Abstract
Three dimensional (3D) culture is a more physiologically relevant method to model cell behavior in vitro than two dimensional culture. Carcinomas, including breast carcinomas, are complex 3D tissues composed of cancer epithelial cells and stromal components, including fibroblasts and extracellular matrix (ECM). Yet most in vitro models of breast carcinoma consist only of cancer epithelial cells, omitting the stroma and, therefore, the 3D architecture of a tumor in vivo. Appropriate 3D modeling of carcinoma is important for accurate understanding of tumor biology, behavior, and response to therapy. However, the duration of culture and volume of 3D models is limited by the availability of oxygen and nutrients within the culture. Herein, we demonstrate a method in which breast carcinoma epithelial cells and stromal fibroblasts are incorporated into ECM to generate a 3D breast cancer surrogate that includes stroma and can be cultured as a solid 3D structure or by using a perfusion bioreactor system to deliver oxygen and nutrients. Following setup and an initial growth period, surrogates can be used for preclinical drug testing. Alternatively, the cellular and matrix components of the surrogate can be modified to address a variety of biological questions. After culture, surrogates are fixed and processed to paraffin, in a manner similar to the handling of clinical breast carcinoma specimens, for evaluation of parameters of interest. The evaluation of one such parameter, the density of cells present, is explained, where ImageJ and CellProfiler image analysis software systems are applied to photomicrographs of histologic sections of surrogates to quantify the number of nucleated cells per area. This can be used as an indicator of the change in cell number over time or the change in cell number resulting from varying growth conditions and treatments.
Keywords: Bioengineering, Issue 111, 3D culture, breast cancer, bioreactor, perfusion, co-culture, tumor microenvironment
Introduction
Three dimensional (3D) culture models that more accurately mimic the tumor architecture and microenvironment in vivo are important for studies aimed to dissect the complex interactions between cells and their microenvironment and to test the efficacy of candidate therapies. Tumor dimensionality impacts oxygen and nutrient gradients, the uniformity of drug exposure, interstitial pressure/blood flow, and 3D architecture1-4. The presence of an appropriate stromal microenvironment contributes to tumor dimensionality and influences cell-ECM signaling and paracrine signaling between stromal cells and malignant epithelial cells. The effects of tumor dimensionality and the microenvironment on cellular function are well established, with both factors altering drug response1,3,5-8. Additionally, cellular growth kinetics, metabolic rates, and cell signaling differ between two dimensional (2D) culture and culture in 3D, with these factors affecting cellular response1,3,8-10.
In vitro, the tumor surrogate microenvironment can be modulated by including representative ECM constituents and stromal cell populations. Malignant epithelial cells are influenced by the ECM and cancer-associated stromal cells either in a synergistic/protective manner to promote tumor progression or in a suppressive manner to inhibit further tumor propagation5,6,10. In either context, the stroma can affect therapeutic response and drug delivery via paracrine signaling and/or by increasing interstitial pressure in the tumor resulting in decreased drug delivery1,6. Therefore, the addition of ECM and stromal cells into preclinical models will help recapitulate aspects of the tumor that cannot be modeled well in 2D culture.
Herein a method to establish breast cancer surrogates that incorporate a recapitulative microenvironment, including ECM constituents and stromal cells, in a 3D volume is described. In breast carcinoma, the stromal cell population is predominately comprised of cancer associated fibroblasts (CAF) and the stromal ECM is largely composed of collagen type I with a smaller proportion of matrix components that are found in the basement membrane, including laminin and collagen type IV1,4,11-13. Therefore, these components of the breast carcinoma microenvironment (i.e., CAF, collagen I, and basement membrane) have been incorporated into the surrogates. This method can be used to generate solid, un-perfused 3D surrogates (Figure 1A) or can be adapted to include perfusion of medium through the surrogate via a bioreactor system (Figure 1B). Both approaches are described here. This method could also be modified to include other stromal elements, such as tumor-associated macrophages, or to model other solid tumors by adjusting the cellular and ECM components, as appropriate.
For the breast carcinoma surrogate described here, we have utilized the MDA-MB-231 (231) breast cancer cell line, CAF previously isolated from human breast carcinoma14, and an ECM composed of 90% collagen I (6 mg/ml) and 10% growth factor reduced basement membrane material (BM). The surrogate is either grown in an 8-well chamber slide (solid surrogate) or a bioreactor system is utilized to provide continuous nutrient perfusion (perfused surrogate). Any perfusion bioreactor system that can accommodate a volume of ECM containing cells can be used15. As an example, we describe the preparation of the tissue surrogates in our bioreactor system. This system was developed in-house and is not commercially available. Because our focus here is on the preparation and analysis of the 3D tissue surrogates, we have not gone into extensive detail regarding the specifics of manufacture and assembly of our bioreactor system. However, a detailed description of this system and its development has been published16. In this bioreactor system, a polydimethylsiloxane (PDMS) flow channel is used to house the surrogate, which is supported by a PDMS foam (formed using methods similar to those described by Calcagnile et al.17). This volume is penetrated by 4 microchannels (each 400 µm in diameter) which are continuously perfused by medium via a microphysiologic pump to supply oxygen and nutrients to the surrogate.
Appropriate analysis of the surrogates is crucial to gain pertinent information regarding cellular function in response to treatment or other manipulations. Surrogates can be analyzed by various methods including direct imaging of intact surrogates using confocal microscopy or other means of non-invasive imaging, indirect cellular analysis by assaying the conditioned media, or perfusate, for secreted products, or analysis on histologic sections after fixation and processing to paraffin. One such parameter that can be evaluated on histologic sections is cell density. We present one approach to measure cell density (i.e., the number of nucleated cells per section area) using semi-automated image processing techniques applied to photomicrographs of surrogate histologic sections stained with hematoxylin and eosin (H&E). The cell density can be used as an indicator of the relative change in cell number over time or that results from varying growth conditions and treatments.
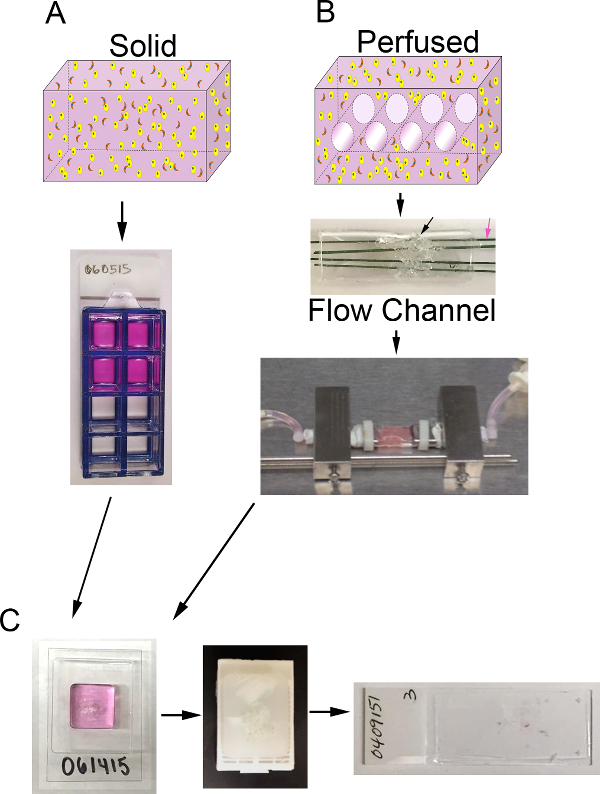 Figure 1. 3D volume and bioreactor system. A) Schematic of the process to generate solid 3D surrogates. Top: cartoon of solid 3D volume containing ECM (pink), epithelial carcinoma cells (yellow), and CAF (orange); Bottom: top view of 8-well chamber slide containing surrogates. B) Schematic of the process to generate perfused 3D surrogates. Top: cartoon of 3D volume with channels to allow for medium perfusion and containing ECM (pink), epithelial carcinoma cells (yellow), and CAF (orange); Middle: image of PDMS flow channel containing PDMS foam (black arrow) to be injected with cell+ECM and penetrated by polymer-coated stainless steel wires (pink arrow) measuring 400 µm in diameter; Bottom: image of the PDMS flow channel containing a surrogate and connected to the bioreactor system to allow for continuous medium perfusion (peristaltic pump and media reservoir not shown). C) Images of processing steps for both solid and perfused surrogates after culture. Left: image of the cryomold containing specimen processing gel and surrogate; Middle: image of a paraffin block containing a fixed and processed surrogate; Right: image of a glass slide with a H&E-stained histologic section of a surrogate. Please click here to view a larger version of this figure.
Figure 1. 3D volume and bioreactor system. A) Schematic of the process to generate solid 3D surrogates. Top: cartoon of solid 3D volume containing ECM (pink), epithelial carcinoma cells (yellow), and CAF (orange); Bottom: top view of 8-well chamber slide containing surrogates. B) Schematic of the process to generate perfused 3D surrogates. Top: cartoon of 3D volume with channels to allow for medium perfusion and containing ECM (pink), epithelial carcinoma cells (yellow), and CAF (orange); Middle: image of PDMS flow channel containing PDMS foam (black arrow) to be injected with cell+ECM and penetrated by polymer-coated stainless steel wires (pink arrow) measuring 400 µm in diameter; Bottom: image of the PDMS flow channel containing a surrogate and connected to the bioreactor system to allow for continuous medium perfusion (peristaltic pump and media reservoir not shown). C) Images of processing steps for both solid and perfused surrogates after culture. Left: image of the cryomold containing specimen processing gel and surrogate; Middle: image of a paraffin block containing a fixed and processed surrogate; Right: image of a glass slide with a H&E-stained histologic section of a surrogate. Please click here to view a larger version of this figure.
Protocol
1. Cell Culture
Thaw BM component overnight at 4 °C, on ice.
Warm medium to 37 °C. To support growth of both 231 cells and CAF, use Dulbecco's Modified Eagle Medium (DMEM) plus 10% fetal bovine serum (FBS). Note: The media used will depend upon the cell type and the experimental goals.
Remove medium from the culture dish (10 cm) of near confluent 231 cells and add 1.5 ml Trypsin/EDTA. Incubate for 1 to 3 min at 37 °C, monitoring for cell detachment. Once cells start to round and come off the plate, stop the reaction by adding 3 ml of the medium containing serum. Pipette the medium and cells into a 15 ml conical tube.
Centrifuge the medium and cells at 150 x g for 5 min. Remove the supernatant, and re-suspend the cell pellet in 2 ml medium.
Count the number of cells per volume using trypan blue and a hemocytometer. Cell viability should be greater than 90%.
Repeat the process with other cell types to be included in the surrogate. For the surrogate described here, the process is repeated with the CAF.
Determine the appropriate volume of each cell suspension to obtain the desired number of cells. Note: For the surrogate described here, a cell density of 2.1 x 106 cells per 100 µl of ECM, with a 2:1 ratio of epithelial cells to fibroblasts (1.4 x 106 231 cells and 7 x 105 CAF per 100 µl ECM), is used. This cell density is a good starting point; however, the optimal cell density will depend on the cell type, the duration of culture, and the objectives of the experiment.
Place the appropriate volume of each cell suspension into a 15 ml conical tube (one tube for each cell type). Centrifuge the medium and cells at 150 x g for 5 min.
Following centrifugation, remove the supernatant, re-suspend one cell type in cell culture grade water (178.8 µl, see Table 1) and the other cell type in 10x DMEM (100 µl, see Table 1, containing phenol red to monitor pH). Place the tubes containing the cells on ice and proceed quickly to preparation of ECM below. Limit the time that the cells remain in water to preserve viability. Note: Both 10x DMEM and cell culture grade water are required components of the ECM; therefore, we have chosen to re-suspend cells in both. Here we arbitrarily chose to re-suspend the 231 cells in cell culture grade water and CAF in 10x DMEM, although either cell type could be re-suspended in either of these two components.
2. Preparation of Cells in ECM (6 mg/ml Bovine Collagen Type I + 10% BM)
Note: An ECM composed of 90% collagen I + 10% BM was chosen to model invasive breast carcinoma because the tumor stroma in this malignancy is composed primarily of collagen I with components of the BM, such as laminin, collagen IV, and entactin, comprising a smaller portion of the ECM12,13,18,19.
On ice, add components listed in Table 1, in order, into a 2 ml microcentrifuge tube. Note: This amount is enough for 8 solid surrogates or, using the bioreactor system described here, 4 perfused surrogates.
| Preparation of cells in ECM (6 mg/ml Bovine Collagen Type I + 10% BM) | |
| 178.8 µl | Cell culture grade water containing the desired number of 231 cells (determined above) |
| 606 µl | Collagen I (10 mg/ml bovine), add drop by drop |
| 100 µl | Basement membrane, thawed |
| 100 µl | 10x DMEM (containing phenol red) with the desired number of CAF (determined above) |
| 15.2 µl | 7.5% (v/v) Sodium Bicarbonate, add drop by drop |
Table 1. Preparation of cells in ECM.
- Gently mix by pipetting, avoiding formation of bubbles. Monitor the pH level using the phenol red in the 10x DMEM. Check that the mixture is an orange/pink color indicating a pH of ~7. If the pH is too low (too yellow), slowly add additional 7.5% sodium bicarbonate one drop at a time (~ 5 µl) until the appropriate color is reached.
- Keep mixture on ice and work quickly to prevent premature ECM polymerization.
3. Surrogate Preparation
- For solid 3D cultures (Figure 1A):
- Working in a cell culture hood using sterile technique, label the lid of a sterile 8-well chamber slide to indicate any experimental variation in surrogates.
- Keeping the chamber slide on ice to prevent premature ECM polymerization, slowly pipette 100 µl of the cell+ECM mixture into each well of the 8-well chamber slide. Note: Pipetting the cell+ECM mixture around the edges of the well first helps to better distribute the cell+ECM mixture in the well.
- Incubate surrogates at 37 °C, 5% CO2 for 45 min to allow ECM polymerization.
- Following ECM polymerization, add 100 µl culture media to each well and incubate at 37 °C, 5% CO2 for the duration of the experiment, changing culture medium every two days.
- For perfused 3D cultures in a bioreactor system (Figure 1B):
- Sterilize all bioreactor components for 3D culture setup (i.e., bioreactor, tubing, forceps, and fittings necessary for bioreactor setup) using the process specific for the bioreactor to be used.
- For the example bioreactor system utilized here, use a combination of autoclaving (e.g., 12 min exposure at 121.1 °C with 15 min drying) and incubating in 70% ethanol for 1 hr.
- Prepare and assemble the portion of the bioreactor system that will house the surrogate.
- For the example bioreactor system used here, insert a PDMS foam backbone into the PDMS flow channel tubing using forceps. Push four (400 µm) polymer-coated stainless steel wires into the PDMS foam to generate parallel microchannels.
- In a cell culture hood, using sterile technique and a 26 gauge needle with syringe, inject the cell+ECM mixture into the area of the perfusion bioreactor designed to contain cells. Proceed quickly to the next step.
- To ensure more even distribution of cells within the surrogates, place the bioreactor component housing the surrogates into a 50 ml conical tube (under the cell culture hood) and continuously rotate at ~18 rpm while incubating at 37 °C for 45 min to allow ECM polymerization. Note: Rotation can be completed using a rotator in the incubator or an oven with a built in rotator, such as a hybridization oven set at 37 °C.
- Connect the bioreactor assembly containing the surrogate to the perfusion pump using manufacturer's instructions. Note: The specifics of this process will vary depending on the bioreactor and pump being used.
- For the example bioreactor system used here, remove the stainless steel wires prior to connecting the bioreactor assembly to the pump.
- Start medium perfusion (bulk flow rate of 167.1 µl/min; microchannel wall shear stress of 1 dyne/cm2) in an incubator at 37 °C, 5% CO2. Note: The rate of medium perfusion can be adjusted, depending on the bioreactor setup and the goals and the design of the experiment.
- Continue medium perfusion for the duration of the experiment, changing culture medium every seven days.
4. Surrogate Fixation and Processing (Figure 1C)
Label cryomolds and plastic tissue cassettes for surrogate fixation and processing.
- Next, encase surrogates in specimen processing gel, which is an aqueous material that is liquid at warm temperatures, but solidifies at room temperature. The specimen processing gel assists in keeping the surrogates intact during processing and facilitates histologic sectioning14,20-22.
- Melt specimen processing gel in a 60 °C water bath to liquefy it, keeping at this temperature until ready to use. Move the bioreactor with the surrogate to a biosafety cabinet.
- Pipette approximately 300 µl specimen processing gel into the bottom of the labeled cryomold (Figure 1C, left panel).
- Using a scalpel blade (No. 10 preferred) and forceps carefully remove the surrogate from the bioreactor or from the well of an 8-well chamber slide and place it into the cryomold containing specimen processing gel. Note: Tissue marking dyes (see Materials/Equipment List for a specific example) of different colors can be used to mark surrogates, thereby allowing multiple samples to be included in one tissue cassette in a distinguishable manner.
- Pipette approximately 300 µl specimen processing gel to cover the surrogate in the cryomold and incubate at 4 °C for 30 min to solidify.
- Once the specimen processing gel has solidified, remove the specimen processing gel containing the surrogate from the cryomold, and place it into a tissue cassette.
Place the tissue cassette containing the surrogate into 10% neutral buffered formalin for 10 to 12 hr at room temperature to allow complete fixation.
Following fixation, move the tissue cassette containing the surrogate to 70% ethanol until processed to paraffin. Note: Transferring the surrogate from formalin to ethanol prevents over-fixation with formalin which can cause loss of immunoreactivity of some epitopes23. The length of time in ethanol is not critical. This change of fixative is important if the surrogate will be used for immunohistochemistry or immunofluorescence. The fixed surrogate is now ready for processing to paraffin (Figure 1C, middle panel). This processing is typically performed in a tissue processor located in an appropriately equipped histology laboratory. A shorter program is recommended due to the size and delicate nature of the surrogates.24
5. Sectioning and H&E Staining (Figure 1C, Right Panel)
Following processing of the surrogates to a paraffin block, section them using a standard microtome for sectioning of formalin-fixed, paraffin-embedded tissues24,25. Note: This can be performed in a qualified histology laboratory, or in a research laboratory, if properly equipped and experienced. The thickness of the histologic sections can vary depending on the intended use of the sections; however, we typically use sections that are 5 µm in thickness. The PDMS foam used here in the perfused surrogates is easily sectioned with a microtome.
Place sections on plain glass histologic slides.
After sectioning, bake the histologic sections at 58 °C for 10-12 hr to prepare for staining with hematoxylin and eosin (H&E). Baking melts the paraffin and also allows better adherence of sections to the glass slide.
- H&E staining:
- Set up the stations described in Table 2 in coplin jars or glass staining dishes, depending on the number of slides to stain. Once reagents are set up, move the sections through each station, in order, incubating for the time indicated below24.
- Mount a coverslip to each slide using mounting media.
- Allow mounting media to dry before imaging.
| H&E Staining | ||
| Station | Solution | Time |
| 1 | Xylene | 5 min |
| 2 | Xylene | 5 min |
| 3 | Xylene | 5 min |
| 4 | 100% Ethanol | 5 min |
| 5 | 100% Ethanol | 5 min |
| 6 | 95% Ethanol | 5 min |
| 7 | 95% Ethanol | 5 min |
| 8 | Tap Water | 5 min |
| 9 | De-ionized Water | 5 min |
| 10 | Hematoxylin 7211 | 5 min |
| 11 | Tap Water | 5 min |
| 12 | Clarifier* | 10 dips |
| *Richard Allan #7401 or 70% Ethanol + 0.5% HCl | ||
| 13 | Tap Water | 5 min |
| 14 | Bluing Reagent | 30 sec |
| 15 | Tap Water | 5 min |
| 16 | 95% Ethanol | 10 dips |
| 17 | Eosin-Y | 1 min |
| 18 | 95% Ethanol | 10 dips |
| 19 | 95% Ethanol | 10 dips |
| 20 | 100% Ethanol | 10 dips |
| 21 | 100% Ethanol | 10 dips |
| 22 | 100% Ethanol | 5 min |
| 23 | Xylene | 10 dips |
| 24 | Xylene | 5 min |
Table 2. H&E Staining.
6. Measuring Cell Density
Image at least one entire H&E stained histologic section of the surrogate using brightfield microscopy at 400X magnification, saving the images as .tif files. Note: The image processing described has only been completed using color images. While untested, we believe the same processing should also be applicable to grayscale images.
Download CellProfiler from the Broad Institute26 (http://cellprofiler.org/download.shtml) and ImageJ from the National Institutes of Health (http://rsb.info.nih.gov/ij/download.html), both of which are publically available at no charge.
To measure the area of the surrogate in each image, open ImageJ, select "Set Measurements" (under the "Analyze" tab), select "Area", and then select "Okay".
Open an image (.tif file) of the surrogate. Using the polygon tool in ImageJ (see Figure 2), outline the area of the surrogate in the image by dragging the mouse and clicking to make anchor points. Use the edges of the ECM as a guide. Once outlined, select "Measure" under the "Analyze" tab.
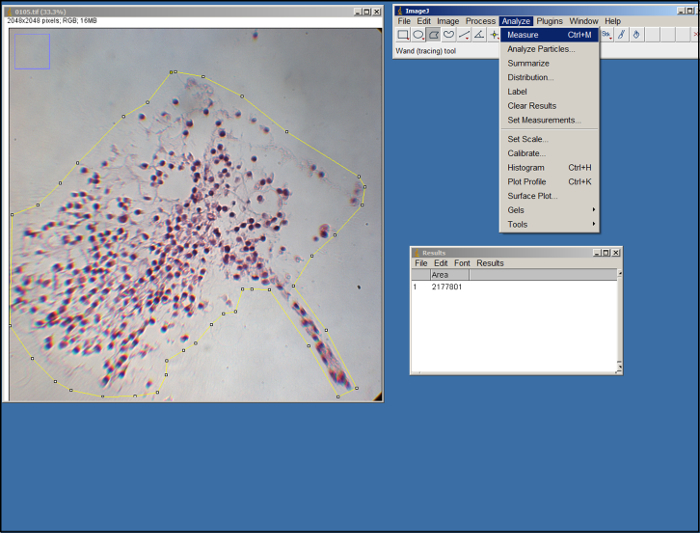 Figure 2. ImageJ analysis. Screenshot of ImageJ processing.
Please click here to view a larger version of this figure.
Figure 2. ImageJ analysis. Screenshot of ImageJ processing.
Please click here to view a larger version of this figure.
Repeat for each image of the tissue surrogate. Save measurements and corresponding image identifiers to a spreadsheet program.
Upload image files used to measure area in ImageJ into CellProfiler by dragging image files to the "File List". Assign a name to the images imported in the "NamesAndTypes" input module and select the image type (i.e., "Color Image").
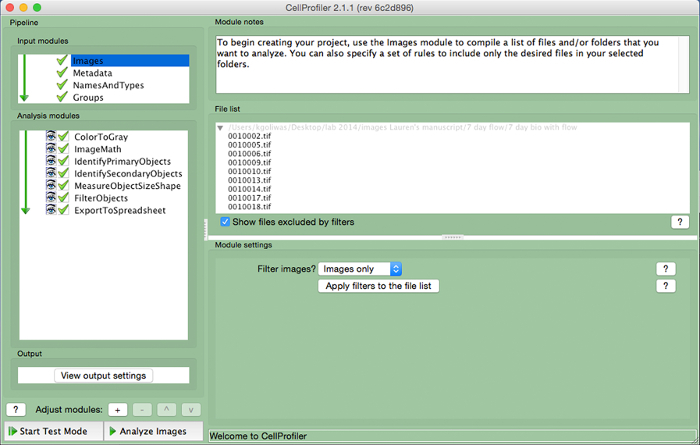 Figure 3. CellProfiler example pipeline. Screenshot of the pipeline designed to measure the number of nucleated cells in CellProfiler. Please click here to view a larger version of this figure.
Figure 3. CellProfiler example pipeline. Screenshot of the pipeline designed to measure the number of nucleated cells in CellProfiler. Please click here to view a larger version of this figure.
- Create an analysis pipeline that includes the following modules to calculate the number of cells per image, by clicking the "+" sign next to "Adjust Module" (Figure 3, at bottom of left panel). Add each of the modules below.
- Select "ColorToGray" (Figure 4).
- Select the input image name from the drop down menu, name the output image, and select the original image type (i.e., RGB if color image input). Note: This module would not be needed if using grayscale images
- Select "ImageMath" (Figure 5).
- Select the "Invert" operation, name the output image, and select the grayscale image in the "select first image" tab.
- Select "IdentifyPrimaryObjects" (Figure 6).
- Select input image (image after math correction), name the primary object to be identified (nuclei), and enter the diameter range for the objects to be measured in pixel units (approximately 25 to 65). Select "adaptive" threshold strategy with "Otsu" thresholding method with "three classes". Do not change any other parameters from the default settings. Note: Optimal range of diameters should be determined by opening an image in the input image module and measuring the diameter of nuclei (i.e., the primary object) using the measure length tool.
- Select "IdentifySecondaryObjects" (Figure 7).
- Select the input image (image after math correction), select the input objects (nuclei), and name the objects to be identified (cells). Choose the "propagation" method to identify secondary objects, use the "adaptive" threshold strategy and "Otsu" method with "two classes" minimizing "weighted variance". Select "no smoothing" and a threshold correction factor of 1, lower and upper bounds of 0 and 1, and a regularization factor of 0.05. Do not change any other parameters from the default settings.
- Select "MeasureObjectSizeShape" (Figure 8).
- Select cells (secondary object) and nuclei (primary object) as the objects to be measured.
- Select "FilterObjects" (Figure 9).
- Name the output objects and select the nuclei (primary object) as the object to filter.
- Keep the next two parameters as per the default settings. Select "AreaShape" as the measurement to filter by category, and "FormFactor" as the measurement.
- Select "Yes" to filter using a minimum measurement value and add a minimum value of 0.52.
- Select "No" to filter using a maximum measurement.
- Select "Yes" to retain the outlines of filtered objects and name the outlined image.
- Select "ExportToSpreadsheet" (Figure 10)
- Select where to save the file and name "Output Files".
- After the analysis pipeline is generated (Steps 6.7.1 to 6.7.7.1), start "Test Mode" in CellProfiler and evaluate each step, including that the nuclei in the test image are appropriately filtered, to ensure optimal parameters were chosen to identify cells. Figure 11 shows an output image from CellProfiler following filtering, where nuclei are correctly identified (circled in green), and the background is filtered out.
- Once parameters are evaluated and determined to be sufficient, save the project and then click "Analyze Images". This project can be used repeatedly for future analysis. Note: Once parameters have been established, the program is set to analyze all the images in the "File List", sequentially. This will result in multiple windows opening for each image analyzed, which causes a longer processing time.
- To avoid a longer processing time, click the eye icon on all of the modules except "ExportToSpreadsheet" under the "Analysis modules" section.
- Once all the surrogate images have been processed by CellProfiler, open the spreadsheet containing the image data generated by CellProfiler and the spreadsheet containing the area measured with ImageJ. Copy the filtered nuclei data (column D in the CellProfiler spreadsheet) and the image identifiers (column R) and paste them into the spreadsheet containing the measured area data.
- Calculate the sum of all of the measurements obtained for the number of nuclei per image of the surrogate.
- Calculate the sum of the measurements obtained for surrogate area from each image of the surrogate. Divide the total area measured by 1x106.
- Divide the total number of nuclei by the total area measurement in the step above to obtain a value for the number of cells per 1x106 pixels2.
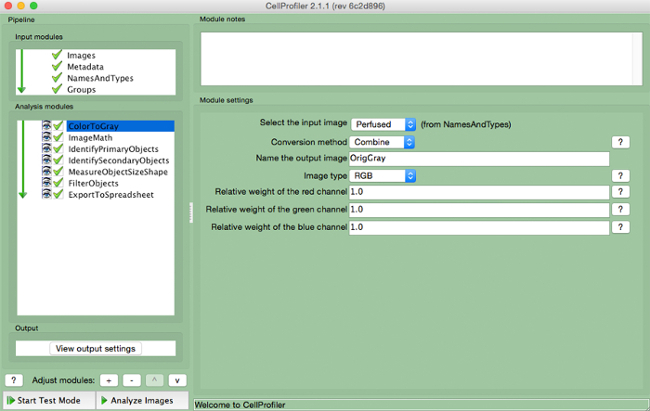 Figure 4. CellProfiler pipeline: changing image to grayscale. Screenshot of "ColortoGray "module. Please click here to view a larger version of this figure.
Figure 4. CellProfiler pipeline: changing image to grayscale. Screenshot of "ColortoGray "module. Please click here to view a larger version of this figure.
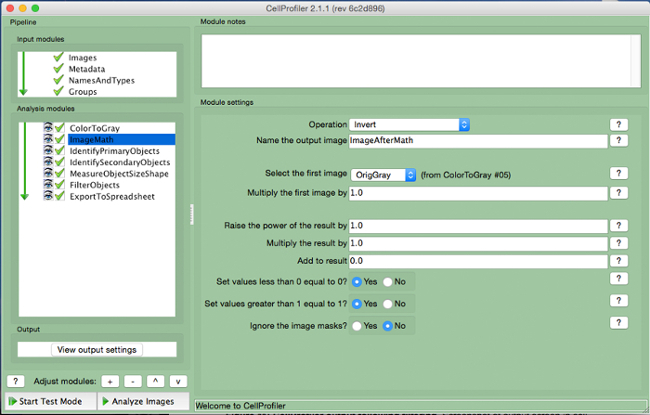 Figure 5. CellProfiler pipeline: inverting image. Screenshot of "ImageMath" module. Please click here to view a larger version of this figure.
Figure 5. CellProfiler pipeline: inverting image. Screenshot of "ImageMath" module. Please click here to view a larger version of this figure.
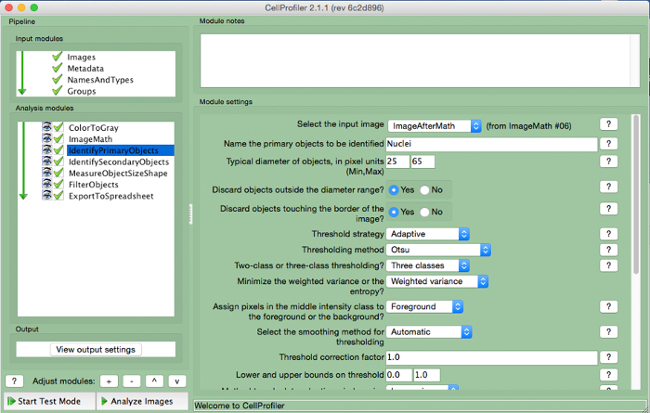 Figure 6. CellProfiler pipeline: identifying nuclei. Screenshot of "IdentifyPrimaryObjects" module. Please click here to view a larger version of this figure.
Figure 6. CellProfiler pipeline: identifying nuclei. Screenshot of "IdentifyPrimaryObjects" module. Please click here to view a larger version of this figure.
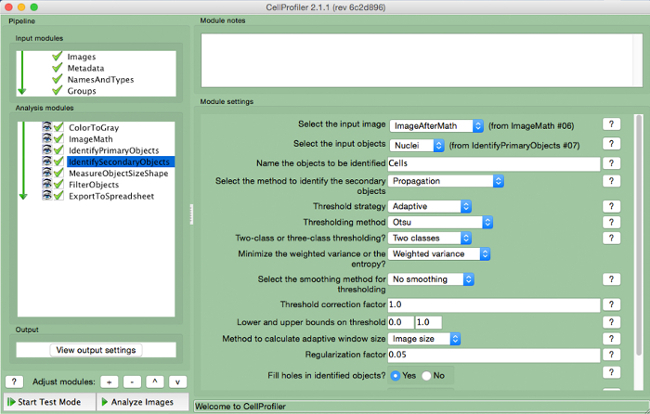 Figure 7. CellProfiler pipeline: identifying cells. Screenshot of "IdentifySecondaryObjects" module. Please click here to view a larger version of this figure.
Figure 7. CellProfiler pipeline: identifying cells. Screenshot of "IdentifySecondaryObjects" module. Please click here to view a larger version of this figure.
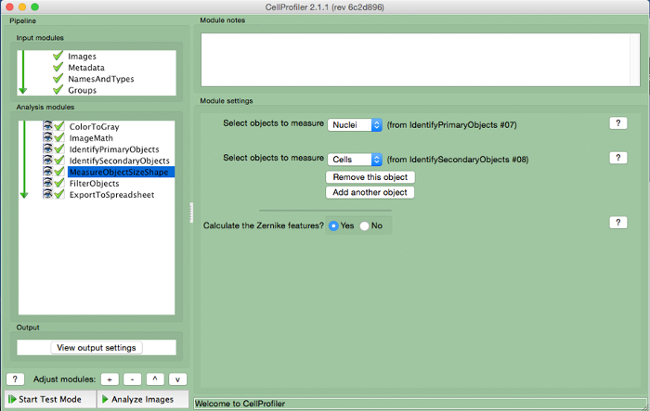 Figure 8. CellProfiler pipeline: measuring objects. Screenshot of "MeasureObjectSizeShape" module. Please click here to view a larger version of this figure.
Figure 8. CellProfiler pipeline: measuring objects. Screenshot of "MeasureObjectSizeShape" module. Please click here to view a larger version of this figure.
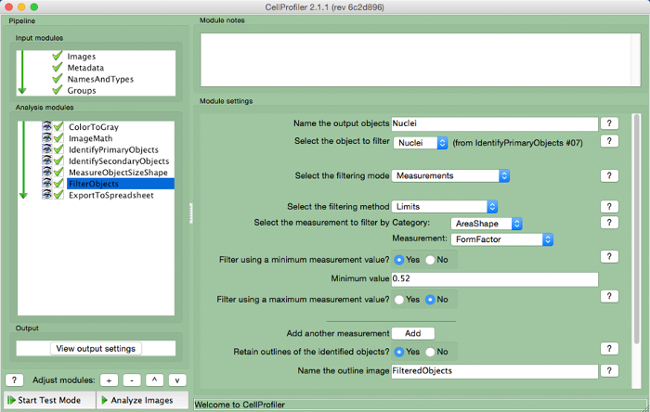 Figure 9. CellProfiler pipeline: filtering objects. Screenshot of "FilterObjects" module. Please click here to view a larger version of this figure.
Figure 9. CellProfiler pipeline: filtering objects. Screenshot of "FilterObjects" module. Please click here to view a larger version of this figure.
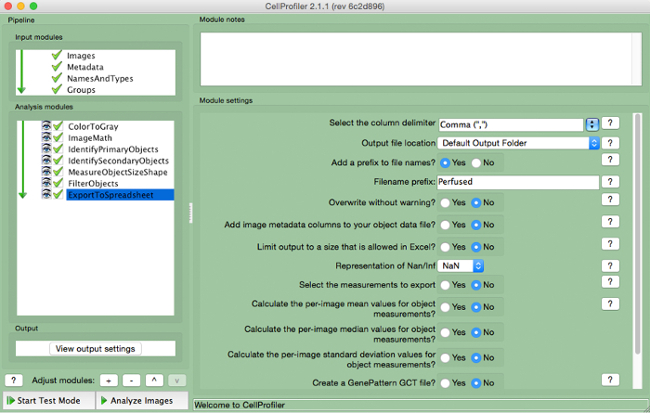 Figure 10. CellProfiler pipeline: exporting data. Screenshot of "ExportToSpreadsheet" module. Please click here to view a larger version of this figure.
Figure 10. CellProfiler pipeline: exporting data. Screenshot of "ExportToSpreadsheet" module. Please click here to view a larger version of this figure.
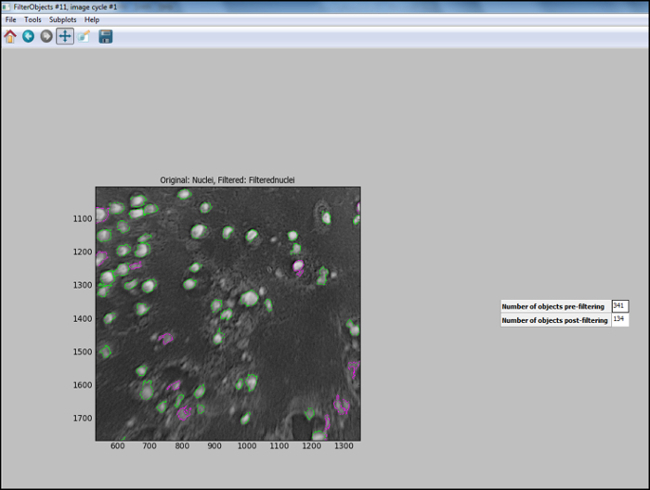 Figure 11. CellProfiler output following filtering. Screenshot of output screen in cell profiler following object filtering. Please click here to view a larger version of this figure.
Figure 11. CellProfiler output following filtering. Screenshot of output screen in cell profiler following object filtering. Please click here to view a larger version of this figure.
Representative Results
Both solid and perfused 3D breast cancer surrogates were prepared as described above and grown for 7 days. Subsequently, surrogates were fixed, processed to paraffin, sectioned, and stained with hematoxylin and eosin, as described above. The number of nucleated cells per area (both 231 cells and CAF) of each surrogate was measured. As can be seen in Figure 12, representative photomicrographs of the H&E-stained sections demonstrate a higher concentration of cells present in the perfused surrogates (Figure 12A) compared to the solid surrogates (Figure 12B), even though the density of cells initially incorporated into the surrogates was the same in both solid and perfused conditions. This visual representation of cell growth is supported by a higher mean number of cells per area (density) in the perfused surrogates (n=3) compared to the solid surrogates (n=3), as determined by the CellProfiler analysis (Figure 12C). To obtain the data in Figure 12C, a complete histologic cross-section of each surrogate was imaged, requiring multiple images for each surrogate, and the images were analyzed as described above. Then, the total number of nuclei for each surrogate was divided by the total area of the surrogate (expressed as the number of nuclei/1 x 106 pixels2), providing a cell density for each surrogate. To validate the use of CellProfiler for nucleated cell quantification, the results using the CellProfiler program were compared with those obtained by simply manually counting the number of cells present per area in each of the 6 surrogates (Table 3). The cell density of each surrogate was calculated as described above. No significant difference was found in the cell densities obtained by manual counting and CellProfiler analysis for either perfused surrogates (p=0.855) or solid surrogates (p=0.553). To support that the difference in cell density between the solid and perfused surrogates correlates with a difference in cell growth, cell proliferation of each surrogate was evaluated via immunohistochemical analysis of Ki-67 labeling (Figure 12D)27. The same trend was found, with a significantly higher percentage of proliferating cells in the perfused surrogates (n=3) compared to the solid surrogates (n=3), indicating a higher growth rate in the perfused surrogates and correlating with the increased cell density of these surrogates. While the cell type (i.e., breast cancer epithelial or CAF) was not evaluated in these measurements, immunostaining for cell type-specific markers could be used to better understand the growth or distribution of a specific cell population. Protocols detailing this process have been previously published 25,28.
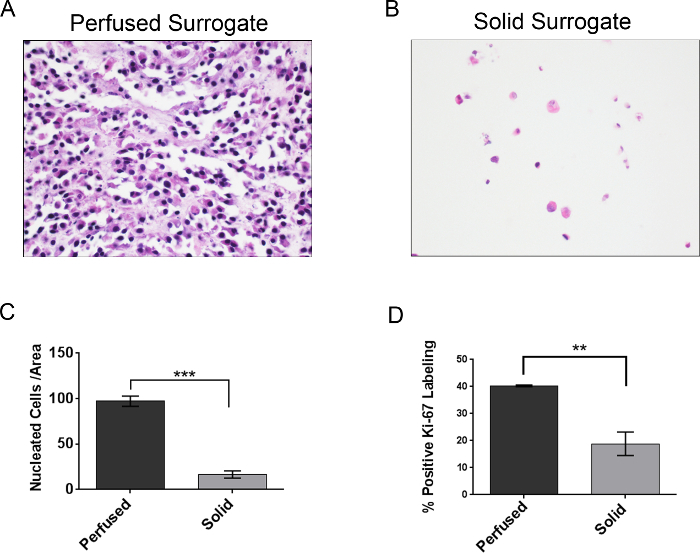 Figure 12. Representative Results. A) Representative image of a H&E-stained section from a perfused surrogate grown for 7 days (400X original magnification). B) Representative image of a H&E-stained section from a solid surrogate grown for 7 days with medium changed every 2 days (400X original magnification). C) Comparison of the mean number of nucleated cells per area, or cell density, following 7 days growth of perfused (no change of media) and solid surrogates (media changed every 2 days). D) Comparison of Ki-67 labeling index (i.e., the percentage of cells in a population that express Ki-67), a measure of cell proliferation, following 7 days growth of perfused and solid surrogates. Data represent the mean ± SEM, n=3 per condition, ** indicates p≤0.01 and *** indicates p≤0.001 (Student's t-test). Please click here to view a larger version of this figure.
Figure 12. Representative Results. A) Representative image of a H&E-stained section from a perfused surrogate grown for 7 days (400X original magnification). B) Representative image of a H&E-stained section from a solid surrogate grown for 7 days with medium changed every 2 days (400X original magnification). C) Comparison of the mean number of nucleated cells per area, or cell density, following 7 days growth of perfused (no change of media) and solid surrogates (media changed every 2 days). D) Comparison of Ki-67 labeling index (i.e., the percentage of cells in a population that express Ki-67), a measure of cell proliferation, following 7 days growth of perfused and solid surrogates. Data represent the mean ± SEM, n=3 per condition, ** indicates p≤0.01 and *** indicates p≤0.001 (Student's t-test). Please click here to view a larger version of this figure.
| Experiment | CellProfiler: nucleated cells per area | Manual: nucleated cells per area | Average CellProfiler | Average Manual | P value (Unpaired t Test) |
| Perfused Surrogate 1 | 88.178 | 75.532 | 97.225 | 99.901 | 0.855 |
| Perfused Surrogate 2 | 107.528 | 117.812 | |||
| Perfused Surrogate 3 | 95.967 | 106.359 | |||
| Solid Surrogate 1 | 19.797 | 17.480 | 16.491 | 12.991 | 0.553 |
| Solid Surrogate 2 | 8.612 | 5.650 | |||
| Solid Surrogate 3 | 21.065 | 15.844 |
Table 3. Cell densities obtained by manual or CellProfiler analysis for 3 solid and 3 perfused surrogates.
Discussion
Herein, a method of 3D culture has been described that incorporates components of the tissue microenvironment, including the extracellular matrix (ECM) and human stromal fibroblasts, in a volume that more closely models human breast cancer to allow for the development of a recapitulative 3D morphology. The 3D culture method described is more representative of human disease than traditional 2D cell culture in that multiple cell types are incorporated into a 3D volume of ECM. It has been noted that these parameters (i.e., multiple cell types, ECM, and 3D volume) provide a more appropriated context to study biological processes because tissue architecture, signals from the microenvironment, and dimensionality are being taken into account 1,8. Other methods of 3D culture have been previously described including, the culture of cells on top of a 3D matrix29, the generation of spheroids30, hanging drop cutlures31, and the use of microfluidic devices32. While each of these systems has unique advantages, overall they fail to include either matrix components, stromal cell populations, or a representative dimensionality. The method of 3D culture described here includes each of these parameters and can be established using a bioreactor system to allow for longer growth periods that are difficult to achieve using solid 3D culture. In addition, perfusion of surrogates provides a greater degree of growth compared with solid surrogates.
Another feature of this approach for in vitro culture is the flexibility in the types of analyses that can be performed using the surrogate "tissues". A variety of endpoints can be monitored indirectly during culture by evaluating the conditioned media, or perfusate, for soluble products (e.g., LDH as an indicator of cell death, specific secreted proteins or metabolites as an indicator of appropriate phenotypic function) or by utilizing non-invasive imaging of fluorescently or luminescently labeled cells to evaluate growth and surrogate architecture overtime. Following growth, the ECM can be digested using an ECM protease and cells can be isolated and used for additional analysis or further experiments, such as flow cytometric quantification33. Lysates of the cells removed from the ECM could also be evaluated genomically, at the mRNA transcript level, or for protein expression. Alternatively, endpoints can be measured directly from the fixed and processed surrogate "tissues" after the experiment has concluded. These include a variety of morphological parameters (e.g., nuclear/cell morphology and the degree of cell aggregation) on H&E-stained histologic sections and other molecular analyses performed on histologic sections using immunohistochemistry (e.g., expression of Ki-67 as an indicator of cell proliferation) or in situ hybridization (e.g., RNA expression of specific gene products). It is also possible to perform additional molecular analyses, such as real-time, quantitative PCR or next generation sequencing, on these formalin-fixed, paraffin-embedded surrogates; however, due to molecular cross-linking induced by fixation and processing, snap-frozen surrogates are preferred for these types of analyses. Such molecular and morphologic evaluations can provide valuable information regarding cell growth, death, or response to pharmacological treatment throughout growth as well as following culture.
The main advantage of using the CellProfiler program for semi-automated quantification of the density of nucleated cells present in histologic sections of surrogates is the time savings that it provides. Although the total amount of time required per surrogate is variable and depends largely on the size of the surrogates and the number of images acquired, it is estimated that arriving at a cell density from the surrogate images requires 4-5 hours of hands-on time per surrogate when manually counting versus half an hour per surrogate when using the CellProfiler program. The majority of the hands-on-time using CellProfiler analysis is attributed to the measurement of the surrogate area in ImageJ. The CellProfiler program does require time to automatically process the images, but does not require hands-on time from the researcher other than importing the images.
There are limitations of the methods and the 3D surrogate cultures described here. As shown in Figure 12, the use of the perfusion bioreactor system provided a greater degree of growth than solid cultures, in which the accumulation of cells and the rate of proliferation is considerably slower. Although the perfusion bioreactor system used here provided a growth advantage, perfusion systems can also have their own limitations. For example, PDMS is often used in the manufacture of bioreactors due to its ease of use, inert properties, and reproducibility. However, this material is known to absorb hydrophobic molecules from cell culture media and may take up lipophilic drugs34-36. While other materials can be used instead of PDMS if these limitations are of concern, other materials available also come with their own drawbacks that must be considered. Because CellProfiler analysis is dependent on the parameters input for the diameter and circularity of the nuclei, optimization of these inputs for each surrogate/cell type may be necessary. It is also of note that the cell density measurement presented here is not a direct measurement of cell viability. It has previously been noted that a direct measurement of cell viability is challenging in 3D culture systems37. However, nuclear degradation is a part of both apoptosis and necrosis, the two predominate forms of cell death. In apoptosis, the nucleus fragments, a process called karyorrhexis, following chromatin condensation, or pyknosis38,39. This differs from necrosis, where the dying cell swells causing the cell membrane to rupture and release the contents of its cytoplasm. Histologically, necrosis is characterized by karyolysis (i.e., dissolution of a nucleus), followed by pyknosis and karyorrhexis40. Nuclei that have undergone these changes can be seen on H&E-stained histologic sections but will not be recognized as intact nuclei by the CellProfiler program. These nuclear changes occur relatively late in the process of cell death however. Therefore, quantifying the number of nucleated cells per area may include cells that are in the earlier stages of cell death, but will nonetheless provide useful information regarding cell viability at the time of surrogate fixation. Other methods that could be used to indicate viability or growth include analyses of perfusates (e.g., Alamar Blue or LDH assays) or evaluation of proliferation in the surrogate "tissues" (e.g., via Ki-67 labeling by immunohistochemistry).
Critical steps in the protocol for surrogate setup include ECM polymerization, where temperature, pH, and time all must be monitored to ensure complete polymerization prior to bathing the surrogate in medium. In addition, keeping the cell+ECM mixture on ice prior to plating in an 8-well chamber slide or injection into a bioreactor system is crucial to preventing premature polymerization. Also to prevent polymerization during surrogate setup, the sodium bicarbonate used to increase the pH to 7 should be added to the mixture last. Finally, adequate incubation time at 37 °C must be given to ensure complete ECM polymerization; 45 minutes has proven to be sufficient. Another important factor in surrogate preparation is the avoidance of bubble formation. In both the solid and perfused systems, it is best to avoid the formation of bubbles in the cell+ECM mixture as to not create a block to the flow of nutrients during culture. It is also important to avoid bubbles during medium introduction in the perfusion system where a bubble may function as an air embolism, blocking medium flow.
Proper fixation is important for downstream analysis; therefore, attention to standardized and consistent fixation (including the type of fixation and the amount of time in fixative) and processing is necessary to obtain replicative results. The duration and type of fixation is less of an issue for the H&E staining described here, but these parameters may affect other types of molecular analysis of the surrogates. This is because formalin fixation, in particular, causes the cross-linking of molecules, including protein, DNA, and RNA in a somewhat time-dependent and molecule-specific manner41,42. This is of particular importance in immunohistochemical analysis, when antibody recognition may be hindered by cross-linking.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
The University of Alabama at Birmingham Center for Metabolic Bone Disease performed the histologic processing and sectioning of surrogates. Southern Research (Birmingham, AL) provided support for the manufacture of the bioreactor system. Funding was provided by the United States Department of Defense Breast Cancer Research Program (BC121367).
References
- Hakanson M, Textor M, Charnley M. Engineered 3D environments to elucidate the effect of environmental parameters on drug response in cancer. Integr Biol (Camb) 2011;3(1):31–38. doi: 10.1039/c0ib00074d. [DOI] [PubMed] [Google Scholar]
- Horning JL, et al. 3-D tumor model for in vitro evaluation of anticancer drugs. Mol Pharm. 2008;5(5):849–862. doi: 10.1021/mp800047v. [DOI] [PubMed] [Google Scholar]
- Dhiman HK, Ray AR, Panda AK. Three-dimensional chitosan scaffold-based MCF-7 cell culture for the determination of the cytotoxicity of tamoxifen. Biomaterials. 2005;26(9):979–986. doi: 10.1016/j.biomaterials.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Place AE, Jin Huh S, Polyak K. The microenvironment in breast cancer progression: biology and implications for treatment. Breast Cancer Res. 2011;13(6):227. doi: 10.1186/bcr2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y, Keller ET, Garfield DH, Shen K, Wang J. Stromal cells in tumor microenvironment and breast cancer. Cancer Metastasis Rev. 2013;32(1-2):303–315. doi: 10.1007/s10555-012-9415-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsson J, Micke P. Prognostic relevance of cancer-associated fibroblasts in human cancer. Semin Cancer Biol. 2014;25:61–68. doi: 10.1016/j.semcancer.2014.02.006. [DOI] [PubMed] [Google Scholar]
- Roskelley CD, Desprez PY, Bissell MJ. Extracellular matrix-dependent tissue-specific gene expression in mammary epithelial cells requires both physical and biochemical signal transduction. Proc. Natl. Acad. Sci. 1994;91:12378–12382. doi: 10.1073/pnas.91.26.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickl M, Ries CH. Comparison of 3D and 2D tumor models reveals enhanced HER2 activation in 3D associated with an increased response to trastuzumab. Oncogene. 2008;28(3):461–468. doi: 10.1038/onc.2008.394. [DOI] [PubMed] [Google Scholar]
- Ivascu A, Kubbies M. Rapid generation of single-tumor spheroids for high-throughput cell function and toxicity analysis. J Biomol Screen. 2006;11(8):922–932. doi: 10.1177/1087057106292763. [DOI] [PubMed] [Google Scholar]
- Lovitt CJ, Shelper TB, Avery VM. Advanced cell culture techniques for cancer drug discovery. Biology (Basel) 2014;3(2):345–367. doi: 10.3390/biology3020345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergamaschi A, et al. Extracellular matrix signature identifies breast cancer subgroups with different clinical outcome. J Pathol. 2008;214(3):357–367. doi: 10.1002/path.2278. [DOI] [PubMed] [Google Scholar]
- Oskarsson T. Extracellular matrix components in breast cancer progression and metastasis. The Breast. 2013;22:S66–S72. doi: 10.1016/j.breast.2013.07.012. [DOI] [PubMed] [Google Scholar]
- Kelley LC, Lohmer LL, Hagedorn EJ, Sherwood DR. Traversing the basement membrane in vivo: A diversity of strategies. JBC. 2014;204(3):291–301. doi: 10.1083/jcb.201311112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadlonova A, et al. Breast fibroblasts modulate epithelial cell proliferation in three-dimensional in vitro co-culture. Breast Cancer Res. 2004;4 doi: 10.1186/bcr949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt D, Marsano A, Jakob M, Heberer M, Martin I. Oscillating perfusion of cell suspensions through three-dimensional scaffolds enhances cell seeding efficiency and uniformity. Biotechnol Bioeng. 2003;84(2):205–214. doi: 10.1002/bit.10759. [DOI] [PubMed] [Google Scholar]
- Marshall LE, et al. Flow-perfusion bioreactor system for engineered breast cancer surrogates to be used in preclinical testing. J Tissue Eng Regen Med. 2015. [DOI] [PMC free article] [PubMed]
- Calcagnile P, Fragouli D, Mele E, Ruffilli R, Athanassiou A. Polymeric foams with functional nanocomposite cells. RSC Adv. 2014;4:19177–19182. [Google Scholar]
- Naba A, Clauser KR, Lamar JM, Carr SA, Hynes RO. Extracellular matrix signatures of human mammary carcinoma identify novel metastasis promoters. eLife. 2014;3 doi: 10.7554/eLife.01308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochter A, Bissell MJ. Involvement of extracellular matrix constituents in breast cancer. Semin Cancer Biol. 1995;6(3):165–173. doi: 10.1006/scbi.1995.0017. [DOI] [PubMed] [Google Scholar]
- Joiner KS, Spangler EA. Evaluation of HistoGel-embedded specimens for use in veterinary diagnostic pathology. J Vet Diagn Invest. 2012;24(4):710–715. doi: 10.1177/1040638712445763. [DOI] [PubMed] [Google Scholar]
- Varsegi GM, Shidham V. Cell Block Preparation from Cytology Specimen with Predominance of Individually Scattered Cells. J Vis Exp. 2009. p. e1316. [DOI] [PMC free article] [PubMed]
- Sadlonova A, et al. Human Breast Fibroblasts Inhibit Growth of the MCF10AT Xenograft Model of Proliferative Breast Disease. Am J Pathol. 2007;170(3) doi: 10.2353/ajpath.2007.060031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otali D, He Q, Stockard CR, Grizzle WE. Preservation of immunorecognition by transferring cells from 10% neutral buffered formalin to 70% ethanol. Biotech Histochem. 2013;88(0):170–180. doi: 10.3109/10520295.2012.754496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster SS, Jenkins L, Burg KJL. Histological Techniques for Porous, Absorbable, Polymeric Scaffolds, Used in Tissue Engineering. J Histotechnol. 2003;26(1):57–65. [Google Scholar]
- Troy T-C, Arabzadeh A, Enikanolaiye A, Lariviere N, Turksen K. Immunohistochemistry on Paraffin Sections of Mouse Epidermis Using Fluorescent Antibodies. J Vis Exp. 2008. [DOI] [PMC free article] [PubMed]
- Carpenter AE, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7(10):R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y-J, et al. Gli1 enhances migration and invasion via up-regulation of MMP-11 and promotes metastasis in ERa negative breast cancer cell lines. Clin Exp Metastasis. 2011. [DOI] [PMC free article] [PubMed]
- Evilsizor MN, Ray-Jones HF, Lifshitz J, Ziebell J. Primer for Immunohistochemistry on Cryosectioned Rat Brain Tissue: Example Staining for Microglia and Neurons. J Vis Exp. 2015. p. e52293. [DOI] [PMC free article] [PubMed]
- Pal A, Kleer CG. Three dimensional cultures: a tool to study normal acinar architecture vs. malignant transformation of breast cells. J Vis Exp. 2014. p. e51311. [DOI] [PMC free article] [PubMed]
- Hasselbach LA, et al. Optimization of High Grade Glioma Cell Culture from Surgical Specimens for Use in Clinically Relevant Animal Models and 3D Immunochemistry. J Vis Exp. 2014. p. e51088. [DOI] [PMC free article] [PubMed]
- Foty R. A Simple Hanging Drop Cell Culture Protocol for Generation of 3D Spheroids. J Vis Exp. 2011. p. e2720. [DOI] [PMC free article] [PubMed]
- Materne E-M, et al. The Multi-organ Chip - A Microfluidic Platform for Long-term Multi-tissue Coculture. J Vis Exp. 2015. p. e52526. [DOI] [PMC free article] [PubMed]
- Sadlonova A, et al. Identification of Molecular Distinctions Between Normal Breast-Associated Fibroblasts and Breast Cancer-Associated Fibroblasts. Cancer Microenviron. 2009;2:9–21. doi: 10.1007/s12307-008-0017-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JD, Douville NJ, Takayama S, ElSayed M. Quantitative analysis of molecular absorption into PDMS microfluidic channels. Ann Biomed Eng. 2012;40(9):1862–1873. doi: 10.1007/s10439-012-0562-z. [DOI] [PubMed] [Google Scholar]
- Halldorsson S, Lucumi E, Gomez-Sjoberg R, Fleming RM. Advantages and challenges of microfluidic cell culture in polydimethylsiloxane devices. Biosens Bioelectron. 2015;63:218–231. doi: 10.1016/j.bios.2014.07.029. [DOI] [PubMed] [Google Scholar]
- Regehr KJ, et al. Biological implications of polydimethylsiloxane-based microfluidic cell culture. Lab Chip. 2009;9(15):2132–2139. doi: 10.1039/b903043c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdett E, Kasper FK, Mikos AG, Ludwig JA. Engineering tumors: a tissue engineering perspective in cancer biology. Tissue Eng Part B Rev. 2010;16(3):351–359. doi: 10.1089/ten.TEB.2009.0676. [DOI] [PubMed] [Google Scholar]
- Caruso RA, et al. Mechanisms of coagulative necrosis in malignant epithelial tumors (Review) Oncol Lett. 2014;8(4):1397–1402. doi: 10.3892/ol.2014.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore S. Apoptosis: A Review of Programmed Cell Death. Toxicol Pathol. 2007;35(4):495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majno G, Joris I. Apoptosis, oncosis, and necrosis. An overview of cell death. Am J Pathol. 1995;146(1):3–15. [PMC free article] [PubMed] [Google Scholar]
- Ogino S, et al. Molecular pathological epidemiology of epigenetics: emerging integrative science to analyze environment, host, and disease. Mod Pathol. 2013;26(4):465–484. doi: 10.1038/modpathol.2012.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otali D, et al. Combined effects of formalin fixation and tissue processing on immunorecognition. Biotech Histochem. 2009;84(5):223–247. doi: 10.3109/10520290903039094. [DOI] [PMC free article] [PubMed] [Google Scholar]


