Abstract
We have developed an inducible system to visualize gene expression at the levels of DNA, RNA and protein in living cells. The system is composed of a 200 copy transgene array integrated into a euchromatic region of chromosome 1 in human U2OS cells. The condensed array is heterochromatic as it is associated with HP1, histone H3 methylated at lysine 9, and several histone methyltransferases. Upon transcriptional induction, HP1α is depleted from the locus and the histone variant H3.3 is deposited suggesting that histone exchange is a mechanism through which heterochromatin is transformed into a transcriptionally active state. RNA levels at the transcription site increase immediately after the induction of transcription and the rate of synthesis slows over time. Using this system, we are able to correlate changes in chromatin structure with the progression of transcriptional activation allowing us to obtain a real-time integrative view of gene expression.
Introduction
Genes are expressed as a result of the concerted processes of transcription, pre-mRNA processing, messenger ribonucleoprotein particle (mRNP) export and translation (reviewed, Maniatis and Reed 2002 and Orphanides and Reinberg 2002). Genetic and biochemical analyses have identified a large number of factors required for the execution of these processes (reviewed, Lemon and Tjian 2000 and Rappsilber et al. 2002), but how their functions are spatially and temporally coordinated is not well understood. Additionally, the transcriptional status of a gene is tightly linked to the structure of its chromatin, but how chromatin proteins are organized and how their dynamics change during the induction of transcription is not well characterized within the context of the living cell.
Efforts to label specific regions of chromatin in vivo have utilized interactions between DNA binding proteins and their target sequences. The introduction of bacterial lac operator repeats into the genomes of eukaryotic cells and expression of a green fluorescent protein (GFP) lac repressor fusion protein is a noninvasive means of identifying and studying specific regions of chromatin (reviewed, Belmont 2001 and Janicki and Spector 2003). Using this approach, the large-scale unfolding of a chromatin structure induced by the VP16 acidic activation domain (Tumbar et al., 1999) and the induction of a tetracycline regulatable array of transcription units (Tsukamoto et al., 2000) have been visualized in living cells. Additionally, the affinity of a GFP glucocorticoid receptor fusion protein for a tandem array of the mouse mammary tumor virus (MMTV) driving a ras reporter was used not only to identify a region of transcriptionally active chromatin in living cells but also to show that transcriptional activators assemble into dynamic complexes at transcription sites McNally et al. 2000 and Muller et al. 2001.
Gene expression is initiated by transcriptional activators that recruit both ATP-dependent nucleosome remodeling complexes and enzymes that posttranslationally modify histone tail domains (reviewed, Emerson 2002 and Jenuwein and Allis 2001). The covalent modification of histone tails (i.e., acetylation, phosphorylation, ubiquitylation, and methylation) is believed to create a “code” that regulates the transcriptional status of chromatin by both modulating nucleosomal structure and promoting and/or preventing the binding of regulatory factors (reviewed, Fischle et al. 2003, Jenuwein and Allis 2001 and Strahl and Allis 2000). For example, methylation of histone H3 at lysine 9 (H3 K9), a modification associated with silent chromatin, creates a binding site for the chromodomain (Jacobs and Khorasanizadeh, 2002) of heterochromatin protein 1 (HP1) Bannister et al. 2001, Jacobs et al. 2001 and Lachner et al. 2001. Lysine residues in histone tails can be either mono-, di- or trimethylated (reviewed, Fischle et al., 2003) and a number of histone H3 K9 methyltransferases (HMTases) have been identified (reviewed, Marmorstein, 2003).
As biochemical analyses of histone lysine methylation suggest that the modification is permanent in nature and a histone demethylase has not yet been identified (reviewed, Bannister et al. 2002 and Jenuwein and Allis 2001), it is not clear how the H3 K9 methyl mark is removed during gene activation. Of particular interest in this regard is the histone H3 variant, H3.3, which has been shown to deposit in active ribosomal DNA genes in a replication-independent manner (Ahmad and Henikoff, 2002). Therefore, histone exchange may be a general mechanism through which histone methylation is removed during transitions in the transcriptional state of chromatin.
The direct readout of a gene template is produced by the synthesis of the encoded mRNA. Although specific mRNAs can be localized to their transcription sites by RNA fluorescence in situ hybridization (RNA FISH) (reviewed, Huang and Spector, 1997), the kinetics of RNA synthesis at a specific transcription site in a single living cell have not been evaluated. In order to track the movement of specific RNAs in living cells, several studies have utilized the MS2 bacteriophage viral replicase translational operator (a 19 nucleotide RNA stem loop) and a GFP MS2 coat fusion protein Bertrand et al. 1998, Forrest and Gavis 2003, Fusco et al. 2003 and Rook et al. 2000. The GFP-MS2 coat protein binds to the stem loop of the translational operator as a dimer (Beckett and Uhlenbeck, 1988) and allows the RNA to be visualized. This labeling approach was used to track the asymmetrical movement of ASH1 mRNA in dividing yeast cells (Bertrand et al., 1998), the movement of RNA particles in the cytoplasm of hippocampal neurons and COS cells Fusco et al. 2003 and Rook et al. 2000 and the dynamic localization of endogenous nanos RNA in Drosophila oocytes (Forrest and Gavis, 2003). Thus far, it has not been used to examine the dynamics of mRNA synthesis at a single specific transcription site.
We have developed a cell line that allows us to investigate how gene expression events are coordinated spatially and temporally in vivo. By combining the lac operator/lac repressor, tetracycline inducible, and MS2 translational operator/coat protein systems, we are able to visualize DNA, RNA, and protein in living cells. Using this system, we show that prior to transcriptional activation a stably integrated 200 copy array of inducible transcription units is highly condensed and heterochromatic and we characterize the dynamic loss of HP1α and the recruitment of the histone variant H3.3 during the induction of transcription. Furthermore, we correlate changes in chromatin structure to RNA synthesis and the recruitment of components of the gene expression machinery. This system allows us to obtain a real-time integrative view of gene expression by simultaneously following the dynamics of a specific region of chromatin and its RNA and protein products in living cells.
Results
Development of a Cell Line to Study Gene Expression in Living Cells
In order to study the dynamics of gene expression in vivo, we constructed a plasmid, containing a transcription unit, that allows the DNA, RNA, and translated protein product to be directly visualized in living cells (Figure 1A). This plasmid, based upon one previously generated in our laboratory (Tsukamoto et al., 2000), was stably integrated into human U2OS cells. 256 copies of lac operator sequence are present at the 5′ end of the construct such that when the lac repressor binding protein is expressed as a cyan fluorescent protein (CFP) or a yellow fluorescent protein (YFP) fusion protein (CFP-lac repressor/YFP-lac repressor), the integration site/transgene array can be visualized. Immediately downstream, 96 copies of the tetracycline response element (TRE) allow the transcription units to be regulated. When the tetracycline-controlled transactivator (rtTA) (rTetR fused to the VP16 activation domain; pTet-On), is expressed in the presence of doxycycline (dox), it binds to the TREs and activates transcription from the minimal CMV promoter.
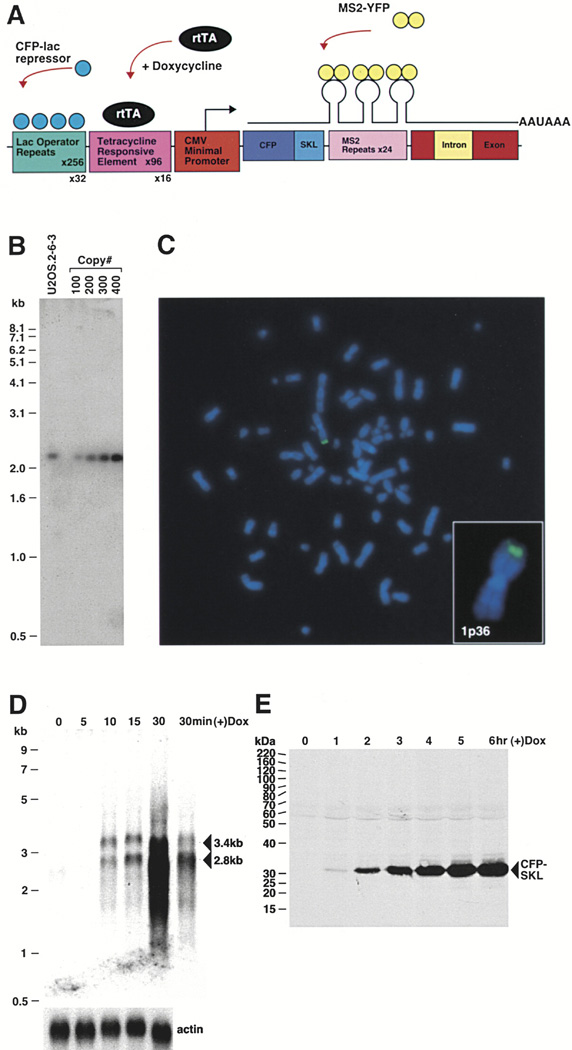
Figure 1. Characterization of Human U2OS 2-6-3 Cells.
(A) Schematic representation of the gene expression plasmid, p3216PECMS2β. The plasmid is composed of 256 copies of the lac operator, 96 tetracycline response elements, a minimal CMV promoter, CFP fused to the peroxisomal targeting signal SKL, 24 MS2 translational operators (MS2 repeats), a rabbit β-globin intron/exon module, and a cleavage/polyadenylation signal. Expression of CFP-lac repressor allows the DNA to be visualized and expression of pTet-On (rtTA) in the presence of doxycycline (dox) drives expression from the CMV minimal promoter. When MS2-YFP (YFP fused to the MS2 coat protein) dimerizes and interacts with the stem loop structure of the translational operator, it allows the transcribed RNA to be visualized.
(B) Quantitative Southern blot of clone 2-6-3 genomic DNA. A 2.4 kb fragment is produced when clone 2-6-3 genomic DNA and p3216PECMS2β are digested with NcoI which cuts at the 5′ end of CFP and within the β-globin intron. Comparison of known quantities of plasmid DNA equal to 100, 200, 300, and 400 copies per cell showed that 2-6-3 cells contain ~200 stably integrated copies of p3216PECMS2β.
(C) DNA fluorescence in situ hybridization (DNA FISH) of 2-6-3 cells shows that there is a single integration site in the euchromatic region of chromosome 1p36.
(D) Northern blot time course analysis of RNA isolated 0, 5, 10, 15, and 30 min after the induction of transcription. The last lane shows a lighter exposure of the 30 min time point. Pre-mRNA transcripts run at 3.4 kb and spliced mRNA at 2.8 kb. The probe recognizes the MS2 repeats. Actin was probed as a loading control.
(E) Immunoblot time course analysis of CFP-SKL expression 0, 1, 2, 3, 4, 5, and 6 hr after the addition of doxycycline.
The transcribed RNA encodes CFP with peroxisomal targeting signal-1 [Ser-Lys-Leu (SKL)] fused to its carboxyl terminus, 24 tandem MS2 translational operators (MS2 repeats), a β-globin intron/exon module, and a cleavage/polyadenylation signal (Figure 1A). Expression of the MS2 coat-YFP fusion protein (MS2-YFP) allows the transcribed RNA to be visualized and the intron/exon module and cleavage/polyadenylation signal allow the recruitment of the pre-mRNA processing machinery to the transcription site to be studied. When the mRNA is exported from the nucleus and translated in the cytoplasm, the CFP-SKL protein product is targeted to peroxisomes and serves as a reporter confirming that all processes required for gene expression have been successfully completed.
The 20 kb gene expression plasmid (p3216PECMS2β) was cotransfected with the hygromycin resistance plasmid, pTK-Hyg, into U2OS cells and drug resistant colonies were isolated and screened for the presence of a single integration site (visualized by the transient expression of YFP-lac repressor) and the ability to be transcriptionally activated by rtTA expression (+) dox (determined by the presence of CFP-labeled peroxisomes in the cytoplasm). Cell line U2OS 2-6-3 was selected for further characterization because of its easily detectable integration site and the high proportion of transcriptionally activated cells.
In order to determine the relative copy number of the integrated plasmid, genomic DNA from U2OS 2-6-3 cells was isolated and used for Southern blotting. This analysis showed that cell line U2OS 2-6-3 contains ~200 copies of p3216PECMS2β (Figure 1B).
As the chromosomal location of transgene loci has been shown to influence their expression as a result of position effects (Spradling and Rubin, 1983), we were interested in determining the chromosomal integration site of the p3216PECMS2β transgene. DNA fluorescence in situ hybridization (DNA FISH) showed that there is a single integration site in an euchromatic region of chromosome 1p36 (Figure 1C).
In order to characterize the induction of RNA synthesis upon transcriptional activation, Northern blot analysis was performed after a time-course induction. U2OS 2-6-3 cells were transiently transfected with pTet-On, dox was added 2.5 hr posttransfection and RNA was isolated 0, 5, 10, 15, and 30 min later (Figure 1D). RNA was first detected 10 min after the addition of dox with levels increasing dramatically between the 15 and 30 min time points. The fact that RNA transcripts were not detected at 0 min demonstrates that transcription is not “leaky” when pTet-On is expressed in the absence of dox.
At the 10 min time point, there was slightly more unspliced RNA compared to spliced mRNA (1.0: 0.9375) (Figure 1D). Over time, the spliced product predominated (15 min, 1.3-fold increase; 30 min > 3-fold increase). As the level of the spliced RNA increased dramatically between 15 and 30 min postinduction, this suggests that the changes that occur at the locus between these time points increase the efficiency of both transcription and pre-mRNA processing. This lag time may reflect the time required for the chromatin remodeling machinery to decondense the chromatin and hence make more of the transcription units accessible in addition to the time required for the pre-mRNA processing machinery to be recruited to the transcription site and the splicing reaction to occur. At later time points, both higher and lower molecular weight species of RNA were detected (Supplemental Data, Supplemental Figure S1 available at http://www.cell.com/cgi/content/full/116/5/683/DC1; 120 min) suggesting that aberrant products may be produced when transcription is elevated to high levels from a strong promoter.
Immunoblotting of protein lysates collected between 0 and 6 hr after the induction of transcription showed that the CFP-SKL protein can be detected at the 1 hr time point with expression levels increasing steadily over time (Figure 1E).
Visualization of Gene Expression and the Recruitment of the Gene Expression Machinery to a Transcription Site
To simultaneously analyze chromatin and RNA dynamics at a transcription site in individual cells, U2OS 2-6-3 cells were transiently cotransfected with CFP-lac repressor, rtTA and MS2-YFP and visualized 2.5 hr posttransfection. In the absence of dox, the chromatin of the transgene locus, marked by CFP-lac repressor, appears as a highly condensed small dot (Figure 2A) (0.89 ± 0.12 µm in diameter; n = 13) that is DAPI dense (data not shown). MS2-YFP is diffusely distributed in a smooth pattern throughout the nucleoplasm without any particular enrichment at the locus (Figure 2B).
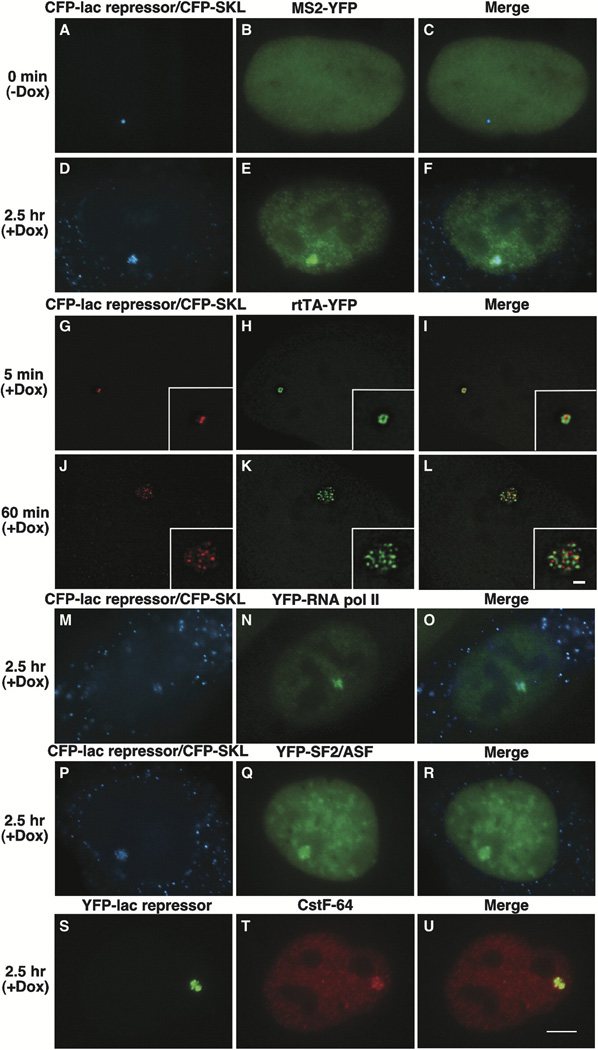
Figure 2. Visualization of DNA, RNA, and Protein in Living Cells.
(A–F) U2OS 2-6-3 cells were transiently transfected with pSV2-CFP-lac repressor, pTet-ON (rtTA) and MS2-YFP, and imaging was begun 2.5 hr posttransfection.
(A–C) At 0 min (−) dox, CFP-lac repressor marks the locus (A) and MS2-YFP is diffusely distributed throughout the nucleus (B).
(D–F) 2.5 hr after the addition of Dox, the locus is highly decondensed and CFP-SKL is seen in the cytoplasmic peroxisomes (D). MS2-YFP accumulates at the site of the decondensed locus and is present in a particulate pattern throughout the nucleoplasm (E).
(G–L) Image stacks of cells expressing pSV2-CFP-lac repressor (pseudocolored red) and EYFP-rtTA-N1 (pseudocolored green) were collected and deconvolved in cells fixed 5 min (G–I) and 60 min (J–L) after the induction of transcription. Single sections from deconvolved stacks are shown.
(M–U) Factors involved in gene expression colocalize with the decondensed locus. YFP-RNA polymerase II (M–O), YFP-SF2/ASF (P–R), and Cstf64 (S–U) are present at the active locus. Scale bar is equal to 5 µm. Scale bar in enlarged insets is equal to 1 µm.
Interestingly, mRNA transcripts from the constitutively active hygromycin resistance plasmid (pTK-Hyg) were detected at the condensed locus by RNA FISH (Supplemental Data, Supplemental Figure S2 available on Cell website) confirming that pTK-Hyg integrated into the array with the gene expression plasmid (p3216PECMS2β). As pTK-Hyg transcripts were generally on the periphery of the locus, it is possible that it integrated throughout the array but that only those copies accessible to the transcription machinery are transcribed. Although pTK-Hyg is constitutively active, the locus is condensed when p3216PECMS2β is inactive suggesting that the higher order structure of the array is dictated by the transcriptional state of the construct in the higher proportion (9:1).
When transcription is induced by the addition of dox to the medium and cells examined 2.5 hr later, the locus appears decondensed and CFP-SKL is localized to the cytoplasmic peroxisomes (Figure 2D). Decondensed loci range in size from ~2.0–4.5 µm in diameter with the variability likely resulting from the transient expression of rtTA. Additionally, MS2-YFP concentrates at the decondensed locus and is seen in a particulate pattern throughout the nucleoplasm that is consistent with the transcribed RNAs being packaged into mRNPs (Figure 2E). RNA in situ hybridization with a probe that recognizes sequence in the β-globin intron confirmed that nascent transcripts are present at the site of the decondensed locus (Supplemental Data, Supplemental Figure S3 available on Cell website).
In order to visualize the association of the transcriptional activator with the lac repressor, we expressed CFP-lac repressor (pseudocolored red) and rtTA as a YFP fusion protein (pEYFP-rtTA-N1) (pseudocolored green) (Figures 2G–2L) and collected and deconvolved image stacks 5 and 60 min after the addition of dox. Though the locus is still compact 5 min postinduction, (Figures 2G–2I, single sections from deconvolved image stacks), the regions bound by CFP-lac repressor and rtTA-YFP are easily distinguishable and minimally overlapping. In fact, rtTA-YFP only binds to a region of the condensed locus suggesting that a small subset of the repeated templates are transcribed at early time points—a result consistent with the low levels of RNA detected in the northern analysis (Figure 1D). In a cell fixed 60 min postinduction (Figures 2J–2L), the locus is highly decondensed and CFP-lac repressor and rtTA-YFP also appear in distinct foci that partially overlap. As we did not observe 200 foci at the decondensed locus, this suggests that the transcription units assemble into repetitive substructures within the array or that the array does not fully decondense.
We next examined the recruitment of factors required for transcription and pre-mRNA processing (Supplemental Data, Supplemental Figure S4 available on Cell website; immunoblot of constructs). YFP-RNA polymerase II (Sugaya et al., 2000)(Figures 2M–2O), the pre-mRNA splicing factor YFP-SF2/ASF (Misteli et al., 1997)(Figures 2P–2R) and the 3′ end processing factor Cstf64 (Takagaki et al., 1990)(Figures 2S–2U) all colocalized with the decondensed locus confirming that gene expression factors from different nuclear domains are recruited to the transcription site.
The Kinetics of Chromatin Decondensation and RNA Synthesis
We were next interested in evaluating the kinetics of RNA synthesis in individual cells during transcriptional activation. Cells were transiently cotransfected with CFP-lac repressor, MS2-YFP, and rtTA, and 2 hr posttransfection placed in a FCS2 live-cell chamber (37 °C) and visualized. Medium containing dox was perfused into the chamber ~0.5 hr later, and cells were imaged every 2.5 min for 4–6 hr in both the YFP and CFP channels (Supplemental Data, Supplemental Movie S1 available on Cell website).
An increase in RNA levels was detected immediately after the addition of dox (i.e., Figure 3E, 5 min; 3H, 7.5 min; 3P). The highly condensed state of the chromatin at the early time points suggests that only a subset of the repeats are being transcribed which is supported by the limited amount of rtTA-YFP detected at the locus 5 min post dox (Figure 2H). By 17.5 min when noticeable changes in higher order chromatin structure begin to be detected (Figures 3J–3L), the RNA coats the entire region of the locus. By ~130 min when RNA levels at the transcription site are at their peak (Figure 3P), the locus is maximally decondensed and CFP-SKL is concentrated in the peroxisomes (Figures 3M–3O). The CFP-SKL protein can first be detected in the cytoplasmic peroxisomes ~1 hr after the addition of dox (data not shown).
Figure 3. Kinetics of RNA Synthesis.
(A–O) Still images from a time series of U2OS 2-6-3 cells during transcriptional activation showing the relationship between the chromatin of the locus, marked by CFP-lac repressor, and the RNA, marked by MS2-YFP. Also see Supplemental Data and Supplemental Movie S1 available on Cell website. Scale bar is equal to 5 µm.
(P) Quantitative analysis and modeling of RNA levels at the locus. The intensity of the MS2-YFP signal at the locus as it associates with the synthesized RNA was measured every 2.5 min. There is an initial linear increase after the induction of transcription which slows over time (following the form A*(1−exp(−B*t))). The fit shows a deviation from this increase at 140 min after which a decrease predominates. There is a delay in the increase at 50 min.
By measuring RNA levels at the transcription site over time, we found that they increase, peak, and subsequently decrease (Figure 3P). Modeling of this data showed that the rate of increase slows with time suggesting that increases in RNA production from this transgene array inhibit further synthesis (Figure 3P). A delay in the increase was also often observed before the peak. It is possible that this delay reflects the slower decondensation of a region of chromatin within the array that forms a specific substructure. Though we found that there is variability in the timing of these features between cells, likely resulting from the transient expression of rtTA, the general features are consistent (data not shown).
The Condensed Locus Is Heterochromatic
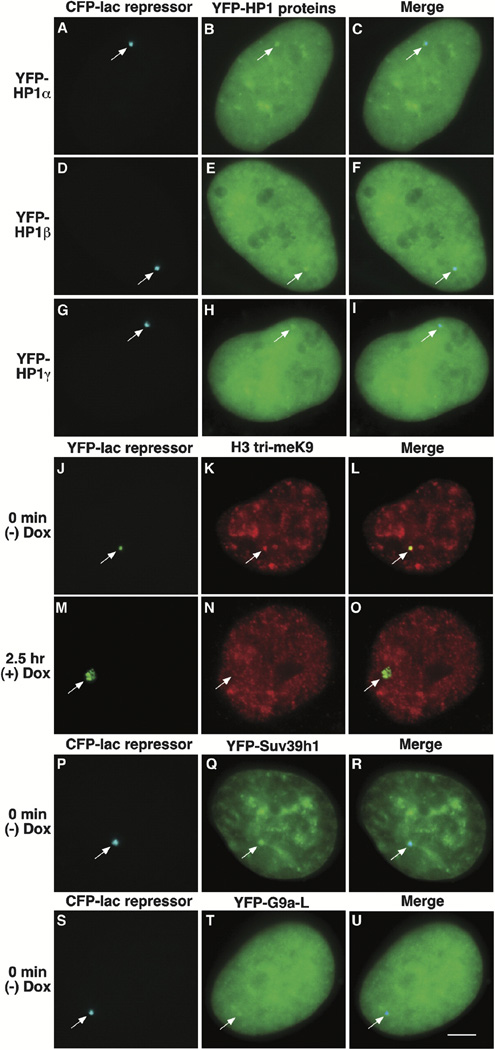
As the ~200 copies of the gene expression plasmid formed a highly condensed structure in interphase cells, we also wanted to characterize the locus in this state. When U2OS 2-6-3 cells expressing YFP-lac repressor were independently labeled with antibodies to HP1α, β, and γ, all three isoforms were detected at the condensed locus (Supplemental Data, Supplemental Figure S5 available on Cell website). Similarly, YFP-HP1α, β and γ fusion proteins colocalized with CFP-lac repressor (Figures 4A–4I; Supplemental Data, Supplemental Figure S4 available on Cell website; Western blot of constructs).
Figure 4. Characterization of the Condensed Heterochromatic Locus.
YFP-HP1α (A–C), YFP-HP1β (D–F), and YFP-HP1γ (G–I) colocalize with the condensed locus, marked by CFP-lac repressor and the histone H3 is trimethylated on lysine 9 (H3 tri-meK9) (J–L). The H3 lysine 9 modification is not detected after the induction of transcription (M–O; 2.5 hr postdox). The histone H3 K9 methyltransferases YFP-Suv39h1 (P–R) and YFP-G9a-L (S–U) are present at the condensed locus. Scale bar is equal to 5 µm.
The association of HP1 with the condensed locus suggested that the histone H3 might be methylated at lysine 9 as this modification serves as a binding site for HP1 Bannister et al. 2001, Jacobs et al. 2001 and Lachner et al. 2001. When cells were labeled with an antibody made to a branched histone H3 peptide methylated at lysine 9 (Peters et al., 2001) which had been affinity purified for antibodies with a preference for trimethylated H3 lysine 9 (H3 tri-meK9) Rice et al. 2003 and Peters et al. 2003, this modification was detected at the condensed locus (Figures 4J–4L) but not at decondensed loci in transcriptionally activated cells (Figures 4M–4O, 2.5 hr postinduction).
As we found all three HP1 isoforms and histone H3 tri-meK9 at the inactive locus, we also wanted to see if we could visualize an association with H3 K9 HMTases. When we expressed Suv39h1, a H3 K9-specific HMTase associated with constitutive heterochromatin (Rea et al., 2000) as a YFP fusion protein, it colocalized with CFP-lac repressor (Figures 4P–4R; Supplemental Data, Supplemental Figure S4 available on Cell website, Western blot). Additionally, YFP-G9a-L, the longer G9a splice variant shown to methylate both H3 K9 and K27 in vitro and to localize to euchromatin Tachibana et al. 2001 and Tachibana et al. 2002, colocalized with CFP-lac repressor (Figures 4S–4U; Supplemental Data, Supplemental Figure S4 available on Cell website, Western blot).
HP1 Isoforms, H3 K9 Methylation, and Eu-HMTase 1 Differentially Associate with the Gene Expression Plasmid by ChIP Analysis
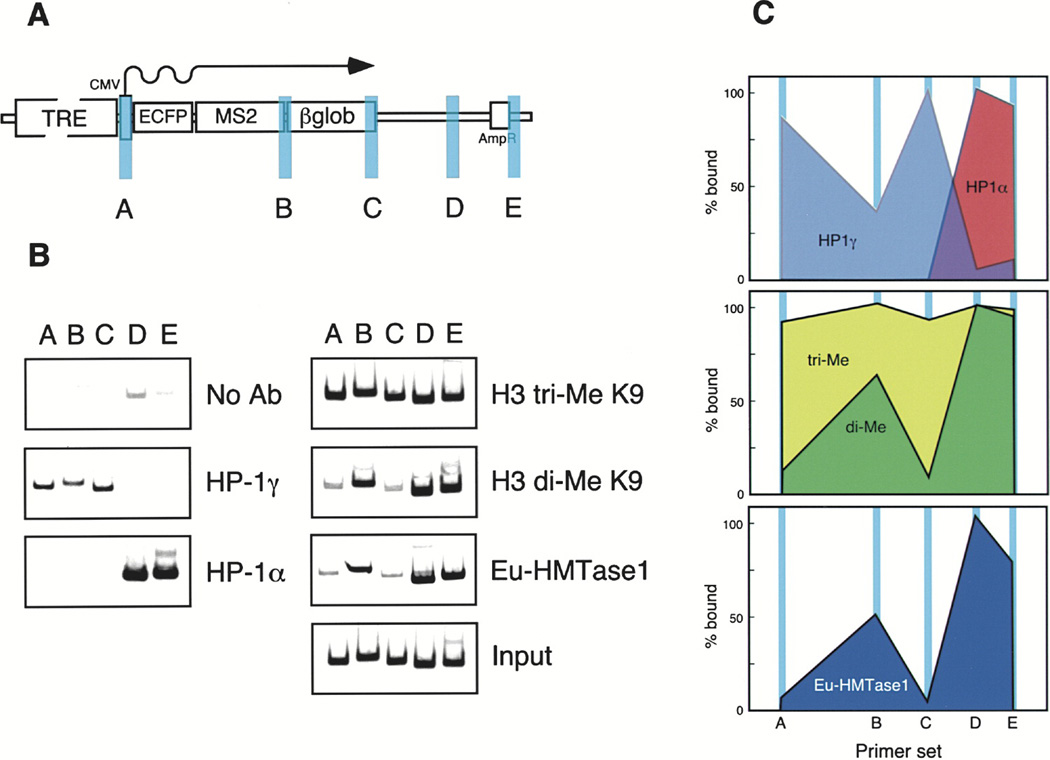
As we were able to visually detect HP1 and H3 tri-meK9 at the condensed locus, we wanted to analyze the specific associations of HP1α and γ, H3 di- and tri-meK9, and H3 K9 HMTases by chromatin immunoprecipitation (ChIP) (Figure 5). Strikingly, HP1γ displayed a distinct preference for the promoter and the transcribed regions of the gene in contrast to HP1α, which was only found downstream on the bacterial plasmid sequence (Figures 5B and 5C). H3 tri-meK9 was present throughout the assayed regions while di-meK9 was enriched on the bacterial plasmid sequence and at the beginning of the intron/exon module in a region immediately after the MS2 repeats (Figures 5B and 5C). Interestingly, euchromatic histone methyltransferase1 (Eu-HMTase 1) colocalized with the H3 K9 dimethylated regions of the gene expression plasmid. As G9a has been reported to be a H3 K9 di-HMTase Rice et al. 2003 and Peters et al. 2003 and Eu-HMTase1 is 63% homologous to G9a and possesses the same basic structure (Ogawa et al., 2002), this suggests that Eu-HMTase1 is the H3 K9 di-HMTase responsible for the pattern of H3 K9 di-methylation at the locus.
Figure 5. Chromatin Immunoprecipitation (ChIP) Analysis of Heterochromatin Proteins and Modifications on the Inactive Gene Expression Plasmid.
Diagram (A) depicts the gene expression plasmid and the location of the primer pairs used: (a) promoter, (b) beginning of β-globin intron/exon module, (c) end of β-globin intron/exon module, (d) and (e) bacterial plasmid sequence.
(B) Results of the ChIP analysis showing the localization of HP1α, HP1γ, histone H3 di- and tri-MeK9, and Eu-HMTase1 on the gene expression plasmid.
(C) The levels of the associated factors and modifications are plotted.
HP1α Is Depleted from the Locus during Transcriptional Activation
As HP1 is a protein critical to the stability of transcriptional silencing, we wanted to examine the kinetics of the association of YFP-HP1α with the locus during transcriptional activation and to relate these dynamics to changes in chromatin structure (Supplemental Data, Supplemental Movie S2 available on Cell website). YFP-HP1α was detected at the locus before the addition of dox (Figures 6A–6C; 0 min) and during the early time points postinduction (Figures 6D–6F; 17.5 min). At the 30 min time point when significant chromatin decondensation had occurred (Figure 6G), YFP-HP1α was seen in punctate structures which were possibly regions of chromatin not yet remodeled by transcriptional activity (Figure 6H). By ~50 min, accumulations of YFP-HP1α could no longer be detected on the locus (Figures 6K–6L) and between 100–140 min, YFP-HP1α became progressively depleted from this region (data not shown) with levels subsequently decreasing further over time. The presence of this dark region, depleted of YFP-HP1α below background levels (Figures 6M–6O; 180 min), suggests that HP1α binding sites are no longer present at the transcriptionally active locus.
Figure 6. Dynamics of YFP-HP1α Depletion from the Locus during Transcriptional Activation.
YFP-HP1α colocalizes with the condensed locus (0 min, A–C) and the condensed regions during the early time points of transcriptional activation (D–F, 17.5 min). It is seen in punctate structures at the 30 min time point (G–I) and appears smooth and diffuse by 50 min (J–L). 180 min postinduction, a dark region (HP1α depleted) that colocalizes with the decondensed locus, is seen in the YFP-HP1α image. Also see Supplemental Data and Supplemental Movie S2 available on Cell website. Scale bar is equal to 5 µm.
The Histone H3 Variant, H3.3, Is Deposited at the Locus during Transcriptional Activation
As YFP-HP1α was dynamically depleted during the induction of transcription and we could not detect the histone H3 tri-MeK9 modification at transcriptionally active loci (2.5 hr postinduction), we were interested in understanding how this modification is removed from the chromatin during transcriptional activation. A histone demethylase has not yet been identified (reviewed, Bannister et al., 2002) but the histone H3 variant, H3.3, has been shown to be deposited into active ribosomal DNA in a replication-independent manner (Ahmad and Henikoff, 2002). Therefore, we were interested in determining whether histone exchange is a general mechanism through which a heterochromatic region is transformed into the active state.
Before the induction of transcription, H3.3-YFP did not show any specific deposition at the locus (Figures 7A–7C; Supplemental Data, Supplemental Movie S3 available on Cell website). A small spot of H3.3-YFP was seen, however, ~7.5 min after the addition of dox adjacent to the region marked by CFP-lac repressor (pseudocolored red) (Figures 7D–7F). Significant incorporation of H3.3-YFP appeared around the periphery of the locus (Figures 7G–7I; 40 min) and subsequently progressed into the interior (Figures 7J–7L; 75 min). Over time, H3.3-YFP deposition became highly concentrated at the locus and did not completely overlap with the regions marked by CFP-lac repressor (Figures 7M–7O) suggesting that deposition may specifically occur in the regions associated with the transcription machinery. Nucleoli were also labeled by H3.3-YFP, as confirmed by double labeling with an antibody to fibrillarin (data not shown), which is consistent with the deposition of H3.3 into rDNA (Ahmad and Henikoff, 2002). Therefore, histone exchange appears to be part of the mechanism by which heterochromatin can be converted to active chromatin.
Figure 7. Analysis of the Deposition of the Histone Variant, H3.3, at the Locus during Transcriptional Activation.
H3.3-YFP is not enriched at the condensed locus (A–C). A small spot adjacent to the locus is seen immediately after induction (D–F; 7.5 min). Significant deposition begins around the periphery of the decondensing locus (G–I, 40 min; J–L, 75 min) and eventually H3.3-YFP appears in a concentrated region that does not completely colocalize with CFP-lac repressor (pseudocolored red) (M–O; 180 min). Also see Supplemental Data and Supplemental Movie S3 available on Cell website. Scale bar is equal to 5 µm.
Discussion
Due to the complexity of the gene expression pathway, the individual processes have long been studied as discrete events. However, as the interconnectedness of chromatin remodeling, transcription, mRNA processing, and mRNP export becomes increasingly apparent, it is necessary to understand how they are coordinated in vivo. We have developed a cell system that allows us to visualize an inducible array of transcription units and their RNA and protein products in living cells and thus, to evaluate dynamic changes in chromatin structure, RNA synthesis, and factor dissociation/association during the induction of transcription. Using this system, we have provided significant insight into the dynamic spatial and temporal changes that occur as chromatin transitions from a heterochromatic to a euchromatic state. This system has extensive potential to address a broad range of questions relating to gene expression, DNA replication, and chromatin stability.
mRNA Dynamics at a Transcription Site
We have been able to evaluate the dynamics of RNA synthesis in real-time by measuring MS2-YFP levels at the site of transcription. Thus far, mRNA levels at a single specific transcription site have not been evaluated over time in living cells although the diffusive properties of the total endogenous population of nuclear poly(A+) RNA have been studied using fluorescently tagged oligodeoxynucleotides labeled with caged flurochromes and fluorescence correlation spectroscopy Politz et al. 1998 and Politz et al. 1999 and by examining the dynamics of mRNP proteins (Calapez et al., 2002). We have observed that RNA levels begin increasing immediately after the induction of transcription, peak, and subsequently decrease. A delay in the increase was often seen at a time point prior to the peak. It is possible that this delay reflects the decondensation of a substructure of the locus exposed after the initial large-scale unfolding. Interestingly, independently isolated clones containing an amplified region of DHFR cDNA labeled withlac operator repeats showed distinct but reproducible arrangements of chromosome segments within the metaphase chromatid axis suggesting that different chromatin regions acquire distinct folding characteristics (Dietzel and Belmont, 2001). In the case of our locus, the juxtaposition of the plasmids within the array could create different substructures that exhibit distinct activation profiles. High-resolution analysis of the locus during transcriptional activation will be required to determine the effects of subchromosomal features on the kinetics of RNA synthesis.
Modeling the rate of increase of RNA at this transgene array locus upon the induction of transcription showed that it slows with time, suggesting that as more RNA is synthesized it may have a negative feedback effect on further transcription. A similar trend was seen in both a cell line containing a tandem array of ras reporters driven by MMTV promoters activated by the glucocorticoid receptor Archer et al. 1994 and Muller et al. 2001 and at the endogenous β-actin gene upon serum induction (Femino et al., 1998). By run-on assay, transcription peaked in the MMTV reporter cell line within the first 2 hr after hormone treatment and declined by 6 hr (Archer et al., 1994). The arrays recondensed 3–8 hr after induction with array size correlated to decreasing amounts of transcript as determined by RNA FISH analysis (Muller et al., 2001). β-actin RNA levels peaked 15 min after serum induction after which they diminished until the 1.5 hr time point when they were indistinguishable from those seen before induction (Femino et al., 1998).
Although we do not know the mechanism(s) through which the rate of transcription is slowed in our cell line, it is possible that the accumulation of RNA at the transcription site sterically hinders the transcription machinery. A number of pathways that monitor mRNA quality in the nucleus have been identified and mRNA that is too slowly or improperly spliced, polyadenylated, or transported is retained at the transcription site and degraded in the nucleus (reviewed, Vasudevan and Peltz 2003 and Wilkinson and Shyu 2002). Northern blotting of RNA from the 120 min time point (Supplemental Data, Supplemental Figure S1 available on Cell website) identified species with molecular weights both higher and lower than the unspliced and spliced products. It is possible that they are truncated, degraded, or read-through products produced when high levels of RNA are transcribed from the strong CMV promoter. Although we do not know the cellular location of these aberrant transcripts, it is possible that they are retained at the transcription site and affect the rate of transcription. In the case of the β-actin gene, transcription was shown to be inhibited at initiation (Femino et al., 1998). The mechanism(s) through which the cell regulates the transcription cycle of a gene is not well understood, but the emerging importance of RNA as a signaling molecule (reviewed, Gottesman 2002 and Nelson et al. 2003) suggests that understanding the kinetics of its production may provide insight not only into transcriptional regulation but possibly also into chromatin structure.
Characterization and Dynamics of the Condensed Locus and Its Transition to the Active State
We have shown that the 200 copies of the gene expression plasmid that integrated into the genome of the U2OS cell line formed a heterochromatic structure. As transcription can be activated at this array in an experimentally regulated manner, we were able to study in living cells the dynamic changes that occur as heterochromatin transits to the active state. By examining YFP-HP1α dynamics during the induction of transcription, we showed that it is depleted from the locus as it decondenses suggesting that H3 K9 methylation is removed during gene activation. We also found that the histone H3 variant, H3.3, is deposited during activation. Therefore, histone exchange is likely the mechanism through which H3 K9 methylation is removed.
By ~50 min postinduction, there are no longer accumulations of YFP-HP1α at the locus but because YFP-HP1α does not become depleted until 100–140 min, this suggests that at least a population of histone H3 is still methylated at K9 prior to these time points. As we see RNA coating the decondensing chromatin 20–25 min postinduction, it is possible that transcription can occur in the presence of HP1 and H3 K9 trimethylation. It is also possible that these silencing marks are removed immediately from specific sequence elements (i.e., promoters) upon induction and that we do not have the resolution to detect these changes or to determine which of the multiple repeats are activated first. Interestingly, HP1 has been shown to be required for expression of genes located in heterochromatin in Drosophila Clegg et al. 1998, Hearn et al. 1991 and Lu et al. 2000 and to be recruited to heat shock and ecdysone-activated puffs on polytene chromosomes(Piacentini et al., 2003). Future studies are required to address how the activational and repressive roles of HP1 may be modulated and coordinated in our system.
As significant deposition of H3.3 at the locus is not seen until later time points (75–150 min postinduction), it is possible that histone exchange is a late event in the transition of heterochromatin to the active state—perhaps playing more of a role in changing and maintaining the epigenetic state of a gene than in aiding in the progression of transcription. It is also possible that in order for transcription to begin, histone exchange must first occur in specific sequence elements, such as promoters, and that we are not able to detect deposition in such small regions. Subsequently, histones may be exchanged throughout the entire coding region resulting in the significant accumulation visualized at later time points. As H3.3-YFP did not completely colocalize with CFP-lac repressor (Figures 7M–7O), it is likely that exchange occurs specifically in actively transcribed areas.
Heterochromatin has been reported to have both dynamic and stable qualities. Its high degree of condensation is believed to contribute to the inhibition of transcription by preventing transcription factors from accessing their binding elements yet it is not intractable to transcriptional activation. Photobleaching studies have shown HP1 to be a dynamic component of heterochromatin Cheutin et al. 2003 and Festenstein et al. 2003in contrast to the core histones, which show little turnover Kimura and Cook 2001, Lever et al. 2000 and Phair and Misteli 2000. Likewise, biochemical analyses of histone methylation suggest that this modification is highly stable (reviewed, Bannister et al. 2002 and Jenuwein and Allis 2001). Therefore, the combination of both dynamic and durable elements may be what makes heterochromatin simultaneously stable and flexible. As histone H3 has been shown to be tightly associated with chromatin (Kimura and Cook, 2001), it will be of particular interest to characterize the histone chaperones and chromatin remodeling proteins that function to remove H3 and exchange it for H3.3 as they may be important regulators of epigenetic states. In fact, the histone chaperone HIRA was recently shown to be required for DNA synthesis-independent nucleosome assembly (Tagami et al., 2004) and Swr1, a Swi2/Snf2-related ATPase was identified in a complex that catalyzes the exchange of H2A with H2AZ in nucleosome arrays Krogan et al. 2003 and Mizuguchi et al. 2003.
Interestingly, we found, by ChIP analysis, that HP1α and γ, di-and trimethyl H3 K9 and Eu-HMTase1 are differentially localized along the inactive gene expression plasmid. It is unlikely that the HP1 isoforms associate in a sequence specific manner, as they have not been reported to show any DNA motif preferences (Zhao et al., 2000). The distinct associations of HP1α and γ with the chromatin are, therefore, more likely determined by complex interactions with other factors involved in establishing and maintaining heterochromatin. It has been suggested that HP1γ associates with euchromatin while HP1α associates with constitutive heterochromatin (reviewed, Li et al., 2002). As the bacterial plasmid sequence is composed of elements that are noneuchromatic as well as noneukaryotic, it may therefore be recognized as constitutive heterochromatin by HP1α. Likewise, the promoter, the coding region and the intron/exon module may recruit HP1γ because they are recognized as euchromatic elements.
Interestingly, there was a strong coincidence between H3 di-meK9 and Eu-HMTase1 suggesting that Eu-HMTase1 is the histone H3 HMTase that dimethylates the locus. As Suv39h1 and G9a-L were also detected, more than one H3 K9 HMTase seems to be responsible for the H3 K9 methylation patterns suggesting that the regulation of the chromatin at the condensed locus is complex. We also observed that the association of HP1γ is inversely related to the H3 di-meK9 and Eu-HMTase1 patterns as it is not detected in the downstream region of the bacterial plasmid sequence and is reduced in the region immediately after the MS2 repeats which is another noneukaryotic sequence element. As Eu-HMTase1 was originally isolated in a complex with E2F-6 that included HP1γ but not HP1α and β (Ogawa et al., 2002), it seems that that Eu-HMTase1 does not preferentially associate with a specific HP1 isoform or alone specify associations of the HP1 isoforms with DNA sequences. Therefore, other as yet unidentified factors may be antagonistic to HP1γ binding. Additionally, the relationship between the strong peaks of H3 K9 dimethylation and HP1α on the bacterial plasmid sequence is not clear. H3 di-meK9 has been reported to localize in a pattern consistent with euchromatin Heard et al. 2001, Rice et al. 2003 and Peters et al. 2003 and constitutive heterochromatin is characteristic of HP1α.
These results, therefore, suggest that there are no simple rules to explain how heterochromatin proteins and modifications are targeted to DNA sequences. Perhaps the high degree of complexity revealed in the studies of heterochromatin structure and regulation (reviewed, Grewal and Moazed 2003 and Richards and Elgin 2002) suggest that cells employ multiple complex interactions in order to package the diverse range of sequence elements in the eukaryotic genome such that heritable gene expression patterns are ensured.
Heterochromatin and Transgene Arrays
When the ~200 copies of the gene expression plasmid integrated into a euchromatic region of the U2OS cell genome, they formed a structure that is highly condensed in interphase cells and heterochromatic. Many regions of constitutive heterochromatin, such as centromeres, telomeres, and transposons, are composed of repetitive sequence elements. In addition, multicopy transgene arrays as small as two copies inserted into euchromatic regions of the genome are often silenced, DNA methylated and, in Drosophila polytene chromosomes, seen as DAPI dense structures enriched with HP1 Fanti et al. 1998, Hsieh and Fire 2000 and Lau et al. 1999.
Transgene silencing is increased in arrays with inverted repeats which are likely a potent source of double-stranded RNA (dsRNA) Dorer and Henikoff 1994, Pal-Bhadra et al. 1997, Pal-Bhadra et al. 1999 and Sabl and Henikoff 1996 and small interfering RNAs (siRNAs) generated by the RNA interference pathway (RNAi) (reviewed, Hannon, 2002) have been detected in Drosophila cells with transgene arrays (Pal-Bhadra et al., 2002). RNAi is also involved in the establishment and maintenance of heterochromatin Hall et al. 2002 and Volpe et al. 2002. Therefore, it will be interesting to look for the presence of siRNAs and inverted repeats in 2-6-3 cells. As we have a cell line in which we can visualize the conversion of a region of heterochromatin to a transcriptionally active state, it may be a useful system in which to study the mechanisms of RNAi-based gene silencing in vivo.
To date, it is not well understood how primary DNA sequence content regulates higher order chromatin structure, how distinct functional domains are established and how these structures regulate gene expression. Repetitive arrays occur naturally in the human genome and include transposons, ribosomal DNA, centromeres, and telomeres (reviewed, Li et al. 2001 and Spector 1993). Recently, a reduction in the number of copies of a repetitive element at 4q35 below a certain threshold was shown to cause Facioscapulohumeral muscular dystrophy (FSHD) with copy number a critical determinant of the age of onset as well as the clinical severity of the disease (reviewed,Bickmore and Van Der Maarel 2003, Jiang et al. 2003 and Ricci et al. 1999). Therefore, an understanding of the packaging and regulation of repetitive elements in the genome will provide important insight into the mechanisms of both gene regulation and disease.
Experimental Procedures
Plasmids
p3216PECMS2β was constructed by modifying p3216PCβ (Tsukamoto et al., 2000)(Supplemental Data available on Cell website). pSV2-YFP-lac repressor-C1 and pSV2-YFP-C1 were previously described (Tsukamoto et al., 2000). pSV2-CFP-lac repressor-C1 was constructed by replacing the coding region of YFP with CFP.
pTet-On and pTK-Hyg were purchased from BD Biosciences Clontech (Palo Alto, CA). pEYFP-rtTA-N1 was constructed by PCR amplifying the rtTA coding region from pTet-On and inserting it into pEYFP-N1. MS2-YFP was expressed from plasmids with either CMV or RNA polymerase II large subunit promoters (Fusco et al., 2003) (Supplemental Data available on Cell website). pSV2-EYFP-Suv39 h1-C1 (mouse) and pSV2-EYFP-G9a-L-C1 (mouse) were constructed from their respective GFP plasmids (gifts of Y. Shinkai)Tachibana et al. 2001 and Tachibana et al. 2002. SV2-YFP-HP1α-C2, EYFP-HP1α-C2 and SV2-YFP-HP1β-C2, and SV2-YFP-HP1γ-C2 were constructed from the respective GFP constructs (gift of S. Lowe). pSV2-YFP-SF2/ASF was constructed from pGFP-SF2/ASF (Misteli et al., 1997) and pSV2-YFP-RNA polymerase II from EGFP-RNA polymerase II (gift of M. Vigneron) (Sugaya et al., 2000). CMV-histone H3.3-YFP was a gift of K. Ahmad and S. Henikoff.
Stable Cell Line Construction
U2OS cells, grown in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% FBS and penicillin-streptomycin (Invitrogen, Carlsbad, CA), were transfected with 18 µg of p3216PECMS2β and 2 µg of pTK-Hyg by calcium phosphate transfection (Chen and Okayama, 1987). Stable transformants were selected in 100 µg/ml hygromycin B (Sigma, St. Louis, MO) for 7–10 days and isolated colonies were maintained in selection medium. Cell lines were screened for insertion of p3216PECMS2β and the ability to be transcriptionally activated by transfecting SV2-YFP-lac repressor and pTet-On in the presence of 1 µg/ml doxycycline and looking for cells with YFP-lac repressor binding sites in the nucleus and CFP-SKL labeled peroxisomes in the cytoplasm 24 hr later. Cells from positive colonies were plated as single cells in conditioned medium and the amplified populations were again transfected with SV2-YFP-lac repressor to determine whether they all contained single integration sites. Copy number and message size were determined by Southern and Northern blotting as described in Supplemental Data available on Cell website.
Transfection
For imaging and Northern and immunoblotting studies, cells were grown in DMEM + 10% tet system approved FBS (BD Biosciences Clontech, Palo Alto, CA) and transfected by electroporation. Cells were grown to 70%–80% confluency, trypsinized, and the cell pellet from a 10 cm dish was resuspended in ~600 µl of medium. 200 µl of cells were added to 4-mm gap cuvettes containing plasmids and 40 µg of sheared salmon sperm DNA (Amresco, Solon, OH) and electroporated using a Gene Pulser II (BioRad, Hercules, CA) (170 V, 950 µF). Electroporated cells were transferred to 2 mL of medium, centrifuged, resuspended, and plated on BD Cell-Tak (BD Biosciences, Palo Alto, CA) coated coverslips. 2.5–5 µl of Cell-Tak was spread until dry onto coverslips with a sterile cell scraper and subsequently rinsed with 100% EtOH followed by sterile water. Hygromycin B was not added after electroporation.
DNA and RNA Fluorescence In Situ Hybridization
For DNA FISH, probe labeling was performed via nick translation of p3216PECMS2β (detailed protocol, http://www.riedlab.nci.nih.gov/). Tran-scripts from p3216PECMS2β were localized by RNA FISH using CY3-labeled oligo nucleotide probes (Femino et al., 1998). pTK-Hyg was nick translated using the VYSIS, Inc. nick translation kit (Downers Grove, IL) and transcripts were localized using standard procedures (Tsukamoto et al., 2000).
Immunoblotting
Cells were electroporated with pTet-On (2 µg) or YFP constructs (5 µg) and sheared salmon sperm DNA (40 µg). For time course analysis, dox (1 µg/ml) was added to the medium 2.5 hr post transfection and cell lysates were harvested in 1× gel loading buffer and blotted according to standard procedures using a monoclonal anti-GFP antibody (1: 1500) (Roche, Indianapolis, IN).
Imaging Transcriptional Activation
For time-lapse imaging, cells were transfected with pSV2-CFP-lac repressor (2 µg), pTet-On (2 µg), sheared salmon sperm DNA (40 µg), and 0.5 µg of either pEYFP-MS2, pEYFP-HP1α, or CMV-H3.3-YFP and plated on Cell-Tak-coated coverslips. Cells were placed in an FCS2 live-cell chamber (Bioptechs Inc., Butler, PA) 2 hr posttransfection and phenol-red free Leibovitz's L15 medium (Invitrogen) was perfused into it. The chamber and objective lens were maintained at 37°C (Bioptechs Inc., Butler, PA). Images were acquired with an Olympus IX-70 inverted microscope with a 100×/1.4 NA objective lens using a TILL Photonics Polychrome II monochromator with a xenon light source and TILLvisION imaging software with a TILL IMAGO-SVGA camera (T.I.L.L. Photonics GmbH).
Stacks of 3–5 images 0.5 µm apart were taken at both the 430 and 510 nm wavelengths with a dual band pass CFP/YFP filter (51017bs +10 nm) (Chroma Technology, Brattleboro, Vermont) every 2.5 min using 2 × 2 binning (640 × 512 pixels). Approximately 2.5 hr posttransfection, medium containing dox was perfused into the chamber. In focus images were selected for each time point using the CollectFrames macro and then combined into a single image series using the Tcombine macro in TILLvisION.
For measurement of RNA levels at the transcription site, image files were exported from TILLvisION as TIFF files and custom software written in Matlab (The Mathworks, Inc., Boston MA) was used to extract the amplitude of light emanating from the regions of interest. The locus was identified in the CFP channel as the brightest spot and a 20 × 20 pixel area around it was correlated to the MS2-YFP image in order to measure the amount of RNA at the locus.
For still images of live cells, cells were transfected with 2 µg pSV2-CFP-lac repressor, 2 µg pTet-On, and 5 µg of the respective YFP expression construct. Images were taken using the TILL imaging system (1 × 1 binning; 1280 × 1024 pixels).
Deconvolution Microscopy
Image stacks at 0.2 µm intervals were collected and deconvolved using softWoRx 2.50 software (Deltavision by Applied Precision, Issaquah, WA) on an Olympus IX70 microscope with a 100×/1.35 NA PlanApo objective lens using CFP and YFP filters (Chroma, Brattleboro, VT).
Immunofluorescence
Cells were transfected with 2 µg SV2-YFP lac repressor, 2 µg pTet-On, and 40 µg sheared salmon sperm DNA. Cells were fixed either 2.5 hr posttransfection (0 min time point) or 2.5 hr after the addition of dox to the medium (see Supplemental Data available on Cell website). For labeling with HP1 β and γ antibodies, cells were preextracted with CSK buffer (100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 10 mM PIPES [pH 6.8]) + 0.5% Triton on ice for 5 min then immediately fixed in 2% formaldehyde in 1× PBS for 10 min. Washes, blocking, and antibody incubation are as described in Supplemental Data available on Cell website. The following antibodies were used: HP1α, 1:100; HP1 β, 1:700; HP1 γ, 1:1000, mouse monoclonals (Chemicon, Temecula, CA); histone H3 tri-methyl K9, 1:500, rabbit polyclonal (gift of D. Allis and T. Jenuwein) Rice et al. 2003 and Peters et al. 2003; Cstf64, 1:60, mouse monoclonal IgG (Takagaki et al., 1990). Secondary antibodies were Texas-red antimouse IgG and antirabbit IgG (1:1000) (Jackson Labs, Bar Harbor, ME). Images were acquired on a Zeiss Axioplan 2i fluorescence microscope with a plan-APO 100×/1.4 NA objective lens using Openlab Software (Improvision, Lexington, MA) and an Orca CCD camera (Hammamatsu, Middlesex, NJ).
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays were performed using the Upstate Biotechnology protocol (http://www.upstate.com/misc/protocols.q.prot.e.chips/). Antibodies against the following antigens were used: HP1-γ and HP1-α (gift of F. Rauscher) (Ayyanathan et al., 2003); histone H3 di-methyl K9 (Upstate Biotechnology, Charlottesville, VA); histone H3 tri-methyl K9 (gift of D. Allis and T. Jenuwein) Rice et al. 2003 and Peters et al. 2003; and Eu-HMTase1 (gifts of Y. Nakatani) (Ogawa et al., 2002). Specific DNA-sequences present in the immune precipitates were detected by quantitative PCR using five sets of primers (A–E) designed to amplify ~200 bp fragments from the plasmid (p3216PECMS2β), centered at the CMV promoter (set A) and 2.5 kb (B), 3.6 kb (C), 4.8 kb (D), and 5.6 kb (E) downstream. Primer sequences are available upon request. PCR products, labeled with 33P-dCTP, were detected, and quantified by phosphorimaging.
Supplementary Material
Cells expressing CFP-lac repressor, MS2-YFP, and rtTA were imaged every 2.5 min for a total of 4 hr and 22.5 min.
Cells expressing CFP-lac repressor, YFP-HP1a, and rtTA were imaged every 2.5 min for a total of 3 hr.
Cells expressing CFP-lac repressor (pseudocolored red), H3.3-YFP, and rtTA were imaged every 2.5 min for a total of 5 hr and 16 min.
Acknowledgments
We would like to thank Carolyn Dong for designing the plasmid schematic in Figure 1; Supriya Prasanth for help with the Northern blot analysis; Steve Henikoff for insightful discussions about histone exchange; David Allis, Thomas Jenuwein, Frank Rauscher, Yoshihiro Nakatani, and James Manley for their generous gifts of antibodies. We would also like to thank Edith Heard and members of the Spector laboratory for critical review of the manuscript. Supported by grant 42694 from NIGMS/NIH to D.L.S. and 067728 to W.P.T. W.P.T. is a Leukemia and Lymphoma Society Scholar.
References
- Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- Archer TK, Lee HL, Cordingley MG, Mymryk JS, Fragoso G, Berard DS, Hager GL. Differential steroid hormone induction of transcription from the mouse mammary tumor virus promoter. Mol. Endocrinol. 1994;8:568–576. doi: 10.1210/mend.8.5.8058066. [DOI] [PubMed] [Google Scholar]
- Ayyanathan K, Lechner MS, Bell P, Maul GG, Schultz DC, Yamada Y, Tanaka K, Torigoe K, Rauscher FJ., 3rd Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: a mammalian cell culture model of gene variegation. Genes Dev. 2003;17:1855–1869. doi: 10.1101/gad.1102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Schneider R, Kouzarides T. Histone methylation: dynamic or static? Cell. 2002;109:801–806. doi: 10.1016/s0092-8674(02)00798-5. [DOI] [PubMed] [Google Scholar]
- Beckett D, Uhlenbeck OC. Ribonucleoprotein complexes of R17 coat protein and a translational operator analog. J. Mol. Biol. 1988;204:927–938. doi: 10.1016/0022-2836(88)90052-6. [DOI] [PubMed] [Google Scholar]
- Belmont AS. Visualizing chromosome dynamics with GFP. Trends Cell Biol. 2001;11:250–257. doi: 10.1016/s0962-8924(01)02000-1. [DOI] [PubMed] [Google Scholar]
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol. Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- Bickmore WA, Van Der Maarel SM. Perturbations of chromatin structure in human genetic disease: recent advances. Hum. Mol. Genet. 2003;12(Suppl 2):R207–R213. doi: 10.1093/hmg/ddg260. [DOI] [PubMed] [Google Scholar]
- Calapez A, Pereira HM, Calado A, Braga J, Rino J, Carvalho C, Tavanez JP, Wahle E, Rosa AC, Carmo-Fonseca M. The intranuclear mobility of messenger RNA binding proteins is ATP dependent and temperature sensitive. J. Cell Biol. 2002;159:795–805. doi: 10.1083/jcb.200203046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheutin T, McNairn AJ, Jenuwein T, Gilbert DM, Singh PB, Misteli T. Maintenance of Stable Heterochromatin Domains by Dynamic HP1 Binding. Science. 2003;299:721–725. doi: 10.1126/science.1078572. [DOI] [PubMed] [Google Scholar]
- Clegg NJ, Honda BM, Whitehead IP, Grigliatti TA, Wakimoto B, Brock HW, Lloyd VK, Sinclair DA. Suppressors of position-effect variegation in Drosophila melanogaster affect expression of the heterochromatic gene light in the absence of a chromosome rearrangement. Genome. 1998;41:495–503. [PubMed] [Google Scholar]
- Dietzel S, Belmont AS. Reproducible but dynamic positioning of DNA in chromosomes during mitosis. Nat. Cell Biol. 2001;3:767–770. doi: 10.1038/35087089. [DOI] [PubMed] [Google Scholar]
- Dorer DR, Henikoff S. Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell. 1994;77:993–1002. doi: 10.1016/0092-8674(94)90439-1. [DOI] [PubMed] [Google Scholar]
- Emerson BM. Specificity of gene regulation. Cell. 2002;109:267–270. doi: 10.1016/s0092-8674(02)00740-7. [DOI] [PubMed] [Google Scholar]
- Fanti L, Dorer DR, Berloco M, Henikoff S, Pimpinelli S. Heterochromatin protein 1 binds transgene arrays. Chromosoma. 1998;107:286–292. doi: 10.1007/s004120050310. [DOI] [PubMed] [Google Scholar]
- Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]
- Festenstein R, Pagakis SN, Hiragami K, Lyon D, Verreault A, Sekkali B, Kioussis D. Modulation of heterochromatin protein 1 dynamics in primary Mammalian cells. Science. 2003;299:719–721. doi: 10.1126/science.1078694. [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- Forrest KM, Gavis ER. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr. Biol. 2003;13:1159–1168. doi: 10.1016/s0960-9822(03)00451-2. [DOI] [PubMed] [Google Scholar]
- Fusco D, Accornero N, Lavoie B, Shenoy SM, Blanchard JM, Singer RH, Bertrand E. Single mRNA molecules demonstrate probabilistic movement in living Mammalian cells. Curr. Biol. 2003;13:161–167. doi: 10.1016/s0960-9822(02)01436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S. Stealth regulation: biological circuits with small RNA switches. Genes Dev. 2002;16:2829–2842. doi: 10.1101/gad.1030302. [DOI] [PubMed] [Google Scholar]
- Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- Hall IM, Shankaranarayana GD, Noma K, Ayoub N, Cohen A, Grewal SI. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Heard E, Rougeulle C, Arnaud D, Avner P, Allis CD, Spector DL. Methylation of histone H3 at Lys-9 is an early mark on the X chromosome during X inactivation. Cell. 2001;107:727–738. doi: 10.1016/s0092-8674(01)00598-0. [DOI] [PubMed] [Google Scholar]
- Hearn MG, Hedrick A, Grigliatti TA, Wakimoto BT. The effect of modifiers of position-effect variegation on the variegation of heterochromatic genes of Drosophila melanogaster. Genetics. 1991;128:785–797. doi: 10.1093/genetics/128.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J, Fire A. Recognition and silencing of repeated DNA. Annu. Rev. Genet. 2000;34:187–204. doi: 10.1146/annurev.genet.34.1.187. [DOI] [PubMed] [Google Scholar]
- Huang S, Spector DL. Pre-mRNA splicing: nuclear organization of factors and substrates. In: Krainer AR, editor. Eukaryotic Messenger RNA Processing. Oxford: IRL Press; 1997. pp. 33–67. [Google Scholar]
- Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Taverna SD, Zhang Y, Briggs SD, Li J, Eissenberg JC, Allis CD, Khorasanizadeh S. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 2001;20:5232–5241. doi: 10.1093/emboj/20.18.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki SM, Spector DL. Nuclear choreography: interpretations from living cells. Curr. Opin. Cell Biol. 2003;15:149–157. doi: 10.1016/s0955-0674(03)00012-7. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Jiang G, Yang F, van Overveld PG, Vedanarayanan V, van der Maarel S, Ehrlich M. Testing the position-effect variegation hypothesis for facioscapulohumeral muscular dystrophy by analysis of histone modification and gene expression in subtelomeric 4q. Hum. Mol. Genet. 2003;12:2909–2921. doi: 10.1093/hmg/ddg323. [DOI] [PubMed] [Google Scholar]
- Kimura H, Cook PR. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 2001;153:1341–1353. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Lau S, Jardine K, McBurney MW. DNA methylation pattern of a tandemly repeated LacZ transgene indicates that most copies are silent. Dev. Dyn. 1999;215:126–138. doi: 10.1002/(SICI)1097-0177(199906)215:2<126::AID-DVDY5>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- Lemon B, Tjian R. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 2000;14:2551–2569. doi: 10.1101/gad.831000. [DOI] [PubMed] [Google Scholar]
- Lever MA, Th'ng JP, Sun X, Hendzel MJ. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature. 2000;408:873–876. doi: 10.1038/35048603. [DOI] [PubMed] [Google Scholar]
- Li WH, Gu Z, Wang H, Nekrutenko A. Evolutionary analyses of the human genome. Nature. 2001;409:847–849. doi: 10.1038/35057039. [DOI] [PubMed] [Google Scholar]
- Li Y, Kirschmann DA, Wallrath LL. Does heterochromatin protein 1 always follow code? Proc. Natl. Acad. Sci. USA. 2002;99(Suppl. 4):16462–16469. doi: 10.1073/pnas.162371699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu BY, Emtage PC, Duyf BJ, Hilliker AJ, Eissenberg JC. Heterochromatin protein 1 is required for the normal expression of two heterochromatin genes in Drosophila. Genetics. 2000;155:699–708. doi: 10.1093/genetics/155.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- Marmorstein R. Structure of SET domain proteins: a new twist on histone methylation. Trends Biochem. Sci. 2003;28:59–62. doi: 10.1016/S0968-0004(03)00007-0. [DOI] [PubMed] [Google Scholar]
- McNally JG, Muller WG, Walker D, Wolford R, Hager GL. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science. 2000;287:1262–1265. doi: 10.1126/science.287.5456.1262. [DOI] [PubMed] [Google Scholar]
- Misteli T, Caceres JF, Spector DL. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 1997;387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-Driven Exchange of Histone H2AZ Variant Catalyzed by SWR1 Chromatin Remodeling Complex. Science. 2003;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- Muller WG, Walker D, Hager GL, McNally JG. Large-scale chromatin decondensation and recondensation regulated by transcription from a natural promoter. J. Cell Biol. 2001;154:33–48. doi: 10.1083/jcb.200011069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson P, Kiriakidou M, Sharma A, Maniataki E, Mourelatos Z. The microRNA world: small is mighty. Trends Biochem. Sci. 2003;28:534–540. doi: 10.1016/j.tibs.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Ogawa H, Ishiguro K, Gaubatz S, Livingston DM, Nakatani Y. A complex with chromatin modifiers that occupies E2F- and Myc-responsive genes in G0 cells. Science. 2002;296:1132–1136. doi: 10.1126/science.1069861. [DOI] [PubMed] [Google Scholar]
- Orphanides G, Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M, Bhadra U, Birchler JA. Cosuppression in Drosophila: gene silencing of Alcohol dehydrogenase by white-Adh transgenes is Polycomb dependent. Cell. 1997;90:479–490. doi: 10.1016/s0092-8674(00)80508-5. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M, Bhadra U, Birchler JA. Cosuppression of nonhomologous transgenes in Drosophila involves mutually related endogenous sequences. Cell. 1999;99:35–46. doi: 10.1016/s0092-8674(00)80060-4. [DOI] [PubMed] [Google Scholar]
- Pal-Bhadra M, Bhadra U, Birchler JA. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol. Cell. 2002;9:315–327. doi: 10.1016/s1097-2765(02)00440-9. [DOI] [PubMed] [Google Scholar]
- Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schofer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin Mol. Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- Phair RD, Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- Piacentini L, Fanti L, Berloco M, Perrini B, Pimpinelli S. Heterochromatin protein 1 (HP1) is associated with induced gene expression in Drosophila euchromatin. J. Cell Biol. 2003;161:707–714. doi: 10.1083/jcb.200303012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz JC, Browne ES, Wolf DE, Pederson T. Intranuclear diffusion and hybridization state of oligonucleotides measured by fluorescence correlation spectroscopy in living cells. Proc. Natl. Acad. Sci. USA. 1998;95:6043–6048. doi: 10.1073/pnas.95.11.6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz JC, Tuft RA, Pederson T, Singer RH. Movement of nuclear poly(A) RNA throughout the interchromatin space in living cells. Curr. Biol. 1999;9:285–291. doi: 10.1016/s0960-9822(99)80136-5. [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Ryder U, Lamond AI, Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Ricci E, Galluzzi G, Deidda G, Cacurri S, Colantoni L, Merico B, Piazzo N, Servidei S, Vigneti E, Pasceri V, et al. Progress in the molecular diagnosis of facioscapulohumeral muscular dystrophy and correlation between the number of KpnI repeats at the 4q35 locus and clinical phenotype. Ann. Neurol. 1999;45:751–757. doi: 10.1002/1531-8249(199906)45:6<751::aid-ana9>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, Hunt DF, Shinkai Y, Allis CD. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell. 2003;12:1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- Richards EJ, Elgin SC. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell. 2002;108:489–500. doi: 10.1016/s0092-8674(02)00644-x. [DOI] [PubMed] [Google Scholar]
- Rook MS, Lu M, Kosik KS. CaMKIIalpha 3′ untranslated region-directed mRNA translocation in living neurons: visualization by GFP linkage. J. Neurosci. 2000;20:6385–6393. doi: 10.1523/JNEUROSCI.20-17-06385.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabl JF, Henikoff S. Copy number and orientation determine the susceptibility of a gene to silencing by nearby heterochromatin in Drosophila. Genetics. 1996;142:447–458. doi: 10.1093/genetics/142.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL. Macromolecular domains within the cell nucleus. Annu. Rev. Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- Spradling AC, Rubin GM. The effect of chromosomal position on the expression of the Drosophila xanthine dehydrogenase gene. Cell. 1983;34:47–57. doi: 10.1016/0092-8674(83)90135-6. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Sugaya K, Vigneron M, Cook PR. Mammalian cell lines expressing functional RNA polymerase II tagged with the green fluorescent protein. J. Cell Sci. 2000;113:2679–2683. doi: 10.1242/jcs.113.15.2679. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- Tachibana M, Sugimoto K, Nozaki M, Ueda J, Ohta T, Ohki M, Fukuda M, Takeda N, Niida H, Kato H, Shinkai Y. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone h3.1 and h3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL, MacDonald CC, Wilusz J, Shenk T. A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes Dev. 1990;4:2112–2120. doi: 10.1101/gad.4.12a.2112. [DOI] [PubMed] [Google Scholar]
- Tsukamoto T, Hashiguchi N, Janicki SM, Tumbar T, Belmont AS, Spector DL. Visualization of gene activity in living cells. Nat. Cell Biol. 2000;2:871–878. doi: 10.1038/35046510. [DOI] [PubMed] [Google Scholar]
- Tumbar T, Sudlow G, Belmont AS. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J. Cell Biol. 1999;145:1341–1354. doi: 10.1083/jcb.145.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Peltz SW. Nuclear mRNA surveillance. Curr. Opin. Cell Biol. 2003;15:332–337. doi: 10.1016/s0955-0674(03)00051-6. [DOI] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Wilkinson MF, Shyu AB. RNA surveillance by nuclear scanning? Nat. Cell Biol. 2002;4:E144–E147. doi: 10.1038/ncb0602-e144. [DOI] [PubMed] [Google Scholar]
- Zhao T, Heyduk T, Allis CD, Eissenberg JC. Heterochromatin protein 1 binds to nucleosomes and DNA in vitro. J. Biol. Chem. 2000;275:28332–28338. doi: 10.1074/jbc.M003493200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cells expressing CFP-lac repressor, MS2-YFP, and rtTA were imaged every 2.5 min for a total of 4 hr and 22.5 min.
Cells expressing CFP-lac repressor, YFP-HP1a, and rtTA were imaged every 2.5 min for a total of 3 hr.
Cells expressing CFP-lac repressor (pseudocolored red), H3.3-YFP, and rtTA were imaged every 2.5 min for a total of 5 hr and 16 min.