The swabs were weighed before and after collection to determine virus titers per 100 mg nasal secretions. To determine the extent of replication of the sw/IA/04 challenge virus in the respiratory tract, 4 pigs per group were euthanized at 4 DPCh. Samples of the upper (nasal mucosa, tonsil and trachea) and lower (left and right lung) respiratory tract were collected for virus titration. Blood samples for serological examinations were collected at the start of the experiment and 4 weeks after the initial inoculation, i.e. at the time of challenge with sw/IA/04. The remaining pigs were also bled at 14 and 28 DPCh.
Virus titration
Nasal swab samples from both nostrils were suspended in 1 ml of phosphate‐buffered saline (PBS) supplemented with 10% fetal bovine serum, 100 IU/ml penicillin and 100 μg/ml streptomycin and mixed vigorously for 1 hour. The medium was collected, clarified by centrifugation and used for titration. Tissue samples were weighed and ground in PBS containing 10 IU/ml penicillin and 10 μg/ml streptomycin to obtain 10% or 20% (w/v) tissue homogenates. The homogenates were clarified by centrifugation and used for titration. All samples were titrated on Madin–Darby canine kidney (MDCK) cells in serum‐free medium with trypsin (10 μg/ml from porcine pancreas). Briefly, MDCK cells were seeded in 96‐well cell culture plates at a concentration of 250 000 cells per ml. After 3 days of incubation, the cells were 100% confluent and were inoculated with 10‐fold dilutions of the samples using 4 wells per dilution. MDCK cells were observed daily for cytopathic effect until 7 days after inoculation. Virus titers were calculated by the method of Reed and Muench. 19
Serological assays
Antibody titers against sw/B/98, sw/IA/04 and Calif/09 were determined in all sera by hemagglutination inhibition (HI), virus neutralization (VN) and neuraminidase inhibition (NI) assays, according to standard methods. 20 , 21 Before use, all sera were heat inactivated (56°C, 30 minutes). In the HI assay, sera were pre‐treated with receptor‐destroying enzyme from Vibrio cholerae and adsorbed onto chicken erythrocytes. Twofold serum dilutions were incubated [1 hour, room temperature (RT)] with four hemagglutinating units of the respective viruses. Finally, 0·5% chicken erythrocytes were added, and the assay was read after 1 h at RT. In the VN assay, twofold serum dilutions were incubated (1 hour, 37°C) with 100 TCID50 of MDCK‐adapted virus in microtiter plates. MDCK cells were added at a concentration of 600000 cells per ml. After incubation (24 hour, 37°C), the cells were fixed with 4% paraformaldehyde. Virus‐positive cells were demonstrated by an immunoperoxidase staining using mouse monoclonal antibodies against influenza A virus nucleoprotein (HB65) and peroxidase‐conjugated goat anti‐mouse IgG. In the NI assay, standard virus doses were selected by an assay of NA activity based on the colorimetric analysis of sialic acid release from fetuin substrate. Twofold serum dilutions were incubated with the standard virus dilution in microtiter plates, and the reduction of NA activity in each serum dilution was compared with that in controls without serum. All sera were tested in duplicate, and antibody titers were expressed as the reciprocal of the highest serum dilution that completely inhibited hemagglutination or virus replication in MDCK cells, or that gave a 50% inhibition of NA activity. Starting dilutions were 1:2 in the VN test, and 1:10 in the HI and NI tests.
Statistical analysis
Differences in serum HI, VN and NI antibody titers were compared between groups in two‐sample Student’s t‐tests. Samples that tested negative in the serological assays were given a value corresponding to half of the minimum detectable antibody titer. P < 0·05 was taken as the level of statistical significance.
Results
Genetic and antigenic relationships between sw/B/98, sw/IA/04 and Calif/09
Percentages of nucleotide and aa identity between the 4 analyzed genes of sw/B/98 and sw/IA/04 or Calif/09 are shown in Table 1. Nucleotide sequence identity of the HA gene of sw/B/98 and the 2 viruses with the classical swine‐lineage HA (sw/IA/04 and Calif/09) was low (74–75%), whereas both classical HAs were more similar (91%). Nucleotide sequences of the NA and M1 genes of sw/B/98 were more similar to Calif/09 (92% and 95%, respectively) than to sw/IA/04 (79% and 88%, respectively), consistent with the Eurasian virus phylogenetic lineage of these 2 genes in the 2009 pandemic viruses. All viruses were equally similar in the NP gene (83%). Similar but generally higher relationships were observed at the aa level.
Table 1.
Percent identity of the nucleotide and amino acid sequences of the hemagglutinin (HA), neuraminidase (NA), matrix (M) and nucleoprotein (NP) genes of sw/B/98 with those of sw/IA/04 or a prototype pandemic (H1N1) 2009 virus
| % identity compared to sw/B/98 | ||||||||
|---|---|---|---|---|---|---|---|---|
| HA | NA | M1 | NP | |||||
| N | aa | N | aa | N | aa | N | aa | |
| Sw/IA/04 | 75 | 75 | 79 | 82 | 88 | 94 | 83 | 98 |
| Calif/09 | 74 | 72 | 92 | 93 | 95 | 98 | 83 | 97 |
N, nucleotide; aa, amino acid.
Amino acid changes at presumed antigenic sites of the HA are shown in Figure 1. The HA1 segment of sw/B/98 contained 76 aa differences compared to sw/IA/04 and 84 aa differences with Calif/09, and respectively 13 and 14 changes were in aa residues at putative antigenic sites. The HA1 regions of the viruses with the classical swine‐lineage HA genes were more closely related (39 aa differences, with 8 at putative antigenic sites). The NA gene of sw/B/98 was more closely related to Calif/09 (28 aa differences, with 12 at putative antigenic sites) than to sw/IA/04 (82 aa differences, with 27 at putative antigenic sites). The NA of sw/IA/04 and Calif/09 was different in 83 aa residues, of which 31 were located at putative antigenic sites.
Figure 1.

Alignment of deduced amino acid sequences at antigenic sites, as defined by Brownlee and Fodor, 17 of the hemagglutinins of sw/B/98, sw/IA/04 and Calif/09. Only the amino acids different from those in the sw/B/98 sequence are indicated, and conserved residues are shown as dashes.
Virus excretion and serological response after initial infection with sw/B/98 or sw/IA/04
Mild fever (40·4–40·8°C) was seen in all pigs 1 and 2 days after inoculation with sw/B/98 (clinical scores 0·30 and 0·20 on 1 and 2 DPI, respectively), and in most pigs after initial inoculation with sw/IA/04 (clinical scores 0·20 and 0·10 on 1 and 2 DPI, respectively), but respiratory signs were absent. All inoculated pigs excreted the SIVs used for inoculation for 6 consecutive DPI. Mean virus titers in nasal swabs are shown in Figure 2. The challenge control pigs remained virus negative.
Figure 2.
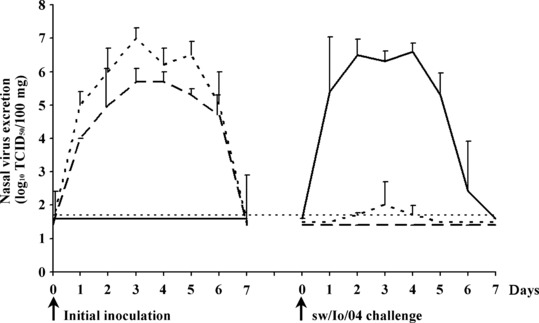
Nasal virus excretion after initial inoculation and after challenge with sw/IA/04. Mean virus titers in nasal swabs of each group are given. Sw/B/98‐sw/IA/04, sw/IA/04‐sw/IA/04 and sw/IA/04‐control groups are represented by dotted, dashed and full lines, respectively. The thin dashed line represents the detection limit (<1·7 log10 TCID50/100 mg).
All pigs were negative in antibody assays at the start of the experiments against all three viruses. Serological findings at the time of challenge with sw/IA/04, i.e. 4 weeks after initial inoculation, are shown in Table 2. As expected, all control pigs were seronegative at the time of challenge, whereas all pigs inoculated previously with either sw/IA/04 or sw/B/98 had HI, VN and NI antibodies to the SIV used for inoculation. In addition, most pigs inoculated with sw/IA/04 had HI antibodies that cross‐reacted with sw/B/98 and/or Calif/09, and all these pigs had cross‐reactive VN and NI antibodies to the other two viruses. Cross‐reactive HI, VN and NI antibody titers to both heterologous viruses were significantly lower than those to the homologous virus (P < 0·05). All pigs inoculated with sw/B/98 lacked cross‐reactive HI antibodies to sw/IA/04 and Calif/09, but most of them had cross‐reactive VN antibodies to both viruses, though at lower titers than to the homologous virus (P < 0·05). Cross‐reactive NI antibodies against both viruses were found in all pigs, and they were higher to Calif/09 than to sw/IA/04 (P < 0·05).
Table 2.
Serological findings at the time of challenge with sw/IA/04 and 2 weeks later in naïve control pigs and sw/B/98 or sw/IA/04‐immune pigs
| Group | Test | Mean antibody titers of positive pigs ± SEM* (number of positive pigs/total number of pigs) | |||||
|---|---|---|---|---|---|---|---|
| Time of challenge | 14 DPCh | ||||||
| sw/B/98 | sw/IA/04 | Calif/09 | sw/B/98 | sw/IA/04 | Calif/09 | ||
| sw/IA/04‐ control | HI | <† | < | < | 18 ± 2 (5/5) | 68 ± 12 (5/5) | 10 ± 0 (2/5) |
| VN | < | < | < | 46 ± 15 (5/5) | 154 ± 33 (5/5) | 12 ± 3 (5/5) | |
| NI | < | < | < | 22 ± 7 (5/5) | 288 ± 93 (5/5) | 16 ± 6 (5/5) | |
| sw/IA/04‐sw/IA/04 | HI | 16 ± 2 (7/8) | 88 ± 22 (8/8) | 15 ± 2 (4/8) | 15 ± 3 (4/4) | 45 ± 13 (4/4) | 15 ± 4 (2/4) |
| VN | 44 ± 13 (8/8) | 180 ± 26 (8/8) | 9 ± 2 (8/8) | 75 ± 40 (4/4) | 208 ± 31 (4/4) | 27 ± 14 (4/4) | |
| NI | 25 ± 6 (8/8) | 355 ± 91 (8/8) | 16 ± 4 (8/8) | 30 ± 6 (4/4) | 280 ± 40 (4/4) | 23 ± 6 (4/4) | |
| sw/B/98‐sw/IA/04 | HI | 135 ± 31 (8/8) | < | < | 120 ± 23 (4/4) | 40 ± 14 (4/4) | 17 ± 3 (3/4) |
| VN | 416 ± 82 (8/8) | 8 ± 2 (7/8) | 3 ± 1 (7/8) | 1216 ± 352 (4/4) | 128 ± 23 (4/4) | 32 ± 6 (4/4) | |
| NI | 230 ± 70 (8/8) | 29 ± 4 (8/8) | 68 ± 15 (8/8) | 800 ± 160 (4/4) | 400 ± 139 (4/4) | 720 ± 201 (4/4) | |
*Standard error of the mean; †antibody titers below detection limits, i.e. <10 in HI and NI tests and <2 in VN test. HI, hemagglutination inhibition; NI, neuraminidase inhibition; VN, virus neutralization.
Clinical and virological protection after challenge with sw/IA/04
After subsequent challenge with sw/IA/04, mild fever (40·6–40·1°C), depression and respiratory signs were seen in the previously uninfected control pigs at 1 (clinical score 0·44) and 2 (clinical score 0·17) DPCh only. Mean virus titers in nasal swabs are shown in Figure 2. All previously uninfected control pigs excreted the sw/IA/04 challenge virus at high titers (up to 7·5 log10 TCID50/100 mg) for 6 consecutive DPCh or until euthanasia at 4 DPCh, whereas disease and virus excretion were undetectable in the sw/IA/04‐sw/IA/04 group. Interestingly, only 1 of 8 pigs inoculated previously with sw/B/98 had mild fever (40·0–40·1°C) at 1 and 2 DPCh with sw/IA/04 (clinical score 0·03 on both days). Likewise, only 2 of 8 pigs of the sw/B/98‐sw/IA/04 group had detectable nasal virus excretion, and nasal virus titers were lower and detectable for a shorter duration than in the previously uninfected control group. One pig tested positive at 3 DPCh (3·0 log10 TCID50/100 mg) only, and one pig (nr V‐09‐949) on 2, 3 and 4 DPCh (1·7, 3·0 and 2·3 log10 TCID50/100 mg, respectively).
Sw/IA/04 virus titers in the respiratory tract of the pigs euthanized at 4 DPCh are shown in Table 3. Sw/IA/04 was recovered from the nasal mucosa, tonsil, trachea, left lung and right lung of all 4 previously uninfected control pigs. Virus titers ranged from 4·0 to 6·6 log10 TCID50/g in the upper respiratory tract, and from 6·3 to 7·2 log10 TCID50/g in the lungs. All four pigs of the sw/IA/04‐sw/IA/04 group were completely virus negative. In the sw/B/98‐sw/IA/04 group, only pig nr V‐09‐949 tested positive, and this was only in the tracheal sample and only at a low titer of 2·0 log10 TCID50/g. Areas of lung consolidation, involving 5–38% of the lung surface, were present in the four previously uninfected control pigs euthanized, but absent in the pigs from the other groups.
Table 3.
Virus titers in the respiratory tract 4 days after challenge with sw/IA/04 in naïve control pigs and sw/B/98 or sw/IA/04‐immune pigs
| Group | Range of virus titers (log10 TCID50/g) of positive pigs (number of positive pigs/total number of pigs) | |||||
|---|---|---|---|---|---|---|
| Nasal mucosa respiratory part | Nasal mucosa olfactory part | Tonsil | Trachea | Left lung | Right lung | |
| sw/IA/04‐control | 4·1–6·6 (4/4) | 4·3–6·0 (4/4) | 4·0–5·0 (4/4) | 5·5–6·5 (4/4) | 6·3–6·7 (4/4) | 6·2–7·2 (4/4) |
| sw/IA/04‐sw/IA/04 | <* | < | < | < | < | < |
| sw/B/98‐sw/IA/04 | < | < | < | 2·0 (1/4) | < | < |
*Virus titers below the detection limit (1·9 log10 TCID50/g for nasal mucosa, tonsil and trachea; 1·7 log10TCID50/g for lung) in all pigs.
Serological profile after challenge with sw/IA/04
Serological findings at 14 DPCh are shown in Table 2. All previously uninfected control pigs seroconverted (i.e. ≥4‐fold increase in antibody titer) to sw/IA/04 after challenge infection and developed cross‐reactive HI, VN and NI antibody titers to both other viruses, though at lower titers than the homologous sw/IA/04 titers. Only two pigs, however, developed HI antibodies to Calif/09. In the sw/IA/04‐sw/IA/04 group, HI, VN and NI antibody titers to the three viruses were comparable to those before challenge (P > 0·05). In the sw/B/98‐sw/IA/04 pigs, antibody titers to sw/B/98 did not change after challenge (P > 0·05). Antibody titers to sw/IA/04 and sw/Calif/09 increased, but only the increases in VN antibody titers to sw/IA/04, and VN and NI antibody titers to Calif/09 were significant (P < 0·05). Antibody titers at 28 DPCh were similar to those at 14 DPCh (P > 0·05).
Discussion
This study shows a stronger than expected protection against infection with a North American triple reassortant H1N1 SIV in pigs infected with a phylogenetically distinct European avian‐like SIV 4 weeks earlier. It has long been known that the HA1 of both virus lineages shows considerable antigenic and genetic differences. 13 , 14 , 22 Cross‐reactive HI antibodies against viruses with a classical H1 are usually only detectable in high titered or hyperimmune antisera against avian‐like H1N1 SIV, and not in lower titered post‐infection swine sera. 3 , 16 , 23 The VN test detects a broader range of neutralizing antibodies than the HI test, 24 which likely explains the cross‐reactive VN antibody titers in our study. Dürrwald et al. 25 detected cross‐reactive VN antibodies against German isolates of the pandemic 2009 (H1N1) virus in pigs experimentally infected with the avian‐like H1N1 SIV A/swine/Haselünne/IDT2617/2003, which is in agreement with our findings. The cross‐protection between European and North American H1N1 SIVs was clearly stronger than that observed between European H1N1 and H1N2 SIVs in previous experimental studies. 20 , 26 , 27 In those prior studies, the pigs were first inoculated with sw/B/98 followed 4 weeks later by sw/Gent/7625/99, a typical European H1N2 virus. One of these studies used the same intranasal inoculation route and dose as in the present experiment. 27 This resulted in high H1N2 virus titers in nasal swabs of all challenge control pigs during 6 consecutive DPCh. All pigs that had been previously infected with sw/B/98 also shed high amounts of H1N2 virus in nasal swabs, and the duration of excretion was only 1 or 2 days shorter than in the control group. This contrasts with the excretion of the North American H1N1 SIV in the present study, which was largely blocked in sw/B/98‐immune pigs. The inferior protection against the H1N2 virus may relate to its more distant relationship to both the NA and HA of sw/B/98 when compared with the North American H1N1 SIV. Indeed, the HA of sw/Gent/7625/99 (H1N2) shows only 70·5% sequence homology with sw/B/98 (H1N1) and as much as 28 aa differences in antigenic sites, versus 75% homology and 13 aa changes in antigenic sites for sw/IA/04 (H1N1). This translates into detectable cross‐reactive VN and NI antibodies to the antigenically more closely related H1N1 virus, but not to H1N2. 20 , 27 The NP and M proteins, which are major targets for T cells, show more than 95% nucleotide identity between European H1N1 and H1N2 SIVs, 26 but they remain relatively conserved in the North American H1N1 SIV. Based on these findings and on general knowledge of the cross‐protective immune response between influenza viruses, 28 we believe that the observed cross‐protection between antigenically distinct H1N1 SIVs in the present study results from a combination of antibody‐ and cell‐mediated immune (CMI) responses to multiple viral proteins. In addition, mucosal as well as systemic immunity is likely involved. Most important, our data provide further proof of the concept that cross‐protection can occur between influenza viruses with multiple aa differences in three of four antigenic sites of the HA, and in the absence of detectable cross‐reactive serum HI antibody.
Our data further support the concept advanced previously 16 that the immune response after infection with avian‐like European H1N1 SIV may also protect pigs against the pandemic (H1N1) 2009 virus. Our genetic data and serologic results agree with previous findings about the swine ancestry of the pandemic virus. 10 , 12 The HA and NP of the pandemic Calif/09 isolate were closely related to the North American sw/IA/04 SIV, and both viruses shared many aa changes at antigenic sites of the HA, compared to the European sw/B/98 SIV. The NA and M1 of the pandemic virus, in contrast, were phylogenetically derived from Eurasian‐lineage, avian‐like SIV. 12 This was reflected in higher NI antibody titers to the pandemic virus than to the North American challenge H1N1 SIV in pigs immune to sw/B/98. It is therefore rational to expect an even more solid cross‐protection against the pandemic virus than to sw/IA/04‐like North American swine viruses in response to prior infection with sw/B/98‐like European viruses, though this assumption needs to be tested by further challenge studies. In experimental studies, in influenza naive pigs, the pandemic virus was as infectious for pigs as the endemic SIVs, and it was readily transmitted between pigs. 29 , 30 , 31 , 32 At this time of writing, however, cases of pandemic (H1N1) 2009 virus in pigs in Europe have been reported in only 10 countries, and the virus is far less widespread than the endemic SIVs. 11 However, it is striking that most cases have been reported in countries that were previously free of SIV, such as Norway, 33 and in countries with a low‐to‐moderate SIV prevalence, like Ireland and the United Kingdom.
The extent of cross‐protection between any two influenza viruses may appear greater under experimental than under natural conditions, because of many possible differences between the experimental and field situations. As an example, the short time interval between the primary influenza virus inoculation and challenge in our and other studies represents an artificial situation and may optimize the outcome of the experiment. On the other hand, fattening pigs in swine‐dense regions of Europe are frequently exposed to multiple SIV subtypes within their 26‐week short lifetime. For instance, as many as 84·5% of fattening swine farms in Belgium, France, Italy and Spain showed serologic evidence of infection with 2 or 3 SIV subtypes during 2006–2008. 6 Such consecutive or co‐infections will likely increase cross‐protection against viruses with a classical H1 HA. Experimental consecutive infections with European H1N1 and H1N2 viruses were also shown to induce cross‐reactive HI antibody to North American SIVs as well as pandemic (H1N1) 2009 virus, whereas the respective single infections failed to do so. 16 Such combined H1N1‐H1N2 infections even afforded protection against challenge with H3N2 SIV, despite no or minimal cross‐reactive HI antibody. 27 Recent serological investigations demonstrated cross‐reactive HI antibodies to the pandemic 2009 (H1N1) virus in as much as 52% of 1559 pig serum samples from 195 German pig herds. 25 The sera had been collected in the mid‐2009, before the first reports of the pandemic virus in European swine populations. Furthermore, vaccines based on the endemic European SIVs are licensed in the main pig‐producing Member States of Europe, but not in countries with lower pig numbers and SIV prevalences like Norway or Ireland. These vaccines seem to offer partial cross‐protection against the pandemic virus. 25 All these data further support the idea that pigs in swine‐dense regions of Europe may experience protection against influenza viruses with a classical swine‐lineage H1.
Acknowledgements
We thank Ruben Donis (Centers for Disease Control and Prevention) for providing the Calif/09 virus, and Lieve Sys, Melanie Bauwens, and Philippe Elskens for excellent technical assistance. This study was supported by the 6th Framework Research Program of the European Commission (FLUINNATE SSPE‐CT‐2006‐044161).
References
- 1. Olsen CW, Brown IH, Easterday BC, Van Reeth K. Swine influenza; in Straw B, D’Allaire S, Zimmerman J, Taylor D. (eds): Diseases of Swine. Iowa: Iowa State University Press, 2006; 469–482. [Google Scholar]
- 2. Olsen CW. The emergence of novel swine influenza viruses in North America. Virus Res 2002; 85:199–210. [DOI] [PubMed] [Google Scholar]
- 3. Pensaert M, Ottis K, Vandeputte J, Kaplan MM, Bachmann PA. Evidence for the natural transmission of influenza A virus from wild ducks to swine and its potential importance for man. Bull World Health Organ 1981; 59:75–78. [PMC free article] [PubMed] [Google Scholar]
- 4. Brown IH. The epidemiology and evolution of influenza viruses in pigs. Vet Microbiol 2000; 74:29–46. [DOI] [PubMed] [Google Scholar]
- 5. Van Reeth K, Brown IH, Dürwald R et al. Seroprevalence of H1N1, H3N2 and H1N2 influenza viruses in pigs in seven European countries in 2002‐2003. Influenza Other Respi Viruses 2008; 2:99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kyriakis CS. Surveillance and control of influenza in pigs. Doctoral thesis, 2009. ISBN 978‐90‐5864‐177‐9.
- 7. Guan Y, Shortridge KF, Krauss S, Li PH, Kawaoka Y, Webster RG. Emergence of avian H1N1 influenza viruses in pigs in China. J Virol 1996; 70:8041–8046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Takemae N, Parchariyanon S, Damrongwatanapokin S et al. Genetic diversity of swine influenza viruses isolated from pigs during 2000 to 2005 in Thailand. Influenza Other Respi Viruses 2008; 2:181–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kingsford C, Nagarajan N, Salzberg SL. 2009 Swine‐origin influenza A (H1N1) resembles previous influenza isolates. PLoS ONE 2009; 4:e6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Smith GJ, Vijaykrishna D, Bahl J et al. Origins and evolutionary genomics of the 2009 swine‐origin H1N1 influenza A epidemic. Nature 2009; 459:1122–1125. [DOI] [PubMed] [Google Scholar]
- 11. ProMedmail . http://www.promedmail.org/pls/apex/f?p=2400:1001:4495432574027595::NO::F2400_P1001_BACK_PAGE,F2400_P1001_PUB_MAIL_ID:1000,82366 .
- 12. Garten RJ, Davis CT, Russell CA et al. Antigenic and Genetic Characteristics of Swine‐Origin 2009 A(H1N1) Influenza Viruses Circulating in Humans. Science 2009; 325:197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hinshaw VS, Alexander DJ, Aymard M et al. Antigenic comparisons of swine‐influenza‐like H1N1 isolates from pigs, birds and humans: an international collaborative study. Bull World Health Organ 1984; 62:871–878. [PMC free article] [PubMed] [Google Scholar]
- 14. Brown IH, Ludwig S, Olsen CW et al. Antigenic and genetic analyses of H1N1 influenza A viruses from European pigs. J Gen Virol 1997; 78:78553–78562. [DOI] [PubMed] [Google Scholar]
- 15. Van Reeth K, Braeckmans D, Cox E et al. Prior infection with an H1N1 swine influenza virus partially protects pigs against a low pathogenic H5N1 avian influenza virus. Vaccine 2009; 27:6330–6339. [DOI] [PubMed] [Google Scholar]
- 16. Kyriakis CS, Olsen CW, Carman S et al. Serologic cross‐reactivity with pandemic (H1N1) 2009 virus in pigs, Europe. Emerg Infect Dis 2010; 16:96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brownlee GG, Fodor E. The predicted antigenicity of the haemagglutinin of the 1918 Spanish influenza pandemic suggests an avian origin. Philos Trans R Soc Lond B Biol Sci 2001; 356:1871–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maurer‐Stroh S, Ma J, Lee RT, Sirota FL, Eisenhaber F. Mapping the sequence mutations of the 2009 H1N1 influenza A virus neuraminidase relative to drug and antibody binding sites. Biol Direct 2009; 4:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reed LJ, Münch H. A simple method of estimating fifty per cent endpoints. Am J Hygiene 1938; 27:493–497. [Google Scholar]
- 20. Van Reeth K, Gregory V, Hay A, Pensaert M. Protection against a European H1N2 swine influenza virus in pigs previously infected with H1N1 and/or H3N2 subtypes. Vaccine 2003; 21:1375–1381. [DOI] [PubMed] [Google Scholar]
- 21. Sandbulte MR, Gao J, Straight TM, Eichelberger MC. A miniaturized assay for influenza neuraminidase‐inhibiting antibodies using reverse genetics‐derived antigens. Influenza Other Respi Viruses 2009; 3:233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Scholtissek C, Bürger H, Bachmann PA, Hannoun C. Genetic relatedness of hemagglutinins of the H1 subtype of influenza A viruses isolated from swine and birds. Virology 1983; 129:521–523. [DOI] [PubMed] [Google Scholar]
- 23. Brown IH, Manvell RJ, Alexander DJ, Chakraverty P, Hinshaw VS, Webster RG. Swine influenza outbreaks in England due to a new H1N1 virus. Vet Rec 1993; 132:461–462. [DOI] [PubMed] [Google Scholar]
- 24. Miller E, Hoschler K, Hardelid P, Stanford E, Andrews N, Zambon M. Incidence of 2009 pandemic influenza A H1N1 infection in England: a cross‐sectional serological study. Lancet 2010; 375:1100–1108. [DOI] [PubMed] [Google Scholar]
- 25. Dürrwald R, Krumbholz A, Baumgarte S et al. Swine influenza A vaccines, pandemic (H1N1) 2009 virus, and cross‐reactivity. Emerg Infect Dis 2010; 16:1029–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Van Reeth K, Brown I, Essen S, Pensaert M. Genetic relationships, serological cross‐reaction and cross‐protection between H1N2 and other influenza A virus subtypes endemic in European pigs. Virus Res 2004; 103:115–124. [DOI] [PubMed] [Google Scholar]
- 27. Van Reeth K, Labarque G, Pensaert M. Serological profiles after consecutive experimental infections of pigs with European H1N1, H3N2, and H1N2 swine influenza viruses. Viral Immunol 2006; 19:373–382. [DOI] [PubMed] [Google Scholar]
- 28. Grebe KM, Yewdell JW, Bennink JR. Heterosubtypic immunity to influenza A virus: where do we stand? Microbes Infect 2008; 10:1024–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brookes SM, Núñez A, Choudhury B et al. Replication, pathogenesis and transmission of pandemic (H1N1) 2009 virus in non‐immune pigs. PLoS ONE 2010; 5:e9068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lange E, Kalthoff D, Blohm U et al. Pathogenesis and transmission of the novel swine‐origin influenza virus A/H1N1 after experimental infection of pigs. J Gen Virol 2009; 90:2119–2123. [DOI] [PubMed] [Google Scholar]
- 31. Weingartl HM, Berhane Y, Hisanaga T et al. Genetic and pathobiologic characterization of pandemic H1N1 2009 influenza viruses from a naturally infected swine herd. J Virol 2010; 84:2245–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Itoh Y, Shinya K, Kiso M et al. In vitro and in vivo characterization of new swine‐origin H1N1 influenza viruses. Nature 2009; 460:1021–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hofshagen M, Gjerset B, Er C et al. Pandemic influenza A(H1N1)v: human to pig transmission in Norway? Euro Surveill 2009; 14:19406. [DOI] [PubMed] [Google Scholar]


