Abstract
Whole slide digital imaging technology enables researchers to study pathologists’ interpretive behavior as they view digital slides and gain new understanding of the diagnostic medical decision-making process. In this study, we propose a simple yet important analysis to extract diagnostically relevant regions of interest (ROIs) from tracking records using only pathologists’ actions as they viewed biopsy specimens in the whole slide digital imaging format (zooming, panning, and fixating). We use these extracted regions in a visual bag-of-words model based on color and texture features to predict diagnostically relevant ROIs on whole slide images. Using a logistic regression classifier in a cross-validation setting on 240 digital breast biopsy slides and viewport tracking logs of three expert pathologists, we produce probability maps that show 74 % overlap with the actual regions at which pathologists looked. We compare different bag-of-words models by changing dictionary size, visual word definition (patches vs. superpixels), and training data (automatically extracted ROIs vs. manually marked ROIs). This study is a first step in understanding the scanning behaviors of pathologists and the underlying reasons for diagnostic errors.
Keywords: Digital pathology, Medical image analysis, Computer vision, Region of interest, Whole slide imaging
Introduction
Whole slide imaging (WSI) technology has revolutionized histopathological image analysis research, yet most automated systems analyze only hand-cropped regions of digital WSIs of tissue biopsies. The fully automated analysis of digital whole slides remains a challenge. A digital whole slide can be quite large, often larger than 100,000 pixels in both height and width, depending on the tissue and the biopsy type. In clinical practice, a trained pathologist examines the full image, discards most of it after a quick visual survey and then spends the remainder of the interpretive time viewing small regions within the slide that contain diagnostic features that seem most significant [1, 2]. The first challenge for any image analysis system is the localization of these regions of interest (ROIs) in order to reduce the computational load and improve the diagnostic accuracy by focusing on diagnostically important areas.
Histopathological image analysis research tackles many problems related to diagnosis of the disease, including nucleus detection [3–7], prediction of clinical variables (diagnosis [8–12], grade [13–18], survival time [19–21]), identification of genetic factors controlling tumor morphology (gene expression [20, 22], molecular subtypes [20, 23]), and localization of ROIs [24–28]. One of the major research directions in histopathological image analysis is to develop image features for different problems and image types. Commonly used image features include low-level features (color [9, 10, 15, 16, 18, 21, 27–31], texture [10–14, 18, 28]), object level features (shape [32–37], topology [8, 11, 14, 18, 26, 31]), and semantic features (statistics [19, 26], histograms [28, 32], bag-of-words [28]).
The majority of the literature on ROI localization considers ROIs manually marked by experts. Gutierrez et al. used functions inspired by human vision to combine over-segmented images and produce an activation map for relevant ROIs [25]. Their method is based on human perception of groupings, also known as Gestalt law. Using a supervised machine-learning method, they merge relevant segments with the help of an energy function that quantifies similarity between two image partitions. They evaluate their findings with pathologist drawn ROIs. Their method outperforms the standard saliency detection models [38].
Bahlmann et al. employed a supervised model to detect ROIs using expert annotations of ROIs to train a linear SVM classifier [24]. They make use of color features to differentiate diagnostically relevant and irrelevant regions on a WSI. However, their evaluation considers only manually marked positive and negative samples and does not apply to the complete digital slide.
The experimental setting of Romo et al. is the most relevant to our work [27]. They calculated grayscale histograms, local binary pattern (LBP) histograms [39], Tamura texture histograms [40], and Sobel edge histograms for 70 × 70 pixel tiles. In a supervised setting, they classify all tiles in a WSI as diagnostically relevant or not. They evaluate their predictions against image regions that are visited longer by the pathologists during their interpretations. Our methodology to extract ground truth ROIs is different from theirs, since we take all actions of the pathologists into account, not only pathologist viewing duration. Thus, we provide a first validation of automated ROI extraction that uses a broader range of pathologist image search behavior, including zooming, panning, and fixating.
The problem we attempt to solve is to locate diagnostically important ROIs on a digital slide using image features such as color and texture. We designed a system that produces a probability map of diagnostic importance given a digital whole slide. In our previous work, we showed the usefulness of the visual bag-of-words representation for ROI localization using a small subset of 20 images [28]. In this paper, we compare different visual word representations and dictionary sizes using 240 whole slide images and report on the results of a larger and more comprehensive study.
Materials and Methods
Human Research Participants Projection
The study was approved by the institutional review boards at Dartmouth College, Fred Hutchinson Cancer Research Center, Providence Health and Services Oregon, University of Vermont, University of Washington, and Bilkent University. Informed consent was obtained electronically from pathologists.
Dataset
The breast pathology (B-Path) and digital pathology (digiPATH) study [41–44] aims are to understand the diagnostic patterns of pathologists and evaluate the accuracy and efficiency of interpretations using glass slides and digital whole slide images. For this purpose, three expert pathologists were invited to interpret a series of 240 breast biopsies on glass or digital media. Cases included benign without atypia (30 %), atypical ductal hyperplasia (30 %), ductal carcinoma in situ (30 %), and invasive breast cancer (10 %). The methods of case development and data collection from pathologists have been previously described [41–44] and will be summarized here briefly.
The 240 core needle and excisional breast biopsies were selected from pathology registries in Vermont and New Hampshire using a random sampling stratified according to woman’s age (40–49 vs. ≥50), parenchymal breast density (low vs. high), and interpretation of the original pathologist. After initial review by an expert, new glass slides were created from the original tissue blocks to ensure consistency in staining and image quality.
The H&E stained biopsy slides were scanned using an IScan Coreo Au® digital slide scanner in 40× magnification, resulting in an average image size of 90,000 × 70,000 pixels. Digital whole slide images of the 240 cases were independently reviewed by three experienced breast pathologists using a web-based virtual slide viewer that was developed specifically for this project using HD View SL, Microsoft’s open source Silverlight gigapixel image viewer. The viewer provides similar functionality to the industry sponsored WSI image viewers. It allows users to pan the image and zoom in and zoom out (up to 40× actual and 60× digital magnification). The expert pathologists are internationally recognized for research and continuing medical education on diagnostic breast pathology. Each of our experts has had opportunities to utilize digital pathology as a tool for research and teaching, yet none of our experts use digital pathology as a tool for the primary diagnosis of breast biopsies. Each expert pathologist independently provided a diagnosis and identified a supporting ROI for each case. On completion of independent reviews, several consensus meetings were held to reach a consensus diagnosis and define consensus ROIs for each case. Detailed tracking data were collected while the expert pathologists interpreted digital slides using the web-based virtual slide viewer. Our dataset for this paper contains the tracking data and ROI markings from the three expert breast pathologists as they independently interpreted all 240 cases.
Viewport Analysis
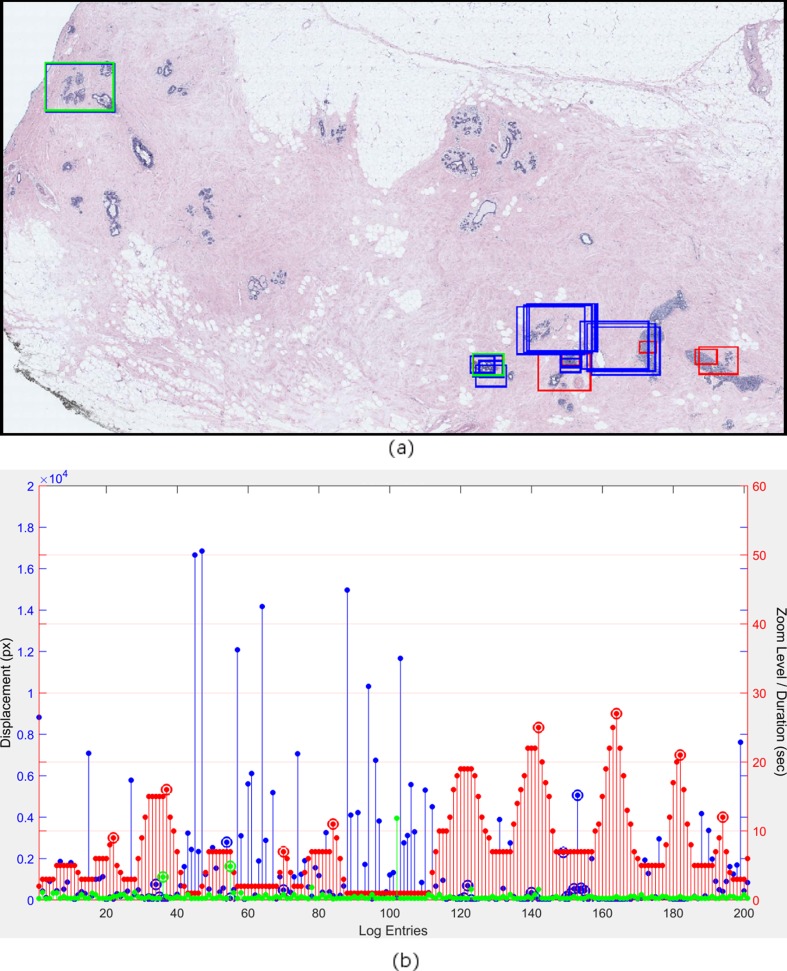
A viewport log provides a stream of screen coordinates and zoom levels with timestamps indicating the location of the pathologists’ screen in the digital whole slide. We used a graph to visualize a pathologist’s reading of a digital whole slide (see Fig. 1a) and defined three actions over the viewport tracking data that are used to extract regions to which pathologists focused their attention:
Zoom peaks are the log entries where the zoom level is higher than the previous and the next entries. A zoom peak identifies a region where the pathologist intentionally zoomed to look at a higher magnification. During the diagnostic process, low magnification views are also very important in terms of planning the search strategy and seeing the big picture. In low magnification, the pathologists determine the areas of importance to zoom into (see the circled red bars in Fig. 1a). They are the local maximal points of the zoom level series plotted in red.
Slow pannings are the log entries where the zoom level is the same as the previous entry, and the displacement is small. We used a 100 pixel displacement threshold on the screen level (100× zoom on the actual image) to define slow pannings. The quick pans intended for moving the viewport to a distant region result in high displacement values (more than 100 pixels). In comparison, slow pannings are intended for investigating a slightly larger and closer area without completely moving the viewport (see the circled blue bars in Fig. 1a). The zoom level represented by the red bars is constant, and the displacement represented by blue bars is small at these points.
Fixations are the log entries where the duration is longer than 2 seconds. Fixations identify the areas to which a pathologist focused extra attention by looking at them longer (see circled green bars in Fig. 1a). In eye-tracking studies, the fixation is defined as maintaining the visual gaze for more than 100 ms, but this definition is not suitable for our mouse tracking data. A much higher threshold than 100 ms is picked, because the mouse cursor movements are much slower than the gaze movements.
Fig. 1.
Viewport analysis. a The selected viewports (rectangular image regions visible on the pathologist’s screen) are shown in colored rectangles on the actual image. A zoom peak noted with a red circle in b corresponds to red rectangles in a. Similarly, slow pannings and fixations which are noted with blue and green circles in b correspond to blue and green rectangles in a. b An example visualization of the viewport log for an expert pathologist interpreting the image in a. The x-axis shows the log entry numbers (not the time). The red bars represent the zoom level, the blue bars represent the displacement, and the green bars represent the duration at each entry. The y-axis on the right shows the zoom level and duration values while the y-axis on the left shows the displacement values. Zoom peaks, slow pannings, and fixations are marked with red, blue and green circles, respectively
The viewports (rectangular image regions) that correspond to one of the above three actions are extracted as diagnostically relevant ROIs (see Fig. 1b for example viewports). Note that these image regions are not necessarily related to the final diagnosis given to a case by the expert; the regions on the image can be distracting regions as well as diagnostic regions.
ROI Prediction in Whole Slide Images
We represent diagnostically relevant ROIs at which the pathologists are expected to look with a visual bag-of-words model. The bag-of-words (BoW) model is a simple yet powerful representation technique commonly used in document retrieval and computer vision [45]. The BoW represents documents (or images) as collections of words in which each bag is different in terms of the frequency of each word in a pre-determined dictionary. In this framework, a visual word is a 120 × 120 pixel image patch cut from a whole slide image whereas a bag represents a 3600 × 3600 pixel image window, and each bag is a collection of words. We considered the sizes of biological structures at 40× magnification in the selection of visual word and bag sizes. A visual word is constructed to contain more than one epithelial cell. A visual bag, on the other hand, may contain bigger structures such as breast ducts.
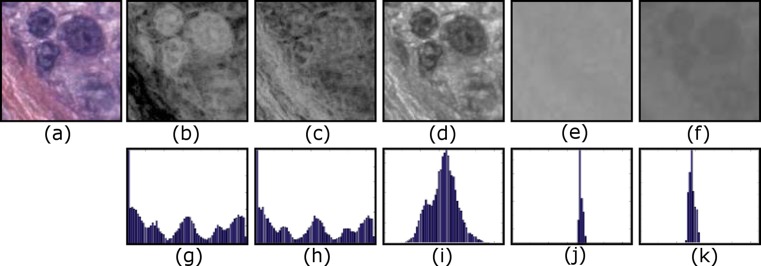
A visual vocabulary is a collection of distinct image patches that can be used to build images. The visual vocabulary is usually obtained by collecting all possible words (120 × 120 pixel patches) from all images and clustering them to reduce the number of distinct words. We selected two commonly used low-level image features for representing visual words: Local binary pattern (LBP) [39] histograms for texture and L*a*b* histograms for color. For LBP histograms, instead of using grayscale as is usually done, we used a well-known color deconvolution algorithm [46] to obtain two chemical dye channels, hematoxylin and eosin (H&E), and calculated LBP feature on these two color channels (for example, images of RGB to H&E conversion, see Fig. 2). Each visual word is represented by a feature vector that is the concatenation of the LBP and L*a*b* histograms. Both LBP and L*a*b* features have values ranging from 0 to 255, and we used 64 bins for each color and texture channel resulting in a feature vector of length 320.
Fig. 2.
a 120 × 120 image patch (visual word) in RGB, b deconvolved hematoxylin color channel that shows nuclei, c deconvolved eosin color channel that shows stromal content, d, e LBP histograms of deconvolved H and E channels, f–h L, a, and b channels of the L*a*b* color space, i–k color histogram of L, a, and b color channels
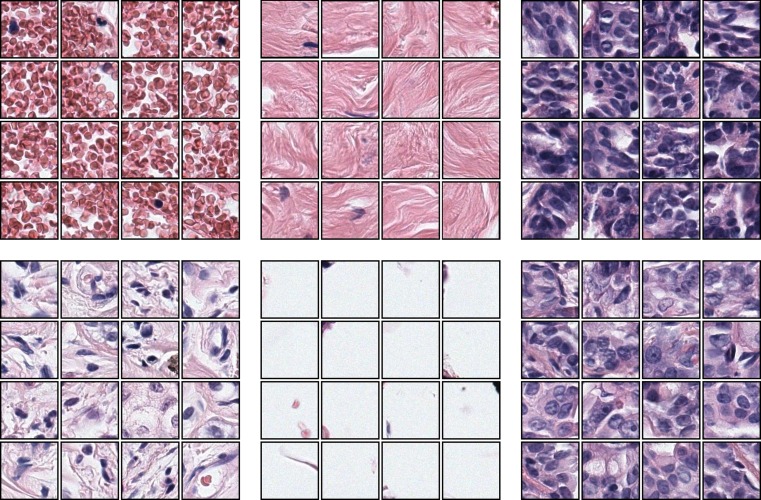
We used k-means clustering to obtain a visual vocabulary that can be represented as the cluster centers. Any 120 × 120 pixel image patch is assigned to the most similar visual word, the one with the smallest Euclidean distance between the feature vector of the patch and the cluster center that represents the visual word. This enables us to represent image windows (bags) as histograms of visual words (see Fig. 3 for some example clusters). Since the cluster center is not always a sample point in the feature space, we show the closest 16 image patches to cluster centers for 6 visual words.
Fig. 3.
Example results from the k-means clustering. Each set show the closest 16 image patches to a cluster center
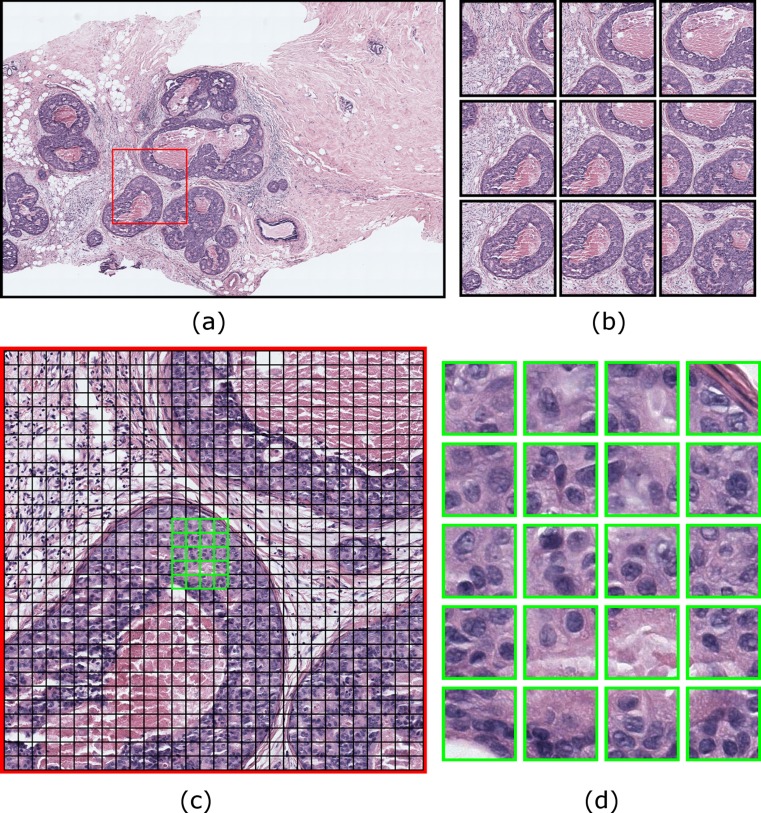
We used a sliding window approach for extracting visual bags that are 3600 × 3600 pixel image windows overlapping by 2400 pixels both horizontally and vertically. Overlapping the sliding windows is a common technique to ensure that at least one window contains an object, if all others fail to encompass it. We picked a two-thirds overlap between sliding windows for performance purposes, since a higher overlap would increase the number of sliding windows and hence the sample size for the classification. Each sliding window contains 30 × 30 = 900 image patches, which are then represented as color and texture histograms and assigned to visual words by calculating distances to cluster centers. In this framework, each sliding window is represented as a histogram of visual words. Figure 4 shows an example sliding window and visual words computed from it.
Fig. 4.
Sliding window and visual bag-of-words approach: a A 3600 × 3600 pixel sliding window is shown with a red square on an image region. b The sliding window from a is shown in the center with neighboring sliding windows overlapping 1200 pixels horizontally and vertically. c 120 × 120 pixel visual words are shown with black borders on the same sliding window from a. Visual words do not overlap. d A group of visual words are shown in higher magnification. They are identified with green borders in c
Results
We formulated the detection of diagnostically relevant ROIs as a classification problem where the samples are sliding windows, the features are visual bag-of-words histograms and the labels are obtained through viewport analysis. We labeled sliding windows that overlap with the diagnostically relevant ROIs as positive samples and everything else as negative samples. We employed tenfold cross-validation experiments using logistic regression.
We conducted several experiments to understand the visual characteristics of ROIs. We compared different dictionary sizes, different visual word definitions (square patches vs. superpixels), and different training data (automatically extracted viewport ROIs vs. manually marked ROIs). The classification accuracies we are reporting are calculated as the percentage of sliding windows that are correctly classified as diagnostically relevant or not over all possible sliding windows of size 3600 × 3600 pixels.
Dictionary Size
The dictionary size corresponds to the number of clusters and the length of the feature vector (as the histogram of visual words) calculated for each image window. For this reason, the dictionary size can determine the representative power of the model, yet large dictionaries present a computational challenge and introduce redundancy. Since the dictionary is built in an unsupervised manner, we tested different visual vocabulary sizes to understand the effect of dictionary size on model predictions. For this purpose, we applied k-means clustering to obtain the initial 200 clusters from millions of image patches and reduced the number of clusters by using hierarchical clustering.
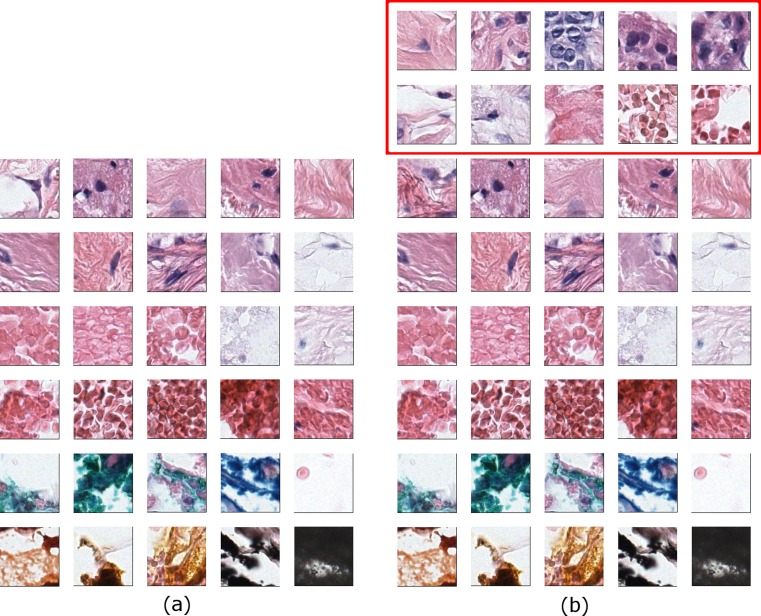
The classification accuracy (74 %) does not change when the dictionary size is reduced from 200 to 40 but drops from 74 to 46 % when the dictionary contains only 30 words. This trend is present in all experiments with different visual words (superpixels) and different training data. We compared the visual dictionaries with 40 words and 30 words to discover critical visual words in the ROI representation. Figure 5 shows the visual dictionaries; the missing words in the 30-word vocabulary are framed in the 40-word dictionary. The missing words include some blood cells, stroma with fibroblast, and, in particular, epithelial cells in which the ductal carcinoma or pre-invasive lesions present abnormal features.
Fig. 5.
Visual dictionaries with b 40 words and a 30 words. Note that visual words that represent epithelial cells are missing in a while present in b. This difference causes classification accuracy to drop from 74 to 46 %. The visual words that represent epithelial cells are absolutely necessary for the diagnostically relevant regions, since all the structures in a are discarded by pathologists during the screening process
Superpixels
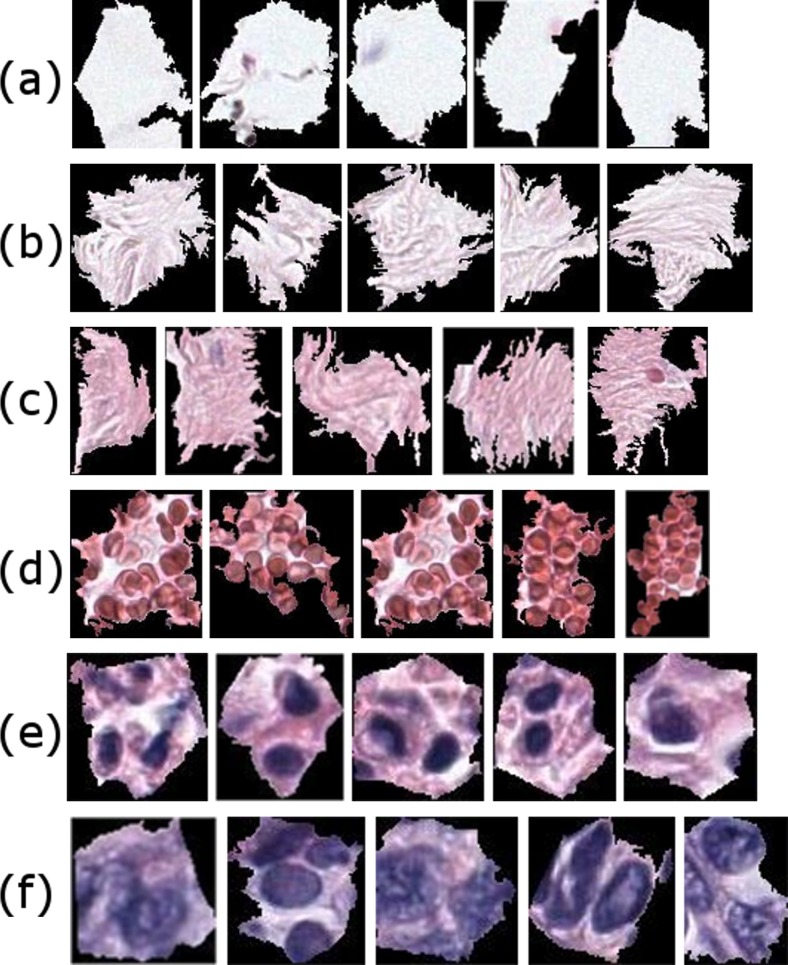
Superpixel [47] segmentation is a very popular method in computer vision. There has been successful work in histopathological image analysis in which superpixels are used as building blocks of the tissue analysis [19, 48]. We tried replacing 120 × 120 pixel image patches with superpixels that are obtained by the efficient SLIC algorithm [49]. Similar to image patches, we calculated color and texture features from all superpixels from all images and built our visual vocabulary by k-means clustering. Figure 6 shows the closest 6 superpixels to cluster centers.
Fig. 6.
Some superpixel clusters as visual words from a dictionary of superpixels. Most superpixel clusters can be named by expert pathologists although they are discovered through unsupervised methods. Some of the superpixel clusters as identified by pathologists: a empty space (e.g., areas on the slide with no tissue), b loose stroma, c stroma, d blood cells, e epithelial nuclei, and f abnormal epithelial nuclei
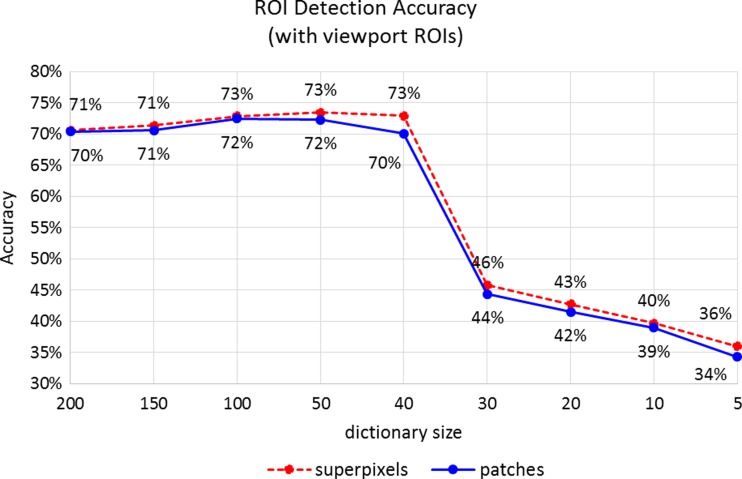
Superpixel segmentation is formulated as an optimization problem that is computationally expensive to calculate. Using superpixels instead of square patches did not improve diagnostically relevant ROI detection significantly. Figures 7 and 8 give a comparison of ROI classification accuracy for superpixel-based visual words and square patch visual words.
Fig. 7.
Classification accuracies with different-sized visual dictionaries and different representations of visual words. The accuracies obtained by tenfold cross-validation experiments using ROIs extracted through viewport analysis of three expert pathologists on 240 digital slides
Fig. 8.
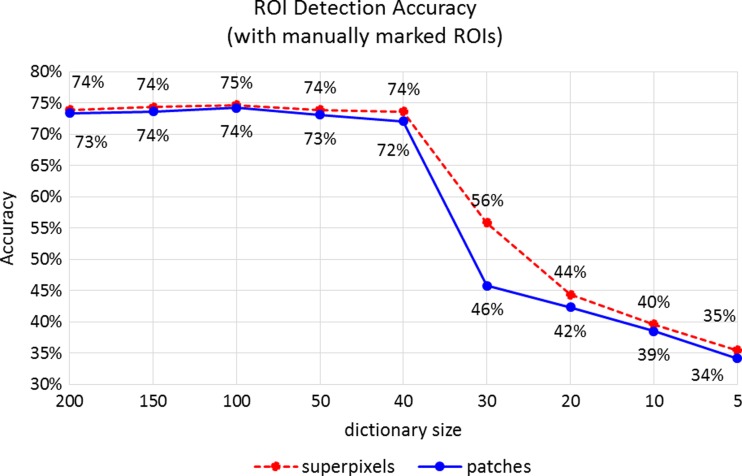
Classification accuracies with different-sized visual dictionaries and different representations of visual words. The accuracies obtained by tenfold cross-validation experiments using manually marked ROIs as training and viewport-extracted ROIs as test data
Training Using Manually Marked ROIs
The viewport analysis produces a set of ROIs that are potentially diagnostically relevant even though not included in the diagnostic ROI that is drawn by the pathologist. These areas include those that are zoomed in, slowly panned around, or fixated by pathologists with the intention of detailed assessment of these regions on the slide. However, due to the nature of viewing software or human factors, some of these areas are incorrectly depicted as diagnostically relevant because their zoom, duration, or displacement characteristics are matched to our criteria. This situation introduces noise in training data by labeling some negative samples as positive. We retrained our model by using the consensus ROI for each case that are agreed upon by three experts and show diagnostic features specific to the diagnosis of the slide as training data. Although comparatively very small and very expensive to collect, hand-drawn ROIs provide very controlled training data but increase detection accuracy very little. Figure 8 shows that the classification accuracy for manually marked ROIs is only slightly higher to that of viewport ROIs as shown in Fig. 7.
Comparison of Computer-Generated Regions to Human Viewport Regions
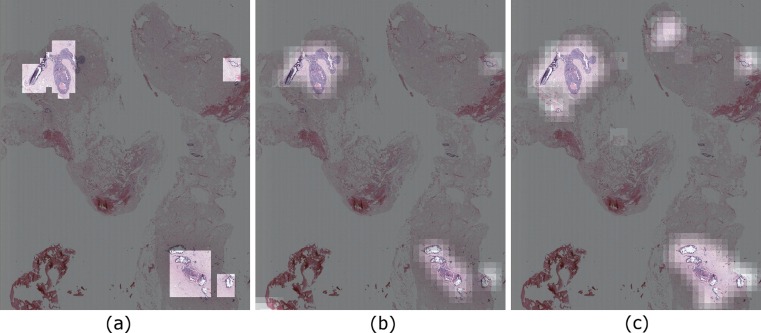
We evaluated our ROI detection framework in a classification setting where each instance is an image region extracted by the sliding window approach. In addition to these quantitative evaluations, we produced probability maps that show the regions detected as ROIs by the computer. Figure 9 shows a comparison of viewport-extracted ROIs (ground truth) and predictions of the two different models. A visual evaluation reveals that our detection accuracy is affected by the rectangular ground truth regions, but in fact our system is able to capture most of the areas the pathologist focused on.
Fig. 9.
a Ground truth calculated by analyzing the viewport logs for a case. b Probability map showing the predictions made by using manually marked ROIs as training data and image patches as visual words. c Probability map showing the predictions made by using viewport-extracted ROIs as training data and image patches as visual words
Discussion
Whole slide digital images provide researchers with an unparalleled opportunity to study the diagnostic process of pathologists. This work presents a simple yet important first step in understanding the visual scanning and interpretation process of pathologists. In the “Viewport Analysis” section, we introduced a novel representation and analysis of the pathologists’ behavior as they viewed and interpreted the digital slides. By defining three distinct behaviors, we can extract diagnostically important areas from the whole slide images. These areas include not only the final diagnostic ROIs that support the diagnosis but also the distracting areas that pathologists may focus attention on during the interpretation process.
The other contribution of this paper is an image analysis approach to understand the visual characteristics of ROIs that attract pathologists’ attention. We used a visual bag-of-words model to represent diagnostically important regions as a collection of small image patches. In classification experiments, we were able to detect ROIs from unseen images with 75 % accuracy. Further analyzing the dictionary size, we were able to identify the important visual words for detecting diagnostically important ROIs.
In additional experiments, we analyzed the model with different-sized visual vocabularies. The dictionary size does not have an impact on the accuracy as long as the dictionary is large enough to include basic building blocks of tissue images. Since breast histopathology images have less variability in comparison to everyday images, the dictionary size needed for a high detection accuracy is around 40 words—much smaller than general computer vision practices for the bag-of-words model. We also discovered that the words representing the epithelial cells are the most important words in representation of ROIs. When the dictionary size is decreased to 30 words where hierarchical clustering merges all epithelial cell clusters to others, the accuracy drops significantly. This is very intuitive since the breast cancer presents diagnostic features especially around epithelial structures of the tissue such as breast ducts and lobules.
We also experimented with a different visual word definition, superpixels. Using superpixels instead of square patches does not increase classification accuracy significantly. Furthermore, superpixels are computationally very expensive and slow in comparison to simple square patches.
A factor in our evaluation that should be considered is the nature of viewport-extracted ROIs. Because the tracking software records the portions of the digital slide visible on the screen, the viewports are always rectangular. Although this simple data collection allowed us to obtain a large dataset that is unique in the field, it has its shortcomings. In lower resolutions that correspond to small zoom levels, the viewports include a lot of surrounding uninteresting tissue (like background white space or tissue stroma) but there is no way to understand, outside of eye tracking, where the pathologist actually focused in these rectangular image regions. Our predictions, on the other hand, can be quite precise in ROI shapes.
Conclusions
With the increasing integration of digital slides into education, research, and clinical practice, the localization of ROIs is even more important. In this work, we explored the use of detailed tracking data in localization of ROIs. This study is a step toward developing computer-aided diagnosis tools with which an automated system may help pathologists locate diagnostically important regions and improve their performance.
We showed that image characteristics of specific regions on digital slides attract the attention of the pathologists, and basic image features, such as color and texture, are very useful in identifying these regions. We applied the bag-of-words model to predict diagnostically relevant regions in unseen whole slide images and achieved a 75 % detection accuracy. Our analysis of the viewport logs is novel and extracts the regions on which the pathologists focused during their diagnostic review process. This analysis enabled us to use a large dataset that consists of interpretations of three expert pathologists on 240 whole slide images.
This study is a first step in understanding the diagnostic process and may contribute to understanding how errors are made by pathologists when screening slides. In future work, we intend to analyze scanning behavior with the help of image analysis techniques and uncover the reasons underlying misdiagnosis.
Acknowledgments
The research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award numbers R01 CA172343, R01 CA140560, and KO5 CA104699. The content is solely the responsibility of the authors and does not necessarily represent the views of the National Cancer Institute or the National Institutes of Health. The authors wish to thank Ventana Medical Systems, Inc., a member of the Roche Group, for the use of iScan Coreo Au™ whole slide imaging system and HD View SL for the source code used to build our digital viewer. For a full description of HD View SL, please see http://hdviewsl.codeplex.com/. Selim Aksoy is supported in part by the Scientific and Technological Research Council of Turkey Grant 113E602.
Compliance with Ethical Standards
The study was approved by the institutional review boards at Dartmouth College, Fred Hutchinson Cancer Research Center, Providence Health and Services Oregon, University of Vermont, University of Washington, and Bilkent University. Informed consent was obtained electronically from pathologists.
References
- 1.Brunyé TT, Carney PA, Allison KH, Shapiro LG, Weaver DL, Elmore JG: Eye Movements as an Index of Pathologist Visual Expertise: A Pilot Study. van Diest PJ, ed. PLoS One 98: e103447, 2014 [DOI] [PMC free article] [PubMed]
- 2.Lesgold A, Rubinson H, Feltovich P, Glaser R, Klopfer D, Wang Y: Expertise in a complex skill: Diagnosing x-ray pictures. Nat Exp 311–342: 1988
- 3.Vink JP, Van Leeuwen MB, Van Deurzen CHM, De Haan G. Efficient nucleus detector in histopathology images. J Microsc. 2013;2492:124–135. doi: 10.1111/jmi.12001. [DOI] [PubMed] [Google Scholar]
- 4.Cireşan DC, Giusti A, Gambardella LM, Schmidhuber J: Mitosis detection in breast cancer histology images with deep neural networks. Lecture Notes in Computer Science (including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), 8150 LNCS., 411–418, 2013 [DOI] [PubMed]
- 5.Irshad H, Jalali S, Roux L, Racoceanu D, Hwee LJ, Le NG, Capron F. Automated mitosis detection using texture, SIFT features and HMAX biologically inspired approach. J Pathol Inform. 2013;4(Suppl):S12. doi: 10.4103/2153-3539.109870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Irshad H, Roux L, Racoceanu D: Multi-channels statistical and morphological features based mitosis detection in breast cancer histopathology. Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS, 6091–6094, 2013 [DOI] [PubMed]
- 7.Wan T, Liu X, Chen J, Qin Z: Wavelet-based statistical features for distinguishing mitotic and non-mitotic cells in breast cancer histopathology. Image Processing (ICIP), 2014 I.E. International Conference on, 2290–2294, 2014
- 8.Chekkoury A, Khurd P, Ni J, Bahlmann C, Kamen A, Patel A, Grady L, Singh M, Groher M, Navab N, Krupinski E, Johnson J, Graham A, Weinstein R: Automated malignancy detection in breast histopathological images. Pelc NJ, Haynor DR, van Ginneken B, Holmes III DR, Abbey CK, Boonn W, Bosch JG, Doyley MM, Liu BJ, Mello-Thoms CR, Wong KH, Novak CL, Ourselin S, Nishikawa RM, Whiting BR, eds., SPIE Medical Imaging, 831515–831515 - 13, 2012
- 9.DiFranco MD, O’Hurley G, Kay EW, Watson RWG, Cunningham P. Ensemble based system for whole-slide prostate cancer probability mapping using color texture features. Comput Med Imaging Graph. 2011;357–8:629–645. doi: 10.1016/j.compmedimag.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 10.Dong F, Irshad H, Oh E-Y, Lerwill MF, Brachtel EF, Jones NC, Knoblauch NW, Montaser-Kouhsari L, Johnson NB, Rao LKF, Faulkner-Jones B, Wilbur DC, Schnitt SJ, Beck AH. Computational Pathology to Discriminate Benign from Malignant Intraductal Proliferations of the Breast. PLoS One. 2014;912 doi: 10.1371/journal.pone.0114885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doyle S, Agner S, Madabhushi A, Feldman M, Tomaszewski J: Automated grading of breast cancer histopathology using spectral clustering with textural and architectural image features. 2008 5th IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Proceedings, ISBI, 496–499, 2008
- 12.Doyle S, Feldman M, Tomaszewski J, Madabhushi A. A boosted Bayesian multiresolution classifier for prostate cancer detection from digitized needle biopsies. IEEE Trans Biomed Eng. 2012;595:1205–1218. doi: 10.1109/TBME.2010.2053540. [DOI] [PubMed] [Google Scholar]
- 13.Jafari-Khouzani K, Soltanian-Zadeh H. Multiwavelet grading of pathological images of prostate. IEEE Trans Biomed Eng. 2003;506:697–704. doi: 10.1109/TBME.2003.812194. [DOI] [PubMed] [Google Scholar]
- 14.Khurd P, Grady L, Kamen A, Gibbs-Strauss S, Genega EM, Frangioni J V.: Network cycle features: Application to computer-aided Gleason grading of prostate cancer histopathological images. Proceedings - International Symposium on Biomedical Imaging, 1632–1636, 2011 [DOI] [PMC free article] [PubMed]
- 15.Kong J, Shimada H, Boyer K, Saltz J, Gurcan M: Image analysis for automated assessment of grade of neuroblastic differentiation. 2007 4th IEEE International Symposium on Biomedical Imaging: From Nano to Macro - Proceedings, 61–64, 2007
- 16.Kong J, Sertel O, Shimada H, Boyer KL, Saltz JH, Gurcan MN. Computer-aided evaluation of neuroblastoma on whole-slide histology images: Classifying grade of neuroblastic differentiation. Pattern Recognit. 2009;426:1080–1092. doi: 10.1016/j.patcog.2008.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sertel O, Kong J, Catalyurek UV, Lozanski G, Saltz JH, Gurcan MN. Histopathological image analysis using model-based intermediate representations and color texture: Follicular lymphoma grading. J Signal Process Syst. 2009;551–3:169–183. doi: 10.1007/s11265-008-0201-y. [DOI] [Google Scholar]
- 18.Basavanhally A, Ganesan S, Feldman M, Shih N, Mies C, Tomaszewski J, Madabhushi A. Multi-Field-of-View Framework for Distinguishing Tumor Grade in ER #002B; Breast Cancer from Entire Histopathology Slides. Biomed Eng IEEE Trans. 2013;608:2089–2099. doi: 10.1109/TBME.2013.2245129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beck AH, Sangoi AR, Leung S, Marinelli RJ, Nielsen TO, van de Vijver MJ, West RB, van de Rijn M, Koller D. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci Transl Med. 2011;3108:108ra113. doi: 10.1126/scitranslmed.3002564. [DOI] [PubMed] [Google Scholar]
- 20.Cooper LAD, Kong J, Gutman DA, Dunn WD, Nalisnik M, Brat DJ. Novel genotype-phenotype associations in human cancers enabled by advanced molecular platforms and computational analysis of whole slide images. Lab Invest. 2015;954:366–376. doi: 10.1038/labinvest.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuchs TJ, Wild PJ, Moch H, Buhmann JM. Computational pathology analysis of tissue microarrays predicts survival of renal clear cell carcinoma patients. Med Image Comput Comput Assist Interv. 2008;11(Pt 2):1–8. doi: 10.1007/978-3-540-85990-1_1. [DOI] [PubMed] [Google Scholar]
- 22.Kong J, Cooper LAD, Wang F, Gao J, Teodoro G, Scarpace L, Mikkelsen T, Schniederjan MJ, Moreno CS, Saltz JH, Brat DJ: Machine-based morphologic analysis of glioblastoma using whole-slide pathology images uncovers clinically relevant molecular correlates. PLoS One. 811: 2013 [DOI] [PMC free article] [PubMed]
- 23.Chang H, Fontenay GV, Han J, Cong G, Baehner FL, Gray JW, Spellman PT, Parvin B. Morphometic analysis of TCGA glioblastoma multiforme. BMC Bioinforma. 2011;121:484. doi: 10.1186/1471-2105-12-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bahlmann C, Patel A, Johnson J, Chekkoury A, Khurd P, Kamen A, Grady L, Ni J, Krupinski E, Graham A, Weinstein R: Automated detection of diagnostically relevant regions in H&E stained digital pathology slides. Prog Biomed Opt Imaging - Proc SPIE 8315: 2012
- 25.Gutiérrez R, Gómez F, Roa-Peña L, Romero E: A supervised visual model for finding regions of interest in basal cell carcinoma images. Diagn Pathol 626, 2011 [DOI] [PMC free article] [PubMed]
- 26.Huang CH, Veillard A, Roux L, Loménie N, Racoceanu D. Time-efficient sparse analysis of histopathological whole slide images. Comput Med Imaging Graph. 2011;357–8:579–591. doi: 10.1016/j.compmedimag.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 27.Romo D, Romero E, González F. Learning regions of interest from low level maps in virtual microscopy. Diagn Pathol. 2011;6(Suppl 1):S22. doi: 10.1186/1746-1596-6-S1-S22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mercan E, Aksoy S, Shapiro LG, Weaver DL, Brunye T, Elmore JG: Localization of Diagnostically Relevant Regions of Interest in Whole Slide Images. Pattern Recognit (ICPR), 2014 22nd Int Conf 1179–1184, 2014
- 29.Kothari S, Phan JH, Young AN, Wang MD: Histological image feature mining reveals emergent diagnostic properties for renal cancer. Proceedings - 2011 I.E. International Conference on Bioinformatics and Biomedicine, BIBM 2011, 422–425, 2011 [DOI] [PMC free article] [PubMed]
- 30.Tabesh A, Teverovskiy M, Pang HY, Kumar VP, Verbel D, Kotsianti A, Saidi O. Multifeature prostate cancer diagnosis and gleason grading of histological images. IEEE Trans Med Imaging. 2007;2610:1366–1378. doi: 10.1109/TMI.2007.898536. [DOI] [PubMed] [Google Scholar]
- 31.Gunduz-Demir C, Kandemir M, Tosun AB, Sokmensuer C. Automatic segmentation of colon glands using object-graphs. Med Image Anal. 2010;141:1–12. doi: 10.1016/j.media.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 32.Yuan Y, Failmezger H, Rueda OM, Ali HR, Gräf S, Chin S-F, Schwarz RF, Curtis C, Dunning MJ, Bardwell H, Johnson N, Doyle S, Turashvili G, Provenzano E, Aparicio S, Caldas C, Markowetz F. Quantitative image analysis of cellular heterogeneity in breast tumors complements genomic profiling. Sci Transl Med. 2012;4157:157ra143. doi: 10.1126/scitranslmed.3004330. [DOI] [PubMed] [Google Scholar]
- 33.Lu C, Mahmood M, Jha N, Mandal M. Automated segmentation of the melanocytes in skin histopathological images. IEEE J Biomed Heal Inform. 2013;172:284–296. doi: 10.1109/TITB.2012.2199595. [DOI] [PubMed] [Google Scholar]
- 34.Martins F, de Santiago I, Trinh A, Xian J, Guo A, Sayal K, Jimenez-Linan M, Deen S, Driver K, Mack M, Aslop J, Pharoah PD, Markowetz F, Brenton JD: Combined image and genomic analysis of high-grade serous ovarian cancer reveals PTEN loss as a common driver event and prognostic classifier. Genome Biol 1512: 2014 [DOI] [PMC free article] [PubMed]
- 35.Naik S, Doyle S, Agner S, Madabhushi A, Feldman M, Tomaszewski J: Automated gland and nuclei segmentation for grading of prostate and breast cancer histopathology. 2008 5th IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Proceedings, ISBI, 284–287, 2008
- 36.Mokhtari M, Rezaeian M, Gharibzadeh S, Malekian V: Computer aided measurement of melanoma depth of invasion in microscopic images. Micron 6140–48: 2014 [DOI] [PubMed]
- 37.Lu C, Mandal M: Automated segmentation and analysis of the epidermis area in skin histopathological images. Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS 5355–5359, 2012 [DOI] [PubMed]
- 38.Itti L, Koch C. Computational modelling of visual attention. Nat Rev Neurosci. 2001;23:194–203. doi: 10.1038/35058500. [DOI] [PubMed] [Google Scholar]
- 39.He DC, Wang L. Texture unit, texture spectrum, and texture analysis. IEEE Trans Geosci Remote Sens. 1990;284:509–512. [Google Scholar]
- 40.Tamura H, Mori S, Yamawaki T: Textural Features Corresponding to Visual Perception. IEEE Trans Syst Man Cybern 86: 1978
- 41.Oster NV, Carney PA, Allison KH, Weaver DL, Reisch LM, Longton G, Onega T, Pepe M, Geller BM, Nelson HD, Ross TR, Tosteson ANA, Elmore JG. Development of a diagnostic test set to assess agreement in breast pathology: practical application of the Guidelines for Reporting Reliability and Agreement Studies (GRRAS) BMC Womens Health. 2013;131:3. doi: 10.1186/1472-6874-13-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng S, Weaver D, Carney P, Reisch L, Geller B, Goodwin A, Rendi M, Onega T, Allison K, Tosteson A, Nelson H, Longton G, Pepe M, Elmore J. A Framework for Evaluating Diagnostic Discordance in Pathology Discovered During Research Studies. Arch Pathol Lab Med. 2014;1387:955–961. doi: 10.5858/arpa.2013-0263-OA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allison KH, Reisch LM, Carney PA, Weaver DL, Schnitt SJ, O’Malley FP, Geller BM, Elmore JG. Understanding diagnostic variability in breast pathology: Lessons learned from an expert consensus review panel. Histopathology. 2014;652:240–251. doi: 10.1111/his.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elmore JG, Longton GM, Carney PA, Geller BM, Onega T, Tosteson ANA, Nelson HD, Pepe MS, Allison KH, Schnitt SJ, O’Malley FP, Weaver DL. Diagnostic concordance among pathologists interpreting breast biopsy specimens. JAMA. 2015;31311:1122–1132. doi: 10.1001/jama.2015.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sivic J, Zisserman A. Efficient visual search of videos cast as text retrieval. IEEE Trans Pattern Anal Mach Intell. 2009;314:591–606. doi: 10.1109/TPAMI.2008.111. [DOI] [PubMed] [Google Scholar]
- 46.Ruifrok AC, Johnston DA. Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol. 2001;234:291–299. [PubMed] [Google Scholar]
- 47.Ren X, Malik J: Learning a classification model for segmentation. Proc Ninth IEEE Int Conf Comput Vis 2003
- 48.Bejnordi BE, Litjens G, Hermsen M, Karssemeijer N, van der Laak JAWM: A multi-scale superpixel classification approach to the detection of regions of interest in whole slide histopathology images. 9420., 94200H - 94200H - 6, 2015
- 49.Achanta R, Shaji A, Smith K, Lucchi A, Fua P, Süsstrunk S. SLIC superpixels compared to state-of-the-art superpixel methods. IEEE Trans Pattern Anal Mach Intell. 2012;3411:2274–2281. doi: 10.1109/TPAMI.2012.120. [DOI] [PubMed] [Google Scholar]