Abstract
Obesity is a risk factor for asthma, but obese subjects with asthma respond poorly to standard asthma drugs. Obesity also alters gut bacterial community structure. Obesity-related changes in gut bacteria contribute to weight gain and other obesity-related conditions, including insulin resistance and systemic inflammation. Here, we review the rationale for the hypothesis that obesity-related changes in gut bacteria may also play a role in obesity-related asthma. The metabolomes of the liver, serum, urine, and adipose tissue are altered in obesity. Gut bacteria produce a large number of metabolites, which can reach the blood and circulate to other organs, and gut bacteria–derived metabolites have been shown to contribute to disease processes outside the gastrointestinal tract, including cardiovascular disease. Here, we describe the potential roles for two such classes of metabolites in obesity-related asthma: short-chain fatty acids and bile acids. Greater understanding of the role of microbiota in obesity-related asthma could lead to novel microbiota-based treatments for these hard-to-treat patients.
Keywords: airway, immune system, bile acids, short-chain fatty acids
Obesity is a global epidemic. The World Health Organization estimates that, in 2014, more than 0.5 billion adults were obese and another 1.9 billion were overweight. Obesity is a risk factor for type 2 diabetes, hypertension, and atherosclerosis. Obesity has also emerged as a risk factor for asthma. The mechanistic basis for this relationship has yet to be fully elucidated. Here, we describe evidence supporting the hypothesis that obesity-related changes in the gut microbiome may contribute to the etiology of obese asthma.
Obesity and Asthma
Obesity increases the prevalence and incidence of asthma in both children and adults (1–3). Obesity is also common in severe asthma. Indeed, within the The Epidemiology and Natural History of Asthma: Outcomes and Treatment Regimes (TENOR) cohort, a U.S. cohort of subjects with severe asthma, 30.7% of the children and 69.3% of the adults were obese versus obesity rates of approximately 20% for children and 35% for adults in the general U.S. population (4). Similarly, in the British Thoracic Society Difficult Asthma Registry, a cohort of adult subjects with severe asthma in the United Kingdom, 48.3% were obese, nearly double the obesity prevalence observed in the general adult U.K. population (25%) (5). The observations that, in obese subjects with asthma, weight loss causes substantial reductions in asthma symptoms, improves asthma control, and reduces airway hyperresponsiveness (6, 7) indicates that, at least for some obese subjects with asthma, obesity is not just a comorbidity, but a causal factor. Data from obese mice also support a causal role for obesity in the development of asthma: obese mice display airway hyperresponsiveness even in the absence of any other inciting stimulus (8–12).
Many obese subjects with asthma have difficulty controlling their asthma (13, 14). Indeed, steroids are less effective in obese than in lean subjects with asthma (15). Conceivably, aspects of the obese state, for example, the systemic inflammation of obesity, reduce steroid efficacy by interfering with corticosteroid signaling pathways (15). However, it is also possible that obese subjects with asthma have a phenotype that is not responsive to steroids: steroids target the immune responses typical of allergic asthma, but many obese subjects with asthma are non-atopic (16, 17). Understanding the mechanistic basis for obese asthma may allow for the development of other therapeutic options that have greater efficacy in this population.
Obesity-Related Changes in the Gut Microbiome
Estimates are that more than 100 trillion bacteria from over 1,000 different species colonize the human gastrointestinal (GI) tract. The collective genome of these bacteria includes at least 150-fold more genes than are present in the human genome, and has functional capacities that humans lack (18). For example, gut bacteria are capable of metabolizing polysaccharides and dietary starches that are otherwise indigestible. This metabolism results in the production of short-chain fatty acids (SCFAs), such as butyrate, propionate, and acetate (19, 20), that can then be used by host cells, especially enterocytes and hepatocytes, for ATP synthesis or conversion to triglycerides or glucose. SCFAs also act as signaling molecules, resulting in effects both inside and outside the GI tract (see subsequent discussion). Gut microbiota participate in the synthesis and absorption of some vitamins and minerals, the conversion of primary to secondary bile acids, and the detoxification of some xenobiotics, and contribute to proper intestinal epithelial functioning (21–29).
After birth, there are marked changes in the human gut bacterial community structure that vary depending on Caesarian versus vaginal birth, formula versus breast feeding, host genetics, and especially with the introduction of solid food (30). The community structure begins to stabilize by the age of 2–3 years, but can still be substantially impacted by diet, antibiotic use, changes in geography, age, and environmental exposures (30). Consequently, there are marked interindividual differences in the gut microbiota—differences that are less marked between related individuals, but nevertheless greatly surpass intraindividual differences assessed across time (31).
Impact of Obesity on Gut Microbiota
Obesity alters the distal gut microbiota. In mice, both genetic obesity and high-fat diet (HFD)–induced obesity alter the two most abundant bacterial phyla in the mammalian GI tract, increasing the ratio of Firmicutes to Bacteroidetes (32–36). The composition of the diet itself has a profound effect on gut community structure. In mice fed an HFD, but food restricted to reduce body weight, the gut bacterial community structure is more similar to that in obese HFD-fed mice with unrestricted access to food than to that in mice fed a low-fat diet ad libitum, even though the latter have body weights more similar to the HFD-fed food-restricted mice (35). The nature of the dietary fat also matters, as changes in gut microbiota induced by HFD differ depending on whether the fat is derived from lard versus fish oil (37).
The marked interindividual variability in the microbial composition of the human gut makes it more difficult to identify obesity-related effects on gut bacterial community structure in humans than in genetically inbred mice (20). Nevertheless, obesity has been shown to reduce the diversity of gut bacteria in human subjects (31, 38). Studies of diet-induced weight loss also indicate that the Firmicutes-to-Bacteroidetes ratio declines with weight loss in humans (39, 40), consistent with the increased Firmicutes-to-Bacteroidetes ratio observed in obese versus lean mice. Compared with obese subjects matched for initial weight who did not undergo surgery, obese subjects who have undergone Roux-en-Y gastric bypass surgery for weight loss also exhibit reductions in several species of Firmicutes, along with increases in γ proteobacteria (40). Importantly, it may be the functional capacities of gut bacteria, rather than their phylogenetic or taxonomic composition, that is important: compositional differences at the taxonomic level do not necessarily result in differences at the functional level (41). Furthermore, key bacteria can have major health effects, even when those bacteria are present at such low abundance that substantial alterations in their prevalence have minimal impact on overall phylogenetic ratios (42). In this respect, it is important to note that the obese and lean human gut microbiomes do differ with respect to genes involved in carbohydrate, lipid, and amino acid metabolism (31). Similarly, diet-induced weight loss in children with genetic obesity (Prader Willi syndrome) also results in changes in microbial genes related to metabolism (43).
Functional Effects of Obesity-Related Changes in Gut Microbiota
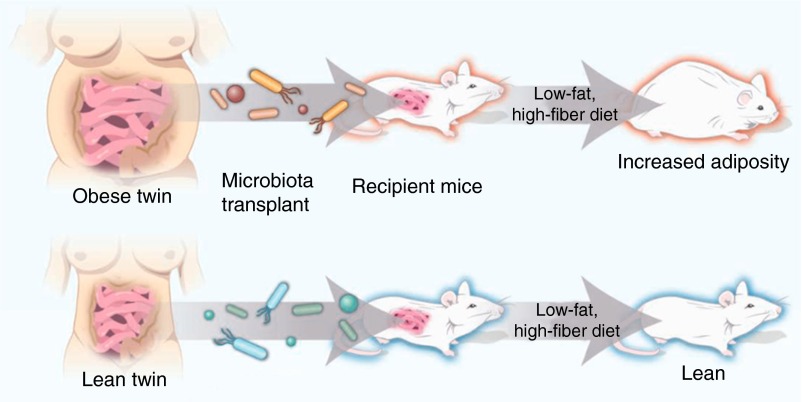
Obesity-related changes in the gut microbiota have important functional consequences for weight gain. Germ-free (GF) mice, mice that are born and raised without exposure to microbes, weigh less than age-, strain-, and sex-matched, conventionally raised (CONV) mice despite greater food consumption and reduced metabolism (44), likely because of reduced microbiota-associated energy harvest from the diet (34, 45). Moreover, GF mice do not gain weight when placed on HFD, but do after bacterial reconstitution by transplant of feces from CONV mice (45, 46). Furthermore, GF mice gain more weight after transplant with fecal contents from CONV obese versus CONV lean mice (34) or after transplant with fecal contents of obese human subjects versus their lean monozygotic twins (47, 48) (Figure 1). Similarly, GF mice colonized with bacteria from feces of obese human subjects who have undergone either dietary or surgically induced weight loss gain less weight than those colonized with bacteria from feces of obese subjects without weight loss intervention (40, 43). In contrast, GF mice transplanted with feces from obese human subjects supplemented with Christensenellaceae resulted in reduced weight compared to GF mice transplanted with feces from obese human subjects without Christensenellaceae supplementation (49). Finally, probiotics (a source of live micro-organisms that provides health benefits to the host) reduce body mass and body fat in mice with diet-induced obesity (50).
Figure 1.
Inoculating germ-free (GF) mice with microbiota from an obese or lean human twin donor results in the GF mice taking on the microbial and adiposity characteristics of the donor. Reproduced from Ref. 48 by permission from the American Association for the Advancement of Science.
There is less evidence for a role of the gut microbiota in regulation of body weight in human subjects. However, a recent large-scale analysis of glycemic responses to a variety of foods in 800 individuals indicated an association between obesity and the gut community structure (51). In addition, a recent case study noted the development of obesity in a previously lean individual who had undergone a fecal transplantation for the management of Clostridium difficile. Notably, the donor was overweight (52). Similarly, Cani and colleagues (53) noted that administration of a prebiotic (a nondigestible food ingredient that stimulates growth of certain gut bacteria, conferring a health benefit to the host) caused increases in several Bifidobacterium, Lactobacillus, Roseburia, and Faecalibacterium species, and also caused a significant increase in meal-induced satiety along with increases in the gut-derived hormones (glucagon-like peptide-1 [GLP-1] and peptide YY [PYY]), which are known to cause satiety in humans (53). Similarly, a report from the Nurses’ Health Study indicates that, among the various foods associated with weight loss, yogurt, a probiotic, has the most profound effect (54).
Although differences in energy harvest from the diet account for at least part of the impact of microbiota on body weight (34, 45), the study by Cani and colleagues (53) indicates that microbes can also impact host eating behavior via effects on satiety hormones. However, some satiety hormones can also impact energy harvest. For example, PYY is reduced in GF versus CONV mice (45). PYY inhibits gut motility, and reductions in PYY in GF mice were associated with an increased rate of food transit through the intestines and, hence, less time for energy harvest. Colonization of GF mice also induces central nervous system resistance to leptin (55), another satiety-inducing hormone, whereas prebiotics that reduce the Firmicutes-to-Bacteroidetes ratio increase leptin sensitivity (56). The production of metabolites that activate cannabinoid receptors (which impact eating) is also affected by gut bacteria (57). Finally, taste receptors for fat are altered in the tongues and intestines of GF versus CONV mice (58), indicating that microbes also impact food preferences.
Microbiome-dependent changes in metabolism may also contribute to the role of the microbiome in obesity. Gut microbiota regulate the intestinal epithelial expression of Fiaf, an angiopoietin-like protein. Fiaf alters lipoprotein lipase activity (44) in adipose tissue, and also impacts expression of genes that regulate mitochondrial oxidation of fatty acids (59).
Obesity-related changes in gut bacteria also impact other aspects of the obese phenotype. For example, antibiotics reverse the insulin resistance caused by high-fat feeding (60), suggesting that obesity-related changes in the microbiome may contribute to type 2 diabetes. Indeed, a small study performed in human subjects indicates that transfer of gut microbiota from nondiabetic into diabetic individuals results in improved insulin sensitivity (61). Furthermore, individual glycemic responses to a large variety of foods can be predicted by components of the gut microbiome assessed in fecal samples (51). Consistent with these observations, probiotic treatments also impact the insulin resistance associated with obesity in mice (62).
Chronic, low-grade systemic inflammation is another consequence of obesity. The adipose tissue of obese mice and obese human subjects is infiltrated with activated macrophages producing a variety of proinflammatory cytokines and chemokines that enter the systemic circulation. Increasing evidence points to a role for the microbiota in these events. For example, HFD feeding results in an increase in the proportion of bacteria containing LPS (endotoxin) in the gut and alters the permeability of the intestinal epithelium, resulting in systemic endotoxemia that contributes to the adipose tissue inflammation. Thus, proinflammatory cytokine levels are elevated by HFD in wild-type, but not Toll-like receptor 4–deficient or myeloid differentiation primary response gene 88 (MyD88)-deficient, mice that lack effective LPS signaling capacities (37, 63, 64). In addition, treatment with Akkermansia muciniphila, bacteria that are typically attenuated in the obese, reverses both endotoxemia and adipose tissue inflammation, in HFD-fed mice (65). Other probiotic treatments also affect the systemic inflammation associated with obesity in mice (66). Given that the systemic inflammation of obesity may also contribute to obesity-associated asthma (67), such results suggest the potential efficacy of probiotic therapeutics in obese subjects with asthma.
Mechanistic Basis for Functional Consequences of Obesity-Related Changes in Gut Microbiota: the Metabolome
As described previously here, endotoxemia may mediate some of the effects of obesity on outcomes such as systemic inflammation and insulin resistance. Given the potential for endotoxin to impact asthma-related outcomes (68), obese microbiome–related changes in endotoxin might also impact asthma. The microbiome also has broad effects on the immune system, including the generation of regulatory T cells (Tregs) and the development of IL-17A–expressing cells (69). As recently reviewed, such effects could also impact obesity-related asthma (70). However, in obesity, there are also profound changes in the metabolome, the set of small-molecule metabolites present in a given biological fluid, cell type, or tissue. Importantly, accumulating evidence suggests a key role for the microbiome in such metabolic changes. Whether and how such changes might contribute to obese asthma remains to be established.
The Metabolome Is Altered in Obesity
Both in humans and in rodents, the metabolomes of the liver, serum, urine, and adipose tissue are altered by obesity (71–80). Given that insulin resistance is common in obesity, it is perhaps not surprising that glucose, lactate, glycerol, fatty acids, and β-hydroxybutyrate are increased in the blood of obese versus lean subjects. Blood metabolomics also consistently indicate obesity-related alterations in branched-chain amino acid metabolites (80–82). Because receptors for many fatty acids (G protein–coupled receptor [GPR] 40, GPR41, GPR49, GPR84, and GPR120), for lactate (hydroxycarboxylic acid [HCA1]/GPR81), and for β-hydroxybutyrate (HCA2/GPR109A) exist (83, 84), and because such small metabolites have the capacity to diffuse across pulmonary capillaries, obesity-related changes in these moieties could impact lung function directly via activation of these receptors.
Gut Microbiota Impact the Metabolome
Gut microbiota metabolize dietary foodstuffs to produce a huge variety of small metabolites that can diffuse across the gut into the circulation, where some are further metabolized by host enzymes, resulting in bacterial–mammalian cometabolites. Thus, gut bacterial–derived metabolites can affect not only the GI tract, but also other target organs, leading to the description of the gut microbiota as an endocrine organ (85). For example, trimethylamine, a microbial-dependent metabolite derived from dietary choline, is oxidized in the liver to produce trimethylamine N-oxide (TMAO). Serum concentrations of TMAO are linked to atherosclerosis and cardiovascular disease risk (86, 87). Similarly, when the cecal contents of atherosclerosis-prone mice are transplanted into antibiotic-treated mice, the mice develop enhanced choline diet–induced atherosclerosis and TMAO (88). Even the brain is impacted by bacterial metabolites. Serum concentrations of the bacterial-dependent metabolite, 4-ethylphenylsulfate, are markedly elevated in mouse models of autism. Importantly, treatment with Bacteroides fragilis in the food reverses these elevations in 4-ethylphenylsulfate and also improves autism-like behavior (89).
Microbiota affect the metabolomes of the intestines, urine, liver, brain, and kidney (27, 90–97). The blood metabolome is also affected. For example, studies using GF mice and studies using antibiotic treatment indicate effects of gut microbiota on serum levels of many bacterial-derived metabolites, including SCFA, pipecolate, choline, phenol sulfate, and hippurate (90, 98, 99). Thus, the altered gut microbiome of obesity may affect metabolites that circulate to the lungs and affect airway function. Subsequently here, we discuss the role of the microbiota in the generation of two groups of metabolites: SCFAs and bile acids, as well as the potential for these metabolites to impact obese asthma.
SCFAs
As discussed previously here, gut bacteria are capable of fermenting polysaccharides and dietary starches that are otherwise indigestible, leading to the production of the SCFAs: butyrate, propionate, and acetate (19, 20). In the GI tract, SCFAs are produced from dietary polysaccharides via bacterial metabolism. Indeed, compared with CONV mice, GF mice have reduced intestinal SCFAs and excrete significantly more calories in their feces in the form of indigestible polysaccharides (20). Consistent with these observations, colonizing GF mice with Bacteroides thetaiotaomicron increases production of SCFAs in GF rodents (100). Similarly, mice fed a high-fiber diet have increased gut Bifidobacterium, a bacterium that ferments dietary fiber to form SCFAs, and increased circulating SCFAs (99). Most SCFA production occurs in the distal colon and cecum, and both the makeup of the microbiota present and the transit time through the colon impact the amount of SCFAs produced (101).
Increasing data support a relationship between the microbiome, SCFAs, and obesity. SCFAs are higher in the cecal contents of genetically obese mice. Increased fecal SCFAs are also observed in human obesity (101). In twins discordant for obesity, the microbiome of the obese twin is enriched for genes involved in carbohydrate fermentation to SCFAs (31). Given the ability of SCFAs to provide an energy source to the host, one might expect that elevated SCFA production in obesity would contribute to weight gain. Nevertheless, exogenously administered SCFAs actually reduce weight gain in mice (102). The ability of SCFAs to increase the production of satiety hormones, including GLP-1, PYY, and leptin (103, 104), and to increase energy expenditure (105), likely explains this apparent conundrum.
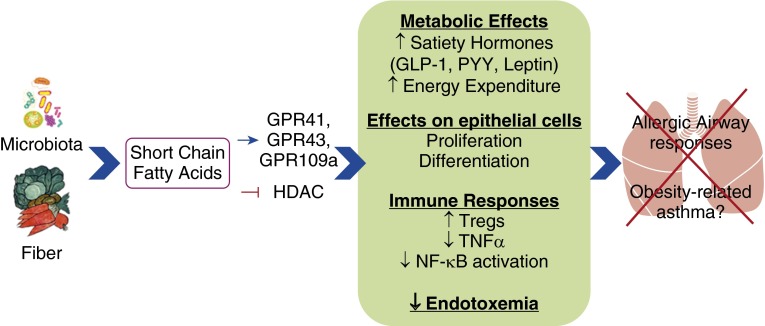
SCFAs have multiple effects that could impact obese asthma (Figure 2). First, SCFAs play a role in the regulation of T cells, both in the GI tract and in peripheral tissues. For example, exogenous administration of SCFA promotes the development of Tregs (106). Reductions of Tregs in adipose tissue are thought to contribute to the systemic inflammation of obesity (107). Second, SCFAs stimulate intestinal epithelial proliferation and differentiation (108), and could also contribute to repair of the epithelial cell damage that is typical of asthma. SCFA-mediated changes in epithelial barrier function could also impact the systemic endotoxemia of obesity. Antiinflammatory effects of SCFAs could also counter the effects of this endotoxemia, as SCFAs inhibit LPS-induced NF-κB activation and increases in TNF-α in neutrophils and macrophages (101, 109). Effects of SCFAs are mediated both by binding of SCFAs to GPRs, including GPR41, GPR43, and GPR109A, which vary in their sensitivity to acetate, propionate, and butyrate, and by inhibition of histone deacetylase (101, 110–112).
Figure 2.
Schematic representation of ways in which short-chain fatty acids from microbiota metabolism of dietary fiber may prevent allergic airway responses and obesity-related asthma. GLP-1, glucagon-like peptide-1; HDAC, histone deacetylase; PYY, peptide YY; Tregs, regulatory T cells.
Although there are, as yet, no studies of the role of SCFAs in obesity-related asthma, data do suggest a role for the gut microbiome, and for SCFAs in particular, in modulating allergic asthma. GF mice, which have a reduced ability to produce SCFAs (20), develop greater allergic airways responses than CONV mice (113, 114). Furthermore, both elevations in circulating SCFAs induced by high-fiber feeding and exogenously administered propionate protect against allergic airways inflammation in mice, and this protective effect is lost in GPR41-deficient mice (99). Similarly, low fiber–fed mice with circulating levels of SCFAs have increased allergic airways responses (99). These effects of SCFAs appear to occur at the level of the bone marrow and dendritic cell precursors that impact the immune response to allergen.
Bile acids
Primary bile acids, such as cholate and chenodeoxycholate, are synthesized from cholesterol and conjugated with either glycine (humans) or taurine (mice) in the liver. Bile is then secreted via the bile duct into the duodenum in response to hormonal signals initiated by eating. In the intestines, bile acids contribute to the digestion of dietary fat by acting as emulsifiers. In the lower GI tract, most of the bile acids are absorbed back into the circulation and returned to the liver, where they are taken up and re-excreted (the enterohepatic circulation), although some escape reuptake and circulate in the systemic blood. The enterohepatic circulation thus prevents loss of cholesterol-containing moieties in the feces (40, 115, 116).
Gut microbes both modify and are modified by bile acids (40, 115, 116). Gut bacteria deconjugate and dehydroxylate bile acids, resulting in the formation of secondary bile acids. Consequently, there is an increase in the ratio of conjugated to unconjugated bile acids in GF and antibiotic-treated mice (117). Bile acids themselves have bactericidal properties via their detergent properties on bacterial membranes, although certain bacteria (e.g., Bilofilia wadsorthia) actually thrive in environments rich in bile acids (118). Thus, changes in the release of bile acids, for example, in response to HFDs, can alter the community structure of the gut microbiome. Indeed there are substantial changes in the gut microbiome in rats fed a diet containing high levels of cholate (119).
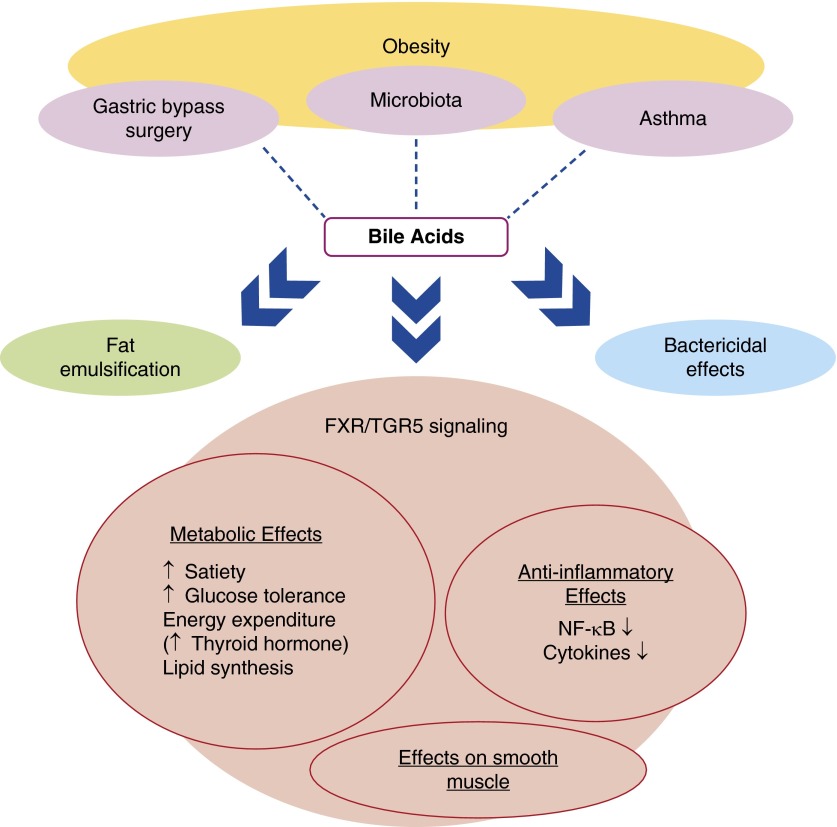
In addition to their bacteriostatic and emulsifying actions, bile acids also have a signaling role. Bile acid binding to two receptors, farnesoid X receptor (FXR), a nuclear receptor, and to a GRP, Takeda G protein-coupled receptor 5 (TGR5), mediate these effects. FXR- and TGR5-mediated signaling events contribute to beneficial effects of bile acids against obesity and obesity-related conditions (Figure 3). For example, bile acids increase energy expenditure through TGR5-mediated changes in thyroid hormone synthesis (120). Bile acids also cause TGR5-dependent secretion of the satiety hormone, PYY (121). In the liver, bile acid–induced activation of FXR results in reduced fatty acid synthesis and decreases circulating triglycerides (120). FXR activation also inhibits hepatic gluconeogenesis. Indeed, FXR-deficient mice are insulin resistant (122). A major role for a bile/microbiome axis in obesity was also revealed by a study showing that GF mice receiving the fecal microbiota from an obese twin displayed not only a greater fat mass than mice receiving gut microbes from the lean twin, but also reduced levels of several bile acids and reduced FXR-dependent gene transcription in the ileum and the liver (47).
Figure 3.
Schematic representation of ways in which bile acid modifications in obesity may impact obesity-related asthma. FXR, farnesoid X receptor; TGR5, Takeda G protein–coupled receptor 5.
Bariatric surgery results in increased circulating bile acids (115) and, in mice with HFD-induced obesity, these bile acids appear to mediate reduced eating, subsequent weight loss, and improved glucose tolerance, because these effects are reduced in FXR-deficient mice (123). Bariatric surgery alters gut microbial communities, and there are also differential effects of bariatric surgery on the gut microbiome in FXR versus wild-type mice (123). Bariatric surgery also improves obesity-associated asthma (6), but whether or not bile acid– and/or microbiome-mediated changes are involved in these events remains to be determined. However, it is interesting to note that a recent metabolomic profiling of plasma from subjects with asthma versus healthy subjects identified bile acids (taurocholate and glycodeoxycholate) among the metabolites that were affected by asthma (124).
Bile acids also have antiinflammatory effects that may be relevant for asthma (125). For example, in macrophages, TGR5 activation causes elevations in cyclic adenosine monophosphate (cAMP) that inhibits NF-κB–mediated induction of proinflammatory cytokines by LPS (126). The observation that TGR5 agonists attenuate atherosclerosis in TGR5-sufficient, but not TGR5-deficient, mice (127) indicates that there are important functional consequences of such antiinflammatory effects of bile acids.
Finally, bile acids, via TGR5 signaling, promote relaxation of gastric smooth muscle (128). TGR5-dependent activation of Gαs and consequent increases in cAMP mediate this relaxation. Because airway smooth muscle also relaxes in response to elevations in cAMP, it is conceivable that changes in bile acids could also impact the bronchoconstriction of asthma.
Conclusions
Obesity alters the gut bacterial community structure. Such changes could play a role in obesity-related asthma via alterations in production of bacterial-derived or modified metabolites, such as SCFAs or bile acids. A role for the microbiome in obesity-related asthma has both public health and therapeutic implications. The gut microbiome is shaped by early life events, including mode of delivery, breastfeeding, diet, and antibiotic use. Understanding the impact of these factors on the development of both obesity and asthma could alter early life decisions that impact long-term disease development. Greater understanding of the role of microbiota in obesity-related asthma could also pave the way for the development of novel microbiota-based treatments for this difficult-to-treat group. For example, it is conceivable that altering the gut microbiota with probiotics, prebiotics, or even fecal transplants could ameliorate obesity-related asthma or improve the ability of obese subjects with asthma to respond to standard asthma therapeutics. Indeed, such interventions are effective against other obesity-related conditions (61, 129–131). Microbiota-based therapies that impact weight might also prove effective, given the efficacy of weight loss in obese subjects with asthma (6). To the extent that obesity-related changes in SCFAs or bile acids are important, interventions that promote the survival of SCFA-producing bacteria, high-fiber diets, or even direct administration of SCFAs, or GRP41/43, TGR5, or FXR ligands might also prove effective.
Footnotes
This work was supported by National Institutes of Environmental Health and Safety grants: ES000002, ES013307, and ES024032.
Author Contributions: S.A.S. and Y.C. both contributed to the drafting of the manuscript for important intellectual content
Originally Published in Press as DOI: 10.1165/rcmb.2016-0052PS on March 7, 2016
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Ford ES. The epidemiology of obesity and asthma. J Allergy Clin Immunol. 2005;115:897–909, quiz 910. doi: 10.1016/j.jaci.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 2.Shore SA, Johnston RA. Obesity and asthma. Pharmacol Ther. 2006;110:83–102. doi: 10.1016/j.pharmthera.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 3.Sutherland ER. Obesity and asthma. Immunol Allergy Clin North Am. 2008;28:589–602, ix. doi: 10.1016/j.iac.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schatz M, Hsu JW, Zeiger RS, Chen W, Dorenbaum A, Chipps BE, Haselkorn T. Phenotypes determined by cluster analysis in severe or difficult-to-treat asthma. J Allergy Clin Immunol. 2014;133:1549–1556. doi: 10.1016/j.jaci.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 5.Gibeon D, Batuwita K, Osmond M, Heaney LG, Brightling CE, Niven R, Mansur A, Chaudhuri R, Bucknall CE, Rowe A, et al. Obesity-associated severe asthma represents a distinct clinical phenotype: analysis of the British Thoracic Society Difficult Asthma Registry Patient cohort according to BMI. Chest. 2013;143:406–414. doi: 10.1378/chest.12-0872. [DOI] [PubMed] [Google Scholar]
- 6.Dixon AE, Pratley RE, Forgione PM, Kaminsky DA, Whittaker-Leclair LA, Griffes LA, Garudathri J, Raymond D, Poynter ME, Bunn JY, et al. Effects of obesity and bariatric surgery on airway hyperresponsiveness, asthma control, and inflammation. J Allergy Clin Immunol. 2011;128:508–15.e1, 2. doi: 10.1016/j.jaci.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boulet LP, Turcotte H, Martin J, Poirier P. Effect of bariatric surgery on airway response and lung function in obese subjects with asthma. Respir Med. 2012;106:651–660. doi: 10.1016/j.rmed.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 8.Shore SA. Obesity and asthma: lessons from animal models. J Appl Physiol (1985) 2007;102:516–528. doi: 10.1152/japplphysiol.00847.2006. [DOI] [PubMed] [Google Scholar]
- 9.Shore SA. Obesity, airway hyperresponsiveness, and inflammation. J Appl Physiol (1985) 2010;108:735–743. doi: 10.1152/japplphysiol.00749.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arteaga-Solis E, Zee T, Emala CW, Vinson C, Wess J, Karsenty G. Inhibition of leptin regulation of parasympathetic signaling as a cause of extreme body weight–associated asthma. Cell Metab. 2013;17:35–48. doi: 10.1016/j.cmet.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim HY, Lee HJ, Chang YJ, Pichavant M, Shore SA, Fitzgerald KA, Iwakura Y, Israel E, Bolger K, Faul J, et al. Interleukin-17–producing innate lymphoid cells and the NLRP3 inflammasome facilitate obesity-associated airway hyperreactivity. Nat Med. 2014;20:54–61. doi: 10.1038/nm.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leiria LO, Arantes-Costa FM, Calixto MC, Alexandre EC, Moura RF, Folli F, Prado CM, Prado MA, Prado VF, Velloso LA, et al. Increased airway reactivity and hyperinsulinemia in obese mice are linked by ERK signaling in brain stem cholinergic neurons. Cell Reports. 2015;11:934–943. doi: 10.1016/j.celrep.2015.04.012. [DOI] [PubMed] [Google Scholar]
- 13.Dixon AE, Shade DM, Cohen RI, Skloot GS, Holbrook JT, Smith LJ, Lima JJ, Allayee H, Irvin CG, Wise RA American Lung Association-Asthma Clinical Research Centers. Effect of obesity on clinical presentation and response to treatment in asthma. J Asthma. 2006;43:553–558. doi: 10.1080/02770900600859123. [DOI] [PubMed] [Google Scholar]
- 14.Pradeepan S, Garrison G, Dixon AE. Obesity in asthma: approaches to treatment. Curr Allergy Asthma Rep. 2013;13:434–442. doi: 10.1007/s11882-013-0354-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sutherland ER, Goleva E, Strand M, Beuther DA, Leung DY. Body mass and glucocorticoid response in asthma. Am J Respir Crit Care Med. 2008;178:682–687. doi: 10.1164/rccm.200801-076OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Veen IH, Ten Brinke A, Sterk PJ, Rabe KF, Bel EH. Airway inflammation in obese and nonobese patients with difficult-to-treat asthma. Allergy. 2008;63:570–574. doi: 10.1111/j.1398-9995.2007.01597.x. [DOI] [PubMed] [Google Scholar]
- 17.Wenzel SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat Med. 2012;18:716–725. doi: 10.1038/nm.2678. [DOI] [PubMed] [Google Scholar]
- 18.Rosenbaum M, Knight R, Leibel RL. The gut microbiota in human energy homeostasis and obesity. Trends Endocrinol Metab. 2015;26:493–501. doi: 10.1016/j.tem.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD, Gordon JI. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 20.Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature. 2012;489:242–249. doi: 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- 21.Hill MJ. Intestinal flora and endogenous vitamin synthesis. Eur J Cancer Prev. 1997;6:S43–S45. doi: 10.1097/00008469-199703001-00009. [DOI] [PubMed] [Google Scholar]
- 22.Kau AL, Ahern PP, Griffin NW, Goodman AL, Gordon JI. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldsmith JR, Sartor RB. The role of diet on intestinal microbiota metabolism: downstream impacts on host immune function and health, and therapeutic implications. J Gastroenterol. 2014;49:785–798. doi: 10.1007/s00535-014-0953-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsai F, Coyle WJ. The microbiome and obesity: is obesity linked to our gut flora? Curr Gastroenterol Rep. 2009;11:307–313. doi: 10.1007/s11894-009-0045-z. [DOI] [PubMed] [Google Scholar]
- 25.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson CH, Patterson AD, Idle JR, Gonzalez FJ. Xenobiotic metabolomics: major impact on the metabolome. Annu Rev Pharmacol Toxicol. 2012;52:37–56. doi: 10.1146/annurev-pharmtox-010611-134748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holmes E, Li JV, Marchesi JR, Nicholson JK. Gut microbiota composition and activity in relation to host metabolic phenotype and disease risk. Cell Metab. 2012;16:559–564. doi: 10.1016/j.cmet.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 28.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. Host–gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 29.Sommer F, Bäckhed F. The gut microbiota—masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 30.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3:213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 35.Ravussin Y, Koren O, Spor A, LeDuc C, Gutman R, Stombaugh J, Knight R, Ley RE, Leibel RL. Responses of gut microbiota to diet composition and weight loss in lean and obese mice. Obesity (Silver Spring) 2012;20:738–747. doi: 10.1038/oby.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010;328:228–231. doi: 10.1126/science.1179721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caesar R, Tremaroli V, Kovatcheva-Datchary P, Cani PD, Bäckhed F. Crosstalk between gut microbiota and dietary lipids aggravates wat inflammation through TLR signaling. Cell Metab. 2015;22:658–668. doi: 10.1016/j.cmet.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy S, et al. MetaHIT Consortium. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 39.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 40.Tremaroli V, Karlsson F, Werling M, Ståhlman M, Kovatcheva-Datchary P, Olbers T, Fändriks L, le Roux CW, Nielsen J, Bäckhed F. Roux-en-Y gastric bypass and vertical banded gastroplasty induce long-term changes on the human gut microbiome contributing to fat mass regulation. Cell Metab. 2015;22:228–238. doi: 10.1016/j.cmet.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, Kuzeljevic B, Gold MJ, Britton HM, Lefebvre DL, et al. CHILD Study Investigators. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7:307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 43.Zhang C, Yin A, Li H, Wang R, Wu G, Shen J, Zhang M, Wang L, Hou Y, Ouyang H, et al. Dietary modulation of gut microbiota contributes to alleviation of both genetic and simple obesity in children. EBioMedicine. 2015;2:966–982. doi: 10.1016/j.ebiom.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samuel BS, Shaito A, Motoike T, Rey FE, Backhed F, Manchester JK, Hammer RE, Williams SC, Crowley J, Yanagisawa M, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein–coupled receptor, Gpr41. Proc Natl Acad Sci USA. 2008;105:16767–16772. doi: 10.1073/pnas.0808567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bäckhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host–bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 47.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walker AW, Parkhill J. Microbiology: fighting obesity with bacteria. Science. 2013;341:1069–1070. doi: 10.1126/science.1243787. [DOI] [PubMed] [Google Scholar]
- 49.Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, Blekhman R, Beaumont M, Van Treuren W, Knight R, Bell JT, et al. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee HY, Park JH, Seok SH, Baek MW, Kim DJ, Lee KE, Paek KS, Lee Y, Park JH. Human originated bacteria, Lactobacillus rhamnosus PL60, produce conjugated linoleic acid and show anti-obesity effects in diet-induced obese mice. Biochim Biophys Acta. 2006;1761:736–744. doi: 10.1016/j.bbalip.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 51.Zeevi D, Korem T, Zmora N, Israeli D, Rothschild D, Weinberger A, Ben-Yacov O, Lador D, Avnit-Sagi T, Lotan-Pompan M, et al. Personalized nutrition by prediction of glycemic responses. Cell. 2015;163:1079–1094. doi: 10.1016/j.cell.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 52.Alang N, Kelly CR. Weight gain after fecal microbiota transplantation. Open Forum Infect Dis. 2015;2:ofv004. doi: 10.1093/ofid/ofv004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cani PD, Lecourt E, Dewulf EM, Sohet FM, Pachikian BD, Naslain D, De Backer F, Neyrinck AM, Delzenne NM. Gut microbiota fermentation of prebiotics increases satietogenic and incretin gut peptide production with consequences for appetite sensation and glucose response after a meal. Am J Clin Nutr. 2009;90:1236–1243. doi: 10.3945/ajcn.2009.28095. [DOI] [PubMed] [Google Scholar]
- 54.Mozaffarian D, Hao T, Rimm EB, Willett WC, Hu FB. Changes in diet and lifestyle and long-term weight gain in women and men. N Engl J Med. 2011;364:2392–2404. doi: 10.1056/NEJMoa1014296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schéle E, Grahnemo L, Anesten F, Hallén A, Bäckhed F, Jansson JO. The gut microbiota reduces leptin sensitivity and the expression of the obesity-suppressing neuropeptides proglucagon (Gcg) and brain-derived neurotrophic factor (Bdnf) in the central nervous system. Endocrinology. 2013;154:3643–3651. doi: 10.1210/en.2012-2151. [DOI] [PubMed] [Google Scholar]
- 56.Everard A, Lazarevic V, Derrien M, Girard M, Muccioli GG, Neyrinck AM, Possemiers S, Van Holle A, François P, de Vos WM, et al. Responses of gut microbiota and glucose and lipid metabolism to prebiotics in genetic obese and diet-induced leptin-resistant mice. Diabetes. 2011;60:2775–2786. doi: 10.2337/db11-0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cani PD, Plovier H, Van Hul M, Geurts L, Delzenne NM, Druart C, Everard A. Endocannabinoids—at the crossroads between the gut microbiota and host metabolism. Nat Rev Endocrinol. 2016;12:133–143. doi: 10.1038/nrendo.2015.211. [DOI] [PubMed] [Google Scholar]
- 58.Duca FA, Swartz TD, Sakar Y, Covasa M. Increased oral detection, but decreased intestinal signaling for fats in mice lacking gut microbiota. PLoS One. 2012;7:e39748. doi: 10.1371/journal.pone.0039748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci USA. 2007;104:979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, Burcelin R. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet–induced obesity and diabetes in mice. Diabetes. 2008;57:1470–1481. doi: 10.2337/db07-1403. [DOI] [PubMed] [Google Scholar]
- 61.Vrieze A, Van Nood E, Holleman F, Salojärvi J, Kootte RS, Bartelsman JF, Dallinga-Thie GM, Ackermans MT, Serlie MJ, Oozeer R, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143:913–6.e7. doi: 10.1053/j.gastro.2012.06.031. [DOI] [PubMed] [Google Scholar]
- 62.Cano PG, Santacruz A, Trejo FM, Sanz Y. Bifidobacterium CECT 7765 improves metabolic and immunological alterations associated with obesity in high-fat diet–fed mice. Obesity (Silver Spring) 2013;21:2310–2321. doi: 10.1002/oby.20330. [DOI] [PubMed] [Google Scholar]
- 63.Kim KA, Gu W, Lee IA, Joh EH, Kim DH. High fat diet–induced gut microbiota exacerbates inflammation and obesity in mice via the TLR4 signaling pathway. PLoS One. 2012;7:e47713. doi: 10.1371/journal.pone.0047713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, Neyrinck AM, Fava F, Tuohy KM, Chabo C, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes. 2007;56:1761–1772. doi: 10.2337/db06-1491. [DOI] [PubMed] [Google Scholar]
- 65.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA. 2013;110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cani PD, Possemiers S, Van de Wiele T, Guiot Y, Everard A, Rottier O, Geurts L, Naslain D, Neyrinck A, Lambert DM, et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2–driven improvement of gut permeability. Gut. 2009;58:1091–1103. doi: 10.1136/gut.2008.165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shore SA. Obesity and asthma: possible mechanisms. J Allergy Clin Immunol. 2008;121:1087–1093, quiz 1094–1095. doi: 10.1016/j.jaci.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 68.Heederik D, von Mutius E. Does diversity of environmental microbial exposure matter for the occurrence of allergy and asthma? J Allergy Clin Immunol. 2012;130:44–50. doi: 10.1016/j.jaci.2012.01.067. [DOI] [PubMed] [Google Scholar]
- 69.Burkett PR, Meyer zu Horste G, Kuchroo VK. Pouring fuel on the fire: Th17 cells, the environment, and autoimmunity. J Clin Invest. 2015;125:2211–2219. doi: 10.1172/JCI78085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cho Y, Shore SA. Obesity, asthma, and the microbiome. Physiology (Bethesda) 2016;31:108–116. doi: 10.1152/physiol.00045.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Griffin JL, Nicholls AW. Metabolomics as a functional genomic tool for understanding lipid dysfunction in diabetes, obesity and related disorders. Pharmacogenomics. 2006;7:1095–1107. doi: 10.2217/14622416.7.7.1095. [DOI] [PubMed] [Google Scholar]
- 72.Connor SC, Hansen MK, Corner A, Smith RF, Ryan TE. Integration of metabolomics and transcriptomics data to aid biomarker discovery in type 2 diabetes. Mol Biosyst. 2010;6:909–921. doi: 10.1039/b914182k. [DOI] [PubMed] [Google Scholar]
- 73.Kim HJ, Kim JH, Noh S, Hur HJ, Sung MJ, Hwang JT, Park JH, Yang HJ, Kim MS, Kwon DY, et al. Metabolomic analysis of livers and serum from high-fat diet induced obese mice. J Proteome Res. 2011;10:722–731. doi: 10.1021/pr100892r. [DOI] [PubMed] [Google Scholar]
- 74.Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17:448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Morris C, O’Grada C, Ryan M, Roche HM, Gibney MJ, Gibney ER, Brennan L. The relationship between BMI and metabolomic profiles: a focus on amino acids. Proc Nutr Soc. 2012;71:634–638. doi: 10.1017/S0029665112000699. [DOI] [PubMed] [Google Scholar]
- 76.Oresic M. Obesity and psychotic disorders: uncovering common mechanisms through metabolomics. Dis Model Mech. 2012;5:614–620. doi: 10.1242/dmm.009845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shah SH, Kraus WE, Newgard CB. Metabolomic profiling for the identification of novel biomarkers and mechanisms related to common cardiovascular diseases: form and function. Circulation. 2012;126:1110–1120. doi: 10.1161/CIRCULATIONAHA.111.060368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xie B, Waters MJ, Schirra HJ. Investigating potential mechanisms of obesity by metabolomics. J Biomed Biotechnol. 2012;2012:805683. doi: 10.1155/2012/805683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang A, Sun H, Wang X. Power of metabolomics in biomarker discovery and mining mechanisms of obesity. Obes Rev. 2013;14:344–349. doi: 10.1111/obr.12011. [DOI] [PubMed] [Google Scholar]
- 80.She P, Olson KC, Kadota Y, Inukai A, Shimomura Y, Hoppel CL, Adams SH, Kawamata Y, Matsumoto H, Sakai R, et al. Leucine and protein metabolism in obese Zucker rats. PLoS One. 2013;8:e59443. doi: 10.1371/journal.pone.0059443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, Haqq AM, Shah SH, Arlotto M, Slentz CA, et al. A branched-chain amino acid–related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9:311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Batch BC, Shah SH, Newgard CB, Turer CB, Haynes C, Bain JR, Muehlbauer M, Patel MJ, Stevens RD, Appel LJ, et al. Branched chain amino acids are novel biomarkers for discrimination of metabolic wellness. Metabolism. 2013;62:961–969. doi: 10.1016/j.metabol.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Oh DY, Lagakos WS. The role of G-protein–coupled receptors in mediating the effect of fatty acids on inflammation and insulin sensitivity. Curr Opin Clin Nutr Metab Care. 2011;14:322–327. doi: 10.1097/MCO.0b013e3283479230. [DOI] [PubMed] [Google Scholar]
- 84.Davenport AP, Alexander SP, Sharman JL, Pawson AJ, Benson HE, Monaghan AE, Liew WC, Mpamhanga CP, Bonner TI, Neubig RR, et al. International Union of Basic and Clinical Pharmacology. LXXXVIII. G protein–coupled receptor list: recommendations for new pairings with cognate ligands. Pharmacol Rev. 2013;65:967–986. doi: 10.1124/pr.112.007179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brown JM, Hazen SL. The gut microbial endocrine organ: bacterially derived signals driving cardiometabolic diseases. Annu Rev Med. 2015;66:343–359. doi: 10.1146/annurev-med-060513-093205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung YM, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bennett BJ, de Aguiar Vallim TQ, Wang Z, Shih DM, Meng Y, Gregory J, Allayee H, Lee R, Graham M, Crooke R, et al. Trimethylamine-N-oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation. Cell Metab. 2013;17:49–60. doi: 10.1016/j.cmet.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gregory JC, Buffa JA, Org E, Wang Z, Levison BS, Zhu W, Wagner MA, Bennett BJ, Li L, DiDonato JA, et al. Transmission of atherosclerosis susceptibility with gut microbial transplantation. J Biol Chem. 2015;290:5647–5660. doi: 10.1074/jbc.M114.618249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Martin FP, Dumas ME, Wang Y, Legido-Quigley C, Yap IK, Tang H, Zirah S, Murphy GM, Cloarec O, Lindon JC, et al. A top-down systems biology view of microbiome-mammalian metabolic interactions in a mouse model. Mol Syst Biol. 2007;3:112. doi: 10.1038/msb4100153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Holmes E, Li JV, Athanasiou T, Ashrafian H, Nicholson JK. Understanding the role of gut microbiome–host metabolic signal disruption in health and disease. Trends Microbiol. 2011;19:349–359. doi: 10.1016/j.tim.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 92.Chuang HL, Huang YT, Chiu CC, Liao CD, Hsu FL, Huang CC, Hou CC. Metabolomics characterization of energy metabolism reveals glycogen accumulation in gut-microbiota–lacking mice. J Nutr Biochem. 2012;23:752–758. doi: 10.1016/j.jnutbio.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 93.Mestdagh R, Dumas ME, Rezzi S, Kochhar S, Holmes E, Claus SP, Nicholson JK. Gut microbiota modulate the metabolism of brown adipose tissue in mice. J Proteome Res. 2012;11:620–630. doi: 10.1021/pr200938v. [DOI] [PubMed] [Google Scholar]
- 94.Matsumoto M, Kibe R, Ooga T, Aiba Y, Kurihara S, Sawaki E, Koga Y, Benno Y. Impact of intestinal microbiota on intestinal luminal metabolome. Sci Rep. 2012;2:233. doi: 10.1038/srep00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Matsumoto M, Kibe R, Ooga T, Aiba Y, Sawaki E, Koga Y, Benno Y. Cerebral low-molecular metabolites influenced by intestinal microbiota: a pilot study. Front Syst Neurosci. 2013;7:9. doi: 10.3389/fnsys.2013.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bajaj JS, Heuman DM, Sanyal AJ, Hylemon PB, Sterling RK, Stravitz RT, Fuchs M, Ridlon JM, Daita K, Monteith P, et al. Modulation of the metabiome by rifaximin in patients with cirrhosis and minimal hepatic encephalopathy. PLoS One. 2013;8:e60042. doi: 10.1371/journal.pone.0060042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Marcobal A, Kashyap PC, Nelson TA, Aronov PA, Donia MS, Spormann A, Fischbach MA, Sonnenburg JL. A metabolomic view of how the human gut microbiota impacts the host metabolome using humanized and gnotobiotic mice. ISME J. 2013;7:1933–1943. doi: 10.1038/ismej.2013.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, Blanchard C, Junt T, Nicod LP, Harris NL, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014;20:159–166. doi: 10.1038/nm.3444. [DOI] [PubMed] [Google Scholar]
- 100.Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host–archaeal-bacterial mutualism. Proc Natl Acad Sci USA. 2006;103:10011–10016. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Canfora EE, Jocken JW, Blaak EE. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat Rev Endocrinol. 2015;11:577–591. doi: 10.1038/nrendo.2015.128. [DOI] [PubMed] [Google Scholar]
- 102.den Besten G, Bleeker A, Gerding A, van Eunen K, Havinga R, van Dijk TH, Oosterveer MH, Jonker JW, Groen AK, Reijngoud DJ, et al. Short-chain fatty acids protect against high-fat diet–induced obesity via a PPARγ-dependent switch from lipogenesis to fat oxidation. Diabetes. 2015;64:2398–2408. doi: 10.2337/db14-1213. [DOI] [PubMed] [Google Scholar]
- 103.Tolhurst G, Heffron H, Lam YS, Parker HE, Habib AM, Diakogiannaki E, Cameron J, Grosse J, Reimann F, Gribble FM. Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein–coupled receptor FFAR2. Diabetes. 2012;61:364–371. doi: 10.2337/db11-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xiong Y, Miyamoto N, Shibata K, Valasek MA, Motoike T, Kedzierski RM, Yanagisawa M. Short-chain fatty acids stimulate leptin production in adipocytes through the G protein–coupled receptor GPR41. Proc Natl Acad Sci USA. 2004;101:1045–1050. doi: 10.1073/pnas.2637002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gao Z, Yin J, Zhang J, Ward RE, Martin RJ, Lefevre M, Cefalu WT, Ye J. Butyrate improves insulin sensitivity and increases energy expenditure in mice. Diabetes. 2009;58:1509–1517. doi: 10.2337/db08-1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, Garrett WS. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nishimura S, Manabe I, Nagasaki M, Eto K, Yamashita H, Ohsugi M, Otsu M, Hara K, Ueki K, Sugiura S, et al. CD8+ effector T cells contribute to macrophage recruitment and adipose tissue inflammation in obesity. Nat Med. 2009;15:914–920. doi: 10.1038/nm.1964. [DOI] [PubMed] [Google Scholar]
- 108.Frankel WL, Zhang W, Singh A, Klurfeld DM, Don S, Sakata T, Modlin I, Rombeau JL. Mediation of the trophic effects of short-chain fatty acids on the rat jejunum and colon. Gastroenterology. 1994;106:375–380. doi: 10.1016/0016-5085(94)90595-9. [DOI] [PubMed] [Google Scholar]
- 109.Steinmeyer S, Lee K, Jayaraman A, Alaniz RC. Microbiota metabolite regulation of host immune homeostasis: a mechanistic missing link. Curr Allergy Asthma Rep. 2015;15:24. doi: 10.1007/s11882-015-0524-2. [DOI] [PubMed] [Google Scholar]
- 110.Davie JR. Inhibition of histone deacetylase activity by butyrate. J Nutr. 2003;133(7) suppl:2485S–2493S. doi: 10.1093/jn/133.7.2485S. [DOI] [PubMed] [Google Scholar]
- 111.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, Schilter HC, Rolph MS, Mackay F, Artis D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kimura I, Ozawa K, Inoue D, Imamura T, Kimura K, Maeda T, Terasawa K, Kashihara D, Hirano K, Tani T, et al. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat Commun. 2013;4:1829. doi: 10.1038/ncomms2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Herbst T, Sichelstiel A, Schär C, Yadava K, Bürki K, Cahenzli J, McCoy K, Marsland BJ, Harris NL. Dysregulation of allergic airway inflammation in the absence of microbial colonization. Am J Respir Crit Care Med. 2011;184:198–205. doi: 10.1164/rccm.201010-1574OC. [DOI] [PubMed] [Google Scholar]
- 114.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Penney NC, Kinross J, Newton RC, Purkayastha S. The role of bile acids in reducing the metabolic complications of obesity after bariatric surgery: a systematic review. Int J Obes. 2015;39:1565–1574. doi: 10.1038/ijo.2015.115. [DOI] [PubMed] [Google Scholar]
- 116.Barlow GM, Yu A, Mathur R. Role of the gut microbiome in obesity and diabetes mellitus. Nutr Clin Pract. 2015;30:787–797. doi: 10.1177/0884533615609896. [DOI] [PubMed] [Google Scholar]
- 117.Swann JR, Want EJ, Geier FM, Spagou K, Wilson ID, Sidaway JE, Nicholson JK, Holmes E. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc Natl Acad Sci USA. 2011;108:4523–4530. doi: 10.1073/pnas.1006734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B, Chang EB. Dietary-fat–induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Islam KB, Fukiya S, Hagio M, Fujii N, Ishizuka S, Ooka T, Ogura Y, Hayashi T, Yokota A. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology. 2011;141:1773–1781. doi: 10.1053/j.gastro.2011.07.046. [DOI] [PubMed] [Google Scholar]
- 120.Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. Targeting bile-acid signalling for metabolic diseases. Nat Rev Drug Discov. 2008;7:678–693. doi: 10.1038/nrd2619. [DOI] [PubMed] [Google Scholar]
- 121.Ullmer C, Alvarez Sanchez R, Sprecher U, Raab S, Mattei P, Dehmlow H, Sewing S, Iglesias A, Beauchamp J, Conde-Knape K. Systemic bile acid sensing by G protein–coupled bile acid receptor 1 (GPBAR1) promotes PYY and GLP-1 release. Br J Pharmacol. 2013;169:671–684. doi: 10.1111/bph.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.de Aguiar Vallim TQ, Tarling EJ, Ahn H, Hagey LR, Romanoski CE, Lee RG, Graham MJ, Motohashi H, Yamamoto M, Edwards PA. MAFG is a transcriptional repressor of bile acid synthesis and metabolism. Cell Metab. 2015;21:298–310. doi: 10.1016/j.cmet.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ryan KK, Tremaroli V, Clemmensen C, Kovatcheva-Datchary P, Myronovych A, Karns R, Wilson-Pérez HE, Sandoval DA, Kohli R, Bäckhed F, et al. FXR is a molecular target for the effects of vertical sleeve gastrectomy. Nature. 2014;509:183–188. doi: 10.1038/nature13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Comhair SA, McDunn J, Bennett C, Fettig J, Erzurum SC, Kalhan SC. Metabolomic endotype of asthma. J Immunol. 2015;195:643–650. doi: 10.4049/jimmunol.1500736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sipka S, Bruckner G. The immunomodulatory role of bile acids. Int Arch Allergy Immunol. 2014;165:1–8. doi: 10.1159/000366100. [DOI] [PubMed] [Google Scholar]
- 126.Wang YD, Chen WD, Yu D, Forman BM, Huang W. The G-protein–coupled bile acid receptor, Gpbar1 (TGR5), negatively regulates hepatic inflammatory response through antagonizing nuclear factor κ light-chain enhancer of activated B cells (NF-κB) in mice. Hepatology. 2011;54:1421–1432. doi: 10.1002/hep.24525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pols TW, Nomura M, Harach T, Lo Sasso G, Oosterveer MH, Thomas C, Rizzo G, Gioiello A, Adorini L, Pellicciari R, et al. TGR5 activation inhibits atherosclerosis by reducing macrophage inflammation and lipid loading. Cell Metab. 2011;14:747–757. doi: 10.1016/j.cmet.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rajagopal S, Kumar DP, Mahavadi S, Bhattacharya S, Zhou R, Corvera CU, Bunnett NW, Grider JR, Murthy KS. Activation of G protein–coupled bile acid receptor, TGR5, induces smooth muscle relaxation via both Epac- and PKA-mediated inhibition of RhoA/Rho kinase pathway. Am J Physiol Gastrointest Liver Physiol. 2013;304:G527–G535. doi: 10.1152/ajpgi.00388.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Delzenne NM, Neyrinck AM, Bäckhed F, Cani PD. Targeting gut microbiota in obesity: effects of prebiotics and probiotics. Nat Rev Endocrinol. 2011;7:639–646. doi: 10.1038/nrendo.2011.126. [DOI] [PubMed] [Google Scholar]
- 130.Park DY, Ahn YT, Park SH, Huh CS, Yoo SR, Yu R, Sung MK, McGregor RA, Choi MS. Supplementation of Lactobacillus curvatus HY7601 and Lactobacillus plantarum KY1032 in diet-induced obese mice is associated with gut microbial changes and reduction in obesity. PLoS One. 2013;8:e59470. doi: 10.1371/journal.pone.0059470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yoo SR, Kim YJ, Park DY, Jung UJ, Jeon SM, Ahn YT, Huh CS, McGregor R, Choi MS. Probiotics L. plantarum and L. curvatus in combination alter hepatic lipid metabolism and suppress diet-induced obesity. Obesity (Silver Spring) 2013;21:2571–2578. doi: 10.1002/oby.20428. [DOI] [PubMed] [Google Scholar]