Abstract
A promising therapeutic strategy for diverse genetic disorders involves transplantation of autologous stem cells that have been genetically corrected ex vivo. A major challenge in such approaches is a loss of stem cell potency during stem cell culture. Here we describe a system for maintaining muscle stem cells (MuSCs) in vitro in a potent, quiescent state. Using a machine learning method, we identified a molecular signature of quiescence and used it to screen for factors that could maintain mouse MuSC quiescence, thus defining a quiescence medium (QM). We also designed artificial muscle fibers (AMFs) that mimic the native myofiber of the MuSC niche. Mouse MuSCs maintained in QM on AMFs showed enhanced potential for engraftment, tissue regeneration and self-renewal after transplantation in mice. An artificial niche adapted to human MuSCs showed similarly prolonged quiescence in vitro and enhanced potency in vivo. Our approach for maintaining quiescence may be applicable to stem cells from a range of other tissues.
The development of stem cell therapeutics has been hindered by the inability to manipulate stem cells in vitro without a loss of potency1,2. Emerging data suggest that stem cell potency depends on the cells’ capacity to remain quiescent prior to their activation by regenerative stimuli such as injury3–5. For many stem cell populations, such as skeletal MuSCs1, hematopoietic stem cells6 and neural stem cells 8, the most potent cell in terms of transplantation efficacy and the ability to repair and repopulate a tissue is the long term–quiescent stem cell. It has been estimated that such cells can remain in the quiescent state for months in mice and years in humans 4,10–12. Stem cells reside in tissues in a specialized microenvironment or niche, characterized by a unique combination of biophysical, biochemical and cellular properties. Mechanical properties, such as stiffness, have been shown to play a critical role in regulating stem cell fate19,37. Similarly, direct contact with other cell types that characterize the niche, that also can express molecules such as cytokines or grow factors, play a critical role in regulating stem cells function in their natural environment6,33,8. Such properties have been identified as promoting quiescence in several tissue compartments 8,13–15, 33.
Previous attempts to mimic the endogenous niche in vitro have focused almost exclusively on aspects of the niche that influence the dynamics of cell division, allowing studies of cell replication and cell fate determination 17–22. What has not been well modeled are niche components that promote and maintain stem cell quiescence13,26,33. Even with the best current culture conditions, as soon as quiescent cells are isolated from their in vivo niche and plated, they immediately begin to exit the quiescent state, to activate, a not completely understood dynamic transition from G0 into the cell cycle, and to undergo proliferation and differentiation 1,19,24. The ability to maintain stem cells in a quiescent state in vitro would facilitate study of the biology of quiescence. In the context of cell therapies, it would preserve the potency of stem cells destined for transplantation and reduce the need to expand them ex vivo1,2,24. This would be especially valuable in cases of extended culture, for example, during genetic manipulation before transplantation. Research on MuSCs2, hematopoietic stem cells25 and neural stem cells26, has shown that very small numbers, down to a single cell, of quiescent stem cells can replace vast amounts of tissue; culture systems that maintain stem cell quiescence may allow these findings to be translated to the clinic.
MuSCs, or ‘satellite cells’, reside in a quiescent state under the basal laminae of muscle fibers27,28. Our approach to mimic the biochemical and mechanical properties of the native niche combines a defined culture condition and a 3D microscaffold assembled from extracellular matrix (ECM) proteins found in the MuSC niche. We show that the resulting artificial niche enables sustained quiescence for days, up to a week, of both mouse and human MuSCs and enhanced engraftment and self-renewal after transplantation. Our system provides a tool for studying the biology of MuSC quiescence and may aid the development of stem cell therapeutics for muscle disorders ranging from traumatic injuries to genetic degenerative diseases such as the muscular dystrophies.
Results
A quiescence medium for mouse MuSCs
Consistent with previous reports2, we observed that mouse MuSCs after isolation lose potency after two days in vitro. Using the Pax7CreER and the ROSA26LuSEAP mouse strains to genetically label MuSCs with the Luciferase reporter, we transplanted 10,000 MuSCs as ‘quiescent’ MuSCs (immediately after isolation), ‘activated’ MuSCs (cultured for 3.5 days), or ‘myoblasts’ (cultured for 2 weeks). Non-invasive imaging for 30 days showed that quiescent MuSCs yielded the highest bioluminescence signal (Fig. 1a). Although the bioluminescent signals for MuSC populations clearly increased over time, suggesting active cell proliferation, we cannot exclude the possibility that some of the observed differences between populations were due to differential susceptibility of the cells to apoptosis.
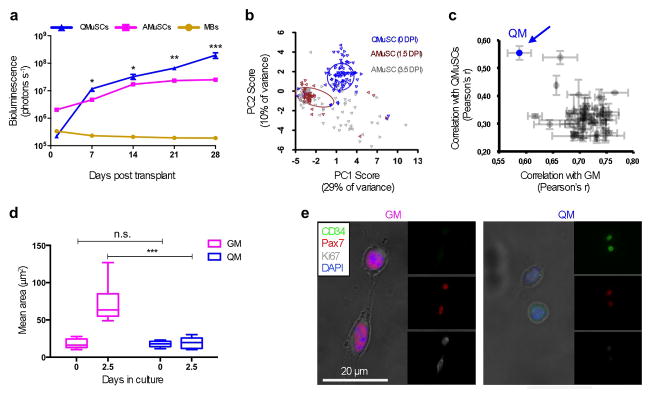
Figure 1. Formulation and functional characterization of “quiescence medium” (QM).
a. Non-invasive bioluminescence of transplanted myogenic cells. Mouse quiescent MuSCs (QMuSCs), activated MuSCs (AMuSCs) or cultured myoblasts (MBs) expressing luciferase were transplanted (10,000 cells per condition) in pre-injured TA muscles and host mice were imaged weekly using an IVIS in vivo bioluminescence imaging system for up to four weeks (n = 6, biological replicates). b. Analysis of single murine MuSC transcriptional profiles. Single freshly isolated MuSCs were isolated by FACS and compared for gene expression profiles using PCA. Single MuSCs were isolated from TA muscles at 0, 1.5, or 3.5 days post injury (DPI). Standard deviational ellipses (radius = 1 SD) are shown for 0 and 3.5 DPI. c. Analysis of combinatorial screening of quiescence-preserving molecules. The graph shows the correlation between transcriptional profiles generated for each group of 500 MuSCs grown in different combinations of the compounds tested. The combination that showed the highest correlation with QMuSCs (Y axis) and the lowest correlation with MuSCs cultured in GM (X axis) was the one chosen for the QM (colored in blue, indicated by the arrow). d. Quantification of the areas, assessed microscopically, of MuSCs cultured in either GM or QM immediately after isolation (0 days) or after 2.5 days in culture (n = 3, biological replicates). e. Representative immunofluorescence images of FACS isolated MuSCs cultured for 2.5 days in GM or QM.
Based on these and previous data suggesting that quiescent cells may be more potent than actively proliferating cells1,2, we sought to define and engineer cell culture conditions that would maintain the quiescent state of freshly isolated MuSCs. To develop a suitable medium, we first identified a molecular signature of quiescent mouse MuSCs and used it to screen various media for their ability to maintain freshly isolated mouse MuSCs in the quiescent state. We selected thirty-nine genes based on their relevance in the myogenic program and we performed a combinatorial Q-RT-PCR array analysis in a microfluidic chip to analyze single, freshly isolated quiescent MuSCs or single activated MuSCs that had been induced to proliferate for either 1.5 or 3.5 days in vivo by the injection of Cardiotoxin into the muscle (Supplementary Table 1). Principal component analysis (PCA) revealed that quiescent and activated MuSCs formed distinct transcriptional clusters (Fig. 1b). A separate analysis of the genes enriched in the two populations revealed clustering of genes highly expressed in quiescent MuSCs, defined here as ‘quiescence genes’ (e.g., Pax7, Notch2, Notch3, Hes1, HeyL, Cd34, and Foxo3) and genes highly expressed in activated MuSCs, defined here as ‘activation genes’ (e.g., Polg, p53, Ezh2, Myod) (Supplementary Fig. 1a). At the single-cell level, topological data analysis (TDA) identified a molecular signature characteristic of quiescent MuSCs distinct from activated MuSCs (Supplementary Fig. 1b).
Next we used combinatorial Q- RT-PCR to screen for molecules that, when added to the medium, would promote expression of the unique quiescence transcriptional signature (Supplementary Table 1). From an initial panel of 50 compounds known or suggested in the literature to positively regulate cell quiescence, we selected the 10 molecules that showed the strongest propensity to prevent quiescent mouse MuSC proliferation (Supplementary Fig. 2). Based on the results of this screening, we chose to further test Elcatonin, a Calcitonin Receptor ligand29; Forskolin, an activator of cAMP30; SB203580, an inhibitor of p3831,32; SU5402, an inhibitor of FGF receptor1,2,33; and TGFβ3–5,34. We also found a novel quiescence-promoting activity for Somatostatin, the c-Met inhibitor MGCD-265 and the CDK/Aurora inhibitor JMJ-7706621 (Supplementary Table 2). We then tested the effect of combinations of these compounds on MuSCs maintained for two days in culture (Supplementary Fig. 3). Using the combinatorial Q-RT-PCR array strategy for a set of 93 genes, we found conditions in which cultured MuSCs were very similar to freshly isolated quiescent MuSCs (Fig. 1c and Supplementary Fig. 3). We thus identified a defined, serum-free “quiescence medium” (QM) formulation (Supplementary Table 3) that maintained the transcriptional signature of quiescence for at least two days in culture (Supplementary Fig. 4a, b).
MuSCs maintained in QM had several characteristics of quiescent cells. One of the most obvious changes in activated MuSCs is a substantial increase in cell size relative to quiescent MuSCs15. We found that MuSCs cultured in QM for 2.5 days remained small, similar to the size of quiescent MuSCs (Fig. 1d). Furthermore, most of the MuSCs cultured in QM continued to express the quiescent marker CD34 and did not express the cell cycle marker Ki67 (Fig. 1e). MuSCs cultured for 2.5 days in QM were able to enter the cell cycle and begin proliferating when switched to growth medium (GM) (Fig. 2a). However, if cultured for one or more days beyond that, they were much less responsive to GM (Fig. 2a).
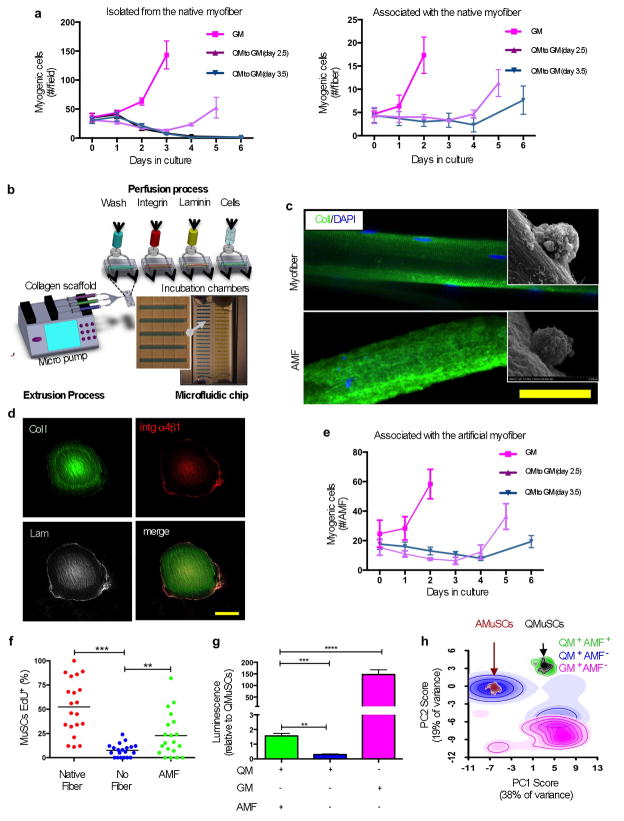
Figure 2. Artificial muscle fibers support functional quiescence of MuSCs in vitro.
a. Cultured mouse MuSCs in different media quantified at different time points. MuSCs that were freshly isolated by FACS (left panel) or isolated associated with single myofiber explants (right panel) were cultured in QM for different days before switching the medium to GM to assess their responsiveness to activate and proliferate. Myogenic cells are cells that stained positive with a cocktail of Pax7 and MyoD antibodies (n = 3, biological replicates). b. Schematic of the fabrication process of AMFs on a microfluidic chip. The design is based on arrays of 20 chambers (500 μm × 300 μm × 7 mm), with media inlet and outlet ports for fluidic lines constituted by 5 parallel channels (50 × 50 μm) used to exchange solutions and to perform cell seeding through the chambers. Monomers of Collagen I are extruded through a nozzle in the chambers to generate AMFs. c. Representative immunostaining of a single murine myofiber (top) and a single AMF (bottom). Scanning electron microscopy images (insets) show MuSCs localized on the fibers. Scale bar = 50 μm. d. Representative confocal immunofluorescence images of a functionalized AMF cross-section. Immunostaining was performed for Collagen I (green), Integrin α4β1 (red) and Laminin (grey). Scale bar = 50 μm. e. Quantification of mouse MuSCs cultured onto AMFs in different media at different time points. Freshly isolated MuSCs were associated with AMFs and cultured in QM or GM as in panel “a”. f. Quantification of MuSCs that were EdU+ve after switching QM to GM at day 3.5. Freshly isolated MuSCs, associated with a native fiber, associated with an AMF, or not associated with a fiber, were cultured in QM in the presence of EdU and switched to GM before being stained (n = 3, biological replicates). g. ATP level quantification measured in a bioluminescence assay. Cultured cells were analyzed and compared to freshly isolated quiescent MuSCs. Freshly isolated MuSCs, with or without being associated with AMFs, were cultured in QM or GM for 2.5 days before being analyzed (10,000 cells per condition from 3 biological replicates). h. PCA of single cell transcriptional profiles. Single MuSCs cultured, with or without being associated with AMFs, in different media were compared with freshly isolated quiescent or activated MuSCs. Clouds represent the densitometry of single cell distributions; colors indicate different cell populations.
AMFs support the maintenance of mouse MuSC quiescence
We hypothesized that the quiescent state could be prolonged by culturing MuSCs in QM on myofibers, a condition that preserves, at least partially, the endogenous niche. Quiescent mouse MuSCs that had not been dissociated from their native myofibers maintained their proliferative responsiveness for a longer time in QM than did cells not associated with myofibers (Fig. 2a). Because MuSCs isolated by fluorescence activated cell sorting (FACS) lack native myofibers, we sought to create an AMF that mimics the endogenous muscle fiber and its surrounding basal lamina. Given the well-established role of substrate elasticity on stem cell fate and function19,27,28,35–37, we tested substrates of different elasticity using a micropost array2,38 with elasticity ranging from 2 to 25 kPa, basing this range on the reported physiological ranges of elasticity of whole muscle and individual myofibers19,29,39,40. We confirmed these values by performing atomic force microscopy (AFM) nanoindentation analysis on individual myofibers (Supplementary Fig. 4a). To determine the elasticity that is most conducive to mouse MuSCs remaining in the quiescent state, we maintained the cells for 2 days in culture in the presence of EdU to test for DNA synthesis and entry into the cell cycle. The propensity of the cells to remain quiescent directly correlated with substrate elasticity over the tested range (Supplementary Fig. 4b, c). Approximately 85% of the cells were quiescent at 2 kPa whereas only ~45% remained quiescent at 25 kPa. Moreover, MuSCs on the most elastic microposts were most similar to quiescent MuSCs in terms of the percentage of cells expressing Pax7 and not expressing MyoD (Supplementary Fig. 4d, e). We concluded that the substrate with greatest elasticity was most conducive to maintaining mouse MuSCs in a quiescent, undifferentiated state.
We also used an alternate method to generate substrates of variable stiffness. The extracellular matrix (ECM) protein Collagen I is biodegradable and biocompatible, and, being a lyotropic material, has uniquely tunable mechanical properties30,41,42. In addition, Collagens are components of the basal lamina, which forms a critical component of the MuSC niche in vivo. We generated Collagen-based hydrogels with elastic moduli ranging from 0.8 to 2.5 kPa (Supplementary Fig. 5a). As with the micropost studies, the entry into the cell cycle was delayed (Supplementary Fig. 5b). However, below ~1.5 kPa (i.e., with a collagen hydrogel formulation at 2.4 mg/ml), the activation of apoptotic programs, as judged by active Caspase staining, increased (Supplementary Fig. 5c). We conclude that there might be an optimal elasticity between 1 and 2 kPa, above which mouse MuSCs tend to activate and differentiate and below which they are more prone to undergo apoptosis. This is in accordance with a recent report showing that increased microenvironment stiffness above 2 kPa promotes myogenic cell proliferation40.
We developed a microfabrication process on a microfluidic chip to generate AMFs with the quiescence-maintaining elastic properties identified in the micropost studies (Fig. 2b). A solution of Collagen I was extruded to generate a scaffold with parallel nanofibrils, resulting from the Collagen cholesteric chemical structure31,32,43, and with a shape and geometry similar to those of a live myofiber (Fig. 2c). The AMFs had an elasticity of 1.33±0.18 kPa (Fig. 6a), consistent with our previous measurements on Collagen I hydrogel films. When mouse MuSCs were plated onto the AMFs (Fig. 2c and Supplementary Movie 1), EdU incorporation was strongly reduced, consistent with maintenance of the quiescent state (Supplementary Fig. 6b). We also tested the mechanical stress capacity of AMFs to evaluate whether they had sufficient structural integrity for transplantation. The mechanical tensile modulus was~27 kPa, which makes them robust to manipulation in a syringe and transplantation similar to a myofiber (Supplementary Fig. 6c).
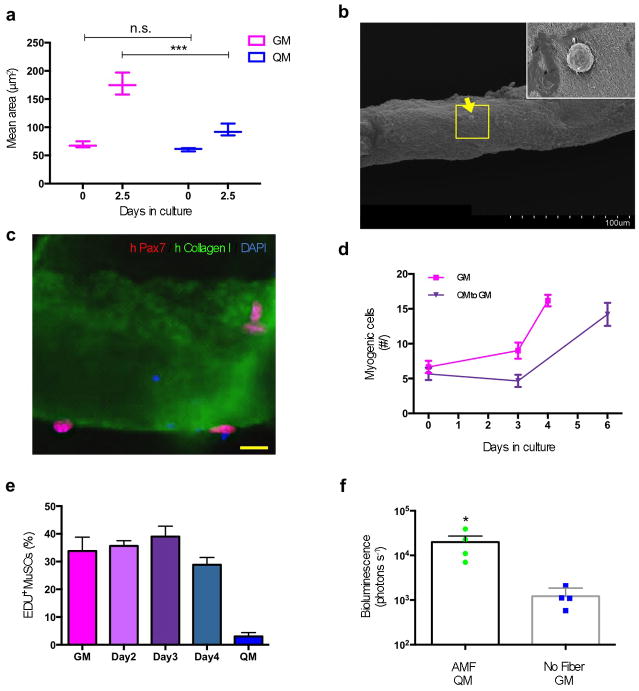
Figure 6. Artificial niche preserves the potency of human MuSCs.
a. Quantification of the areas, assessed microscopically, of hMuSCs cultured in either GM or QM immediately after isolation (0 days) or after 2.5 days in culture (n = 3, biological replicates). b. Scanning electron microscopy images show hMuSC (indicated by yellow arrow) localized on a human AMF. Scale bar = 100 μm. The yellow box is magnified (inset) to show the hMuSC. c. Immunostaining of a single human AMF seeded with hMuSCs. Scale bar = 10 μm. d. Reversible quiescence of hMuSCs cultured on AMFs in QM. Freshly isolated hMuSCs were associated with AMFs and cultured in QM before switching to GM (as in Fig. 2a). e. Quantification of hMuSCs that were EdU+ve after switching QM to GM at 2.5 days. Freshly isolated hMuSCs, associated with AMFs, were cultured in QM and switched to GM, pulsing EdU for 24 hours before being stained (n=3, biological replicates). f. Results of non-invasive in vivo bioluminescence imaging of transduced and transplanted hMuSCs. Similar to experiments of Fig. 5d, isolated hMuSCs were transduced to express Luciferase then either cultured in QM while associated with AMFs or cultured in GM alone for 3.5 days and then transplanted into TA muscles.
MuSCs are regulated by protein components of the niche, such as the ECM protein Collagen VI44. As MuSCs are closely apposed to the muscle fiber membrane in vivo, we identified muscle fiber membrane proteins that might interact with MuSCs to maintain their quiescence. We found that Integrin α4β1 is expressed in the adult mouse niche on the muscle fiber membrane adjacent to quiescent MuSCs (Supplementary Fig. 7a, b). Quiescent MuSCs express VCAM45, and the primary Integrin to which VCAM binds is α4β146. We found that Integrin α4β1 was superior to other Integrin heterodimers, such as α5β1, αVβ1, and α6β1, that have been implicated in MuSC biology46–51, in terms of reducing EdU incorporation and inhibiting cell death in vitro (Supplementary Fig. 7c, d). Laminins are a key component of the basal lamina surrounding MuSCs52,53. It has been suggested that Laminins are not only structural proteins of the basal lamina but are also signaling molecules that are important for the adhesion and localization of MuSCs in their niche53,54.
Based on these considerations, we coated the Collagen-based AMFs with recombinant Integrin α4β1 followed by recombinant Laminin (Fig. 2d). When seeded onto the functionalized AMFs and cultured for three days, mouse MuSCs showed reduced activation as assessed by EdU incorporation, increased viability as assessed by ATP levels, and higher Pax7 and lower MyoD protein expression when compared to AMFs alone or functionalized with Integrin α4β1 only (Supplementary Fig. 8a – c).
Finally, to study the specific role of elastic properties of AMF in maintaining quiescence, we compared fully functionalized AMFs generated either as thick soft gels or as thin coatings on top of a rigid substrate. Consistent with previous experiments (Supplementary Fig. 5), MuSCs cultured for 36 hours on thick functionalized AMFs showed a lower degree of activation on softer substrates (Supplementary Fig. 8d). However, this difference was completely lost when MuSCs were cultured on thin functionalized AMFs, a condition where the different Collagen formulations did not affect the higher stiffness of the rigid substrates to which they were attached. All of these results are consistent with the functionalized AMFs promoting a viable quiescent state by virtue of their biophysical and biochemical properties.
We next tested whether mouse MuSCs cultured on functionalized AMFs could maintain reversible quiescence in QM, as we observed for MuSCs in their native niche but not for isolated MuSCs on a two-dimensional tissue culture substrate (Fig. 2e). Indeed, functionalized AMFs facilitated the maintenance of reversible MuSC quiescence. When maintained in QM for 3.5 days, quiescent MuSCs activated in response to GM with similar kinetics seen when freshly isolated MuSCs were plated in GM (Fig. 2e, f). With time, there was a gradual reduction in cell survival in QM, but even after seven days on maintenance in QM, MuSCs were able to be induced to proliferate upon exposure to GM. We did not test time points beyond seven days. Moreover, levels of ATP showed a sharp reduction when MuSCs were not associated with a fiber but not when associated with AMFs, suggesting that the AMFs promoted a sustained viability of cultured MuSCs in QM (Fig. 2g). We conclude that QM is able to support the survival of MuSCs in the quiescent state without inducing activation and proliferation, and that this is enhanced when MuSCs are cultured on a substrate that mimics their endogenous niche.
Preservation of the transcriptional signature of mouse MuSC quiescence
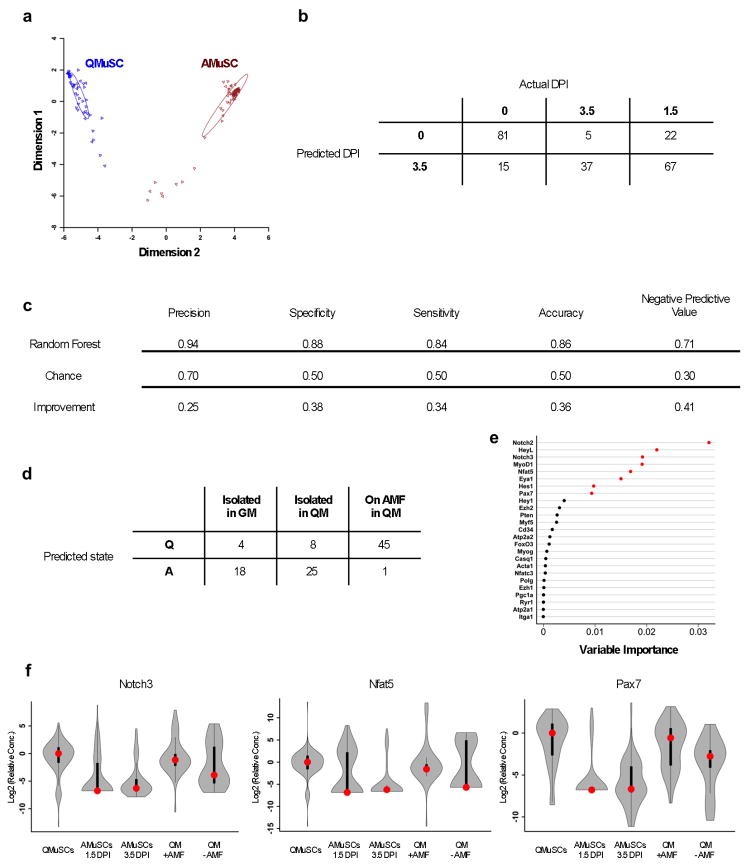
We analyzed the transcriptional profile of single mouse MuSCs under three different in vitro conditions— maintained on AMFs and in QM, maintained on two-dimensional substrate in QM, and maintained on a two-dimensional substrate in GM—and compared these to the profiles of freshly isolated quiescent MuSCs. MuSCs maintained in QM and on AMFs clustered with quiescent MuSCs, whereas MuSCs cultured on two-dimensional substrates did not, regardless of whether they were maintained in QM (Fig. 2h). In a complementary approach, we generated a cell classification model using random forests, a machine learning strategy that uses an ensemble of gene expression decision trees55. In this model, we predicted cell state as either quiescent or activated based on freshly isolated MuSC transcriptional profiles (Fig. 3a). This model performed well, with 86% accuracy during cross-validation on an independent dataset (Fig. 3b, c). Confirming the preservation of quiescence by AMFs, 45 out of 46 MuSC replicates cultured in QM on AMFs were classified as quiescent, in contrast to 8 of 33 MuSCs cultured in QM but on a two-dimensional substrate and only 4 of 22 cultured without either AMFs or QM (Fig. 3d). Analysis of the importance of the genes used to construct this predictive model revealed that the genes most discriminating between quiescence and activation were, in this order of diminishing importance, Notch2, HeyL, Notch3, MyoD, Nfat5, Eya1, Hes1, Pax7, Hey1, Ezh2, Pten, Myf5, Cd34, Atp2a2, and Foxo3 (Fig. 3e). The distribution of expression of single cell levels of these genes in quiescent MuSCs was very similar to that of MuSCs cultured in QM on AMFs, as exemplified by Pax7, Notch3 and Nfat5 (Fig. 3f).
Figure 3. Mouse MuSCs cultured in QM on AMFs are transcriptionally similar to quiescent MuSCs in vivo.
a. Multidimensional scaling representation for the training dataset of single MuSC gene expression. The analysis is based on the proximity matrix of the proportion of trees in which cell pairs share terminal nodes. b. Generation of a random forest model for single MuSCs. After combinatorial Q-RT-PCR on single cells, random forest construction was performed using the gene expression profiles of 48 cells 0 DPI and 68 cells 3.5 DPI. c. Cross-validation dataset. Data are analyzed from a separate experiment, which also included 1.5 DPI cells, to validate the random forest performance in predicting and recognizing the MuSC quiescent or activated state. d. Classification of cells in three conditions (isolated and grown in GM; isolated and grown in QM; associated with AMFs and grown in QM) as being in either a “quiescent” or an “activated” state based on the single cell gene expression profile of individual MuSCs. After this model construction and validation, single cells from one of three culture conditions (GM, QM, or AMF+QM) were classified as 0 DPI (“Q”) or 3.5 DPI (“A”). e. Loadings for genes expressed in single MuSCs. The analysis was performed after combinatorial Q-RT-PCR on cells 0 DPI (for quiescent MuSCs) or on cells 1.5 or 3.5 DPI (for activated MuSCs). The most important genes whose expression is correlated with and predictive of the quiescent state are shown in red. f. Violin plots for selected genes expressed in single MuSCs. The graphs compare the results from single MuSCs obtained in different conditions in vivo (QMuSCs; AMuSCs 1.5 DPI; AMuSCs 3.5 DPI) and in vitro (QM + AMF; QM − AMF). Red dots represent the median. Black bars represent the first and third quartiles. Whiskers represent the minimum and maximum within 1.5 interquartile distances of the first or third quartile.
The artificial niche enhances the potency of transplanted MuSCs
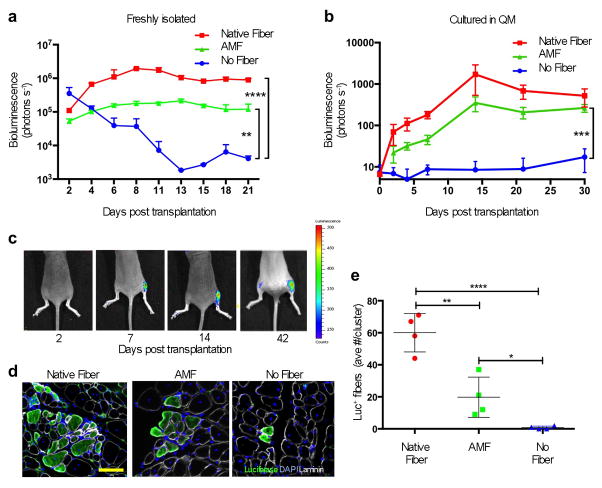
We compared the engraftment potential of freshly isolated MuSCs, MuSCs in the artificial niche (the combination of AMFs and QM) and MuSCs associated with native fibers57,58. In each case, 100 cells were transplanted into tibialis anterior (TA) muscles of immunocompromised mice that had been subjected to Cardiotoxin-induced injury 12 hours prior in order to create an environment that would promote MuSC engraftment. MuSCs were all obtained from Pax7CreER/ROSA26LuSEAP mice that had previously been treated with tamoxifen, so the donor cells expressed Luciferase and their engraftment could be assessed non-invasively. MuSCs associated with AMFs were far more potent than isolated MuSCs and nearly as potent as the same number of MuSCs associated with native fibers (Fig. 4a). These data demonstrate the importance of a niche-like environment in vitro for maintaining MuSC potency.
Figure 4. Transplant potency enhancement via the artificial niche.
a. Results of non-invasive in vivo bioluminescence imaging of freshly isolated, transplanted mouse MuSCs. MuSCs (100 cells per condition) were isolated and immediately transplanted into pre- injured TA muscles: 1) still associated with native myofibers; 2) isolated by FACS and plated onto AMFs in vitro prior to transplantation; or 3) isolated by FACS and in suspension (n ≥ 4, biological replicates). b. Results of non-invasive in vivo bioluminescence of pre-cultured, transplanted MuSCs. MuSCs were cultured for 2.5 days in QM, associated or not with a fiber as in panel “a”, prior to transplantation (50 cells per condition) and imaged weekly for one month (n = 5, biological replicates). c. Representative bioluminescence images of a time course analysis of one of the host mice, quantified as in panel “b”, that received 50 MuSCs transplanted in each TA muscles. The right leg (which is on the right side since the mouse is prone in each image) received MuSCs associated with AMFs; the left leg received MuSCs not associated with any fiber. Images were obtained at different time points as indicated (bioluminescence values are indicated as photons cm−2 s−1 on the scale to the right). d. Representative immunofluorescence immunohistochemistry (IF-IHC) of Luciferase expression in TA muscle cross sections. Muscles of the mice imaged and quantified in panels “b” and “c” were isolated 40 days after transplantation. Scale bars = 100 μm. e. Quantification of IF-IHC staining for Luciferase+ve fibers per cluster in TA muscles that were recipients of transplanted MuSCs. The average number of fibers per cluster per TA is shown; the number of clusters/TA was: Native Fibers 4.6±1.02; AMF 2.6±1.02; No Fiber 0.4±0.48.
One characteristic of stem cell potency is the capacity of the cells to differentiate into new mature tissue15. To assess this, we cultured ~50 mouse MuSCs in the artificial niche for 2.5 days before transplantation. Bioluminescence signals in TAs transplanted with AMFs, similarly to myofibers, increased to a plateau. MuSCs alone were unable to engraft, likely due to the challenging conditions of transplanting only 50 cells in a non-irradiated TA muscle (Fig. 4b, c). We confirmed by immunohistochemical analysis of the same transplanted muscles that the bioluminescence signal corresponded to regenerated muscle fibers (Fig. 4d, e).
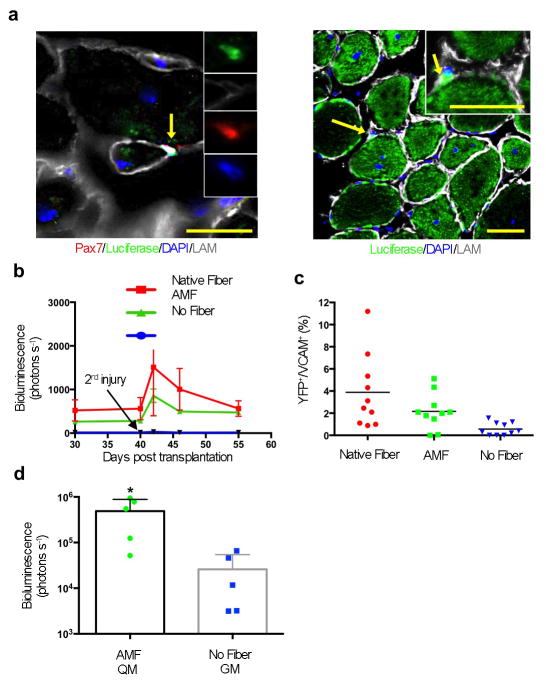
Another important feature of stem cells is the capacity to self-renew15. Having shown that the artificial niche enhanced MuSC engraftment, we tested its effect on self-renewal, again with native fiber-associated MuSCs as controls. First, we examined muscles that had been transplanted with MuSCs cultured in the artificial niche (as in Fig. 4) for evidence of Luciferase-expressing cells in the satellite cell position. Indeed, we could detect Luciferase+ cells beneath the basal lamina of regenerated fibers (Fig. 5a). Second, we performed an injury, 40 days after the initial transplantation, on muscles previously injured and transplanted with Luciferase-expressing MuSCs. In response to the second injury, any self-renewed, transplanted MuSCs, such as those shown in Fig. 4b, would be expected to activate, proliferate and differentiate to form new muscle. Indeed, for native fiber–associated MuSCs cultured in QM and MusCs in the artificial niche, the bioluminescent signal increased after the second injury (Fig. 5b). Cells that had been previously transplanted without any niche not only showed very little steady-state bioluminescence before the second injury, but exhibited almost no increase after it.
Figure 5. AMF maintains MuSC self-renewal capacity after in vitro manipulations.
a. IF-IHC staining of TA muscles transplanted with mouse MuSCs associated with AMFs showing representative images of transplanted Luciferase+ve MuSCs (Luciferase+ve MuSCs localized in between Luciferase+ve fibers are indicated by arrows and magnified in the insets). Scale bars = 100 μm. b. Results of non-invasive in vivo bioluminescence imaging of muscles that were recipient of transplanted Luciferase+ve MuSCs and re-injured after 40 days (indicated by the arrow) after the transplantation. The second injury was performed to test if the bioluminescence signal increased as a consequence of activating and expanding Luciferase+ve MuSCs that were initially transplanted and that had engrafted under the basal lamina (n ≥ 4, biological replicates). c. Quantification of the number of transplanted MuSCs expressing YFP that engrafted as stem cells. Cells were isolated and cultured in QM prior to transplantation in TA muscles. An injury was induced 40 days after transplantation. Ten days later, the percentage of MuSCs (VCAM+ve) that were donor- derived (YFP+ve) was assessed by FACS. d. Results of non-invasive in vivo bioluminescence imaging of transduced and transplanted MuSCs. Isolated MuSCs were either cultured in QM while associated with AMFs or cultured in GM alone for 3.5 days. During culturing, cells were transduced with a lentivirus expressing Luciferase, and 1,000 cells were then transplanted into pre-injured TA muscles. Recipient mice were imaged by bioluminescence 30 days later.
To confirm that mouse MuSCs cultured in the artificial niche for 2.5 days retain self-renewal potential, we transplanted 1,000 quiescent MuSCs expressing the reporter fluorescence protein “enhanced YFP” (derived from a Pax7CreER/ROSA26eYFP mouse) into a pre-injured TA muscle. After 40 days, we performed a second injury with Cardiotoxin, and 10 days later we isolated MuSCs by FACS and analyzed the number of YFP+ MuSCs. TA muscles transplanted with MuSCs associated with either native fibers or AMFs showed the presence of donor-derived MuSCs (Fig. 5c). Conversely, TA muscles transplanted with MuSCs not associated with any niche structure did not have donor-derived MuSCs. We conclude that MuSCs cultured in the artificial niche, similar to MuSCs associated with native fibers, maintain quiescence in culture and retain engraftment and self-renewal potential upon transplantation.
Finally, we tested the effect of our optimized culture conditions on the transplantation potency of MuSCs after genetic manipulation in vitro. We transduced MuSCs maintained in the artificial niche or on a two-dimensional substrate and cultured in GM (standard conditions) with a lentivirus expressing Luciferase and GFP. One day later, we transplanted 1,000 MuSCs from each culture into pre-injured TA muscles of immunocompromised mice. Bioluminescence measurement 30 days later showed that MuSCs maintained in the artificial niche during lentiviral infection yielded, on average, a signal two orders of magnitude higher than MuSCs in standard conditions (Fig. 5d), suggesting that maintenance of a quiescent state by association with AMFs and treatment with QM might allow efficient genetic modification ex vivo without compromising MuSC potency.
The artificial niche supports human MuSCs
We studied whether freshly isolated hMuSCs respond similarly to murine MuSCs in the artificial niche. First, we used FACS, as previously reported 61,62, to isolate hMuSCs from surgical samples. Like murine MuSCs, hMuSCs cultured for 2.5 days in QM modified for human cells remained small (Fig. 6a). We then generated AMFs similar to murine AMFs but based on human proteins, and seeded hMuSCs onto their surface in either GM or QM (Fig. 6b and c). The AMFs facilitated the maintenance of reversible hMuSC quiescence for up to 3.5 days in QM; even at this point, the cells could exit quiescence and begin proliferating when cultured in GM (Fig. 6d, e).
To test the transplantation potency of genetically modified hMuSCs cultured in the artificial niche, we replicated our experiments with mouse MuSCs (Fig. 5d) by transducing Luciferase into 1,000 hMuSCs, cultured either in our optimized conditions or in standard conditions and then transplanting them one day later into TA muscles of immunodeficient mice. 30 days later, hMuSCs cultured in our optimized conditions yielded, on average, a signal one order of magnitude higher than that of hMuSCs cultured in standard conditions (Fig. 6f). Taken together, these results suggest that hMuSCs can also be isolated and genetically manipulated in the artificial niche while retaining much greater potency than hMuSCs maintained under standard conditions.
Discussion
Here we provide evidence that engineering a biomimetic microenvironment enables maintenance of quiescent, potent mouse and human MuSCs. Our optimized condition consists of a defined medium to maintain quiescence of MuSCs, and of a microscaffold in the shape of a muscle fiber, generated with biochemical and biophysical properties of the native niche. The potential utility of our approach for therapeutic applications was shown by the enhanced engraftment and self-renewal potential of both mouse and human MuSCs cultured in the artificial niche compared to traditional conditions. In addition, MuSCs cultured in the artificial niche that were transduced with a lentiviral reporter engrafted after transplantation without loss of potency.
The loss of engraftment ability of cultured MuSCs is a major challenge in developing efficient strategies to manipulate isolated stem cells for cell therapy1,2,57,59,60. The potential to correct genetic mutations in hMuSCs derived from a patient with a genetic disorder, such as a muscular dystrophy, and transplant the corrected cells to replace the pathogenic tissue may enable improved stem cell therapeutics for muscle disorders59,60. Moreover, transplantation of hMuSCs seeded onto microscaffolds may facilitate tissue engineering to treat traumatic injuries, as in the case of volumetric muscle loss63.
Online Methods
Animals
C57BL/6, ROSA26eYFP, and B6.Cg-Foxn1nu/J mice were obtained from Jackson Laboratory. Pax7CreER mouse and ROSA26LuSEAP were provided by Dr. Charles Keller, Oregon Health and Science University, Portland, Oregon, USA. Tamoxifen injections for Cre recombinase activation were performed as described previously 68. To control for tamoxifen injection toxicity, we injected all mice with tamoxifen. All experimental mice employed were 3 to 6 months old. Mice were housed and maintained in the Veterinary Medical Unit at the Veterans Affairs Palo Alto Health Care Systems. Animal protocols were approved by the Administrative Panel on Laboratory Animal Care of Stanford University.
Human skeletal muscle specimens
Subjects ranged in age from 38 to 85 years. All experiments were performed using fresh muscle specimens from operative procedures. Sample processing for cell analysis began within one hour of specimen isolation. In all studies, standard deviation reflects variability in data derived from studies using true biological replicates (i.e., unique donors). Data were not correlated with donor identity.
MuSC Isolation and FACS
MuSCs were isolated as previously described 45. Briefly, muscles were harvested from hind limbs, dissociated to yield a fragmented muscle suspension using a gentleMACS dissociator (Miltenyl Biotec), and digested with Collagenase II (500 units per ml; Invitrogen) in Ham’s F10 medium for 90 minutes. After washing, a second digestion was performed for 30 minutes with Collagenase II (100 units per ml) and Dispase (2 units per ml; Invitrogen). The resulting mononuclear cell suspension was washed, filtered and stained with VCAM-biotin (clone 429; BD Bioscience), CD31-APC (clone MEC 13.3; BD Bioscience), CD45-APC (clone 30-F11; BD Bioscience) and Sca-1-Pacific-Blue (clone D7; Biolegend) antibodies at a dilution of 1:75. Streptavidin-PE-cy7 was used to amplify the VCAM signal (BD Biosciences, 1:75) and FACS grade DAPI dilactate for non-vital cell exclusion (D3571 Invitrogen). Cells isolation was performed on a BD-FACS Aria II or BD FACSAria III equipped with 488-nm, 633-nm and 405-nm lasers to obtain the MuSC population. For single cell sorting, this cell population was sorted a second time to get individual cells. Human MuSCs were purified from fresh operative samples as previously described 61,62. Briefly, operative samples were carefully dissected from adipose and fibrotic tissue and a dissociated muscle suspension was prepared as described for mouse tissue. The resulting cell suspension was then washed, filtered and stained with anti-CD31-Alexa Fluor 488 (clone WM59; BioLegend; #303110), anti-CD45-Alexa Fluor 488 (clone HI30; Invitrogen; #MHCD4520), anti-CD34-FITC (clone 581; BioLegend; #343503), anti-CD29-APC (clone TS2/16; BioLegend; #303008) and anti-NCAM-Biotin (clone HCD56; Biolegend; #318319). Unbound primary antibodies were then washed and the cells incubated for 15 min at 4°C in streptavidin-PE/Cy7 (BioLegend) to detect NCAM-biotin. The machine was carefully optimized for purity, viability, and calibration for single cell sorting. A small fraction of sorted cells was plated and stained for Pax7 and MyoD to assess the purity of the sorted population.
Single Cell Q-RT-PCR
MuSCs were FACS sorted individually into 96 well plates prepared with 9 μl RT-STA buffer (5 μl CellsDirect 2X reaction mix (Life Technologies, PN 11753-500), 0.2 μl Superscript III Platinum Taq mix, 2.7 μl nuclease free water, 0.1 μl SUPERase In RNAse inhibitor (Life Technologies) and 1 μl 10X primer mix (96 × 1 μl of 100 μM primer pairs plus 104 μl DNA suspension buffer)). Plates were spun down and stored at −80°C to ensure complete cell lysis. Next, plates were thawed and subjected to a reverse transcription (RT) (15 minutes at 50°C (RT reaction), 2 minutes at 95°C (to inactivate reverse transcriptase and activate Taq polymerase), and subjected to a pre-amplification reaction (15 seconds at 95°C, 4 minutes at 60°C (for 20 cycles)). Pre-amplified cDNAs were treated with Exonuclease I (2.52 μl water, 0.36 μl buffer, 0.72 μl exonuclease I (New England BioLabs, PN M0293L)) for 30 minutes at 37°C, followed by 15 minutes inactivation at 80°C. Exonuclease treated cDNAs were diluted 5X in DNA suspension buffer (TEKnova, PN T0221) and stored at −80°C. To perform the Q-RT-PCR on the Fluidigm Biomark HD, sample mixes were prepared with 2.7 μl of diluted cDNAs mixed with 3.0 μl of 2X SsoFast EvaGreen Supermix with low ROX (Bio-Rad, PN 172-5211) and 0.3 μl of 20X DNA Binding Dye Sample loading reagent (Fluidigm, PN 100-0388), vortexed and spun down. Separately, to create the assay mix, 0.3 μl of 100 μM primer mix was combined with 2.5 μl 2X Assay Loading Reagent (Fluidigm, PN 8500073) and 2.25 μl DNA suspension buffer, vortexed and spun down. A 96.96 Dynamic Array IFC (Fluidigm) was primed on an IFC Controller HX (Fluidigm). The primed Dynamic Array was loaded with the assay and sample mixes using an IFC Controller HX. Finally the PCR was performed on a Biomark HD, using protocol GE 96×96 PCR+Melt v2 (2400 seconds at 70°C, 30 seconds at 60°C, 60 seconds at 95°C hot start, 30 cycles (5 seconds at 95°C, 20 seconds at 60°C), followed by a melt curve). Biomark data was analyzed using Q-RT-PCR Analysis software (Fluidigm). Data were exported into Microsoft Excel for further analysis. For each amplicon, a limit of detection (LOD) was determined by running 10-fold serial dilutions of embryonic cDNA in 6 replicates. Limit of detection was set to the lowest dilution for which all six replicates give a PCR signal. This value was set to denote the amplification of 2–10 copies of transcript per reaction chamber. A list of primers is attached in Table S4.
Single Cell Data Analysis and Molecular Signature
Raw Ct values were filtered and normalized according to the manufacturer’s instructions and are represented in Table S4. Log2 (expression values) were used for all analyses. For analyses involving multiple experiments (random forests and violin plots), only genes with primers common to all experiments were used, and datasets were first batch-corrected using COMBAT 69. For PCA, genes were mean-centered and unit variance-scaled. K-medians clustering with k-means++ seeding using Manhattan distance was performed for genes, using k = 2. Random forest construction was done using 8000 trees and 25 genes with 5 random genes selected at each node 55. Conditional variable importance for the genes involved in random forest construction was calculated as the mean decrease in out-of-bag (OOB) accuracy of the trees after permuting that gene within sample subsets split by correlated genes with an association p<0.2. P-values for these conditional variable importance measures are based on 1000 permutations of the response variable to mimic the distribution of variable importance when no gene has predictive value. Hierarchical clustering used Manhattan distance and complete linkage after mean-centering and UV-scaling genes. P-values are approximately unbiased values calculated using multiscale bootstrap resampling with 10,000 resamples at ten sizes 70. Scripts generated in R computational language to analyze the data are posted at http://www.stanford.edu/~jbrett/R_Scripts/Quarta2014/. Topological data analysis (TDA) methods were used to identify the quiescent population signature in an unsupervised fashion using the Ayasdi Iris software (Ayasdi Inc., Menlo Park CA). TDA benefits from properties inherent in the classic topological methods useful for characterizing shape, as applied to point cloud space. These properties include deformation invariance in the form of relative noise insensitivity; compressed representation, as reified by the partitioning and clustering steps; and coordinate freeness that relies only on the notion of similarity between points. While mathematical details regarding the method of construction for the topological network is described in complete form elsewhere 73, we summarize the approach here briefly. Two key parameters are needed for generating a topological analysis: a notion of similarity, or metric, and filter(s). For each point X, in this case a single cell, a filter function f is applied yielding some real-valued output Y. Based on the value of Y for each X, the points are binned with some overlap, the span of which is variable. Then, for each X and for all bins, a metric is used to determine the distance d(X,X′) in the original space. A partial clustering scheme is finally used to aggregate points within bins and form connections between points across multiple bins. In the topological network, the aggregated points form the nodes (and thus collections of points), while the edges appear when two aggregations in the clustering contain one or more of the same points. The method described above essentially generates Reeb graphs from the data, the result of which can be used for identification of natural segmentations of points for noisy data, and has been useful in identifying patterns in complex, high-dimensional datasets 73. In the present case, the data is based on population signatures across 35 genes over the single cells. For this dataset, the metric used was correlation, and the functions applied were principal and secondary components (metric embedded). The resulting representation revealed a distinct segregation of the quiescent population. A non-parametric statistical test (Kolmogorov-Smirnov) was used to identify the most significant markers for the quiescent population relative to the entire dataset.
Single-fiber Explants
Extensor digitorum longus (EDL) or flexor digitorum brevis (FDB) muscles were excised and digested in Collagenase II (500 units per ml in Ham’s F10 medium) as previously described 75. Fibers were then washed extensively and cultured in medium containing Ham’s F10, 10% horse serum and 0.05% chick embryo extract. Every 24 hours, 50% of the medium was replaced with Ham’s F10 medium with 20% fetal bovine serum (FBS). EDL fibers were cultured in suspension. Fixed fibers were stained and the number of MuSCs was quantified per fiber.
Micropost-arrays
Elastomeric micropost-arrays were generated with PDMS by replica-molding as published elsewhere 76. Briefly, PDMS prepolymer was poured over a template containing an array of holes, degassed under vacuum, cured at 110°C for 20 hours and peeled off the template. Microposts were fabricated 1 μm in diameter and 3 μm of center-to-center distance; elasticity of the microposts was changed by modulating the amount of cross-linker and their length 38 to obtain nominal spring constants of 1.9223 nN/μm, 7.22106 nN/μm, 18.1905 nN/μm, 32.0312 nN/μm for effective moduli of approximately 2, 6, 12, 25 kPa respectively.
Injections
Mice were anaesthetized using isoflurane through a nose cone. Muscle injury was induced by injecting 20 μl of Cardiotoxin (Invitrogen) into TA muscles.
Transduction
Luciferase and GFP protein reporters were subcloned into a third generation HIV-1 lentiviral vector and used to transduce freshly isolated MuSCs.
Histology and Immunohistochemistry
For hematoxylin and eosin staining, TA muscles were dissected and directly frozen in OCT (Tissue-Tek). For immunohistochemistry, TA muscles were fixed for 5 hours using 0.5% electron-microscopy-grade paraformaldehyde and subsequently transferred to 20% sucrose overnight. Muscles were then frozen in OCT, cryosectioned at a thickness of 6 μm and stained using an M.O.M. kit (Vectorlabs) or a Zenon labelling kit (Invitrogen) according to the manufacturers’ instructions.
Atomic Force Microscopy
AFM was used to analyze Collagen-based hydrogel properties. Gels were made from bovine tendon Collagen I (Cellmatrix I-A, Nitta Gelatin, Osaka, Japan). The Collagen concentrations that were tested ranged from 1.8 to 2.7 mg/ml. Varying Collagen concentrations were achieved by reducing the volume of Collagen stock solution (3.0 mg/ml) and replacing that volume with an equivalent amount of 10X Ham’s F12 media (Life Technologies, Carlsbad, CA) without sodium bicarbonate. Reconstitution buffer volumes were kept constant across all conditions (Table 1). Reconstitution buffer was comprised of 2.2 g NaHCO3 (Sigma-Aldrich, St. Louis, MO) in 100 ml of 0.05N NaOH and 200 mM HEPES.
Table 1.
Collagen matrix compositions utilized to generate encapsulating matrices.
| Concentration [mg/ml] | Collagen stock [μl] | Ham’s F12 [μl] | Reconstitution buffer [μl] | Collagen:media:buffer ratio |
|---|---|---|---|---|
| 2.7 | 450 | 0 | 50 | 9:0:1 |
| 2.4 | 400 | 50 | 50 | 8:1:1 |
| 1.8 | 400 | 150 | 50 | 6:3:1 |
Gel films were generated according to protocol and kept on ice prior to casting onto glass surfaces for testing. Samples were given 30 minutes to gel at 37°C and then rehydrated with phosphate buffer solution (PBS, Life Technologies, Carlsbad, CA) for a minimum of 15 minutes prior to testing. Similarly, individual muscle fibers were adhered to glass surfaces prior to testing and submerged in PBS. Gel and muscle fiber samples were tested while remaining submerged in PBS. Short silicon contact mode probes (SHOCON, Applied NanoStructures, Mountain View, CA) were used to collect individual force measurements from all samples. Probes were rinsed with FBS (Life Technologies, Carlsbad, CA) prior to testing to prevent non-specific adsorption of samples to the probe tip. Force measurements were collected using 1 μm force distances and 1 Hz scanning rates with a 0.5 V trigger point upon contact. The integral gain was set to 10 due to the submerged testing conditions. A Hertz indentation model for cone tip geometry was used to fit the data. Poisson ratios of 0.4 and 0.33 were assumed for Collagen gels and primary tissue, respectively. All AFM data were collected using an Asylum Research MFP-3D-BIO.
AMF Tensile Testing Analysis
Collagen fibers were extruded according to protocol and transferred to 1.5 ml Eppendorf tubes filled with PBS. Eppendorf tubes were rinsed once with FBS prior to use to prevent non-specific adsorption of fibers to the tube walls. Tubes were then sealed, placed on ice, and transferred to the CellScale Biomaterials Testing facility. All tensile testing was performed on-site at the CellScale facility using the Microsquisher device. AMF samples were tested in a hydrated state. One end of the AMF fiber was fixed in place and the other was attached to the moving force transducer wire of the Microsquisher device. This enabled collection of mechanical data in a tensile rather than compressive mode. Force versus displacement curves were then converted to stress versus strain curves for calculation of the Young’s moduli.
Master Mold Fabrication
All photo masks were designed using SolidWorks (Solidworks Corp, Waltham, MA) and printed at a resolution of 20,000 dots per inch on a transparency film (CAD/Art Services). Master Mold fabrication was performed as described elsewhere77. Briefly, the flow-layer master was fabricated from a combination of positive and negative photoresists using a three-step lithography process. Channel sections were fabricated from the resist material SU8-2010. SU8 2010 (MicroChem, Newton, MA) was spun onto a silicon wafer (3,000 rpm for 45 seconds), baked before exposure (1 minute at 65°C, then 3 minutes at 95°C), exposed through a negative transparency mask (40 seconds at 7 mW/cm2), baked after exposure (1 minute at 65°C, then 3 minutes at 95°C), and developed in an SU8 nanodeveloper (MicroChem). Channel sections were fabricated using the positive photoresist SJR 5740 (MicroChem). To promote photoresist adhesion, the wafer was first treated with hexamethyldisilazane (Microprime HP-Primer; ShinEtsu MicroSi, Phoenix) (1 minute at 1 atmosphere). The photoresist was spun onto the patterned wafer (2,000 rpm for 60 seconds), soft baked (1 minute 45 seconds at 95°C), aligned to the existing features, exposed (45 seconds at 7 mW/cm2), and developed (20% Microposit 2401 developer; MicroChem). The mold was then annealed (20 minutes at 120°C), and hard baked (2 hours at 170°C). Low-impedance input and output channels were fabricated to allow for the rapid flushing of viscous reagents. A 60 mm layer of SU8 2075 (MicroChem) was spun onto a silicon wafer (3,000 rpm for 60 seconds), baked before exposure (7 minutes at 65°C, then 20 minutes at 95°C), aligned to the primary flow structure, and exposed through a negative transparency mask (40 seconds at 7 mW/cm2), baked after exposure (1 minute at 65°C, then 15 minutes at 95°C), and developed in an SU8 nanodeveloper (MicroChem). Control features (25 mm high) were fabricated on a separate wafer using a single lithographic step. SU8 2025 (MicroChem) was spun onto a silicon wafer (3,000 rpm for 45 seconds), baked before exposure (1 minute at 65°C, then 3 minutes at 95°C), aligned to the primary flow structure, exposed through a negative transparency mask (40 seconds at 7 mW/cm2), baked after exposure (1 minute at 65°C, then 3 minutes at 95°C), and developed in an SU8 nanodeveloper (MicroChem).
Microfluidic Device Fabrication
The microfluidic chip was fabricated as previously described 77. Briefly, using silicone elastomer (General Electric RTV 615) the technique of multilayer soft lithography was applied. To facilitate the release of the elastomer from the mold, all molds were treated with chlorotrimethylsilane (Aldrich). Liquid silicone elastomer (20 parts A:1 part B) was spun onto the control master (2,400 rpm for 60 seconds) and baked in a convection oven at 80°C for 60 minutes. Liquid silicone elastomer (5 parts A:1 part B) was poured on the flow master to a thickness of 7 mm, degassed, and baked at 80°C for 75 minutes. The bonded elastomer was then peeled from the control mold, and access ports were punched at the flow and control inlets using a 0.055-inch punch (Technical Innovations, Gaithersburg, MD). The device was peeled from the silicon wafer, cut to size, and sealed to a glass substrate for mechanical rigidity via plasma bonding. The final design, sized to fit a standard 22 × 50 mm glass coverslip, consisted of two rows of twenty culture chambers (500 μm × 300 μm × 7 mm) each, with media inlet and outlet ports and smaller (50 μm × 50 μm) flow channels connecting the culture chambers of each row. This design was chosen because it allows for the side-by-side comparison of two different culture conditions on the same chip, thus eliminating any undesired variability in handling or manipulation between experimental conditions while maintaining identical conditions within experimental groups. The photomasks, master mold and polydimethylsiloxane (PDMS) chips were fabricated at the Stanford Microfluidics Foundry based using multilayer soft lithography. After that, 18G needles were cut down to approximately 1.5 cm and inserted into the inlet and outlet ports where they were secured with additional PDMS. Tygon tubing (1/16″) was used to connect the inlet needles to a SP220I syringe pump (World Precision Instruments; Sarasota, FL), which was used to control all subsequent fluid manipulations within the device.
Artificial Muscle Fiber Fabrication
The Collagen used was bovine tendon Collagen I (Nitta Gelatin, Japan) or Collagen solution from human fibroblasts (Sigma-Aldrich, USA). All polymerization procedures were carried out using sterile techniques and sterile solutions. Polymerization was achieved by mixing monomeric Collagen solution, F10 medium, and reconstitution buffer at a ratio of 8:1:1, according to manufacturer’s instructions. Lower modulus Collagen was achieved by decreasing the amount of Collagen solution and replacing that volume with F10 medium. All solutions and mixtures were kept on ice to slow down polymerization. Collagen fibers were then produced by extruding through a 26S fine-gauge syringe needle (Hamilton Company; Reno, NV) with an internal needle diameter of 0.127 mm. The syringe plunger was pushed at a constant rate of 5 μl/min by a SP220I syringe pump (World Precision Instruments; Sarasota, FL), volume setting to 10 μl and inner syringe diameter to 0.461 mm. All Collagen fibers were extruded into warm PBS in petri dishes or through the PDMS in microchambers. The chip chambers were pre-coated with 10% horse serum for 10 minutes and rinsed twice with PBS prior to storing the Collagen fibers. The coating step was performed to prevent non-specific adhesion of Collagen fibers to the chambers. For functionalizing the Collagen microfibers, mouse or human recombinant Integrin α4β1 was perfused in the chip and absorbed on the surface of the Collagen fiber by incubating for 1 hour. The scaffold was then washed by perfusing with PBS for 30 minutes. A third functionalization step was performed by perfusing mouse or human Laminin and incubating for 1 hour. A washing step was performed by perfusing with PBS for 1 hour. FACS sorted mouse or human MuSCs were then perfused in the chamber, allowed to adhere to the scaffold, cultured in the chip until fixed with 1% PFA and removed for analysis or employed unfixed for transplantation. Cell density in the media perfused was titrated in order to obtain an average number of 10±5 to 100±20 (according to the experiment) MuSCs per AMF.
Transplantation
Recipient nude mice were pre-injured with Cardiotoxin under anesthesia 12 hours before transplantation. Mice received randomly via intramuscular injection 50 to 1000 MuSCs either in suspension, associated with viable muscle fibers, or associated with AMFs. Live MuSCs (expressing YFP, GFP or immunostained) were counted at a fluorescent microscope prior to transplantation. The injection was performed with a pulled transparent glass needle pre-coated with FBS, that was connected with silicon tubing, pre-loaded with mineral oil, to a Hamilton syringe. The syringe was controlled by an automated micropump (SP220I syringe pump, World Precision Instruments; Sarasota, FL). Cell suspension or individual fibers were carefully loaded in the needle under a microscope. A small opening of the skin was performed to expose the TA muscles. The needle was inserted into the proximal TA muscle and the cells or fibers were slowly injected into the muscle. The needle was left undisturbed for 5 minutes in the muscle. Before extracting the needle, the injection site was sealed with surgical glue (Tiessel, fibrin sealant, Baxter) to prevent leaking of cells or fibers. The skin wound was carefully sutured with one point stich (Coated Vicryl suture, 8-0, Ethicon).
Bioluminescence Imaging
Bioluminescent imaging was performed using the Xenogen IVIS-Spectrum System (Caliper Life Sciences). Mice were anesthetized using 2% isoflurane and 100% oxygen at a flow rate of 2.5 l/min. Then 300 μl of 50 mg/ml sterile D-Luciferin (Biosynth International Inc.) dissolved in PBS was administered by intraperitoneal injection. After 23 minutes from the substrate injection, the mice were imaged for 30 seconds at the maximal light collection (f-stop 1) at the highest resolution (small binning). Each image was saved for subsequent analysis. Imaging was performed in bind: the investigators performing the imaging did not know the identity of the experimental conditions for the transplanted cells.
Bioluminescence Image Analysis
Analysis of each image was performed using Living Image Software, version 4.0 (Caliper Life Sciences). Briefly, a manually-generated circle was placed on top of the region of interest and resized to completely surround the limb or the specified region on the recipient mouse. Imaging was performed in bind: the investigators performing the analysis did not know the identity of the experimental groups.
Bioluminescence Analysis of ATP levels
ATP levels were measured using the CellTiter-Glo (Promega) luminescent assay as described by the manufacturer.
Scanning Electron Microscopy
Single myofibers or AMFs were fixed (4% PFA and 2% glutaraldehyde in 0.1M NaCacodylate buffer, pH 7.3) at 4°C, post-fixed in 1% OsO4, and transferred to microporous (120 μm) cylindrical capsules (Electron Microscopy Sciences, Hatfield, PA) for gradual dehydration in ethanol, followed by critical point drying (Tousimis, Rockville, MD). Samples were then mounted onto aluminum stubs, Au/Pd sputter-coated and imaged with a Zeiss Sigma FESEM (Thornwood, NY) operated at 2–5 kV with SE detection. Hydrated samples were similarly fixed and then mounted fully hydrated onto the chamber in a Coolstage (Deben, UK) for imaging with a Hitachi 3400N Variable Pressure (VP) SEM (Hitachi Northridge, CA) operated at 15 kV and 50 Pa using BSE detection in VP-mode.
Recombinant Proteins
The recombinant proteins used in this study were, for mouse: Integrin α4β1 (R&D cat 7810-A6-050); Integrin α5β1 (R&D cat 7728-A5-050); Integrin αVβ1 (R&D cat 7705-AV-050); Integrin α6β1 (R&D cat 7810-A6-050); TGFβ1 (R&D Systems cat 7666-MB-005); Laminin (Invitrogen cat 23017-015). For human: Integrin α4β1 (R&D cat 5668-A4-050); Laminin (BioLamina cat LN-211); Collagen (Sigma-Aldrich cat C2249).
Immunofluorescence
Immunofluorescence was performed using a Zeiss Observer Z1 fluorescent microscope (Zeiss) equipped with a Hamamatsu Orca-ER camera or a Zeiss confocal system LSM710 (Zeiss). Data acquisition and fiber-diameter measurements were performed using Improvision Volocity software (Perkin Elmer) or Zeiss LSM ZEN software (Zeiss).
Antibodies and Staining
Antibodies used in this study were to the following proteins, with the source of each antibody indicated: Pax7 (DSHB); Ki67 (Abcam 15580 and BD 558615); CD34 (Biolegend 343501 and Biolegend 119213); Laminin (Sigma L9393, Millipore MAB1903 and One World Lab C13071); cleaved Caspase3 (Cell Signaling 9669); MyoD (Dako M3512; Santa Cruz sc-760); green fluorescent protein (GFP) (Invitrogen A11122 and Abcam ab13970); Collagen I (Cedarlane clone 50151AP); Integrin α4β1 (ABcam ab8991-100 and ABcam ab24695). EdU chemical staining was performed as indicated by the manufacturer (Invitrogen).
Statistical Analysis
Unless otherwise noted, all statistical analyses were performed using GraphPad Prism 5 (GraphPad Software). All error bars represent s.e.m.; * P<0.05; ** P<0.001; *** P<0.0001; **** P<0.00001.
Sample size
A minimal number of animals necessary to obtain statistically relevant experimental data was predicted and utilized. The size of groups needed was determined based on previous studies or preliminary data. The number needed per group was estimated based on expected magnitude of any measurable outcome to achieve statistical significance, and was based to achieve a power to 80% and P value of <0.054. The coefficient of variation ranges from <5% to approximately 30%. To detect difference of 20% with a 10% standard deviation (SD) (using the bioluminescence assays as the limiting assay, to calculate the highest variability), 4 mice per group was calculated to be required (2 TA muscles per mouse); with a 20% SD of the assay, 13 (26 muscle) mice per group; with a 30% SD of the assay, 20 mice (40 muscles). Thus, based on the variation used in our previous work, it was expected that 4–13 mice with each group was required.
General Methods
Unless stated otherwise, sample size (n values) are reported as biological replicates of mice and/or SC isolations from separate mice performed on 28 different days. In most cases, the data presented were compiled over the course of 3 years, as mice with the appropriate genotype became available. Therefore the magnitude of the effect and variability in the measurements were primary factors in determining sample size and replication of data. Although samples were not explicitly randomized or blinded, mouse identification numbers were used as sample identifiers and thus the genotypes and experimental conditions of each mouse/sample were not readily known or available to the experimenters during sample processing and data collection. The only criteria used to exclude samples involved the health of the animals, such as visible wounds from fighting. In these cases, the animals were handled in accordance with approved IAUCUC guidelines.
Study Approval
Animals were handled and housed according to the guidelines set forth by the Veterinary Medical Unit of the VA Palo Alto Health Care System, and all procedures were approved by the IACUC prior to being performed. For human subjects, all operative specimens were obtained with appropriate written informed consent according to a protocol approved by the Stanford University Institutional Review Board.
Supplementary Material
Sheet 1. Analysis of MuSCs in different states analyzed at the single cell level for gene expression profiles. Single MuSCs were freshly FACS isolated from uninjured muscles (0 DPI), injured muscles (1.5 and 3.5 DPI), or after having been cultured in different conditions (associated or not with AMFs and cultured in either GM or QM). Single MuSCs were analyzed for gene expression profiles using a Fluidgm Biomark platform. Raw data obtained from Biomark microfluidic chip runs are presented. Sheet 2. Freshly isolated MuSCs were FACS isolated and 500 cells/well were directly loaded in a 96 well plate. In each well, MuSCs were cultured in the presence of candidate molecules at different concentrations and combinations. RNA was then isolated and analyzed for gene expression profiles as in Sheet 1 for single MuSCs. The molecules tested and the different dilutions employed from the working concentrations are indicated as in Table S2. Raw data obtained from Biomark microfluidic chip runs are presented.
Initial panel of 50 compounds known or suggested in the literature to positively regulate cell quiescence. The 10 molecules that showed the strongest propensity to prevent quiescent MuSC proliferation are highlighted in red.
Serum-free “quiescence medium” (QM) formulation that best maintained the transcriptional signature of quiescence for at least two days in culture among all conditions analyzed. Using a combinatorial Q-RT-PCR array strategy for a set of 93 genes (see methods), we found conditions in which cultured MuSCs were very similar to freshly isolated quiescent MuSCs.
Primer sequences of the probes employed in the single cell gene expression array and in the screening for compounds promoting quiescence as indicated in Table S1.
Representative movie of 3D immunofluorescence imaging of MuSCs cultured onto an AMF. Freshly FACS isolated MuSCs were seeded onto an AMF in one of the culture micro-chambers of a microfluidic chip mounted on a glass slide. Cells and AMF were immediately fixed and immunostained by perfusing the culture micro-chamber through microchannels. Imaging with confocal microscopy was performed directly on the microfluidic chip. Immunostaining was performed for Collagen I (green) and Pax7 (red). Nuclei were stained with DAPI.
Acknowledgments
We thank the members of the Rando laboratory for comments and discussions, in particular Mark Hamer and Igor Akimenko for helping with some of the experiments performed with human MuSCs; Lydia-Marie Joubert of the CSIF:EM Stanford Core Facility for the technical support with the SEM imaging; Emre Araci for the technical support at the Stanford Microfluidic Foundry; Caleb Horst of Cellscale Biomaterials Testing for the support in the tensile measurements of the AMF; Maddalena Adorno from Stanford Stem Cell Institute for providing the lentivirus construct expressing Luciferase and GFP; Eithon Cadag of Ayasdi Academic Program for the technical support with the TDA analysis; Michelle Lynch from Dept. of Bioengineering, University of Pennsylvania for the technical support with the micropost array fabrication; Andrew Wang from the Melosh lab for his technical support with the AFM measurements. This work was supported by the Glenn Foundation for Medical Research and by grants from the National Institutes of Health (NIH) (P01 AG036695, R01 AG23806 (R37 MERIT Award), R01 AR062185, and R01 AG047820) the California Institute of Regenerative Medicine, and the Department of Veterans Affairs (BLR&D and RR&D Merit Reviews) to T.A.R.; by NIH P41 program: RESBIO - The Technology Resource for Polymeric Biomaterials (EB001046); and by the CalPoly funding Award # TB1-01175.
Footnotes
Author Contributions:
M.Q. conceived and designed most of the experiments reported. T.A.R. provided guidance throughout. M.Q., J.O.B., A.D.M. and S.C.B. performed experiments and collected data. S.H. provided guidance throughout. M.Q., R.D. and J.S. designed AMF fabrication and performed AMF experiments. J.O.B. analyzed single cell gene expression data and developed the random forests model. M.Q. and R.D. designed and performed AFM experiments and analysis. M.Q., A.D.M. and S.C.B. designed, performed and analyzed single cell gene expression experiments and the screening for factors promoting quiescence of MuSCs. M.Q. and R.C. performed transplant experiments, in vivo imaging and data analysis. M.Q. and M.C.G. designed fabrication and experiments with microfluidic chips for AMFs. M.Q. and J.S. designed the AMF and performed AMFs fabrication experiments and imaging. V.G. performed the experiments with the human MuSCs. J.B.S. collected operative samples during surgeries. M.Q. and T.A.R. analyzed data and wrote the manuscript.
Competing Financial Interests Statement:
The authors declare no competing financial interests.
References
- 1.Montarras D, et al. Direct isolation of satellite cells for skeletal muscle regeneration. Science. 2005;309:2064–2067. doi: 10.1126/science.1114758. [DOI] [PubMed] [Google Scholar]
- 2.Sacco A, Doyonnas R, Kraft P, Vitorovic S, Blau HM. Self-renewal and expansion of single transplanted muscle stem cells. Nature. 2008;456:502–506. doi: 10.1038/nature07384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cerletti M, et al. Highly efficient, functional engraftment of skeletal muscle stem cells in dystrophic muscles. Cell. 2008;134:37–47. doi: 10.1016/j.cell.2008.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327:542–545. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuchs E. The tortoise and the hair: slow-cycling cells in the stem cell race. Cell. 2009;137:811–819. doi: 10.1016/j.cell.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pietras EM, Warr MR, Passegué E. Cell cycle regulation in hematopoietic stem cells. J Cell Biol. 2011;195:709–720. doi: 10.1083/jcb.201102131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ottone C, et al. Direct cell-cell contact with the vascular niche maintains quiescent neural stem cells. Nat Cell Biol. 2014;16:1045–1056. doi: 10.1038/ncb3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung TH, Rando TA. Molecular regulation of stem cell quiescence. Nat Rev Mol Cell Biol. 2013;14:329–340. doi: 10.1038/nrm3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schepers K, Campbell TB, Passegué E. Normal and leukemic stem cell niches: insights and therapeutic opportunities. Cell Stem Cell. 2015;16:254–267. doi: 10.1016/j.stem.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishikawa SI, Osawa M, Yonetani S, Torikai-Nishikawa S, Freter R. Niche required for inducing quiescent stem cells. Cold Spring Harb Symp Quant Biol. 2008;73:67–71. doi: 10.1101/sqb.2008.73.024. [DOI] [PubMed] [Google Scholar]
- 15.Collins CA, et al. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell. 2005;122:289–301. doi: 10.1016/j.cell.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 17.Dellatore SM, Garcia AS, Miller WM. Mimicking stem cell niches to increase stem cell expansion. Curr Opin Biotechnol. 2008;19:534–540. doi: 10.1016/j.copbio.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lutolf MP, Gilbert PM, Blau HM. Designing materials to direct stem-cell fate. Nature. 2009;462:433–441. doi: 10.1038/nature08602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert PM, et al. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 2010;329:1078–1081. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu X, et al. Combination of inflammation-related cytokines promotes long-term muscle stem cell expansion. Cell Res. 2015 doi: 10.1038/cr.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lutolf MP, Doyonnas R, Havenstrite K, Koleckar K, Blau HM. Perturbation of single hematopoietic stem cell fates in artificial niches. Integr Biol (Camb) 2009;1:59–69. doi: 10.1039/b815718a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Csaszar E, et al. Rapid expansion of human hematopoietic stem cells by automated control of inhibitory feedback signaling. Cell Stem Cell. 2012;10:218–229. doi: 10.1016/j.stem.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 25.Busch K, et al. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature. 2015;518:542–546. doi: 10.1038/nature14242. [DOI] [PubMed] [Google Scholar]
- 26.Lugert S, et al. Quiescent and active hippocampal neural stem cells with distinct morphologies respond selectively to physiological and pathological stimuli and aging. Cell Stem Cell. 2010;6:445–456. doi: 10.1016/j.stem.2010.03.017. [DOI] [PubMed] [Google Scholar]
- 27.Schultz E, Gibson MC, Champion T. Satellite cells are mitotically quiescent in mature mouse muscle: an EM and radioautographic study. J Exp Zool. 1978;206:451–456. doi: 10.1002/jez.1402060314. [DOI] [PubMed] [Google Scholar]
- 28.MAURO A. Satellite cell of skeletal muscle fibers. 1961 doi: 10.1083/jcb.9.2.493. at < http://jcb.rupress.org/content/9/2/493.full.pdf>. [DOI] [PMC free article] [PubMed]
- 29.Fukada SI, et al. Molecular signature of quiescent satellite cells in adult skeletal muscle. Stem Cells. 2007;25:2448–2459. doi: 10.1634/stemcells.2007-0019. [DOI] [PubMed] [Google Scholar]
- 30.Xu C, et al. A zebrafish embryo culture system defines factors that promote vertebrate myogenesis across species. Cell. 2013;155:909–921. doi: 10.1016/j.cell.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cosgrove BD, et al. Rejuvenation of the muscle stem cell population restores strength to injured aged muscles. Nat Med. 2014;20:255–264. doi: 10.1038/nm.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernet JD, et al. p38 MAPK signaling underlies a cell-autonomous loss of stem cell self-renewal in skeletal muscle of aged mice. Nat Med. 2014;20:265–271. doi: 10.1038/nm.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chakkalakal JV, Jones KM, Basson MA, Brack AS. The aged niche disrupts muscle stem cell quiescence. Nature. 2012;490:355–360. doi: 10.1038/nature11438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rathbone CR, et al. Effects of transforming growth factor-beta (TGF-β1) on satellite cell activation and survival during oxidative stress. J Muscle Res Cell Motil. 2011;32:99–109. doi: 10.1007/s10974-011-9255-8. [DOI] [PubMed] [Google Scholar]
- 35.Dupont S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 36.Discher DE, Janmey P, Wang Y. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;18:1139–43. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 37.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–89. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 38.Fu J, et al. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat Methods. 2010;7:733–736. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Defranchi E, et al. Imaging and elasticity measurements of the sarcolemma of fully differentiated skeletal muscle fibres. Microsc Res Tech. 2005;67:27–35. doi: 10.1002/jemt.20177. [DOI] [PubMed] [Google Scholar]
- 40.Trensz F, et al. Increased microenvironment stiffness in damaged myofibers promotes myogenic progenitor cell proliferation. Skelet Muscle. 2015;5:5. doi: 10.1186/s13395-015-0030-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brightman AO, et al. Time-lapse confocal reflection microscopy of collagen fibrillogenesis and extracellular matrix assembly in vitro. Biopolymers. 2000;54:222–234. doi: 10.1002/1097-0282(200009)54:3<222::AID-BIP80>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 42.Badylak SF, Freytes DO, Gilbert TW. Extracellular matrix as a biological scaffold material: Structure and function. Acta Biomater. 2009;5:1–13. doi: 10.1016/j.actbio.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 43.Kirkwood JE, Fuller GG. Liquid crystalline collagen: a self-assembled morphology for the orientation of mammalian cells. Langmuir. 2009;25:3200–3206. doi: 10.1021/la803736x. [DOI] [PubMed] [Google Scholar]
- 44.Urciuolo A, et al. Collagen VI regulates satellite cell self-renewal and muscle regeneration. Nat Commun. 2013;4:1964. doi: 10.1038/ncomms2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bjornson CRR, et al. Notch signaling is necessary to maintain quiescence in adult muscle stem cells. Stem Cells. 2012;30:232–242. doi: 10.1002/stem.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dean DC, Iademarco MF, Rosen GD, Sheppard AM. The integrin alpha 4 beta 1 and its counter receptor VCAM-1 in development and immune function. Am Rev Respir Dis. 1993;148:S43–S46. doi: 10.1164/ajrccm/148.6_Pt_2.S43. [DOI] [PubMed] [Google Scholar]
- 47.Jülich D, Geisler R, Holley SA. Tübingen 2000 Screen Consortium. Integrinalpha5 and delta/notch signaling have complementary spatiotemporal requirements during zebrafish somitogenesis. Dev Cell. 2005;8:575–586. doi: 10.1016/j.devcel.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 48.McDonald KA, Lakonishok M, Horwitz AF. Alpha v and alpha 3 integrin subunits are associated with myofibrils during myofibrillogenesis. 1995;108(Pt 3):975–983. doi: 10.1242/jcs.108.3.975. [DOI] [PubMed] [Google Scholar]
- 49.Hirsch E, et al. Alpha v integrin subunit is predominantly located in nervous tissue and skeletal muscle during mouse development. Dev Dyn. 1994;201:108–120. doi: 10.1002/aja.1002010203. [DOI] [PubMed] [Google Scholar]
- 50.Sastry SK, Lakonishok M, Thomas DA, Muschler J, Horwitz AF. Integrin alpha subunit ratios, cytoplasmic domains, and growth factor synergy regulate muscle proliferation and differentiation. J Cell Biol. 1996;133:169–184. doi: 10.1083/jcb.133.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang JT, et al. Genetic analysis of alpha 4 integrin functions in the development of mouse skeletal muscle. J Cell Biol. 1996;135:829–835. doi: 10.1083/jcb.135.3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yin H, Price F, Rudnicki MA. Satellite cells and the muscle stem cell niche. Physiol Rev. 2013;93:23–67. doi: 10.1152/physrev.00043.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.The basement membrane/basal lamina of skeletal muscle. 2003;278:12601–12604. doi: 10.1074/jbc.R200027200. [DOI] [PubMed] [Google Scholar]
- 54.Laminins during muscle development and in muscular dystrophies. 1999;56:442–460. doi: 10.1007/PL00000616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Breiman L. Random Forests. Machine Leraning. 2001;45:5–32. [Google Scholar]
- 56.Strobl C, Boulesteix AL, Kneib T, Augustin T, Zeileis A. Conditional variable importance for random forests. BMC Bioinformatics. 2008;9:307. doi: 10.1186/1471-2105-9-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rando TA. The adult muscle stem cell comes of age. Nat Med. 2005;11:829–831. doi: 10.1038/nm0805-829. [DOI] [PubMed] [Google Scholar]
- 58.Hall JK, Banks GB, Chamberlain JS, Olwin BB. Prevention of muscle aging by myofiber-associated satellite cell transplantation. Sci Transl Med. 2010;2:57ra83. doi: 10.1126/scitranslmed.3001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Konieczny P, Swiderski K, Chamberlain JS. Gene and cell-mediated therapies for muscular dystrophy. Muscle Nerve. 2013;47:649–663. doi: 10.1002/mus.23738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Partridge TA. Impending therapies for Duchenne muscular dystrophy. Curr Opin Neurol. 2011;24:415–422. doi: 10.1097/WCO.0b013e32834aa3f1. [DOI] [PubMed] [Google Scholar]
- 61.Bareja A, et al. Human and mouse skeletal muscle stem cells: convergent and divergent mechanisms of myogenesis. PLoS One. 2014;9:e90398. doi: 10.1371/journal.pone.0090398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Charville GW, et al. Ex Vivo Expansion and In Vivo Self-Renewal of Human Muscle Stem Cells. Stem Cell Rep. 2015;5:621–632. doi: 10.1016/j.stemcr.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shandalov Y, et al. An engineered muscle flap for reconstruction of large soft tissue defects. Proc Natl Acad Sci USA. 2014;111:6010–6015. doi: 10.1073/pnas.1402679111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nishijo K, et al. Biomarker system for studying muscle, stem cells, and cancer in vivo. FASEB J. 2009;23:2681–2690. doi: 10.1096/fj.08-128116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 73.Singh G, Mémoli F, Carlsson GE. Topological Methods for the Analysis of High Dimensional Data Sets and 3D Object Recognition. SPBG. 2007 [Google Scholar]
- 75.Rosenblatt JD, Lunt AI, Parry DJ, Partridge TA. Culturing satellite cells from living single muscle fiber explants. In Vitro Cell Dev Biol Anim. 1995;31:773–779. doi: 10.1007/BF02634119. [DOI] [PubMed] [Google Scholar]
- 76.Rouredu O, et al. Force mapping in epithelial cell migration. Proc Natl Acad Sci USA. 2005;102:2390–2395. doi: 10.1073/pnas.0408482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hansen CL, Sommer MOA, Quake SR. Systematic investigation of protein phase behavior with a microfluidic formulator. Proc Natl Acad Sci USA. 2004;101:14431–14436. doi: 10.1073/pnas.0405847101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sheet 1. Analysis of MuSCs in different states analyzed at the single cell level for gene expression profiles. Single MuSCs were freshly FACS isolated from uninjured muscles (0 DPI), injured muscles (1.5 and 3.5 DPI), or after having been cultured in different conditions (associated or not with AMFs and cultured in either GM or QM). Single MuSCs were analyzed for gene expression profiles using a Fluidgm Biomark platform. Raw data obtained from Biomark microfluidic chip runs are presented. Sheet 2. Freshly isolated MuSCs were FACS isolated and 500 cells/well were directly loaded in a 96 well plate. In each well, MuSCs were cultured in the presence of candidate molecules at different concentrations and combinations. RNA was then isolated and analyzed for gene expression profiles as in Sheet 1 for single MuSCs. The molecules tested and the different dilutions employed from the working concentrations are indicated as in Table S2. Raw data obtained from Biomark microfluidic chip runs are presented.
Initial panel of 50 compounds known or suggested in the literature to positively regulate cell quiescence. The 10 molecules that showed the strongest propensity to prevent quiescent MuSC proliferation are highlighted in red.
Serum-free “quiescence medium” (QM) formulation that best maintained the transcriptional signature of quiescence for at least two days in culture among all conditions analyzed. Using a combinatorial Q-RT-PCR array strategy for a set of 93 genes (see methods), we found conditions in which cultured MuSCs were very similar to freshly isolated quiescent MuSCs.
Primer sequences of the probes employed in the single cell gene expression array and in the screening for compounds promoting quiescence as indicated in Table S1.
Representative movie of 3D immunofluorescence imaging of MuSCs cultured onto an AMF. Freshly FACS isolated MuSCs were seeded onto an AMF in one of the culture micro-chambers of a microfluidic chip mounted on a glass slide. Cells and AMF were immediately fixed and immunostained by perfusing the culture micro-chamber through microchannels. Imaging with confocal microscopy was performed directly on the microfluidic chip. Immunostaining was performed for Collagen I (green) and Pax7 (red). Nuclei were stained with DAPI.