Abstract
The fetal origins of disease hypothesis suggests that variations in the course of prenatal lung development may affect life-long pulmonary function growth, decline, and pathobiology. Many studies support the existence of differences in the developing lung trajectory in males and females, and sex-specific differences in the prevalence of chronic lung diseases, such as asthma and bronchopulmonary dysplasia. The objectives of this study were to investigate the early developing fetal lung for transcriptomic correlates of postconception age (maturity) and sex, and their associations with chronic lung diseases. We analyzed whole-lung transcriptome profiles of 61 females and 78 males at 54–127 days postconception (dpc) from nonsmoking mothers using unsupervised principal component analysis and supervised linear regression models. We identified dominant transcriptomic correlates for postconception age and sex with corresponding gene sets that were enriched for developing lung structural and functional ontologies. We observed that the transcriptomic sex difference was not a uniform global time shift/lag, rather, lungs of males appear to be more mature than those of females before 96 dpc, and females appear to be more mature than males after 96 dpc. The age correlate gene set was consistently enriched for asthma and bronchopulmonary dysplasia genes, but the sex correlate gene sets were not. Despite sex differences in the developing fetal lung transcriptome, postconception age appears to be more dominant than sex in the effect of early fetal lung developments on disease risk during this early pseudoglandular phase of development.
Keywords: lung, embryonic and fetal development, bronchopulmonary dyplasia, asthma, sex dimorphism
Clinical Relevance
This study relates the transcriptomic correlates of lung maturity and sexual dimorphism in the pseudoglandular phase of the developing human fetal lung to genes that are significantly differentially expressed in airway diseases.
The fetal origins of disease hypothesis suggest that in utero exposures can play a role in disease development and progression in later life. In utero exposures, such as smoking, have been shown to influence the developing fetal lung (1). Variations in lung maturity and in genes known to be expressed during lung development, and birth weight, have been associated with childhood asthma, bronchopulmonary dysplasia (BPD), adult lung function, and with death from chronic obstructive airways disease in adult life (2–4). This highlights the need to understand variability in the course of prenatal lung development, as it may contain the imprint for life-long pulmonary function growth/decline, and the developmental origins of chronic lung disease.
Lung development is a complex process divided into embryonic, pseudoglandular, canalicular, saccular, and alveolar phases based on histology. The initial phases of lobation and branching are known to be stereotypic, and are determined by a well conserved, genetically hard-wired program (5). Many genes have been implicated in human lung development; however, its complete transcriptome remains uncharacterized. We know that this process requires the interaction of many molecular processes that are influenced by gene expression. We have previously used genome-wide gene expression profiling to describe characteristic lung development genes in a small set of human lung samples (6). Similarly, animal studies performed by us and others have shown the validity of this approach to identify the key regulators of fetal lung development (6, 7). Elucidating these genomic contributions to lung development will help determine molecular pathways involved in the development of lung disease and identify potential therapeutic targets. For instance, we have previously identified genomic determinants of lung disease using this approach for both asthma and chronic obstructive pulmonary disease (8, 9). In this study, we advance our previous analysis by identifying gene sets associated with lung development during early gestation in a larger set of human fetal lung tissue samples and by assessing their enrichment in asthma and BPD.
Furthermore, epidemiological and experimental data support the existence of differences in the trajectory of lung development in males and females. Sex has previously been shown to influence lung function and the development of neonatal pulmonary disease, such as respiratory distress syndrome and BPD. Response to in utero exposure, such as maternal cigarette smoking, differs in the male and female lung (10). Androgens have been shown to enhance branching morphogenesis (11) and inhibit surfactant production (12) in the developing fetal lung. Male lung development lags behind female lung development in the alveolar phase of development in late gestation, based on the assessment of amniotic fluid indices (13, 14). Thus, it is reasonable to postulate a sexual dimorphism in the developing lung transcriptome and its regulation. Here, we investigate the developing lung transcriptome in males and females from the pseudoglandular and early canalicular phases using both supervised and unsupervised analyses to achieve a more comprehensive understanding of sex differences and similarities in developing fetal lung tissue, and their relationship to chronic lung disease.
Materials and Methods
Developing Human Fetal Whole Lung Tissue Transcriptome Profiles
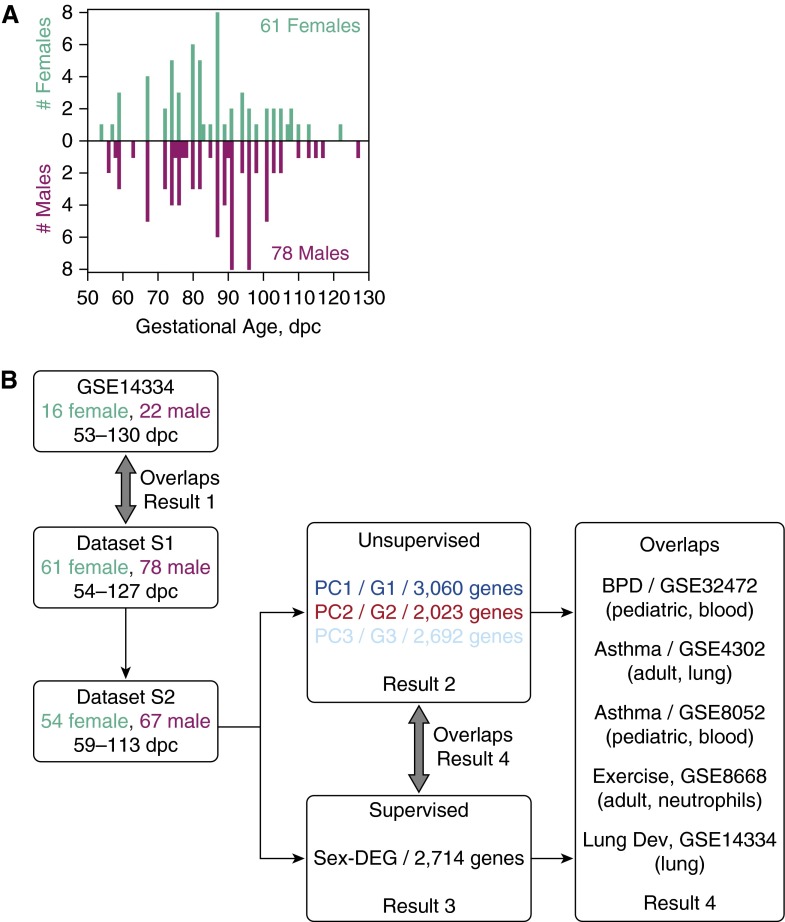
Human fetal lung tissues were obtained from two National Institute of Child Health and Development (Bethesda, MD) programs at the University of Maryland Brain and Tissue Bank for Developmental Disorders (Baltimore, MD), and the Laboratory of Developmental Biology (University of Washington, Seattle, WA) (9). The primary sample population was comprised of 61 females (estimated ages, 54–122 days postconception [dpc]) and 78 males (estimated ages, 56–127 dpc) from 138 nonsmoking mothers with 0 ng/g cotinine in their placental tissue (15) (see Text E1 and Table E2 in the online supplement; Figure 1A). Sex was reassessed using principal component (PC) analysis (PCA) of the corresponding microarray transcriptomic data using 53 microarray probes for genes unique to the Y chromosome.
Figure 1.
Overview of data integration and analyses. (A) Histogram of postconception age in days (dpc) and sex of dataset S1. Females, green; males, magenta. Darker colors mark dataset S2. (B) Overview of the data integration and analyses. GSE numbers are Gene Expression Omnibus accession numbers. BPD, bronchopulmonary dysplasia; Lung Dev, lung development; PCk, the k-th principal component and its characteristic gene set is Gk; Sex-DEG, genes differentially expressed between the sexes.
Total RNA was extracted from lung tissue (RNeasy Mini Kit; Qiagen, Valencia, CA) and profiled using Affymetrix Human Gene 1.0 ST microarrays (Affymetrix, Santa Clara, CA) following the manufacturers’ protocols with 33,297 probes interrogating 19,666 unique genes. The dataset was robust multiarray analysis (RMA) normalized (16) using the oligo package in Bioconductor (http://www.bioconductor.org/) and rendered into a dataset S1 of 33,297 probes × 139 samples with gene expression RMA signal values in log2 scale. To investigate sexual dimorphism, we used an S1 subset of 54 females and 67 males from 19 matching common ages: 59, 67, 72, 74, 76, 80, 82, 85, 87, 89, 91, 94, 96, 98, 101, 103, 105, 110, and 113 dpc. We called this dataset S2. Furthermore, we used an S2 subset of 19 females and 19 males with matching ages representing the 19 common ages and called this dataset S3. To identify the single sample representative for each common age, we applied PCA of S2 samples in transcriptome space, and picked the sample closest to the sample centroid in PCs 1–3 for each common age and sex. The complete dataset is available at the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nih.gov/geo/; GSE68896).
Independent Transcriptome Datasets for Integrative Bioinformatics
Additional details are provided in Text E1. Five other human tissue Gene Expression Omnibus datasets were integrated into the analysis (Figure 1B: GSE14334 [developing lung, positive control]; GSE32472 [leukocytes, BPD]; GSE4302 [airway epithelium, asthma]; GSE8052 [lymphoblastoid cells, asthma]; GSE8668 [neutrophils, postexercise, negative control]).
PCA
Unsupervised PCA of samples in probe/gene space was used to visualize the transcriptome-scale relationship between samples, and to identify the dominant directions of sample variation in transcriptome (probe/gene) space called PCs. Samples transcriptome profiles were standardized to average 0, variance 1 across probes/genes. We focused on PC1–3. Each PC is a linear combination of n probes/genes. For example, the k-th PC (where k is a positive integer e.g., k = 1, 2, 3, …), PCk = ak,1*g1 + ak,2*g2 + … + ak,j*gj + … + ak,n*gn, where the magnitude of the loading coefficient (ak,j) indicates the contribution of gj in PCk. We define the “characteristic genes” of a PC as the probes/genes with the top 5% loading coefficient magnitudes (6, 17), or the probes/genes with loading coefficient magnitude greater than 0.01 of that PC.
Results
Lung Development Trajectory in Transcriptome Space
Here, we verified the concordance between dataset S1 and our previous dataset GSE14334 (6) in terms of the critical turning points of the developmental trajectory in transcriptome space and the overlap between their respective developmental characteristic gene sets. S1 is comprised of whole-lung transcriptome profiles of 61 females (54–122 dpc) and 78 males (56–127 dpc) from 138 nonsmoking mothers (Figure 1A). GSE14334 is limited to 28 samples spanning 53–130 dpc to approximate the age range of the 139 samples in S1.
To find a representative microarray probe for each gene in S1 and GSE14334, respectively, we first identified two samples for each postconception age with replicate sample profiles using PCA. For each age, we picked a pair of samples that were closest to each other in PCs 1–3, and calculated the linear correlation between the first and second replicate age series for each probe. For each gene, we picked the probe with the highest correlation value as its representative profile. Following Ref. 6, we minimized subject- and age-related variations in expression measurement and modeled global gene-specific developmental expression patterns via probe-wise nonparametric regression (Text E1) (18). The normalized datasets represent individual smoothened trajectories for every gene modeled at 35 distinct postconception ages (54–127 dpc) spanned by 139 samples of S1 (19,666 unique genes), and 23 distinct ages (53 to 130 dpc) spanned by 28 samples of GSE14334 (20,361 unique genes). We limited both datasets to 17,929 common genes.
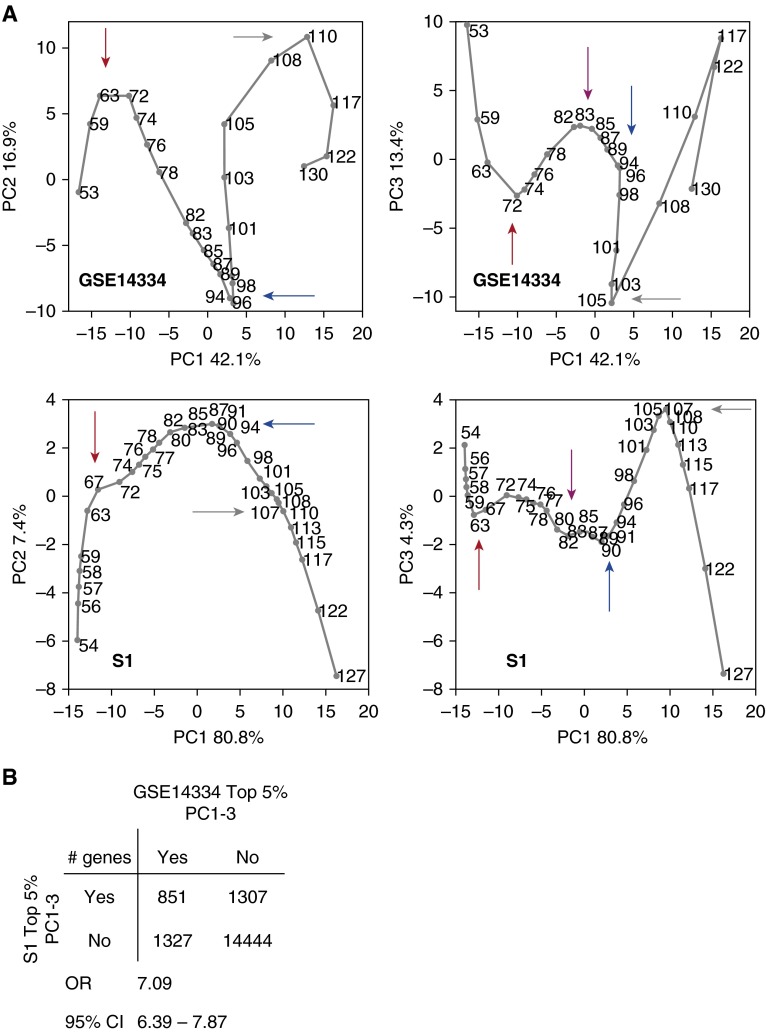
PCA was performed separately on the resulting datasets S1 and GSE14334 to identify the dominant directions of sample variation in their respective transcriptome (gene) spaces, focusing on the three most dominant variations, called PCs 1–3. We studied the sample trajectories in PC1–3 and the overlap in their top 5% contributing genes to PC1–3. The critical turning points of the two trajectories occur at approximately the same age ranges (Figure 2A, colored arrows; 59–67 dpc [red], 80–83 dpc [magenta], 89–96 dpc [blue], and 105–110 dpc [gray]), despite difference in microarray technology and sampling ages. The overlap between their top 5% PC1–3 contributing genes was significant (Figure 2B and Table E3). All subsequent analyses described subsequently here involve transcriptome data that are only RMA normalized, and not further probe-wise normalized by nonparametric regression.
Figure 2.
Comparison of the developing lung datasets GSE14334 and S1 after nonparametric regression normalization. (A) Principal component (PC) analysis (PCA) of GSE14334 (top) and S1 (bottom) in PC1 versus PC2, and PC1 versus PC3 planes. Ages are indicated. Four common, critical turning points (marked by arrows): 59–67 dpc (red); 80–83 dpc (magenta); 89–96 dpc (blue); and 105–110 dpc (gray). (B) Overlap of the top 5% PC1–3 contributing genes to GSE14334 and S1. The odds ratio (OR) and its 95% confidence interval (CI) are shown.
Developing Human Fetal Lung Transcriptomic Correlates for Sex and Age
Here, we investigated the developing lung transcriptome for age and sex correlates. We used the dataset S2 of 54 females and 67 males from 19 matching (common) ages between 59–113 dpc. Each of the 19,666 genes on the microarray was mapped to a representative probe with the highest linear correlation value between the first and second replicate age series, as described in the first result.
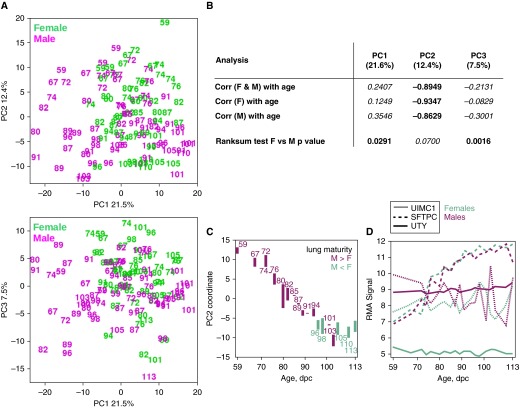
PCA was performed on dataset S2 of 19,666 genes × 121 samples to identify the dominant directions of sample variation in transcriptome space (PC1–3; Figure 3A). We found PC1 and PC3 sample coordinates to be separately associated with sex, and PC2 sample coordinates to be anticorrelated with age (Figure 3B). Regarding PC2 as a transcriptomic proxy for postconception age, where PC2 sample coordinates are inversely proportional to pulmonary maturity, we investigated the difference in PC2 sample coordinates between the sexes at the 19 common ages by first computing the PC2 sample coordinate centroid at each common age and sex (Figure 3C). We observed the male lung to be more mature than the female lung before 96 dpc (i.e., PC2centroid[male, X dpc] < PC2centroid[female, X dpc] for X < 96 dpc). In five of the seven common ages after 94 dpc, the male lung is less mature than the female (i.e., PC2centroid[male, X dpc] > PC2centroid[female, X dpc] for X > 94 dpc). Previously, we had reported a novel transcriptomic turning point at 91–96 dpc within the pseudoglandular phase (6). The genes with the highest PC1–3 loading magnitudes are UIMC1 (EntrezID 51720), SFTPC (EntrezID 6440), and UTY (EntrezID 7404), respectively (Figure 3D).
Figure 3.
Transcriptomic analyses of dataset S2, females (F) (green) and males (M) (magenta) from 19 matching common ages 59–113 dpc. (A) PCA of S2 in PC1 versus PC2, and PC1 versus PC3 planes. Ages are indicated. (B) Two-sided Wilcoxon rank sum tests and linear correlations of PC1–3 sample coordinates with age/sex. PC1 and PC3 are associated with sex difference. PC2 is anticorrelated with postconception age. (C) PC2 sample coordinates of the 19 common age sample centroids for each sex. A bar joins the female–male centroids at each common age, and magenta color indicates if the male centroid is below the female centroid (green otherwise). (D) Temporal expression profiles of genes with the greatest contribution to PC1 (UIMC1, EntrezID 51720), PC2 (SFTPC, EntrezID 6440), and PC3 (UTY, EntrezID 7404) sample averaged at each common age for females (green) and males (magenta). Corr, correlation.
Limiting S2 to unique age- and sex-matched samples (dataset S3), we observed that the gene-wise correlation between the sexes over their 19 common ages had a Gaussian distribution (based on quantile–quantile plot relative to a Gaussian random number generator) centered near 0, even when the time series profile of one sex was shifted up to ±5 common age steps from the other sex, suggesting that the transcriptomic difference between the sexes is not a uniform global time shift (Text E1 and Figure E4).
Sex Differences in the Developing Human Fetal Lung and Their Associations in Chronic Lung Diseases
Here, we further investigated sex differences in the developing human fetal lung transcriptome for developmental and chronic lung disease associations. We considered two gene sets representing sex differences: genes that contribute most to PC1 and PC3 (separately found to be associated with sex) from an unsupervised PCA of dataset S2 described previously here, and genes that are differentially expressed between sexes from a supervised, age-adjusted linear regression model of S2.
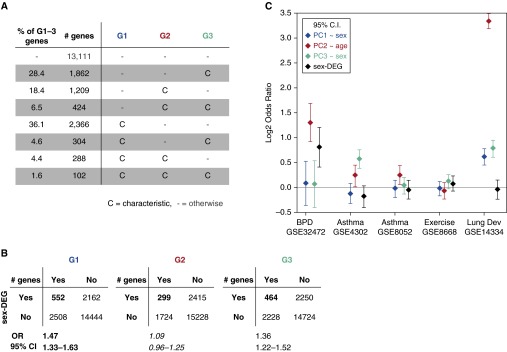
First, we define the characteristic genes of PCk (i.e., genes that contribute most to PCk) as genes with loading magnitude greater than 0.01 in PCk, and call them Gk (Figure 4A and Table E5). Gene ontology (GO) enrichment analyses of G1–G3 show them to be enriched for ontological terms that are associated with developing lung structures and functions, such as branching morphogenesis and gas transport (Tables E6–E8). Second, we used a supervised, age-adjusted linear regression model to identify genes differentially expressed between sexes (Text E1). We found that 3,929 (of 33,297) probes were significant, corresponding to a set of 2,714 unique genes that we call sex-DEG: 1,112 up in females, 1,602 up in males (Table E5). GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://genome.jp/kegg) pathway analysis of sex-DEG both showed three significantly enriched pathways after false discovery rate (FDR) correction: olfactory transduction (FDR-adjusted P = 1.62e−14); DNA replication (FDR-adjusted P = 4.07e−02); and the citric acid cycle (FDR-adjusted P = 5.26e−02). There were significant overlaps between sex-DEG and G1 and G3, respectively (Figure 4B).
Figure 4.
PC1–3 characteristic gene overlaps with genes representing different lung conditions of interest. (A) Tally of PC1–3 characteristic genes, G1–G3. (B) Overlaps between G1–3 and sex-DEG. (C) Overlaps between G1–G3, sex-DEG and genes representing five conditions of interest: differential expression in bronchopulmonary dysplasia (BPD; GSE32472), asthma (GSE4302, GSE8052), and postexercise neutrophils (GSE8668), and characteristic genes of our previous human fetal lung lung dataset (GSE14334). The log2 OR means and their 95% CI lower and upper bounds are shown.
Next, we investigated the relationships between G1–G3 and sex-DEG with genes related to BPD, asthma, fetal lung development (positive control), and postexercise neutrophils (negative control) from other published studies (Text E1). BPD was represented by genes that were differentially expressed in peripheral blood leukocytes from 68 patients with BPD relative to 43 control patients at Days 5, 14, or 28 after birth from GSE32472 (19). GSE32472 and dataset S2 used the same microarray platform, cf (Table E5). Asthma was represented by two datasets: GSE4302 (airway epithelial brushings from 11 healthy subjects and 7 adults with asthma) (20) and GSE8052 (lymphoblastoid B cells from 95 sibling pairs of children with and without asthma) (21). In each asthma dataset, we identified differentially expressed genes between control and asthma groups. The positive control was PC1–3 characteristic genes of our previous developing human fetal lung dataset, GSE14334 (6). The negative control was genes that were differentially expressed after 30 minutes of exercise in 12 healthy male subjects in GSE8668 (22). There were 17,929 common genes between the asthma and control datasets, and our dataset S2 microarray platform, cf (Table E9).
We found G2 (anticorrelated with postconception age, unsupervised) to be consistently enriched for BPD (odds ratio [OR] = 2.46, 95% confidence interval [CI] = 1.88–3.23) and asthma (GSE4302 OR = 1.18, 95% CI = 1.01–1.37; GSE8052 OR = 1.19, 95% CI = 1.04–1.36) genes (Figure 4C; Text E1 and Table E10). G1 and G3 (both associated with sex) did not consistently overlap with BPD or asthma. Sex-DEG was enriched for BPD (OR = 1.75, 95% CI = 1.33–2.29), but not asthma genes.
Discussion
In this evaluation of global gene expression in normal early-phase human fetal lung development, we highlight three findings. First, we confirmed our previous analyses demonstrating a histology-based transcriptomic trajectory of lung development and a common set of genes associated with postconception age (6). Second, we observed that the developmental transcriptome significantly differs by sex. These differences do not appear to be due to uniform global time shift between sexes, as the distributions of gene correlation between sexes remain almost similar after shifting the developmental expression profiles of one sex up to ±5 common age steps from the other sex along the postconception age axis (Figure E4). Finally, for two different lung diseases—asthma and BPD—the genes most tightly correlated with the PC associated with postconception age during lung development were significantly enriched for such disease genes. This points toward the role of developing lung transcriptomic changes in the development of chronic lung disease later in life.
Pathway analyses further enhanced our findings. Overall, our lung developmental gene sets were enriched for GO terms suggestive of developing lung structure and function, such as surfactants, branching morphogenesis, and development of specific tissues, such as epithelium, vasculature, and respiratory tube. That these pathways are enriched supports their role in lung development and reiterates our previously described role of these processes even in an early phase of gestation. Pathway analysis further demonstrates that the differentially expressed genes between the two sexes map to olfactory transduction, DNA replication and citrate cycle (TCA cycle). The olfactory pathway is known to differ between sexes, and ectopic expression of the olfactory receptor genes has been demonstrated (23, 24). Sex-based differential expression of human corneal epithelial cells has also demonstrated that one of the enriched pathways is the DNA replication pathway (25). Similarly, sex differences in the expression of TCA cycle enzymes in skeletal muscle (26), and the excretion of citrate in urine (27), have been demonstrated. Together, the sex-specific pathways enriched in our analysis have previously been shown to differ between sexes, albeit in different tissues.
Transcriptomically, we observed the male lung to be more mature than the female lung before 96 dpc, and the male to be less mature than the female after 96 dpc. Previous studies of amniotic fluid at 28–40 weeks of gestation have shown the female lung to have a higher index of pulmonary maturity than males at this phase of development (14). However, due to the effect of androgens on branching morphogenesis (11), it is possible that, earlier in gestation, the development of the male lung is more advanced than in the female. MicroRNAs that are significantly different between sexes during murine lung development have been identified (28).
We previously determined that genes that are important in lung development might be associated with lung function in subjects with asthma (9). We now add to this result, showing that the gene set correlated with postconception age of the developing human fetal lung (G2) is significantly enriched for asthma- and BPD-associated genes. Although there exist sex-based differences in the natural history of both diseases, we did not see consistent enrichment of the gene sets associated with sex in the developing lung (G1 and G3) for these disease genes. Our results suggest that sex-specific effects previously described in the third trimester (29) are not observed during earlier phases of gestation. Finally, it is possible that these transcriptomic changes are due to a more dominant effect of postconception age when compared with sex differences in the first two trimesters. We hope to investigate these questions using fetal lung samples from the third trimester in a future study.
As would be expected, the enrichment of G2 in BPD was stronger than in asthma. Pathway analysis of genes in the G2 and BPD overlap showed that the most enriched pathways included the p53 signaling pathway and the complement and coagulation cascades. Previous studies have demonstrated a role for p53 in the development of BPD (30). Among the genes identified in this analysis GTSE1, THBS1, and SERPINE1 act downstream from p53. Gene expression analysis of lung samples from short-term ventilated preterm infants was shown to have increased expression of THBS1 (31). Similarly increased levels of plasminogen activator inhibitor 1 (expressed by SERPINE1) in tracheal aspirate have been related to the severity of respiratory distress syndrome (32), and may be related to the development of BPD.
Although these results provide a comprehensive evaluation of the transcriptomic profile of early human lung development, there are several limitations to this study. Because our samples were deidentified tissues from a fetal tissue biorepository, limited phenotypic information was available. We were not able to adjust for differences in race and potential confounders, such as in utero exposures. We attempted to reduce confounders that have been associated with asthma and BPD development, such as in utero smoke exposure, by limiting our samples to those with a measured placental cotinine concentration of 0 ng/g. Furthermore, we used unsupervised PCA to select representative samples for each gestational age and sex, allowing us to study the sex differences in lung tissue. In addition, we recognize that some genes identified in our human lung developmental profile are tissue specific, and may not be identified in other tissues. To address this possibility, we used different tissues to identify the genes associated with asthma and BPD. Our ability to identify a positive enrichment for these developmental genes in the other independent tissue samples suggests that our results are not tissue specific, and may provide additional insight to the developmental origins of disease.
In conclusion, despite sex differences in the transcriptome in the developing human male and female lung tissue, postconception age appears to be a more dominant influence than sex in the effect of early fetal lung development on disease risk. There are clearly fetal lung antecedents of both asthma and BPD identified in our data, but these are not sex specific during this early, pseudoglandular phase of development. The enrichment of asthma and BPD genes in developmental datasets supports the fetal origins of disease hypothesis, and further strengthens the need to study developmental time series transcriptome data to understand the origins of airway-related lung disease.
Footnotes
This work was supported by National Institutes of Health grants R01 HL097144, K08 HL096833, K25 HL091124, T32 HL007427, R21 HL107927, R01 HL092197 and U01 HL65899.
The content of this article does not necessarily represent the official views of the Eunice Kennedy Shriver National Institute of Child Health and Human Development. This manuscript is subject to the National Institutes of Health public access policy.
Author Contributions: A.T.K., D.C., S.S., K.G.T., and S.T.W. designed the study, analyzed, interpreted data, and wrote the manuscript; R.G., C.A.V., and J.S.L. acquired the samples and interpreted the data; W.Q. and V.J.C. provided statistical analyses.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2015-0326OC on November 19, 2015
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Sharma S, Chhabra D, Kho AT, Hayden LP, Tantisira KG, Weiss ST. The genomic origins of asthma. Thorax. 2014;69:481–487. doi: 10.1136/thoraxjnl-2014-205166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker DJ, Godfrey KM, Fall C, Osmond C, Winter PD, Shaheen SO. Relation of birth weight and childhood respiratory infection to adult lung function and death from chronic obstructive airways disease. BMJ. 1991;303:671–675. doi: 10.1136/bmj.303.6804.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Canoy D, Pekkanen J, Elliott P, Pouta A, Laitinen J, Hartikainen AL, Zitting P, Patel S, Little MP, Järvelin MR. Early growth and adult respiratory function in men and women followed from the fetal period to adulthood. Thorax. 2007;62:396–402. doi: 10.1136/thx.2006.066241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orfei L, Strachan DP, Rudnicka AR, Wadsworth ME. Early influences on adult lung function in two national British cohorts. Arch Dis Child. 2008;93:570–574. doi: 10.1136/adc.2006.112201. [DOI] [PubMed] [Google Scholar]
- 5.Warburton D, Bellusci S, Del Moral PM, Kaartinen V, Lee M, Tefft D, Shi W. Growth factor signaling in lung morphogenetic centers: automaticity, stereotypy and symmetry. Respir Res. 2003;4:5. doi: 10.1186/1465-9921-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kho AT, Bhattacharya S, Tantisira KG, Carey VJ, Gaedigk R, Leeder JS, Kohane IS, Weiss ST, Mariani TJ. Transcriptomic analysis of human lung development. Am J Respir Crit Care Med. 2010;181:54–63. doi: 10.1164/rccm.200907-1063OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Y, Wang Y, Besnard V, Ikegami M, Wert SE, Heffner C, Murray SA, Donahue LR, Whitsett JA. Transcriptional programs controlling perinatal lung maturation. PLoS One. 2012;7:e37046. doi: 10.1371/journal.pone.0037046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demeo DL, Mariani TJ, Lange C, Srisuma S, Litonjua AA, Celedon JC, Lake SL, Reilly JJ, Chapman HA, Mecham BH, et al. The SERPINE2 gene is associated with chronic obstructive pulmonary disease. Am J Hum Genet. 2006;78:253–264. doi: 10.1086/499828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharma S, Tantisira K, Carey V, Murphy AJ, Lasky-Su J, Celedón JC, Lazarus R, Klanderman B, Rogers A, Soto-Quirós M, et al. A role for Wnt signaling genes in the pathogenesis of impaired lung function in asthma. Am J Respir Crit Care Med. 2010;181:328–336. doi: 10.1164/rccm.200907-1009OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J, Naeem E, Tian J, Lombardi V, Kwong K, Akbari O, Torday JS, Rehan VK. Sex-specific perinatal nicotine-induced asthma in rat offspring. Am J Respir Cell Mol Biol. 2013;48:53–62. doi: 10.1165/rcmb.2011-0344OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levesque BM, Vosatka RJ, Nielsen HC. Dihydrotestosterone stimulates branching morphogenesis, cell proliferation, and programmed cell death in mouse embryonic lung explants. Pediatr Res. 2000;47:481–491. doi: 10.1203/00006450-200004000-00012. [DOI] [PubMed] [Google Scholar]
- 12.Nielsen HC, Zinman HM, Torday JS. Dihydrotestosterone inhibits fetal rabbit pulmonary surfactant production. J Clin Invest. 1982;69:611–616. doi: 10.1172/JCI110488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fleisher B, Kulovich MV, Hallman M, Gluck L. Lung profile: sex differences in normal pregnancy. Obstet Gynecol. 1985;66:327–330. [PubMed] [Google Scholar]
- 14.Torday JS, Nielsen HC, Fencl MdeM, Avery ME. Sex differences in fetal lung maturation. Am Rev Respir Dis. 1981;123:205–208. doi: 10.1164/arrd.1981.123.2.205. [DOI] [PubMed] [Google Scholar]
- 15.Vyhlidal CA, Riffel AK, Haley KJ, Sharma S, Dai H, Tantisira KG, Weiss ST, Leeder JS. Cotinine in human placenta predicts induction of gene expression in fetal tissues. Drug Metab Dispos. 2013;41:305–311. doi: 10.1124/dmd.112.049999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 17.Kho AT, Bhattacharya S, Mecham BH, Hong J, Kohane IS, Mariani TJ. Expression profiles of the mouse lung identify a molecular signature of time-to-birth. Am J Respir Cell Mol Biol. 2009;40:47–57. doi: 10.1165/rcmb.2008-0048OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loader C.Local regression and likelihood.New York: Springer; 1999 [Google Scholar]
- 19.Pietrzyk JJ, Kwinta P, Wollen EJ, Bik-Multanowski M, Madetko-Talowska A, Günther CC, Jagła M, Tomasik T, Saugstad OD. Gene expression profiling in preterm infants: new aspects of bronchopulmonary dysplasia development. PLoS One. 2013;8:e78585. doi: 10.1371/journal.pone.0078585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woodruff PG, Boushey HA, Dolganov GM, Barker CS, Yang YH, Donnelly S, Ellwanger A, Sidhu SS, Dao-Pick TP, Pantoja C, et al. Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc Natl Acad Sci USA. 2007;104:15858–15863. doi: 10.1073/pnas.0707413104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, Depner M, von Berg A, Bufe A, Rietschel E, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 22.Radom-Aizik S, Zaldivar F, Jr, Leu SY, Galassetti P, Cooper DM. Effects of 30 min of aerobic exercise on gene expression in human neutrophils. J Appl Physiol (1985) 2008;104:236–243. doi: 10.1152/japplphysiol.00872.2007. [DOI] [PubMed] [Google Scholar]
- 23.Feldmesser E, Olender T, Khen M, Yanai I, Ophir R, Lancet D. Widespread ectopic expression of olfactory receptor genes. BMC Genomics. 2006;7:121. doi: 10.1186/1471-2164-7-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, De la Cruz O, Pinto JM, Nicolae D, Firestein S, Gilad Y. Characterizing the expression of the human olfactory receptor gene family using a novel DNA microarray. Genome Biol. 2007;8:R86. doi: 10.1186/gb-2007-8-5-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki T, Richards SM, Liu S, Jensen RV, Sullivan DA. Influence of sex on gene expression in human corneal epithelial cells. Mol Vis. 2009;15:2554–2569. [PMC free article] [PubMed] [Google Scholar]
- 26.Green HJ, Fraser IG, Ranney DA. Male and female differences in enzyme activities of energy metabolism in vastus lateralis muscle. J Neurol Sci. 1984;65:323–331. doi: 10.1016/0022-510x(84)90095-9. [DOI] [PubMed] [Google Scholar]
- 27.Slupsky CM, Rankin KN, Wagner J, Fu H, Chang D, Weljie AM, Saude EJ, Lix B, Adamko DJ, Shah S, et al. Investigations of the effects of gender, diurnal variation, and age in human urinary metabolomic profiles. Anal Chem. 2007;79:6995–7004. doi: 10.1021/ac0708588. [DOI] [PubMed] [Google Scholar]
- 28.Mujahid S, Logvinenko T, Volpe MV, Nielsen HC. miRNA regulated pathways in late stage murine lung development. BMC Dev Biol. 2013;13:13. doi: 10.1186/1471-213X-13-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yunginger JW, Reed CE, O’Connell EJ, Melton LJ, III, O’Fallon WM, Silverstein MD. A community-based study of the epidemiology of asthma: incidence rates, 1964–1983. Am Rev Respir Dis. 1992;146:888–894. doi: 10.1164/ajrccm/146.4.888. [DOI] [PubMed] [Google Scholar]
- 30.Das KC, Ravi D, Holland W. Increased apoptosis and expression of p21 and p53 in premature infant baboon model of bronchopulmonary dysplasia. Antioxid Redox Signal. 2004;6:109–116. doi: 10.1089/152308604771978417. [DOI] [PubMed] [Google Scholar]
- 31.De Paepe ME, Greco D, Mao Q. Angiogenesis-related gene expression profiling in ventilated preterm human lungs. Exp Lung Res. 2010;36:399–410. doi: 10.3109/01902141003714031. [DOI] [PubMed] [Google Scholar]
- 32.Cederqvist K, Sirén V, Petäjä J, Vaheri A, Haglund C, Andersson S. High concentrations of plasminogen activator inhibitor-1 in lungs of preterm infants with respiratory distress syndrome. Pediatrics. 2006;117:1226–1234. doi: 10.1542/peds.2005-0870. [DOI] [PubMed] [Google Scholar]