Abstract
Despite advances in the understanding of head and neck squamous cell carcinomas (HNSCC) progression, the five-year survival rate remains low due to local recurrence and distant metastasis. One hypothesis to explain this recurrence is the presence of cancer stem-like cells (CSCs) that present inherent chemo- and radio-resistance. In order to develop new therapeutic strategies, it is necessary to have experimental models that validate the effectiveness of targeted treatments and therefore to have reliable methods for the identification and isolation of CSCs. To this end, we present a protocol for the isolation of CSCs from human HNSCC cell lines that relies on the combination of two successive cell sortings performed by fluorescence activated cell sorting (FACS). The first one is based on the property of CSCs to overexpress ATP-Binding Cassette (ABC) transporter proteins and thus exclude, among others, vital DNA dyes such as Hoechst 33342. The cells sorted with this method are identified as a "side population" (SP). As the SP cells represent a low percentage (<5%) of parental cells, a growing phase is necessary in order to increase their number before the second cell sorting. The next step allows for the selection of cells that possess two other HNSCC stem cell characteristics i.e. high expression level of the cell surface marker CD44 (CD44high) and the over-expression of aldehyde dehydrogenase (ALDHhigh). Since the use of a single marker has numerous limitations and pitfalls for the isolation of CSCs, the combination of SP, CD44 and ALDH markers will provide a useful tool to isolate CSCs for further analytical and functional assays requiring viable cells. The stem-like characteristics of CSCs was finally validated in vitro by the formation of tumorispheres and the expression of β-catenin.
Keywords: Medicine, Issue 111, Head and neck squamous cell carcinoma, cancer stem cells, fluorescent activated cell sorting, aldehyde dehydrogenase, CD44, cell line
Introduction
Head and neck squamous cell carcinoma (HNSCC) is a common malignancy worldwide and despite progress in current treatments, patients with advanced disease have a poor prognosis. The overall 5 year survival rate of the patient is around 30% despite the combination of therapeutic approaches including surgery, chemo-radiotherapy and targeted-therapies. Recent studies attribute local recurrence and distant metastasis to the survival of cancer stem-like cells (CSCs) following anticancer therapies1. There is accumulating evidence supporting the existence of cells presenting stem cells properties (undifferentiated status, self-renewal and differentiation capacities, and telomerase activity) in various solid tumors including breast, brain, prostate, lung, colon, pancreas, liver and skin2-10. However, the origin of CSCs remains unclear11,12. They may result from the malignant transformation of normal stem cells3,13 or dedifferentiation of tumor cells that acquire CSCs-like features14,15. Therefore, understanding distinctive pathways relating to CSCs will provide insight into early diagnosis and treatment of resistant HNSCC.
It has been proposed that CSCs also possess resistant phenotypes that evade standard chemotherapy and radiotherapy, resulting in tumor relapse compared to the bulk of tumor cells16-19 and are localized into hypoxic niches20. Numerous factors have been proposed to explain these resistances of CSCs, such as propensity to quiescence, enhanced DNA repair, up-regulated cell cycle control mechanisms, and free-radical scavenging21. Moreover, several oncogenic molecular pathways may be specifically activated in CSCs17. In order to improve knowledge of CSCs for further targeted-therapies, we need reliable methods for the identification and isolation of CSCs, owing to the heterogeneity of stem cell-related markers in various types of cancers22.
In HNSCC, stem-like tumor-initiating cells have been isolated from primary patient tumors by sorting cells expressing different CSC biomarkers (such as drug efflux transporters expression23, CD44high, CD24low CD133high, c-Met+ phenotype10,24,25, or ALDHhigh activity26) or cultivating primary patient tumor to form squamospheres that have CSC properties. Nevertheless, the number of squamospheres decreases dramatically after two passages, thus giving a small sample size for further characterization studies27. Therefore, in vitro assays starting from well-established cell lines is an easier solution to design experiments in order to improve knowledge of CSCs.
The aim of this article is to propose a method to isolate CSCs from HNSCC cell lines using multiparametric flow cytometric analysis and cell sorting. The expression of CD44 in correlation with several CSCs properties including ALDH activity, Side Population (SP) phenotype, spheroid formation ability and tumorigenicity are used to isolate and characterize this sub-population of CSCs. CD44, a cell-surface glycoprotein, is involved in cell adhesion and migration. CD44 is highly expressed in many solid tumors CSCs28, including in head and neck cancer models29-31. Moreover, CD44high cells can generate in vivo a heterogeneous tumor whereas CD44low cells cannot10. The SP assay is based on the differential potential of cells to efflux the Hoechst dye22 via the ATP-binding cassette (ABC) family of transporter proteins overexpressed within the CSC membrane. This assay includes the use of ABC transporter inhibitors such as verapamil in control samples. Aldehyde dehydrogenase (ALDH) is an intracellular enzyme that is involved in converting retinol to retinoic acid during early stem cell differentiation25,26. Cells that exhibit high ALDH activity show stem-like cell behavior in HNSCC26 and a very few number of ALDHhigh cells are able to generate tumors in vivo26,32.
The combination of these markers and properties was successfully used by Bertrand et al. to study the resistance in vitro and in vivo of these CSCs to photon and carbon ion radiation19. Their results clearly showed that the combination of various cell markers and properties are more selective for useful studies on HNSCC CSCs populations than single-marker approaches.
Protocol
All animal procedures were performed according to local guidelines on animal care. All the details of this study were approved by the CECCAPP, a French ethics committee.
1. Selection of a Side Population (SP) by the Hoechst Dye Efflux Assay
- Staining 50 million cells with Hoechst 33342 dye.
- Prepare two 15 ml sterile tubes with a conical bottom: one tube labeled "Hoechst" and one labeled "Hoechst and Verapamil". Prepare 10 ml of 5 mM Verapamil hydrochloride solution in sterile water. Prepare the culture medium (CM) for stem cells.
- To prepare CM for CSC (CM-CSC), combine the following: Dulbecco's modified Eagle medium (DMEM):F12 (1:3, v:v), 5% of fetal calf serum (FCS), 0.04 mg/L of hydrocortisone, 100 U/ml of penicillin, 0.1 g/L of streptomycin, and 20 µg/L of Epidermal Growth Factor (EGFR).
- Under a laminar flow hood, trypsinize cells. Remove media, wash with sterile phosphate buffered saline (PBS) and add 1 ml of trypsin-EDTA (0.5 g/L) for a 75 cm² flask. Incubate 3 min at 37 °C. Stop the reaction by adding culture medium used for the parental cell line. Count the number of cells using a cell counter.
- To prepare CM for parental cell line (CM-P), supplement DMEM with 10% FCS, 0.4 mg/L of hydrocortisone, 100 U/ml of penicillin and 0.1 g/L of streptomycin).
- Dilute the cell suspension obtained in the step 1.1.2 to obtain 107 cells/ml in CM-P. Split the cell suspension in the 2 tubes prepared: put 100 µl (106 cells) in the tube "Hoechst and Verapamil" and 4 ml (4 x 107 cells) in the tube "Hoechst".
- In the sample "Hoechst and Verapamil", add 10 µl of 5 mM Verapamil hydrochloride solution (final concentration: 0.5 mM) and mix gently. Add 5 µl of 1 g/L Hoechst solution (final concentration: 0.1 g/L). In the sample "Hoechst", add 5 µl of 1 g/L Hoechst solution per 106 cells (200 µl total). Henceforth, keep samples protected from direct light exposure using aluminum foil.
- Incubate all tubes in a water bath at 37 °C for 1 hr 30 min. Mix gently every 15 min to prevent cells from settling down.
- Centrifuge tubes at 250 x g for 5 min at 4 °C. Remove supernatant and resuspend each pellet with 2 mL of 1x PBS.
- Centrifuge tubes at 250 x g for 5 min at 4 °C. Remove supernatant. Resuspend "Hoechst and Verapamil" pellet in 500 µl of 5 mg/L propidium iodide (PI) diluted in PBS buffer and "Hoechst" pellet in 4 ml of 5 mg/L PI diluted in PBS buffer.
- Transfer sample solutions through a 70 µm cell strainer to remove aggregates and collect single cells in tubes for sample uptake on the flow cytometer. Keep samples on ice and protect from direct light exposure using aluminum foil.
- Isolation of the Side Population Excluding Hoechst dye by Cell Sorting.
- Perform Hoechst negative cell separation on a flow cytometry sorter with the following parameters: UV laser (355 nm), 2 detectors on the UV laser path with blue Hoechst (450/50 BP) and red Hoechst (610/20 BP) filters, and 2 collectors.
- First, analyze sample "Hoechst" that serves as a positive control for staining and flow cytometer set-up.
- Using the cytometer software, on the "Global Worksheet" window, click on "Dot Plot" (fifth top right picture) and create a chart on the global worksheet. On ordinate, with a right click, select FSC-A (forward scatter) and on abscissa, SSC-A (side scatter) (Figure 1A). In the same way, create a SSC-W versus SCC-H dot plot. On this second dot plot, to create a P1 region, click "Polygon gate" (fourteenth top right picture) (Figure 1B)33. Note: The P1 region will encompass single cells and discriminate doublets.
- Optional: Exclude PI positive cells by creating a FSC-A versus PI gated on P1. On this dot plot, select the PI negative population (P2) to exclude PI positive dead cells.
- Using the cytometer software, on the "Global Worksheet" window, click on "Dot Plot" and create a chart on the global worksheet. On ordinate, with a right click, select blue Hoechst-A and on abscissa, red Hoechst-A. With a right click on the population, select "Show population" and P1. On this dot plot, create a region (P2) to select the negative Hoechst dye side population (SP) cells that appears as a side arm on the left of the main population of cells (Figure 1C).
- Analyze the sample "Hoechst" and collect 10,000 events. To ensure that the gate P2, which represents the SP population, is well positioned, analyze the sample "Hoechst and Verapamil" (10,000 events) to observe the disappearance of SP population (Figure 1D).
- Collect the SP Hoechst dye negative cells in a 15 ml tube containing 1 ml of CM-CSC prepared in 1.1.1.1.
- At the end of the cell sorting, centrifuge the cell suspension at 250 x g for 5 min, remove the supernatant and resuspend the pellet with 1 ml of CM-CSC. Count the number of sorted cells using a cell counter and transfer appropriate number of cells to a culture flask (see Table 1). Add CM-CSC and incubate at 37 °C and 5% CO2 atmosphere.
- Maintain cells in culture under the same conditions for a maximum of 2 passages until there is a minimum of 5 x 107 cells (see step 3, "Cell culture method").
2. Selection of the CD44high/ALDHhigh Sub-population in the Side Population Sorted
- Staining 50 million of SP Cells with the ALDH Detection Kit and CD44 Antibody.
- Prepare seven 15 ml sterile tubes labeled as follows: "Unstained" (Tube a), "CD44-APC" (Tube b), "IgG1-APC" (Tube c), "ALDH" (Tube d), "ALDH and DEAB" (Tube e), "ALDH and CD44-APC" (Tube f), "ALDH, DEAB and CD44-APC" (Tube g). Keep all tubes and reagents at 4 °C during the staining.
- Prepare buffer A with 4.5 ml of the buffer 1 (contained in the kit) and 45 µl of CD44-APC (Allophycocyanin) antibody (dilution 1:100). Prepare buffer B with 100 µl of buffer 1 and 1 µl of IgG1-APC antibody (dilution 1:100). Prepare buffer C with 4 ml of buffer 1 and 20 µl of the reagent of the kit.
- Prepare a cell suspension of the previously sorted cells (step 1.2.5) at 107 cells /ml. Add 100 µL (106 cells) of the cell suspension to tubes a, b and c, and 4 ml (4 x 107 cells) to tube f. For the moment do not put cells in tubes d, e and g.
- Centrifuge the tubes containing the cells at 250 x g for 5 min. Remove the supernatant and resuspend the pellets in tubes a, b and c in 100 µl of buffer 1. Add 5 µl of diethylaminobenzaldehyde (DEAB), an ALDH inhibitor to tubes e and g.
- Resuspend cells in tube f with 4 ml reagent C and immediately transfer 100 µl into tubes d, e and g. Incubate all tubes in a water bath at 37 °C for 30 min and protect from direct light exposure using aluminum foil. Mix the cell suspension gently by vortexing after 15 min to prevent cells from settling.
- From this moment, keep the tubes on ice and protected from direct light exposure using aluminum foil. Centrifuge all tubes at 250 x g for 5 min at 4 °C and remove the supernatant.
- Resuspend tube f in 4 ml and tubes b and g with 100 µl of buffer A. Resuspend tube c with 100 µl of buffer B. To the other tubes, add 100 µl of buffer 1. Incubate 10 min at 4 °C.
- Centrifuge all tubes at 250 x g for 5 min at 4 °C and remove the supernatant. Rinse once with 1 ml of buffer 1 (4 ml for tube f) and centrifuge again at 250 x g for 5 min at 4 °C. Remove the supernatant. Re-suspend tube f pellet in 4 ml and all other tubes in 1 ml of buffer 1.
- Transfer the sample solutions through 70 µm cell strainers to remove aggregates and collect single cells in appropriate tubes for sample uptake on flow cytometer.
- Isolation of the CD44high/ALDH high Cell Population by Fluorescence Activated Cell Sorting.
- Perform CD44high/ALDH high cell sorting on a flow cytometry sorter with the following parameters: Blue laser (488 nm) and red laser (633 nm), 1 detector on the blue laser path with a FITC filter (530/30), 1 detector on the red laser path with an APC filter (660/20) and 2 collectors.
- Using the cytometer software, click on "Dot Plot" (the fifth top right picture) to create a FSC-A versus SSC-A dot plot as described in section 1.2.1.2, to check cell morphology and select a population composed of single cells with a SSC-W versus SSC-H dot plot.
- Using the cytometer software, on the "Global Worksheet" window, click on "Dot Plot" and create a chart on the global worksheet. On ordinate, with a right click, select APC-A and on abscissa, FITC-A in order to select the double stained population. Create a gate using tubes d and e to select ALDHhigh cells (Figure 2A). Note: Positive cells disappear in tube e treated with DEAB (Figure 2B).
- On the same chart, create a second gate using tubes b and c in order to select CD44high cells (Figure 2C and 2D). Positive cells disappear in the tube containing IgG1-APC. Analyze tubes f and g and create a third gate that includes CD44high/ALDHhigh cells (Figure 2E and 2F). Note: If positive cells are present in the tube containing IgG1-APC (Figure 2D), the interaction between cells and the APC-stained antibody is not specific.
- Collect CD44high/ALDHhigh cells into a 15 ml tube containing 1 ml of CM-CSC. Collect also CD44low/ALDHlow into a 15 ml tube containing 1 ml of CM19. Note: Prepare CM-CSC as described in step 1.1.1.1.
- Centrifuge the cell suspension at 250 x g for 5 min, remove the supernatant and resuspend the pellet with 1 ml of CM-CSC. Count the number of sorted cells.
3. Cell Culture Method
After the double-sorting of the cells as described above, plate the sorted cells into an appropriate culture flask (Table 1) with CM-CSC at 37 °C with 5 % CO2. After 18-24 hr, check if the cells have adhered and change the culture medium. Change the culture medium every 3 days until the expanding colonies are greater than 50% confluent.
Use the appropriate volume of trypsin 0.5 g/L - EDTA (Table 1) to trypsinize cells from the culture flask. Incubate cells 3 to 5 min at 37 °C. Add the appropriate volume of CM-CSC (Table 1) to stop the action of trypsin.
Count the number of cells obtained and plate 4 x 105 cells in a 175 cm² culture flask with CM-CSC. Incubate at 37 °C and 5 % CO2. At this concentration, cells with a doubling time of 24 hr will be 70% confluent in 7 days.
Use CSCs for in vitro or in vivo experiments before they have undergone 3 passages.
4. Confirmation of Tumor Potential and CSC Characteristics
- Tumor Sphere Formation to Confirm the tumor Potential of the CD44high/ALDHhigh Cells.
- Trypsinize cells as described in 3.2. Add 1 x 106 cells to a 15 ml tube and centrifuge at 250 x g for 5 min. Remove the supernatant and re-suspend the pellet in a DMEM:F12 (3:1) medium FCS free, 20 ng/ml of rhEGF, 4 mg/L of heparin and 1x B27. Incubate cells in a 6 well low anchorage plates culture flask at 37 °C and 5% CO2. Note: Use DMEM:F12 (3:1) medium containing 5% of FCS, 20 ng/ml of rhEGF, 4 mg/L of heparin and 1x B27 if cell do not grow without FCS.
- Observe the tumor sphere formation with an optical microscope from 4 to 10 days after seeding (Figure 3A). Note: Tumor sphere diameter should be more than 35 µm. CD44high/ALDHhigh cells will show a faster and more enhanced tumor sphere formation in terms of number and size compared to CD44low/ALDHlow.
- Evaluation of In Vivo Tumorigenicity After Subcutaneous Injection of CD44high/ALDHhigh Cells in NOD-SCID Mice.
- After sorting, re-suspend cells in PBS at three different concentrations (104 cells/ml, 105 cells/ml, and 106 cells/ml). Inject subcutaneously 100 µl of 104 cells/ml (103 cells) in the right flank region of 6 mice. Do the same for 105 cells/ml (104 cells), and 106 cells/ml (105 cells). Note: Lower dilution than 103 cells can be tested.
- Inject the same concentration of CD44low/ALDHlow cells in the left flank region. Monitor injected mice for up to 10 weeks to see tumor progression19.
Representative Results
The isolation of CSCs from HNSCC cell lines required two successive sorting because of the very low percentage of CSCs in the parental cell line. The first sorting was based on the ability of CSCs to exclude the Hoechst dye due to drug efflux transporters. This resulted in acquisition of 1-5% of the total cell population sorted (Figure 1). During the Hoechst dye negative cell sorting, check the size and granulation of sorted cells by looking at the FSC-A versus SSC-A dot plot (Figure 1A). Then, discriminate doublets and cells fragments by using the SSC-W versus SSC-H dot plot and selecting the population P1 (Figure 1B). On this P1 population, create a Hoechst Red-A versus Hoechst Blue-A dot plot. With the tube labeled "Hoechst", the SP appears as a side arm on the left from the main cells population (Figure 1C). This population must disappear when they are treated with Verapamil (Figure 1D), an inhibitor of ABC transporters. The PI staining allows the exclusion of PI-positive dead cells because this population is under-scale on the Hoechst Red-A scale (Figure 1C and 1D, blue ellipses).
The second cell sorting was based on the high expression of the CD44 receptor and high ALDH enzyme activity which allowed the acquisition of 0.5-2% from the SP cells previously sorted (Figure 2). Before the sorting, various controls were used. The first ones are the "ALDH" and the "ALDH + DEAB" tubes needed in order to place the first gate on FITChigh cells (Figures 2A and 2B): ALDHhigh cells were gated on the FITC-A versus APC-A dot plot using the "ALDH" tube (Figure 2A). The good position of the gate was checked using the "ALDH + DEAB" tube: as DEAB inhibits ALDH, positive cells must disappear from the gate (Figure 2B). If they don't, change the reaction conditions (by increasing the amount of DEAB for example). The second control is the "CD44-APC" and the "IgG1-APC" tubes which allowed to position the second gate on APChigh cells (Figures 2C and 2D) using the cells stained with the CD44-APC antibody (Figure 2C). This population must disappear with the control tube which contained IgG1-APC cells (Figure 2D). If it does not, the bond with the antibody is not specific and BSA 0.5% should be added into buffer 1 from the ALDH detection kit during the antibody reaction. Finally, the third control concerns the "ALDH and CD44-APC" tube and "ALDH, DEAB and CD44-APC" tubes (Figures 2E, 2F, 2G and 2H). The double staining ALDH/CD44 tube is used to position the last gate on the double positive cells (Figure 2E) and the same tube treated with DEAB is a control to verify that ALDHhigh cells disappear (Figure 2F).
This protocol is used to sort CSCs from the SQ20B and the FaDu cell lines. When the sorting is done for the first time on a new cell line, in order to ensure that sorted cells have stem-like cells properties, it is necessary to confirm their tumor potential. One of the stem-like cell property is its ability to form tumorspheres in vitro in a FSC free medium. Under this condition, only cancer stem-like cells can survive and proliferate (Figure 3A). Moreover, qPCR experiments show a high expression of β-catenin (marker of stem-like characteristic) in CD44high/ALDHhigh cells (Figure 3B), as well as Bmi-1 and Notch19. Finally, CD44high/ALDHhigh cells are also able to form tumors when injected in low quantities as compared with CD44low/ALDHlow cells19.
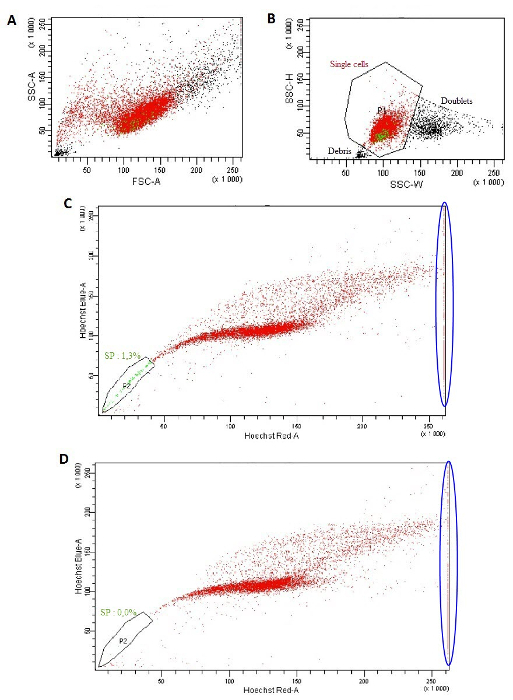 Figure 1: Isolation of a side population excluding the Hoechst dye. (A) FSC-A versus SSC-A dot plot. (B) SSC-W versus SSC-H dot plot. (C, D) Hoechst Red-A versus Hoechst Blue-A dot plot using the "Hoechst" tube (C) or with the "Hoechst and Verapamil" tube (D). Please click here to view a larger version of this figure.
Figure 1: Isolation of a side population excluding the Hoechst dye. (A) FSC-A versus SSC-A dot plot. (B) SSC-W versus SSC-H dot plot. (C, D) Hoechst Red-A versus Hoechst Blue-A dot plot using the "Hoechst" tube (C) or with the "Hoechst and Verapamil" tube (D). Please click here to view a larger version of this figure.
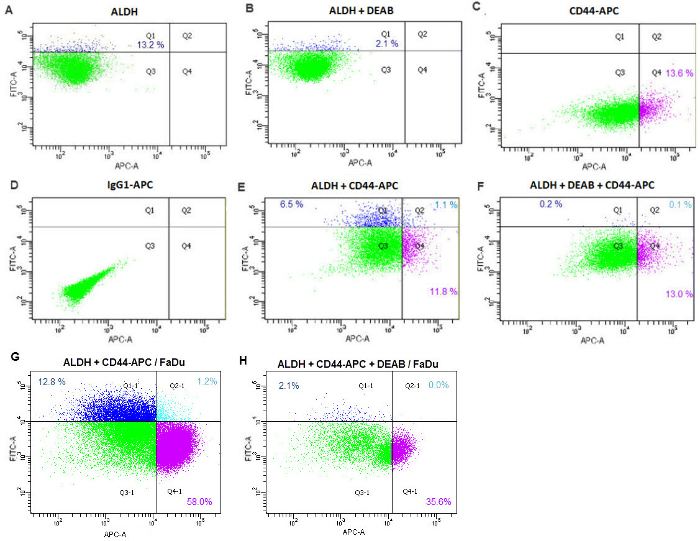 Figure 2: Sorting of CD44high/ALDHhigh cells from Hoechst dye negative cells. FITC-A versus APC-A dot plot using the "ALDH" tube (A), the "ALDH + DEAB" tube (B), the "CD44-APC" tube (C), the "IgG1-APC" tube (D), the "ALDH and CD44-APC" tube (E and G) or the "ALDH, DEAB and CD44-APC" tube (F and H). Figures A, B, C, D, E and F were obtained using SQ20B cell line. Figures G and H obtained using FaDu cell line. Green indicates CD44low/ALDHlow cells (Q3); dark blue indicates CD44low/ALDHhigh cells (Q1); purple indicates CD44high/ALDHlow cells (Q4); light blue indicates CD44high/ALDHhigh cells (Q2). Please click here to view a larger version of this figure.
Figure 2: Sorting of CD44high/ALDHhigh cells from Hoechst dye negative cells. FITC-A versus APC-A dot plot using the "ALDH" tube (A), the "ALDH + DEAB" tube (B), the "CD44-APC" tube (C), the "IgG1-APC" tube (D), the "ALDH and CD44-APC" tube (E and G) or the "ALDH, DEAB and CD44-APC" tube (F and H). Figures A, B, C, D, E and F were obtained using SQ20B cell line. Figures G and H obtained using FaDu cell line. Green indicates CD44low/ALDHlow cells (Q3); dark blue indicates CD44low/ALDHhigh cells (Q1); purple indicates CD44high/ALDHlow cells (Q4); light blue indicates CD44high/ALDHhigh cells (Q2). Please click here to view a larger version of this figure.
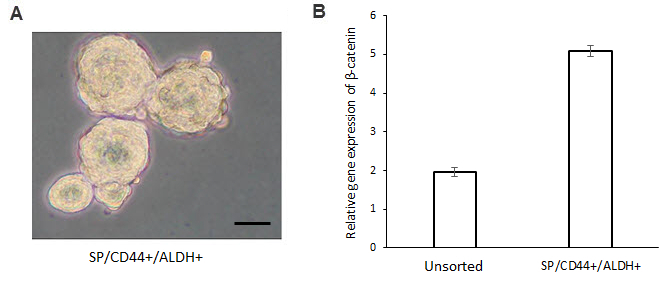 Figure 3: Confirmation of tumor potential of CD44high/ALDHhigh cells. Sorted CD44high/ALDHhigh cells were able to form tumorspheres in vitro (A) in a poor-FCS media. Scale bar, 25 µm. Furthermore, CD44high/ALDHhigh overexpress β-catenin, a marker of tumorigenicity (B). This quantification has been performed by qPCR and error bars represent SD. Please click here to view a larger version of this figure.
Figure 3: Confirmation of tumor potential of CD44high/ALDHhigh cells. Sorted CD44high/ALDHhigh cells were able to form tumorspheres in vitro (A) in a poor-FCS media. Scale bar, 25 µm. Furthermore, CD44high/ALDHhigh overexpress β-catenin, a marker of tumorigenicity (B). This quantification has been performed by qPCR and error bars represent SD. Please click here to view a larger version of this figure.
| Number of cells sorted | Culture flask type | Trypsin Volume (ml) | Culture Medium Volume (ml) |
| 10,000-200,000 | 1 well of a 6-well plate or a 3.5 cm petri dish | 0.5 | 2 |
| 200,000-1,000,000 | 1 T25 culture flask | 1 | 4 |
| >1,000,000 | 1 T75 culture flask | 2 | 10 |
Table 1: Culture flask type to use according to the number of cells sorted. Details of the culture flask size for approximate number of cells sorted are given. The volume of trypsin and volume of medium required for the culture flask type are also provided.
Discussion
This protocol describes a reliable method for the successful isolation of CSCs from a specific cell line that is applicable to other HNSCC cell lines. Isolated head and neck CSCs are then suitable for further molecular characterization in vitro and functional validation by transplantation in immunodeficient mice19. However, some modifications can be tested depending on the side population or the CD44high/ALDHhigh percentages present in the parental cell line. For example, if the percentage of cells in the side population is too low in a particular cell line, the CD44high/ALDHhigh sorting can be performed directly. The markers used in this study may be replaced by other markers appropriate to the cell line studied such as CD133high 34 or CD10high 35.
This protocol is based on two cell sortings. Three color sorting with SP + CD44 + ALDH was not possible with the cell lines tested. First, the parental cell lines tested here present less than 5% of SP and less than 10% CD44high/ALDHhigh cells in the SP. Therefore, SP/CD44high/ALDHhigh represent less than 0.5% of the parental cell line. Hence, a first SP sorting is necessary in order to enrich the population in CD44high and/or ALDHhigh cells. Second, to sort 1% of the parental cell line, it takes 5 hr to obtain approximately 300,000 cells. Therefore, if a 3 color cell sorting is undertaken, the quantity of cells collected will be very low. Furthermore, longer sorting is not recommended as it may affect cell viability.
Owing to the selectivity of this isolation protocol, the main limitation is the small number of CSCs obtained. This could be problematic to perform further experiments, since it is not recommended to use them after 3 passages because of the rapid loss of CD44 and ALDH markers. Furthermore, before each new experiment, the percentage of CD44high/ALDHhigh cells still present in the cell suspension should be tested in order to check the number of CSCs that has differentiated.
It is imperative to prepare Verapamil hydrochloride solution and culture medium containing EGF just before use as these molecules are very unstable. A stock solution of EGF at 20 mg/L is stable for three months at -20 °C. Variations of the Hoechst protocol exist, but the final dye concentration commonly used is 5 µg/ml. Moreover, during the first sorting, different culture media compositions should be tested according to the cell line used. Once the sorting conditions are validated, it is necessary to check the properties of the sorted cell population using the different methods (section 4).
Cell surface markers, ALDH activity and ability to efflux vital dyes have been already used individually in the literature to isolate CSCs from HNSCC. However, the protocol described here has the unique advantage of using combinations of various markers in order to achieve high specificity in CSCs isolation from HNSCC cell lines. Moreover, the CD44 sorting could be realized with an antibody anti-CD44 conjugated with magnetic micro-beads and sorted with a magnetic column36, but this method is only applicable to cell surface markers and cannot be used for an ALDH sorting, preventing double sorting. Another method used to obtain CSCs is the tumorsphere culture from primary tumors37 or xenografts38. However, the acquisition of these primary tumors or xenografts are associated with ethical constraints.
Since this method isolates viable HNSCC CSCs, they can be used (after checking their tumorigenicity) in a number of experiments that measure the physiological function of these cells. It therefore allows assessment of the behavior of CSCs following different therapeutic approaches (radiotherapy, chemotherapy). It also allows the study of various biological parameters such as migration/ invasion, DNA repair, cell signaling, etc. Hence, obtaining CSCs from different cell lines is an attractive choice for investigating CSCs properties.
Disclosures
The authors have nothing to disclose.
Acknowledgments
We thank Thibault Andrieu and Sebastien Dussurgey from the Flow Cytometry Platform of UFR BioSciences Gerland-Lyon-Sud (UMS3444/US8) for their advice and help during our sorting. This work was achieved within the scientific framework of ETOILE and Labex-PRIMES (ANR-11LABX-0063).
References
- Baumann M, Krause M, Hill R. Exploring the role of cancer stem cells in radioresistance. Nat Rev Cancer. 2008;8(7):545–554. doi: 10.1038/nrc2419. [DOI] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65(23):10946–10951. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- Eramo A, et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2008;15(3):504–514. doi: 10.1038/sj.cdd.4402283. [DOI] [PubMed] [Google Scholar]
- Dalerba P, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci USA. 2007;104(24):10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann PC, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1(3):313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Yang ZF, et al. Significance of CD90 cancer stem cells in human liver cancer. Cancer Cell. 2008;13(2):153–166. doi: 10.1016/j.ccr.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Fang D, et al. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res. 2005;65(20):9328–9337. doi: 10.1158/0008-5472.CAN-05-1343. [DOI] [PubMed] [Google Scholar]
- Prince ME, et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proc Natl Acad Sci USA. 2007;104(3):973–978. doi: 10.1073/pnas.0610117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke MF, et al. Cancer stem cells -- Perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66(19):9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- Soltanian S, Matin MM. Cancer stem cells and cancer therapy. Tumor Biol. 2011;32(3):425–440. doi: 10.1007/s13277-011-0155-8. [DOI] [PubMed] [Google Scholar]
- Molyneux G, et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 2010;7(3):403–417. doi: 10.1016/j.stem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- Vermeulen L, et al. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc Natl Acad Sci USA. 2008;105(36):13427–13432. doi: 10.1073/pnas.0805706105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajczak MZ. Cancer stem cells -- Normal stem cells 'Jedi' that went over to the 'dark side.'. Folia Histochem Cytobiol. 2005;43(4):175–181. [PubMed] [Google Scholar]
- Bao S, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Liu G, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncharmont C, et al. Targeting a cornerstone of radiation resistance: Cancer stem cell. Cancer Lett. 2012;322(2):139–147. doi: 10.1016/j.canlet.2012.03.024. [DOI] [PubMed] [Google Scholar]
- Bertrand G, et al. Targeting Head and Neck Cancer Stem Cells to Overcome Resistance to Photon and Carbon Ion Radiation. Stem Cell Rev. 2013;10(1):114–126. doi: 10.1007/s12015-013-9467-y. [DOI] [PubMed] [Google Scholar]
- Das B, Tsuchida R, Malkin D, Koren G, Baruchel S, Yeger H. Hypoxia enhances tumor stemness by increasing the invasive and tumorigenic side population fraction. Stem Cells. 2008;26(7):1818–1830. doi: 10.1634/stemcells.2007-0724. [DOI] [PubMed] [Google Scholar]
- Diehn M, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458(7239):780–783. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZG. The cancer stem cell concept in progression of head and neck cancer. J Oncol. 2009;2009:894064. doi: 10.1155/2009/894064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Zhang Y, Mao L, Zhang Z, Chen W. Side population in oral squamous cell carcinoma possesses tumor stem cell phenotypes. Cancer Lett. 2009;277(2):227–234. doi: 10.1016/j.canlet.2008.12.015. [DOI] [PubMed] [Google Scholar]
- Zhang Q, et al. A subpopulation of CD133(+) cancer stem-like cells characterized in human oral squamous cell carcinoma confer resistance to chemotherapy. Cancer Lett. 2010;289(2):151–160. doi: 10.1016/j.canlet.2009.08.010. [DOI] [PubMed] [Google Scholar]
- Sun S, Wang Z. Head neck squamous cell carcinoma c-Met⁺ cells display cancer stem cell properties and are responsible for cisplatin-resistance and metastasis. Int J Cancer. 2011;129(10):2337–2348. doi: 10.1002/ijc.25927. [DOI] [PubMed] [Google Scholar]
- Chen YC, et al. Aldehyde dehydrogenase 1 is a putative marker for cancer stem cells in head and neck squamous cancer. Biochem Biophys Res Commun. 2009;385(3):307–313. doi: 10.1016/j.bbrc.2009.05.048. [DOI] [PubMed] [Google Scholar]
- Lim YC, et al. Cancer stem cell traits in squamospheres derived from primary head and neck squamous cell carcinomas. Oral Oncol. 2011;47(2):83–91. doi: 10.1016/j.oraloncology.2010.11.011. [DOI] [PubMed] [Google Scholar]
- Yu Q, Stamenkovic I. Cell surface-localized matrix metalloproteinase-9 proteolytically activates TGF-beta and promotes tumor invasion and angiogenesis. Genes Dev. 2000;14(2):163–176. [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy S, et al. Endothelial cell-initiated signaling promotes the survival and self-renewal of cancer stem cells. Cancer Res. 2010;70(23):9969–9978. doi: 10.1158/0008-5472.CAN-10-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikamatsu K, Takahashi G, Sakakura K, Ferrone S, Masuyama K. Immunoregulatory properties of CD44+ cancer stem-like cells in squamous cell carcinoma of the head and neck. Head Neck. 2011;33(2):208–215. doi: 10.1002/hed.21420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YW, et al. Cucurbitacin I suppressed stem-like property and enhanced radiation-induced apoptosis in head and neck squamous carcinoma--derived CD44(+)ALDH1(+) cells. Mol Cancer Ther. 2010;9(11):2879–2892. doi: 10.1158/1535-7163.MCT-10-0504. [DOI] [PubMed] [Google Scholar]
- Clay MR, et al. Single-marker identification of head and neck squamous cell carcinoma cancer stem cells with aldehyde dehydrogenase. Head Neck. 2010;32(9):1195–1201. doi: 10.1002/hed.21315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinelt E, et al. Technical Bulletin: Standardizing Application Setup Across Multiple Flow Cytometers Using BD FACSDiva Version 6 Software. BD Biosciences; 2012. [Google Scholar]
- Zhou L, Wei X, Cheng L, Tian J, Jiang JJ. CD133, one of the markers of cancer stem cells in Hep-2 cell line. Laryngoscope. 2007;117(3):455–460. doi: 10.1097/01.mlg.0000251586.15299.35. [DOI] [PubMed] [Google Scholar]
- Fukusumi T, et al. CD10 as a novel marker of therapeutic resistance and cancer stem cells in head and neck squamous cell carcinoma. Br J Cancer. 2014;111(3):506–514. doi: 10.1038/bjc.2014.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Xiang M, Nie C, Hu H, Ma Y, Wu H. CD44 as a molecular marker to screen cancer stem cells in hypopharyngeal cancer. Acta Otolaryngol. 2013;133(11):1219–1226. doi: 10.3109/00016489.2013.811750. [DOI] [PubMed] [Google Scholar]
- Kanojia D, et al. Proteomic profiling of cancer stem cells derived from primary tumors of HER2/Neu transgenic mice. Proteomics. 2012;12(22):3407–3415. doi: 10.1002/pmic.201200103. [DOI] [PubMed] [Google Scholar]
- Higgins DM, et al. Brain tumor stem cell multipotency correlates with nanog expression and extent of passaging in human glioblastoma xenografts. Oncotarget. 2013;4(5):792–801. doi: 10.18632/oncotarget.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]


