Abstract
Intercellular protein-protein interactions (PPIs) enable communication between cells in diverse biological processes, including cell proliferation, immune responses, infection and synaptic transmission, but they are challenging to visualize because existing techniques1,2,3 have insufficient sensitivity and/or specificity. Here we report split horseradish peroxidase (sHRP) as a sensitive and specific tool for detection of intercellular PPIs. The two sHRP fragments, engineered through screening of 17 cut sites in HRP followed by directed evolution, reconstitute into an active form when driven together by an intercellular PPI, producing bright fluorescence or contrast for electron microscopy. Fusing the sHRP fragments to the proteins neurexin (NRX) and neuroligin (NLG), which bind each other across the synaptic cleft4, enabled sensitive visualization of synapses between specific sets of neurons, including two classes of synapses in the mouse visual system. sHRP should be widely applicable for studying mechanisms of communication between a variety of cell types.
Protein-fragment complementation assays (PCAs) are a powerful strategy for detection of protein-protein interactions (PPIs)5. In these assays, two proteins of interest are fused to complementary fragments of a reporter protein, and reporter activity is reconstituted only if a PPI occurs. Although PCAs have been widely adopted to detect intracellular PPIs, they have rarely been demonstrated for spatially-resolved visualization of extracellular PPIs. One exception is split green fluorescent protein (GFP), which reconstitutes intercellularly to generate fluorescence at cell-cell contacts2,6,7. However, the relatively dim fluorescence of GFP greatly limits the sensitivity of this method, particularly at small cell-cell contacts such as neuronal synapses6.
We envisioned that replacing GFP with a signal-amplifying enzyme could lead to a dramatic improvement in sensitivity. To implement this approach, we required a PCA that: 1) reconstitutes an active enzyme across intercellular contact sites, 2) produces strong enzyme-amplified signal, and 3) generates spatially restricted labeling. Because existing reporters8,9,10,11 did not meet these requirements, we sought to develop a new PCA based on horseradish peroxidase (HRP), which functions in extracellular environments12, generates spatially-restricted fluorescent signal13, and is one of the most sensitive reporter enzymes known14. HRP is a 308 amino acid enzyme that catalyzes the H2O2-dependent oxidation of a wide variety of substrates, and it has been harnessed for diverse applications, including light and electron microscopy12, proximity tagging15 and chemiluminescence16 (Fig. 1A). However, HRP requires a heme cofactor and contains four structurally-essential disulfide bonds, nine N-linked glycosylation sites, and two Ca2+ ions, making it challenging to engineer into a PCA.
Figure 1. Protein engineering of split HRP (sHRP).
A. Schematic overview of the split HRP reporter. Two inactive fragments of HRP reconstitute into an active complex capable of producing a variety of enzymatic reaction products, visible by multiple modalities. The nonfluorescent Amplex Red is converted to fluorescent resorufin. Biotin-phenol is converted to a reactive radical that becomes covalently attached to neighboring proteins13. Tyramide signal amplification (TSA) substrates function analogously, except biotin is replaced with a fluorophore, such as Cy3. Diaminobenzidine (DAB) is converted to a colored and insoluble product that becomes electron-dense upon treatment with osmium tetroxide.
B. Overview of sHRP protein engineering. Structure-guided cut-site screening was followed by two rounds of yeast display directed evolution.
C. Overview of yeast display evolution of sHRPa (top) and sHRPb (bottom).
D. Fluorescence activated cell sorting (FACS) of yeast displaying sHRP fragments at various stages of evolution. Aga1p and 2p are cell surface mating proteins. Re-amplified pools of yeast were labeled and analyzed under matched conditions (1 min biotin-phenol labeling).
E. Crystal structure of HRP (PDB ID 1H5A). The sHRPa fragment is colored blue, and the sHRPb fragment is green.
F. Amplex UltraRed live cell labeling followed by immunostaining of HEK293T cells expressing sHRP fragments in the ER lumen.
We screened 17 HRP cut sites, located on solvent exposed loop regions, by co-expressing complementary fragments in the endoplasmic reticulum (ER) lumen, a subcellular compartment that enables glycosylation and proper folding of disulfide-containing proteins17 (Supplementary Fig. 1A and Supplementary Note 1). Seven of the seventeen cut sites produced enzymatic activity, detected using the fluorogenic substrate Amplex UltraRed, when the protein fragments were fused to the rapamycin-dependent dimerization domains FRB and FKBP18 The cut site after amino acid 213 produced the most active fragment pair, which we call sHRPa′ and sHRPb′ (Supplementary Fig. 1B–E). These HRP fragments reconstituted not only in the ER, but also on the cell surface when driven together by an intercellular protein-protein interaction (PPI) between neurexin (NRX) and neuroligin (NLG), the trans-synaptic binding partners (Supplementary Fig. 1F–G and Supplementary Note 1). Reconstitution in the ER lumen occurred even in the absence of a PPI, indicating that the HRP fragments have substantial affinity for each other, but reconstitution at intercellular contact sites was PPI-dependent. We attribute this discrepancy to the lower effective concentrations of the fragments on the cell surface. However, the reconstituted complex gave much dimmer fluorescence than full-length HRP.
To improve the brightness of sHRP labeling, we utilized yeast display evolution to optimize both fragments in turn (Fig. 1B). We first expressed sHRPb′ stably as a fusion to Aga1P on the yeast surface, then introduced a library of 4 × 107 randomized sHRPa′ mutants fused to Aga2P, the binding partner of Aga1P (Fig. 1C and Supplementary Note 2). We treated the yeast library with biotin-phenol, a substrate that produces spatially restricted labeling13,19, then stained with fluorescent streptavidin, such that yeast cells expressing the most active mutants of sHRPa′ exhibited the brightest fluorescence. We isolated the most active yeast cells using fluorescence activated cell sorting (FACS) (Supplementary Fig. 2A–C). After four rounds of successive labeling, sorting and regrowth, the activity of the yeast pool had increased substantially, indicating enrichment of improved sHRPa′ mutants (Fig. 1D). By sequencing the DNA, validating promising clones in mammalian cells, and combining beneficial mutations, we generated an improved quadruple mutant that we call sHRPa (Fig. 1E and Supplementary Fig. 2D–E). We next performed yeast display evolution of sHRPb′ using the same approach to produce an improved double mutant that we call sHRPb (Supplementary Fig. 3A–D and Supplementary Note 3). To our knowledge, these results represent the first instance in which yeast display has been utilized for evolution of a PCA.
When co-expressed in the ER lumen, the evolved sHRP fragments produced much brighter fluorescence than the original fragment pair, and the reconstituted activity was close to that of full-length HRP (Fig. 1F). Both evolved fragments were completely inactive on their own, and like the original fragment pair, they reconstituted in the ER in the absence of a PPI. Heme supplementation did not boost the signal, suggesting that sHRP reconstitution was not limited by endogenous heme levels (Supplementary Fig. 4A–B). The single intermolecular disulfide bond between the sHRP fragments (Cys97-Cys301) was indispensable for reconstitution (Supplementary Fig. 4A–B). The sHRP fragments were N-glycosylated and not susceptible to proteolytic cleavage (Supplementary Fig. 4C).
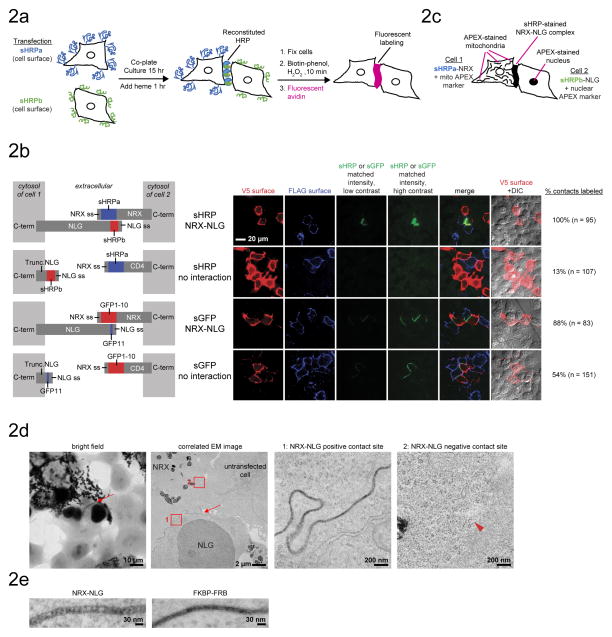
To evaluate sHRP reconstitution at intercellular contacts, we fused the sHRP fragments to NRX and NLG, then transfected the constructs into separate pools of HEK293T cells. Transfected cells were lifted, co-plated, and cultured together for 12 hours, then fixed with formaldehyde, permeabilized with methanol, labeled with biotin-phenol, and stained with fluorescent avidin (Fig. 2A). sHRP activity survived this procedure, and we observed bright fluorescence at 100% of the expected intercellular contact sites (Fig. 2B, Supplementary Fig. 5A–C, and Supplementary Note 4). Heme supplementation to the media was required for intercellular reconstitution (in contrast to intracellular reconstitution), indicating that neither sHRP fragment preloads with intracellular heme (Supplementary Fig. 5D). However, sHRP became active even when heme was added to fixed cells, an advantageous feature for situations in which heme delivery to living cells or tissues is undesirable or impractical.
Figure 2. Intercellular reconstitution of sHRP for fluorescent and EM labeling.
A. Scheme for intercellular sHRP reconstitution across HEK293T cells.
B. Comparison of sHRP and split GFP (sGFP)2,7,6. Split HRP produces brighter fluorescence than split GFP, and its activity is dependent on a protein-protein interaction. The two pools of cells were visualized by surface immunostaining of N-terminal V5 and FLAG epitope tags. V5 and FLAG intensities are normalized between the two sHRP experiments and between the two sGFP experiments, but not across all 4 conditions. Surface expression levels of sHRP constructs and sGFP constructs were similar. Construct designs are shown at left. “ss” is signal sequence.
C. Scheme for EM staining of HEK293T intercellular contacts by sHRP. APEX co-transfection markers provide contrast for both light microscopy and EM23.
D. EM imaging of the intercellular NRX-NLG protein-protein interaction using sHRP (as in C). Arrow points to contact site stained by sHRP. Arrowhead points to a contact site not stained by sHRP. The third and fourth images are zooms of boxed regions in second image.
E. High magnification EM of sHRP staining generated by different protein-protein interactions.
The NRX-NLG protein-protein interaction (PPI) was essential for intercellular sHRP reconstitution, since deleting key interaction domains abolished activity (Fig. 2B and Supplementary Fig. 5A). By contrast, split GFP reconstitution was not dependent on the NRX-NLG PPI, consistent with previous reports that these fragments bind with extremely high affinity and reconstitute spontaneously in extracellular environments20 (Fig. 2B and Supplementary Fig. 5A). Hence sHRP is more accurate and less perturbative than split GFP for readout of intercellular PPIs. We determined that sHRP is broadly applicable for detection of strong intercellular PPIs (i.e., nanomolar dissociation constant) and has a response time of < 10 minutes, much faster than the 4 h reported for split GFP21 (Supplementary Fig. 6A–D). Like split GFP22, sHRP reconstituted irreversibly (Supplementary Fig. 4D–E). This feature could potentially perturb interaction dynamics, but natural mechanisms for clearing trapped intercellular protein complexes mitigate this concern2 (Supplementary Fig. 5C and Supplementary Note 4).
Taking advantage of the ability of peroxidases to convert 3,3′-diaminobenzidine (DAB) into an electron-dense stain12, we asked whether sHRP could be employed for nanoscale imaging of intercellular PPIs by EM. We transfected separate pools of HEK293T cells with sHRPa-NRX and sHRPb-NLG, along with co-transfection markers based on the EM tag APEX23, which were distinguishable by both light microscopy and EM (Fig. 2C). Imaging of thin sections by EM revealed a dark reaction product exclusively at the expected intercellular contact sites (Fig. 2D and Supplementary Note 4). Sensitivity was high, with sHRP staining detectable at all expected contact sites (Supplementary Fig. 7A). At high magnification, a periodic staining pattern was apparent, with individual peg-like structures dispersed along the plasma membrane (Fig. 2E and Supplementary Fig. 7B). These pegs were approximately 10–15 nm in diameter, similar to the dimensions of the NRX-NLG complex (thought to exist as a dimer of dimers24), and were discernible in a 3D tomographic reconstruction (Supplementary Fig. 7C and Supplementary Movies 1 and 2). When we replaced the extracellular domains of NRX and NLG with FRB and FKBP, an unrelated pair of strongly-interacting proteins18, we observed strong and continuous sHRP staining at cell-cell contacts, but the “pegs” were no longer evident (Fig. 2E and Supplementary Fig. 7D).
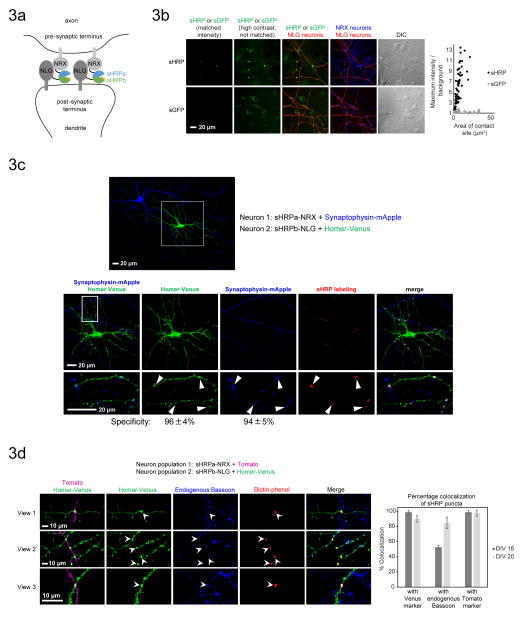
An important challenge in neuroscience is to determine if and where in the brain defined sets of neurons communicate via synapses25,26,27. One methodology called GRASP2,6,7 (“GFP Reconstitution Across Synaptic Partners”) targets split GFP fragments to the pre- and post-synaptic membranes, respectively, of specific sets of neurons, leading to reconstitution of GFP fluorescence exclusively at the synapses of interest. A major limitation of GRASP, however, is its poor sensitivity6, a consequence of the small number of GFP molecules, and thus the low fluorescent signal, per synapse.
To evaluate sHRP as an alternative to GRASP, we transfected sHRPa-NRX and sHRPb-NLG into separate pools of dissociated rat hippocampal neurons (Fig. 3A). After 1–3 days, we fixed and stained the neurons with biotin-phenol, which produced bright punctate sHRP labeling at synapses, as demonstrated by co-localization with both transfected and endogenous synaptic markers (Figs. 3B top panels and 3C–D, Supplementary Fig. 8A–C, and Supplementary Note 5). The sHRP puncta exhibited 94% co-localization with synaptophysin-mApple, a pre-synaptic marker, and 96% co-localization with homer-Venus (post-synaptic). To assess sensitivity, we determined that 62% of mApple-Venus overlay sites were marked by sHRP puncta. The absence of sHRP signal at 38% of these sites could be attributed to these sites not actually being synapses or to a lack of sHRP fragment expression at these particular locations.
Figure 3. Synapse detection in cultured neurons using sHRP.
A. Scheme for reconstitution of sHRP by the trans-synaptic NRX-NLG interaction in the neuronal synaptic cleft. sHRP constructs are expressed under the synapsin promoter to minimize overexpression artifacts.
B. Comparison of sHRP and split GFP2,7,6 for synapse detection. Constructs with synapsin promoter were introduced into separate neuron populations using a two-step transfection procedure. Confocal images are shown at left, and quantitation of maximum signal/noise as a function of contact site area is shown at right. Plots present >70 contact sites across >5 fields of view for each condition. The sHRP-NRX neurons and the sGFP-NLG neurons were marked by a Tomato co-transfection marker, and the sHRP-NLG neurons were marked by a Venus co-transfection marker. sGFP-NRX neurons were detected by anti-V5 staining (AlexaFluor 647 readout). Intensity scales are not normalized for the transfection markers.
C. sHRP labeling is localized to synapses. Confocal fluorescence imaging of sHRP (synapsin promoter) with respect to pre- and post-synaptic markers, synaptophysin-mApple and Homer-Venus, respectively. Arrowheads point to sHRP labeling sites. Specificity is calculated as the fraction of sHRP puncta that overlap with each marker. Values are the mean +/− std dev of 3 independent experiments.
D. Confocal fluorescence imaging of sHRP (synapsin promoter) with respect to endogenous Bassoon, a pre-synaptic marker. The graph presents the % overlap of sHRP puncta. Data in bar graph represent the mean +/− std dev of 3 independent experiments. Experiments were performed at either 16 days in vitro (DIV) or 20 days in vitro.
Detection of authentic synapses required expression of NRX and NLG fusions at low to moderate levels; when we used the strong CAG promoter instead of the weaker synapsin promoter, many of the sHRP-labeled contact sites were oblong and much larger than physiological synapses (Supplementary Fig. 8D). This artifact, which has been reported previously3, was caused by overexpression of NRX and NLG, not the sHRP fragments (Supplementary Figs. 9A–C and 10A–C). EM imaging confirmed that the CAG promoter constructs produced strong sHRP activity at the expected neuron-neuron contact sites, but these sites did not resemble endogenous synapses (Supplementary Figure 11 and Supplementary Note 6). Expressing the sHRP constructs at lower levels removed the artifactual contacts, but also failed to produce unambiguous EM contrast at the expected physiologically appropriate synapses (Supplementary Figure 11 and Supplementary Note 6). This result may reflect the lower sensitivity of EM staining compared to fluorescence.
Synapses labeled by sHRP (expressed under the synapsin promoter) were not enlarged relative to untransfected synapses, suggesting that sHRP is not synaptogenic at low expression levels (Figure 3D and Supplementary Note 5). Heme supplementation for 24 hours did not affect the density of dendritic spines, indicating it was not toxic (Supplementary Fig. 9D).
In a side-by-side comparison to split GFP, sHRP produced much brighter synapse labeling (Fig. 3B and Supplementary Fig. 8A). The fluorescence intensity of split GFP relative to background (i.e., signal to noise ratio) ranged from 1.03 to 1.8 (mean = 1.13), while the corresponding values for sHRP ranged from 1.5 to 13.4 (mean = 5.45). The weaker fluorescence of split GFP was not due to lower surface expression (Supplementary Fig. 10D–F). The previously reported “mGRASP” split GFP constructs7, in which the extracellular domains of NRX and NLG are deleted, were also much dimmer than sHRP, with maximal signal-to-noise values below 1.8 (Supplementary Fig. 8D). Collectively, these results support the conclusion that sHRP is a sensitive and minimally-perturbative reporter of neuronal synapses.
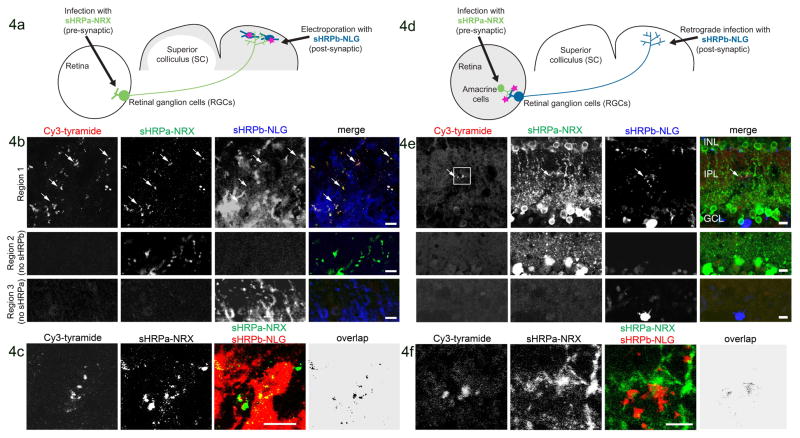
To determine if sHRP could also detect synapses in vivo, we investigated the mouse visual system. Amacrine cells in the retina form synapses on retinal ganglion cells (RGCs), which in turn extend axons through the optic nerve to “retinorecipient” regions in the brain, including the superior colliculus28 (SC). To test whether sHRP could detect synapses between axons of RGCs and dendrites of SC neurons, we introduced sHRPa-NRX to the RGCs, via adeno-associated virus (AAV)29, and sHRPb-NLG into cells of the SC, via electroporation (Fig. 4A). Four weeks later, mice were perfused with heme-containing media, and colliculi were removed, incubated with heme, fixed, sectioned, and stained for sHRP activity and expression of the sHRP fragments. Punctate sites of sHRP activity were present in regions where sHRPa-positive RGC terminals apposed sHRPb-positive SC cells (Fig. 4B–C). As expected, puncta were confined to the superficial retinorecipient layer within which retino-collicular synapses are known to form28. Few puncta were detected, likely reflecting in part the small number of synapses formed between the modest numbers of pre- and postsynaptic cells expressing the sHRP constructs. Nonetheless, puncta were specific in that they were not observed in tissue regions negative for either sHRPa or sHRPb (Fig. 4B).
Figure 4. Detection of reconstituted sHRP in vivo.
A. Scheme for detection of anterograde contacts between RGCs and neurons of the SC.
B. Four weeks after mouse transduction as in (A), the SC was removed by dissection, incubated with heme, fixed, sectioned, and labeled with Cy3-tyramide, a fluorescent sHRP substrate. sHRPa-NRX and sHRPb-NLG expression were detected by staining with anti-HRP and anti-NLG antibodies, respectively (anti-HRP does not recognize sHRPb). The second and third rows show tissue regions lacking sHRPb or sHRPa, respectively. Arrows point to sHRP-stained terminals.
C. Labeled SC from a different animal. “Overlap” is the calculated overlap between sHRPa and sHRPb channels. This experiment was repeated 8 times with similar results.
D. Scheme for detection of retrograde contacts between RGCs and amacrine cells in the mouse retina. sHRPa-NRX expression was restricted to amacrine cells via use of a Cre line (see Methods).
E. One week after transduction as in (D), the retina was dissected and prepared as in (B). Second and third rows show tissue regions lacking sHRPb or sHRPa expression, respectively. Arrow points to sHRP staining. Box indicates the region shown in (F).
F. Zoom region indicated by the box in (E). All scale bars, 10 μm.
We also tested whether sHRP could detect synapses between amacrine cells and RGCs (Fig. 4D). We introduced sHRPa-NRX selectively to GABAergic amacrine cells by injecting a Cre-dependent AAV30 into a targeted Cre-expressing mouse line, and we delivered sHRPb-NLG to RGCs using a modified lentiviral vector optimized for retrograde transport to somata31 (Fig. 4D). Fluorescent sHRP puncta were present exclusively at overlap sites between sHRPa-expressing amacrine cells and sHRPb-expressing RGCs in the inner plexiform layer of the retina, where these synapses are known to form (Fig. 4E–F)32.
These results demonstrate that sHRP can be used to locate synapses between two defined sets of neurons, a challenging task given that synapses are small, with low numbers of trans-synaptic PPIs at a single cleft. Whereas previous technologies1,2,3 detected trans-synaptic PPIs stoichiometrically, sHRP features enzymatic amplification and accepts a wide variety of commercial substrates, enabling fluorescence readout in any color and 3D EM readout with nanometer spatial resolution. Furthermore, sHRP requires a PPI for intercellular reconstitution, in contrast to split GFP, which reconstitutes in a PPI-independent manner (Figure 2B). A potential concern is that fusion constructs of sHRP to NRX and NLG, which possess high binding affinity, could play a role in the formation of the synapses they are designed to detect. However, analogous split GFP (GRASP) NRX-NLG constructs did not perturb synapse size, numbers, location, or specificity in transgenic mouse brain6. sHRP is less perturbative than GRASP because the interaction-dependence of its reconstitution indicates that the sHRP fragments have lower intrinsic affinity for one another.
sHRP requires heme and exogenous labeling reagents, which are straightforward to add to cultured cells, but challenging to deliver in vivo. Further optimization is required to improve the sensitivity of sHRP for synapse detection in vivo. Challenges include inadequate expression or transport of sHRP fragments, difficulties in delivering heme, and tissue background caused by endogenous peroxidase activity. Nevertheless, we have demonstrated that sHRP can detect small synapses involved in long-range (~1 cm) connections in the mouse brain. In cultured cells, sHRP should be immediately applicable for highly sensitive visualization of many different intercellular adhesion complexes beyond NRX-NLG.
ONLINE METHODS
Cloning and mutagenesis
The HRP C gene used for initial cut site screening was amplified from an expression plasmid from Frances Arnold33. For all other constructs, the HRP C gene was custom synthesized (DNA2.0) with codons optimized for expression in mammalian cells. HRP and its fragments were fused to mammalian proteins and localization signals using standard restriction cloning methods and overlap extension PCR34. Mutants of HRP were either generated using QuikChange mutagenesis (Stratagene) or isolated from individual yeast clones and transferred to mammalian expression vectors using standard cloning techniques. Supplementary Table 5 presents an overview of all plasmids used in this study, with detailed descriptions of construct designs, linker orientations, epitope tags, and sequence identities for non-HRP proteins.
HEK293T cell culture and transfection, co-plating, and heme supplementation
HEK 293T cells from ATCC (passage number <25) were cultured as a monolayer in growth media (either MEM (Cellgro) or a 1:1 DMEM:MEM mixture (Cellgro)) supplemented with 10% (w/v) fetal bovine serum (Sigma), 50 units/mL penicillin, and 50 μg/mL streptomycin at 37 °C under 5% CO2. Where indicated, fetal bovine serum (FBS) was replaced with 2% (v/v) B27 supplement (Life Technologies). For confocal fluorescence imaging experiments, cells were grown on 7 × 7 mm glass cover slips in 48-well plates. For EM imaging experiments, cells were grown on poly-d-lysine coated glass-bottom dishes (P35GC-0-14-C, MatTek Corp.). For Western blotting, cells were grown on polystyrene 6-well plates (Corning). To improve adherence of HEK293T cells, growth surfaces were pretreated with 50 μg/mL fibronectin (Millipore) for at least 10 min at 37 °C before cell plating and washed two times with DPBS, pH 7.4. Cells were transfected at 60–90% confluence using Lipofectamine2000 (Life Technologies), typically with 1.0 μL Lipofectamine2000 and 200 ng plasmid per 300,000 cells in serum-free media for 3–4 h, after which time Lipofectamine-containing media was replaced with fresh serum-containing media. Where indicated in Extended Data Figure 1, 400 nM rapamycin (Calbiochem) was supplemented into the media after transfection. Rapamycin was delivered by diluting 1000-fold a 400 μM stock in DMSO, which was maintained at −20 °C for months and thawed as needed. Cells were labeled and/or fixed 18–24 h after transfection. For co-plating of two distinct pools of transfected cells, cells were initially grown on plastic multi-well plates, transfected, switched to fresh serum-containing media, then lifted 4–8 h later by trypsinization and co-plated at 80% density onto fibronectin-coated glass slips. For intercellular sHRP reconstitution experiments, we recommend using B27 (with heme supplementation) or FBS (with or without heme supplementation), but adult calf serum should be avoided, since it inhibited sHRP reconstitution in some experiments. Cells were labeled and/or fixed 12–20 h after co-plating. Where indicated, rapamycin was supplemented into growth media of co-plated cells at a concentration of 400 nM. Where indicated (Extended Data Figure 5C only), DNAseI (Roche) was supplemented at the time of co-plating (0.2 mg/mL final concentration). Where indicated, heme was supplemented at a concentration of 1–7 μM by diluting a 483 μM heme stock solution, which was prepared by dissolving hemin-Cl (Sigma) in 10 mM NaOH with vortexing for 3 min. Heme was typically added at the time of co-plating, but strong sHRP labeling was detected when heme was added only 5 minutes prior to fixation. Heme stock solutions were used fresh (within 40 minutes). Except for a few cases, which are noted explicitly in the text, heme supplementation was used for extracellular sHRP constructs, but was not used for intracellular constructs.
Introduction of sHRP constructs to mice for the anterograde paradigm (Figure 4A–C)
Animals were used in accordance with NIH guidelines and protocols approved by Institutional Animal Use and Care Committee at Harvard University. We infected RGCs with sHRPa-NRX using a recombinant adeno-associated virus (AAV). sHRPa-NRX was cloned into an AAV backbone with a synapsin promoter35 and woodchuck posttranslational response element (WPRE). Viral particles were prepared using a mixture of AAV1 and AAV2 helpers, as described in the protocol below. AAV was injected to one side of adult mouse eyes (P30–40) using 32G Hamilton syringes at 4 weeks before dissection. We overexpressed sHRPb-NLG in neurons of the superior colliculus by lipofection or electroporation. Cerulean-triple F2A-sHRPb-NLG1 was placed between piggyBac-transposon terminal repeats in pXL-CAG36. Plasmid DNAs were prepared using Qiagen Plasmid columns (Valencia, CA) and further treated with an UltraClean Endotoxin Removal Kit (MO BIO Laboratories, Inc., Carlsbad, CA). Lipofection was carried out using Lipofectamine LTX (Life Technologies, Grand Island, NY). Briefly, 5 μg of pXL-CAG-Cerulean-triple F2A-sHRPb-NLG1 and a transposase plasmid (pCAG-PBorf) (10:1 wt ratio) in 50 μL OptiMEM (Life Technologies) were mixed with 5 μL of Lipofectamine PLUS reagent. To this DNA solution, 5 μL Lipofectamine LTX in 50 μL OptiMEM was added, incubated for >15 min, and used as a lipofection mixture for injection to mice. At one week before dissection, 5 μL of the lipofection mixture was injected using 32G Hamilton syringes to the superior colliculus in anesthetized adult mice that had previously been injected with AAV sHRPa-NRX, as described above. In some experiments, 5 μg/2 μL of a 10:1 wt mixture of pXL-CAG-Cerulean-triple F2A-sHRPb-NLG1 and pCAG-PBorf in 1 mM TrisHCl, 0.1 mM EDTA was injected to superior colliculus at postnatal days (P) 1–2, and electroporated using an BTX-ECM830 electroporator and 5 mm electrodes (delivery: 20 pulses, 50 V, 50 ms). To those electroporated pups, presynaptic AAV was injected to an eye at P12, and mice were dissected at P20.
Introduction of sHRP constructs to mice for the retrograde paradigm (Figure 4D–F)
We retrogradely infected RGCs by injecting postsynaptic pseudotyped lentiviral vectors to the superior colliculus and overexpressed sHRPa-NRX in amacrine cells by using a presynaptic Cre-dependent AAV. Cerulean-triple F2A-sHRPb-NLG1 was cloned into a lentiviral plasmid with the CMV promoter. The pseudo-typed lentivirus was generated with FuGB2 (VSV and rabies fusion) envelope glycoprotein, and concentrated by centrifugation as described previously31. A Cre-dependent FLEX-AAV-sHRPa-NRX (AAV-synapsin promoter-DIO-sHRPa-NRX-WPRE) plasmid was designed as described previously30. AAV particles were prepared using an AAV2 helper system, as described below, and injected into the eye ball of adult mice expressing Cre in most if not all amacrine cells (vesicular GABA transporter (Slc32a1)-IRES-Cre line, Jackson laboratories, Bar Harbor, ME). At the same time, pseudotyped lentiviral particles were injected to the superior colliculus, and mice were sacrificed at 7 days after the injections.
Mouse dissection, brain fixation, cryosectioning, and sHRP staining of tissue sections
Each animal was perfused with 10 mL Neurobasal medium (Life Technologies) supplemented with 2 μM heme. Mice were sacrificed, and the superior colliculus or retina were immediately removed from mice and slowly rocked in 2 μM heme and 10% FCS in Neurobasal for 2 h at 4 °C. The tissues were fixed with 4% paraformaldehyde/PBS for 2 h at 4 °C, briefly rinsed with cold PBS, and sunk in 15% (w/v) and 30% (w/v) sucrose/PBS at 4 °C overnight, frozen in Tissue Freezing medium (Electron Microscopy Sciences, Hatfield, PA), and cryosectioned to prepare 20 μm-thick dry sections on Superfrost Plus slides (VWR). Sections were treated in 0.3% H2O2/methanol at −20 °C for 15 h (to reduce endogenous background peroxidase activity), and rinsed with PBS. In some experiments, this step was omitted. Reconstituted HRP activity was detected using a Cy3-tyramide dye (1:100 dilution) from the Tyramide Signal Amplification (TSA) Plus system (PerkinElmer, Waltham, MA) for 2 h at room temperature. Stained sections were blocked with 5% skim milk/PBS supplemented with 0.2% NaN3 for 30 min, then subsequently incubated with primary antibodies, rinsed, and incubated with fluorophore-conjugated secondary antibodies. Mouse α-NLG1 and rabbit α-HRP primary antibodies were used to detect sHRPb-NLG and sHRPa-NRX, respectively. This α-HRP antibody specifically recognizes the larger sHRPa fragment, but does not recognize sHRPb. α-Rabbit 488 and α-mouse 647 secondary antibodies were used. See Supplementary Table 4 for details on antibody vendors and dilutions used. Finally, sections were mounted in Fluoro-Gel (Electron Microscopy Sciences) and imaged using a Zeiss LSM510 confocal microscope. Images were processed using Fiji (ImageJ 1.47d) and Adobe Photoshop CS5.
Rat hippocampal neuron culture, transfection, and heme supplementation
Hippocampal neurons were harvested from rat embryos sacrificed at embryonic day 18 and plated in 24-well plates as previously described3. For electron microscopy (EM) experiments, neurons were plated on poly-d-lysine coated glass-bottom dishes (P35GC-0-14-C, MatTek Corp.) that had been additionally coated and washed as previously described3, and cell suspension was added only to the central glass portion of the dish , with an additional 2 mL media added 2–3 h after plating. Neurons were transfected using Lipofectamine2000 between 10–17 days in vitro using 2 μL Lipofectamine2000 and 400 ng plasmid DNA per 1.91 cm2 well, in a volume of 0.5 mL of a 1:1 DMEM:MEM mixture (Cellgro) without serum. The transfection time was 1–5 h, depending on the age and density of the neuron culture, with more dense and more mature cultures (DIV 15 and older) requiring a longer transfection time for efficient transfection. The original growth medium was preserved, and neurons were placed back into this original medium after transfection. For two-step lipofection experiments, the second transfection was performed 20–48 h after the first transfection, and neurons were fixed and labeled 1–6 days after the second transfection. To avoid the formation of artificially large synapses (as in Extended Data Figure 8E), it was important to use the shortest possible transfection time that still yielded detectable synapses between the two pools of transfected neurons. This transfection time varied depending on the age and density of the culture. Shorter transfection times lowered the transfection efficiency, making sHRP-positive synapses much rarer, but they also maintained lower expression levels on average. Note that this two-step lipofection procedure produces a small percentage of neurons expressing both sHRP fragments, which leads to bright cis signal throughout all processes (see Supplementary Discussion 4). For intercellular sHRP reconstitution experiments in neurons, heme was supplemented into the media from a 483 μM stock (described above) to a concentration of 0.5 – 2 μM. Heme was added 2–18 h prior to fixation.
Split HRP fluorescent labeling of cell-cell contact sites HEK293T cells
In cases when immunostaining was not performed, co-cultured cells were washed in Dulbecco’s phosphate-buffered saline (DPBS) for 4 min at room temperature to remove excess heme. In cases when surface immunostaining was performed, cells were placed into a room temperature solution containing primary antibody and 1% bovine serum albumin (BSA, Fisher Scientific) in DPBS. After 10 minutes, primary antibody solution was removed, and cells were washed 2 × 2 min in room temperature DPBS. After immunostaining and/or washing, cells were submerged in room temperature 4% formaldehyde in phosphate buffered saline (PBS), freshly prepared from a 10% formaldehyde stock (Polysciences), then promptly moved to ice for 20–30 min. Cells were rinsed 3 × 1 min in PBS. In cases where heme and/or serum were supplemented post-fixation, cells were placed in the appropriate media and allowed to rock at 4 °C overnight, followed by rinsing 3 × 5 min in chilled PBS. Cells were treated with methanol at −20 °C for 10 min, rinsed 3 × 1 min in PBS, then submerged in a room temperature solution containing 100 μM biotin-phenol, synthesized as described previously13, and 1 mM H2O2 in PBS. It was important to sonicate the biotin-phenol solution for at least 5 min to dissolve the compound completely. After 10 min, the biotin-phenol solution was removed, and the cells were rinsed 3 × 1 min in PBS, blocked using 1% (w/v) BSA in PBS for 10 min, then placed on ice and stained with a home-made neutravidin-AlexaFluor647 conjugate (1/1000 dilution of the as-synthesized conjugate) in PBS with 1% BSA for 20 min at 4 °C with rocking. In cases where primary immunostaining had been performed, a fluorescent secondary antibody was included in the neutravidin-647 solution. After secondary staining, cells were rinsed 4 × 5 min in PBS, then imaged. For experiments involving >10 samples, cells were fixed a second time using 4% formaldehyde at room temperature for 10 min, then washed 2 × 1 min in room temperature PBS and stored in PBS containing 30 μg/mL kanamycin (IBI Scientific) in the dark at 4 °C prior to imaging. Samples were stable for weeks under these conditions.
Split HRP fluorescent labeling of contact sites between cultured neurons
In cases when immunostaining was not performed, cultured neurons on glass coverslips were washed in Tyrode’s buffer (120 mM NaCl, 2 mM CaCl2, 3 mM KCl, 2 mM MgCl2, 0.5 mM NaH2PO4, 30 mM glucose, 25 mM HEPES, pH 7.4 adjusted with NaOH) for 4 min at room temperature to remove excess heme. In cases when surface immunostaining was performed, neurons were placed into a room temperature solution containing primary antibody and 1% bovine serum albumin (BSA, Fisher Scientific) in Tyrode’s buffer. After 10 min, the antibody solution was removed, and neurons were washed 2 × 2 min in room temperature Tyrode’s buffer. After surface immunostaining and/or washing, neurons were fixed at room temperature for 30 min using 4% formaldehyde with 0.12 M sucrose, 1.8 mM CaCl2, and 1.0 mM MgCl2 in PBS. After fixation, neurons were rinsed 3 × 1 min in PBS at room temperature, then permeabilized with methanol at −20 °C for 10 min. Neurons were rinsed 3 × 1 min in PBS, then submerged in a solution containing 100 μM biotin-phenol and 1 mM H2O2 in PBS for 30–45 min at room temperature. Neurons were rinsed 3 × 1 min in PBS, then blocked using 1% (w/v) BSA for 10 min and stained with a home-made neutravidin-AlexaFluor647 conjugate (1/1000 dilution of the as-synthesized conjugate) in PBS with 1% BSA for 20 min at 4 °C with rocking. In cases where immunostaining was performed, a fluorescent secondary antibody was included in the neutravidin-647 solution. Neurons were rinsed 4 × 5 min in PBS, then imaged. For experiments involving >6 samples, neurons were fixed a second time using 4% formaldehyde prior to imaging.
Amplex UltraRed labeling and immunostaining of HEK293T cells
Amplex UltraRed labeling and immunostaining of HEK293T cells were performed as described previously23. For the live-cell imaging in Extended Data Figure 1, cells were labeled with Amplex UltraRed for 30 min. For the fixed-cell imaging shown in Figure 1F and Extended Data Figure 4A, cells were labeled with Amplex UltraRed for 20 min.
Confocal fluorescence imaging of cultured cells and image analysis
Confocal imaging was performed with a Zeiss AxioObserver inverted confocal microscope as described previously23. All images were collected using SlideBook (Intelligent Imaging Innovations). Acquisition times ranged from 100 ms to 3 s. All image analysis was performed in SlideBook. Fluorophore channels in each experiment were normalized to the same intensity ranges unless explicitly noted otherwise. For the analysis in Figure 3B and Extended Data Figure 8E, a binary mask was created in each field of view for sHRP biotin-phenol labeling and for each of the two transfected pools of neurons, which were marked either by co-transfected fluorescent proteins or surface immunostaining. An intersection mask was created between the two transfection markers, and this intersection mask was then intersected with the sHRP labeling mask to create a new triple intersection mask. Regions within this triple intersection mask with area > 0.42 μm2 were considered contact sites and included in the plots. Within each field of view, “background” was defined as the mean sHRP intensity across 5 manually drawn regions of untransfected neurons with areas of 4–10 μm2. The maximum sHRP signal contained within each contact site was divided by the “background” calculated for the corresponding field of view. Each dataset includes at least 60 contact sites collected across at least 4 independent fields of view. The specificity values in Figure 3C were determined by manual inspection of whether sHRP puncta overlaid with puncta of each synaptic marker (either Homer-Venus or Synaptophysin-mApple). Each experiment examined >40 sHRP puncta across 4 FOV. In some FOV, the Homer-Venus and/or Synaotphysin-mApple synaptic markers failed to display a punctate localization pattern, and in other FOV, the sHRP labeling was so bright and the mApple fluorescence so dim that bleedthrough from sHRP labeling overwhelmed the mApple signal. sHRP puncta in these FOV were excluded from the analysis. Very occasionally, artificially large, oblong sHRP labeling features were observed. These enlarged sHRP labeling sites, which accounted for <5% of labeling features when the weaker synapsin promoter35 and mild transfection conditions were used, were excluded from the analysis. The intersection image presented in Extended Data Figure 9A represents an intersection mask between individual binary masks of the Venus and tdTomato co-transfection markers. The “% contacts labeled” presented in Figure 2B were determined by manual inspection; contact sites were identified as sites of overlap between the V5 and FLAG channels, and a contact site was deemed “labeled” if sHRP signal was visible above background.
DAB staining and preparation of HEK293T cells and cultured neurons for EM
Cells plated on glass-bottom dishes were fixed, stained with 3,3′-diaminobenzidine (DAB), and processed for EM as described previously19, but with the following modifications. After fixation and glycine treatment, HEK293T cells were incubated in 0.5 mg/mL DAB in PBS without H2O2 for 15 min at 4 °C. This solution was replaced with 0.5 mg/mL DAB in PBS with 0.01% H2O2 (3.33 mM) to initiate DAB labeling. DAB staining was performed at 4 °C for 45 min. For cultured neurons, the pre-incubation step with DAB without H2O2 was 30 min, and the DAB staining period was 60–70 min. Neurons were fixed as described previously19 except with 1 mM MgCl2 added to the fixative.
Electron microscopy
DAB-stained areas of embedded cultured cells were identified by transmitted light, and the areas of interest were sawed out using a jeweler’s saw and mounted on dummy acrylic blocks with cyanoacrylic adhesive. The coverslip was carefully removed, the block trimmed, and ultrathin sections (60–80 nm) were cut using an ultramicrotome. Sections were placed on 200 mesh copper grids, and in some cases, grids were coated with 15 nm gold nanoparticles as a reference standard by brief exposure to a diluted solution of colloidal gold (BBI Solutions, Cardiff, UK). Electron micrographs were recorded using a JEOL 1200 TEM operating at 80 keV.
Electron tomography
Semi-thin sections (250 nm thick) were cut from the same material as for the EM imaging of thin sections. A mix of 5 and 10 nm diameter colloidal gold was then deposited on each section side to serve as fiducial markers. Electron micrographs were collected with a 4k×4k detector (Gatan Ultrascan) mounted on a transmitted electron microscope (FEI Titan) operating at 300 kV, while sections were tilted every 0.5° (from −60° to +60°) in a 4-tilt series scheme. The final 3D representations were built from the projection sets following the TxBR alignment protocol37 and using iterative procedures (weighted SIRT). Volumes are displayed with IMOD38, and animations created with AMIRA. Tomograms reported in this paper were all acquired at a magnification of 22,500x, with an effective pixel size being 0.41 nm.
Generation of stable HEK293T NRX and NLG cells
To generate HEK293T cell lines stably expressing sHRPa-NRX and sHRPb-NLG, the constructs were cloned into a piggyBac transposon vector pXL-CAG-Zeocin-3xF2A (NotI/AscI sites) to generate pXL-CAG-Zeocin-3xF2A-sHRPa-NRX and pXL-CAG-Zeocin-3xF2A-sHRPb-NLG, respectively. HEK293T cells were transfected with the appropriate sHRP construct together with a piggyBac transposase vector pCAG-PBorf, trypsinized after 2 days, re-plated into larger plates, and selected with 1 mg/mL Zeocin (Invivogen, San Diego, CA) for 10 days. Surviving colonies were transferred to new plates and screened with antibodies against sHRPa or NLG to select clones with high expression (see Supplementary Table 4).
Split GFP labeling of cell-cell contact sites HEK293T cells
Co-cultured cells on glass coverslips were stained with primary antibodies, fixed, and rinsed as described above for the split HRP samples. Cells were then blocked using 1% (w/v) BSA for 10 min, stained with secondary antibodies, rinsed, and processed for imaging as described for the split HRP samples. A second fixation with 4% formaldehyde did not affect the brightness of GFP (data not shown).
Split GFP fluorescent labeling of contact sites between cultured neurons
Cultured neurons were surface immunostained, rinsed, fixed, and permeabilized with methanol as described above for split HRP in cultured neurons. Neurons were rinsed 3 × 1 min in PBS, blocked using 1% (w/v) BSA in chilled PBS for 10 min, then stained with the appropriate fluorescent secondary antibody in PBS with 1% BSA for 20 min at 4 °C with rocking. Neurons were rinsed 4 × 5 min in PBS, then imaged. In cases when many samples were being imaged (>6), neurons were fixed a second time using 4% formaldehyde prior to imaging.
Split HRP fluorescent labeling in neurons with immunostaining of endogenous bassoon
Cultured neurons on glass coverslips were cooled to room temperature, washed in Tyrode’s buffer for 4 min at room temperature to remove excess heme, then fixed at room temperature for 12 min using 4% formaldehyde with 0.12 M sucrose, 2 mM MgCl2, 10 mM EGTA, 25 mM HEPES, and 60 mM PIPES, pH 7.4. Neurons were rinsed 3 × 1 min in PBS, then permeablized with 0.1% TritonX-100 (Sigma) in PBS for 7 min at room temperature. Neurons were rinsed 3 × 1 min in PBS, then submerged in a solution containing 100 μM biotin-phenol and 1 mM H2O2 in PBS for 45 min at room temperature. Neurons were rinsed 3 × 1 min in PBS, then blocked using 2% (w/v) BSA for 6 h at 4 °C and stained with primary mouse α-Bassoon antibody (Enzo) with 2% BSA in PBS overnight with rocking at 4 °C. Neurons were rinsed 3 × 5 min at room temperature with PBS containing 0.1% TritonX-100, then stained with neutravidin-AlexaFluor647 and fluorescent secondary antibodies using the procedure described above. Neurons were rinsed 4 × 5 min in PBS, then imaged. For experiments involving >6 samples, neurons were fixed a second time using 4% formaldehyde prior to imaging.
Surface immunostaining of cultured neurons
Cultured neurons on glass coverslips were cooled to room temperature, placed into a solution containing primary antibody and 1% bovine serum albumin (BSA, Fisher Scientific) in Tyrode’s buffer, rinsed 2 × 2 min in room temperature Tyrode’s buffer, then fixed at room temperature for 30 min using 4% formaldehyde with 0.12 M sucrose, 1.8 mM CaCl2, and 1.0 mM MgCl2 in PBS. Neurons were rinsed 3 × 1 min in PBS at room temperature, blocked using 1% (w/v) BSA in PBS for 10 min, then stained with the appropriate fluorescent secondary antibody in PBS with 1% BSA for 20 min at 4 °C with rocking. Neurons were rinsed 4 × 5 min in PBS then imaged.
Total immunostaining of cultured neurons
Cultured neurons on glass coverslips were cooled to room temperature, rinsed 1 × 1 min in room temperature Tyrode’s buffer, then fixed at room temperature for 30 min using 4% formaldehyde with 0.12 M sucrose, 1.8 mM CaCl2, and 1.0 mM MgCl2 in PBS. Neurons were rinsed 3 × 1 min in PBS, permeabilized with methanol for 10 min at −20 °C, rinsed again 3 × 1 min in PBS, then blocked using 1% (w/v) BSA in chilled PBS for 10 min and stained with a primary antibody solution with 1% BSA in for 60 min at 4 °C. Neurons were rinsed 4 × 5 min in PBS, stained with the appropriate fluorescent secondary antibody in PBS with 1% BSA for 45 min at 4 °C with rocking, then rinsed 4 × 5 min in PBS and imaged.
Biotin-phenol fluorescent labeling of ER-lumen reconstituted sHRP fragments
Transfected cells were placed under cell culture media containing biotin-phenol (500 μM) and allowed to incubate for 45 min at 37 °C. It was important to sonicate the biotin-phenol solution beforehand for at least 5 min to dissolve the compound. Cells were moved to room temperature, followed immediately by addition of 1 mM H2O2 and gentle manual shaking to mix. After 1 minute, the biotin-phenol solution was removed, and cells were directly fixed with 4% formaldehyde with 5 mM Trolox in PBS at room temperature for 30 min. Cells were rinsed 3 × 1 min in PBS, permeabilized with pre-chilled methanol at −20 °C for 10 min, rinsed 3 × 1 min in PBS, blocked using 1% (w/v) BSA in chilled PBS for 10 min, then stained with a solution containing primary antibodies and 1% (w/v) BSA in PBS with rocking at 4 °C for 45 min. Cells were rinsed 4 × 5 min in PBS, then stained with a solution containing neutravidin647 (1/1000) and the appropriate secondary antibodies with 1% (w/v) BSA in PBS for 20 min at 4 °C. Cells were rinsed 4 × 5 min in PBS and imaged.
Production of lentiviruses
HEK 293T cells in a T25 flask (Corning) were transfected at ~60–70% confluency with 3 mL of serum-free media containing the lentiviral vector containing the gene of interest (2.5 μg), the lentiviral packaging plasmids dR8.91 (2.25 μg) and pVSV-G (0.25 μg)39, and 30 μL of 1 mg/mL Polyethylenimine “Max” (Polysciences) pH 7.1 for 3 h. Transfection media was replaced with fresh serum-containing media, and cell media containing lentivirus was harvested after 48 h and passed through a 0.45 μm filter to produce infectious media that was added directly to neuronal cultures, as described below.
Production of adeno-associated viruses
Adeno-associated viruses (AAVs) with a mixed serotype AAV1/AAV2 were produced according to a published procedure29, except that the HEK293T cells were transfected differently. Three T150 flasks of HEK293T cells were transfected at 60% confluency by addition of (per flask) the AAV vector containing the gene of interest (5.2 μg), AAV1 serotype plasmid (4.35 μg), AAV2 serotype plasmid (4.35 μg), pFdelta6 Adenovirus-helper plasmid (10.4 μg), and 1 mg/mL PEI Max solution (130 μL) dissolved in 26 mL cell culture media (DMEM with high glucose containing 10% FBS (Sigma), 1× GlutaMAX (Life Technologies), and 10 mM sterile HEPES buffer (Life Technologies)). Cells were transfected in this solution for 48 h, after which time infectious media was harvested, passed through a 0.45 μm filter, then used directly for infection of cultured neurons, as described below. For preparation of concentrated AAVs for in vivo experiments, the cells from all three T150 flasks were harvested by scraping and combined, followed by cell lysis and heparin column purification as described29. AAV2 particles were essentially prepared as described previously40. Briefly, HEK293FT cells in ten 15cm plates were transfected with the AAV vector, AAV2 serotype plasmid, and pFdelta6 Adenovirus helper plasmid using calcium phosphate and glycerol shock. Cells were harvested and lysed using chloroform. Viral particles were precipitated with polyethylene glycol 8000, extracted with chloroform, and concentrated using Amicon Ultra15 (Millipore, Billerica, MA).
Infection of cultured neurons with lentiviruses and adeno-associated viruses
For the results presented in Extended Data Figure 9C, hippocampal neurons were transfected using Lipofectamine 2000 at DIV 12 using the procedure described above. At DIV 13, neurons were infected with either lentivirus (400 μL of infectious media) or AAV (25 μL of infectious media). Infectious media was added directly to the existing culture medium, which was normally ~2 mL in volume. At DIV 17, heme was supplemented into the existing culture medium to a concentration of 1 μM. Neurons were fixed and labeled at DIV 18.
Yeast strains, transformation, and cell culture
Aga2P-HRP (full-length) yeast were generated by transformation of the yeast display plasmid pCTCON241 into the S. cerevisiae strain EBY100, as described previously19. All other yeast were generated using a two-step procedure in the S. cerevisiae strain BJ5465, based on a previously described protocol42. First, BJ5465 yeast were transformed with a YIP plasmid that had been linearized by digestion with the restriction enzyme BsiW. The YIP plasmid is designed for integration into the yeast genome42. See Supplementary Table 5 for details on all plasmids used in this study. Transformation was performed using the Frozen E-Z Yeast Transformation II Kit (Zymoprep) according to the manufacturer’s protocols. Transformed yeast were plated on solid media lacking uracil, allowing for selection of individual colonies constitutively expressing the desired Aga1P fusion, as confirmed by FACS analysis of immunostained yeast (see procedure below) and by DNA sequencing of PCR products amplified from genomic DNA, which was extracted using a published procedure43. For introduction of individual, non-mutagenized Aga2P constructs (as opposed to libraries) into yeast constitutively expressing recombinant Aga1P fusions (such as the “template” shown in Extended Data Figure 2A), Aga1P fusion-expressing yeast were made competent for transformation using the Frozen E-Z Yeast Transformation II Kit (Zymoprep), followed by transformation with the appropriate pCTCON2 plasmid using the same kit. Transformed cells containing the Trp1 gene were selected on synthetic dextrose plus casein amino acid (SDCAA) plates. Yeast cell culture and induction of pCTCON2 construct expression were performed as described previously19.
Generation of error-prone libraries for yeast display
Libraries of sHRP fragment mutants were generated using error-prone PCR according to published protocols19,44. In brief, for the sHRPa′ library, 150 ng of the template sHRPa gene in the pCTCON2 vector41 was amplified for 20 rounds with 0.4 μM forward and reverse primers, 2 mM MgCl2, 5 units of Taq polymerase (NEB), and 2μM each of the mutagenic nucleotide analogues 8-oxo-2′-deoxyguanosine-5′triphosphate (8-oxo-dGTP) and 2′-deoxy-p-nucleoside-5′-triphosphate (dPTP). The PCR product was then gel-purified and re-amplified for another 30 cycles under normal PCR conditions. The error-prone PCR product was electroporated along with BamHI-NheI linearized pCTCON2 vector (10 μg insert:1 μg vector) backbone into electrocompetent S. cerevisiae BJ5465 constitutively expressing sHRPb′-Aga1P. Electroporation was performed using a Bio-Rad Gene pulser XCell. Transformation efficiency was 4.0 × 107. DNA sequencing of 19 distinct colonies showed an average of 2.0 nucleotides changed per clone with a range of 0–5. For the sHRPb′ library, the same procedure was used except that 10 rounds of error-prone PCR were performed with 10 μM each of the mutagenic nucleotide analogues, and electroporation was performed into S. cerevisiae BJ5465 constitutively expressing sHRPa-Aga1P. Transformation efficiency was 3.8 × 107. DNA sequencing of 24 distinct colonies showed an average of 1.8 nucleotides changed per clone with a range of 0–6. The electroporated cultures were rescued in 100 mL of SDCAA media supplemented with 50 units/mL penicillin and 50 μg/mL streptomycin for two days at 30°C.
Yeast display selections
Yeast libraries were labeled and sorted following a published protocol19 with a few modifications. Expression of pCTCON2 libraries was induced by growing yeast in 1:9 SDCAA:SGCAA media overnight. Where indicated, 1 mM succinyl acetone (Sigma) was supplemented into the media during this overnight induction period. 10-fold oversampling above the diversity of the yeast pool was employed for all passaging steps. Prior to the first round of sorting, “diversity” was considered to be the number of transformants in the initial library. In subsequent rounds, the “diversity” was considered to be the number of yeast cells collected in the previous round of sorting. Induced yeast were pelleted at 500 × g for 4 min at 4°C and resuspended in an equivalent volume of DPBS with 0.1% BSA (DPBS-B). All subsequent steps were performed in DPBS-B. Cultures were washed once more, and the OD600 for a 1/100 dilution of the culture was measured to determine cell density. An aliquot of yeast culture, containing at least 10-fold more yeast cells than the diversity of the culture (1 OD600 ~ 1 × 107 yeast cells), was pelleted in a microfuge tube at 14,000 × g for 30 seconds. Cells were resuspended in a 1:50 dilution of mouse α-c-Myc (Calbiochem) at a density between 5 × 107 and 1.3 × 108 cells/mL, then incubated at 4°C with rotation for 1 h. Samples were washed twice with DPBS-B, then resuspended in a room temperature solution consisting of 100 μM biotin-phenol and 1 mM H2O2 at a density between 2.5 × 106 and 6.3 × 106 cells/mL. After 1 minute, the yeast suspension was quenched by addition of an equal volume of 20 mM Trolox and 10 mM sodium ascorbate in DPBS-B, followed by rapid vortexing. The cells were pelleted at 14,000 × g for 1 min. Excess solution was removed, and yeast cells were resuspended in a solution containing 1/50 streptavidin-phycoerythrin (Jackson ImmunoResearch) and 1/100 Alexa Fluor 647 goat α-mouse IgG (Life Technologies) at a density between 5 × 107 and 1.3 × 108 cells/mL. After 30 min of incubation with rotation at 4°C, samples were washed twice and resuspended in DPBS-B at a density of 1.3 × 107 cells/mL in the appropriate tube for FACS sorting. Cells were sorted on a Beckman Coulter MoFlo equipped with 488 and 640 nm lasers and appropriate emission filters (580/30 for PE, 657 low-pass for AF647). Trapezoidal gates were drawn to collect yeast cells positive for AF647 signal that also had high PE signal. Gates were drawn to collect the following % of cells: 0.6% for sHRPa′ round 1,1.0% for sHRPa′ round 2, 0.9% for sHRPa′ round 3, 0.5% for sHRPa′ round 4, 1.71% for sHRPb′ round 1, 0.5% for sHRPb′ round 2, 0.57% for sHRPb′ round 3, 0.6 % for sHRPb′ round 4, and 0.35% for sHRPb′ round 4 with succinyl acetone. In each round of sorting, the number of yeast cells passed through the sorter was at least 10-fold larger than the diversity size of the yeast pool, except for round 1, in which case 3-fold oversampling was employed. This lower sampling in round 1 was a result of the very large diversity size and the practical difficulties and expense of labeling large numbers of yeast cells. Sorted cells were regrown for subsequent rounds of sorting as described previously19. Sequence analysis of isolated yeast clones was performed as described previously19.
FACS analysis of yeast
Yeast samples were taken from picked colonies of transformed clones or from samples regrown after sorting. Expression was induced by inoculating 4.5 mL of SGCAA with 500 μL of a saturated SDCAA yeast culture and incubating at 30°C for 20–24 h. Yeast were labeled as described above, except in some cases different primary and secondary antibodies were used for immunostaining (see Supplementary Table 4). Samples were analyzed using a BD LSR II flow cytometer (BD Biosciences) equipped with 488 and 640 nm lasers and appropriate emission filters (515/20 for AlexaFluor488, 582/42 for phycoerythrin, and 670/30 low-pass for AlexaFluor647).
Synthesis of home-made neutravidin-AlexaFluor647 conjugate
A reaction mixture was assembled in a 1.5 mL Eppendorf tube with following components (added in this order): 200 μL of 5 mg/mL Neutravidin (Life Technologies) in PBS, 20 μL of 1 M sodium bicarbonate in water, and 10 μL of 10 mg/mL AlexaFluor647-NHS Ester (Life Technologies) in anhydrous DMSO. The tube was incubated at room temperature with rotation in the dark for 3 h. The neutravidin-AlexaFluor647 conjugate was purified from unreacted dye using a NAP-5 size-exclusion column (GE Healthcare Life Sciences) according to the manufacturer’s instructions. The conjugate was typically eluted from the column in 500 μL cold PBS. Absorbance values, determined using a Nanodrop 2000c UV-vis spectrophotometer (Thermo Scientific), were typically as follows: A280 = ~0.284 and A647 = ~1.625. The conjugate was stable at 4 °C in the dark for at least 4 months and was flash frozen and stored at −80 °C for longer term. For mammalian cell labeling experiments, the conjugate was diluted 1000-fold in PBS containing 1% BSA.
N-linked glycan removal assays and Western blotting
For each condition, HEK293T cells were plated into a single well of a 6-well plate, cultured, and transfected with the appropriate constructs using the procedure described above. Transfected cells were moved to ice, washed twice with 1000 μL pre-chilled DPBS at 4 °C, then scraped and pelleted at 800 × g at 4 °C for 5 min. The supernatant was carefully removed, and the pellet was stored at −80 °C. The pellet was thawed on ice, then submerged in 80 μL cell lysis buffer (1mM HEPES pH 7.5, 5 mM magnesium acetate, 1mM PMSF [phenylmethylsulfonyl fluoride, G-Biosciences] and 1× protease inhibitor cocktail [Sigma]) and resuspended with gentle pipetting. Pellets were subjected to 3 cycles of freezing in liquid nitrogen followed by thawing in a 4 °C water bath. Lysates were clarified by centrifugation at 8000 × g for 10 min at 4 °C. Removal of N-linked glycans was performed using PNGaseF (New England Biolabs) following the manufacturer’s protocol for deglycosylation of denatured proteins. To denature the lysates, 5 μL of clarified lysates were mixed with 4 μL Millipore water and 1 μL 10× glycoprotein denaturing buffer, then heated to 100 °C for 10 min. To the denatured lysates were added 1.5 μL PNGaseF, 2 μL 10× G7 reaction buffer, 2 μL of 10% NP-40, and 4.5 μL Millipore water, followed by gentle pipetting to mix. The resulting reaction mixture was incubated at 37 °C for 1 h. Deglycosylated lysates were separated using 12% SDS-PAGE. Color Prestained Protein Standard (New England Biolabs) was run in parallel for molecular weight calibration. For blotting analysis, gels were transferred to nitrocellulose membrane, stained by Ponceau S (10 min in 0.1% w/v Ponceau S in 5% acetic acid/water), and blocked with 2% BSA in 1.2× TBS-T buffer (24 mM Tris-HCl, 164 mM NaCl, 0.12% Tween, pH 7.6) at 4 °C overnight. Blots were immersed in 1/1000 mouse α-Flag (Agilent) or 1/2500 mouse α-V5 antibody (Life Technologies) in 1.2× TBS-T with 1% BSA for 1 h at room temperature. Blots were rinsed 4 × 5 min with 1.2× TBS-T, then immersed in 1/2000 Goat α-Mouse IgGH + L-HRP Conjugate (Bio-Rad) in 1.2× TBS-T with 1% BSA for 30 min at room temperature. Blots were rinsed 4 × 5 min in 1.2× TBS-T, then rinsed 1 × 2 min in Millipore water before development with Clarity™ reagent (Bio-Rad) and imaging on an Alpha Innotech gel imaging system.
Co-immunoprecipitation to assess sHRP reversibility
For each condition, HEK293T cells were plated into a single well of a 6-well plate, cultured, and transfected with the appropriate constructs using the procedure described above. Transfected cells were lysed, and the lysates were clarified as described above for deglycosylation assays. One quarter of the lysates were diluted to a volume of 115 μL using cell lysis buffer, and 15 μL of the resulting mixture were saved at 4 °C as “input” for subsequent analysis. The remaining 100 μL were mixed with a 10 μL slurry of ANTI-FLAG M2 Affinity Gel (Sigma), which were prepared for binding following the manufacturer’s instructions, and allowed to incubate at 4 °C with rotation for 3 h. Resin was washed 3 × 1 mL using lysis buffer at 4 °C, followed by two successive 25 μL elutions with 100 μg/mL FLAG peptide at room temperature for 15 min. Eluates were combined and analyzed by Western blotting using the procedure described above for deglycosylation assays.
Supplementary Material
Acknowledgments
We thank Jenifer Einstein (MIT) for preparing neuron cultures. Dr. Wenjing Wang (MIT) provided yeast expressing the LAP peptide and gave helpful advice for yeast display and AAV preparation. Dr. Philipp Stawski, Kurt Cox, and Ken Loh (MIT) provided synaptic fluorescent protein fusion plasmids. Dr. Faycal Touti and Dr. Hyun-Woo Rhee (MIT) synthesized biotin-phenol. FACS experiments were performed at the Koch Institute Flow Cytometry Core (MIT). Funding was provided by the US National Institutes of Health (R01-CA186568-1 to A.Y.T.; R37NS029169 to J.R.S.; P41 GM103412 and R01GM086197 to M.H.E.) and the Howard Hughes Medical Institute Collaborative Initiative Award (A.Y.T. and J.R.S.). J.D.M. was supported by NSFGR and NDSEG fellowships.
Footnotes
AUTHORS’ CONTRIBUTIONS STATEMENT
J.D.M. performed all experiments except those explicitly noted below. J.R.S. and M.Y. designed in vivo experiments and analyzed the results. M.Y. performed all in vivo experiments, prepared constructs and viruses for in vivo experiments, and generated stable HEK293T cells. T.J.D. prepared thin sections and performed EM imaging. T.J.D. and S.P. performed electron tomography and processed the data. M.H.E. guided and oversaw EM experiments and analyzed results with J.D.M. and T.J.D. C.G.K. contributed to deglycosylation and HEK293T cell labeling experiments. J.D.M. and A.Y.T. designed the research and analyzed the data. J.D.M., J.R.S., and A.Y.T wrote the paper. All authors edited the paper.
COMPETING INTERESTS STATEMENT
Massachusetts Institute of Technology has filed a patent covering part of the information contained in this article.
References
- 1.Kim SA, Tai CY, Mok LP, Mosser EA, Schuman EM. Calcium-dependent dynamics of cadherin interactions at cell–cell junctions. Proceedings of the National Academy of Sciences. 2011;108:9857–9862. doi: 10.1073/pnas.1019003108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feinberg EH, et al. GFP Reconstitution Across Synaptic Partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron. 2008;57:353–363. doi: 10.1016/j.neuron.2007.11.030. [DOI] [PubMed] [Google Scholar]
- 3.Liu DS, Loh KH, Lam SS, White KA, Ting AY. Imaging trans-cellular neurexin-neuroligin interactions by enzymatic probe ligation. PloS one. 2013;8:e52823. doi: 10.1371/journal.pone.0052823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Craig AM, Kang Y. Neurexin–neuroligin signaling in synapse development. Current opinion in neurobiology. 2007;17:43–52. doi: 10.1016/j.conb.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michnick SW, Ear PH, Manderson EN, Remy I, Stefan E. Universal strategies in research and drug discovery based on protein-fragment complementation assays. Nature reviews Drug discovery. 2007;6:569–582. doi: 10.1038/nrd2311. [DOI] [PubMed] [Google Scholar]
- 6.Yamagata M, Sanes JR. Transgenic strategy for identifying synaptic connections in mice by fluorescence complementation (GRASP) Frontiers in Molecular Neuroscience. 2012;5 doi: 10.3389/fnmol.2012.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim J, et al. mGRASP enables mapping mammalian synaptic connectivity with light microscopy. Nature methods. 2012;9:96–102. doi: 10.1038/nmeth.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Remy I, Michnick SW. A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nature methods. 2006;3:977–979. doi: 10.1038/nmeth979. [DOI] [PubMed] [Google Scholar]
- 9.Galarneau A, Primeau M, Trudeau LE, Michnick SW. β-Lactamase protein fragment complementation assays as in vivo and in vitro sensors of protein–protein interactions. Nature biotechnology. 2002;20:619–622. doi: 10.1038/nbt0602-619. [DOI] [PubMed] [Google Scholar]
- 10.Rossi F, Charlton CA, Blau HM. Monitoring protein–protein interactions in intact eukaryotic cells by β-galactosidase complementation. Proceedings of the National Academy of Sciences. 1997;94:8405–8410. doi: 10.1073/pnas.94.16.8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luker KE, et al. Kinetics of regulated protein–protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:12288–12293. doi: 10.1073/pnas.0404041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, Wang Y, Chiu S-L, Cline HT. Membrane targeted horseradish peroxidase as a marker for correlative fluorescence and electron microscopy studies. Frontiers in neural circuits. 2010;4 doi: 10.3389/neuro.04.006.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhee HW, et al. Proteomic Mapping of Mitochondria in Living Cells via Spatially Restricted Enzymatic Tagging. Science. 2013;339:1328–1331. doi: 10.1126/science.1230593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Porstmann B, Porstmann T, Nugel E, Evers U. Which of the commonly used marker enzymes gives the best results in colorimetric and fluorimetric enzyme immunoassays: horseradish peroxidase, alkaline phosphatase or β-galactosidase? Journal of immunological methods. 1985;79:27–37. doi: 10.1016/0022-1759(85)90388-6. [DOI] [PubMed] [Google Scholar]
- 15.Li XW, et al. New Insights into the DT40 B Cell Receptor Cluster Using a Proteomic Proximity Labeling Assay. Journal of Biological Chemistry. 2014;289:14434–14447. doi: 10.1074/jbc.M113.529578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Azevedo AM, et al. Horseradish peroxidase: a valuable tool in biotechnology. Biotechnology annual review. 2003;9:199–247. doi: 10.1016/s1387-2656(03)09003-3. [DOI] [PubMed] [Google Scholar]
- 17.Wilkinson B, Gilbert HF. Protein disulfide isomerase. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics. 2004;1699:35–44. doi: 10.1016/j.bbapap.2004.02.017. [DOI] [PubMed] [Google Scholar]
- 18.Choi J, Chen J, Schreiber SL, Clardy J. Structure of the FKBP12-rapamycin complex interacting with binding domain of human FRAP. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- 19.Lam SS, et al. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nature methods. 2015;12:51–54. doi: 10.1038/nmeth.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pinaud F, Dahan M. Targeting and imaging single biomolecules in living cells by complementation-activated light microscopy with split-fluorescent proteins. Proceedings of the National Academy of Sciences. 2011;108:E201–E210. doi: 10.1073/pnas.1101929108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsetsenis T, Boucard AA, Araç D, Brunger AT, Südhof TC. Direct Visualization of Trans-Synaptic Neurexin–Neuroligin Interactions during Synapse Formation. The Journal of Neuroscience. 2014;34:15083–15096. doi: 10.1523/JNEUROSCI.0348-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shekhawat SS, Ghosh I. Split-protein systems: beyond binary protein–protein interactions. Current opinion in chemical biology. 2011;15:789–797. doi: 10.1016/j.cbpa.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martell JD, et al. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nature Biotechnology. 2012;30:1143–1148. doi: 10.1038/nbt.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Araç D, et al. Structures of neuroligin-1 and the neuroligin-1/neurexin-1β complex reveal specific protein-protein and protein-Ca 2+ interactions. Neuron. 2007;56:992–1003. doi: 10.1016/j.neuron.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 25.Wickersham IR, Feinberg EH. New technologies for imaging synaptic partners. Current opinion in neurobiology. 2012;22:121–127. doi: 10.1016/j.conb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 26.Jagadish S, Barnea G, Clandinin TR, Axel R. Identifying functional connections of the inner photoreceptors in Drosophila using Tango-Trace. Neuron. 2014;83:630–644. doi: 10.1016/j.neuron.2014.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Y, et al. Cell-type-specific labeling of synapses in vivo through synaptic tagging with recombination. Neuron. 2014;81:280–293. doi: 10.1016/j.neuron.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hong YK, Kim IJ, Sanes JR. Stereotyped axonal arbors of retinal ganglion cell subsets in the mouse superior colliculus. Journal of Comparative Neurology. 2011;519:1691–1711. doi: 10.1002/cne.22595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McClure C, Cole KL, Wulff P, Klugmann M, Murray AJ. Production and titering of recombinant adeno-associated viral vectors. Journal of visualized experiments: JoVE. 2011 doi: 10.3791/3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Atasoy D, Aponte Y, Su HH, Sternson SM. A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. The Journal of neuroscience. 2008;28:7025–7030. doi: 10.1523/JNEUROSCI.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kato S, et al. Selective neural pathway targeting reveals key roles of thalamostriatal projection in the control of visual discrimination. The Journal of Neuroscience. 2011;31:17169–17179. doi: 10.1523/JNEUROSCI.4005-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chalupa LM, Williams RW. Eye, retina, and visual system of the mouse. Mit Press; 2008. [Google Scholar]
- 33.Lin Z, Thorsen T, Arnold FH. Functional expression of horseradish peroxidase in E. coli by directed evolution. Biotechnology progress. 1999;15:467–471. doi: 10.1021/bp990037r. [DOI] [PubMed] [Google Scholar]
- 34.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 35.Kügler S, et al. Neuron-specific expression of therapeutic proteins: evaluation of different cellular promoters in recombinant adenoviral vectors. Molecular and Cellular Neuroscience. 2001;17:78–96. doi: 10.1006/mcne.2000.0929. [DOI] [PubMed] [Google Scholar]
- 36.Yamagata M, Sanes JR. Expanding the Ig superfamily code for laminar specificity in retina: expression and role of contactins. The Journal of Neuroscience. 2012;32:14402–14414. doi: 10.1523/JNEUROSCI.3193-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawrence A, Bouwer JC, Perkins G, Ellisman MH. Transform-based backprojection for volume reconstruction of large format electron microscope tilt series. Journal of structural biology. 2006;154:144–167. doi: 10.1016/j.jsb.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 38.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. Journal of structural biology. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 39.Pagliarini DJ, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo P, et al. Rapid and simplified purification of recombinant adeno-associated virus. Journal of virological methods. 2012;183:139–146. doi: 10.1016/j.jviromet.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chao G, et al. Isolating and engineering human antibodies using yeast surface display. Nature protocols. 2006;1:755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- 42.Chen I, Dorr BM, Liu DR. A general strategy for the evolution of bond-forming enzymes using yeast display. Proceedings of the National Academy of Sciences. 2011;108:11399–11404. doi: 10.1073/pnas.1101046108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lõoke M, Kristjuhan K, Kristjuhan A. Extraction of genomic DNA from yeasts for PCR-based applications. Biotechniques. 2011;50:325. doi: 10.2144/000113672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Colby DW, et al. Engineering antibody affinity by yeast surface display. Methods in enzymology. 2004;388:348–358. doi: 10.1016/S0076-6879(04)88027-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.