Abstract
Orange-red fluorescent proteins (FPs) are widely used in biomedical research for multiplexed epifluorescence microscopy with GFP-based probes, but their different excitation requirements make multiplexing with new advanced microscopy methods difficult. Separately, orange-red FPs are useful for deep-tissue imaging in mammals due to the relative tissue transmissibility of orange-red light, but their dependence on illumination limits their sensitivity as reporters in deep tissues. Here we describe CyOFP1, a bright engineered orange-red FP that is excitable by cyan light. We show that CyOFP1 enables single-excitation multiplexed imaging with GFP-based probes in single-photon and two-photon microscopy, including time-lapse imaging in light-sheet systems. CyOFP1 also serves as an efficient acceptor for resonance energy transfer from the highly catalytic blue-emitting luciferase NanoLuc. An optimized fusion of CyOFP1 and NanoLuc, called Antares, functions as a highly sensitive bioluminescent reporter in vivo, producing substantially brighter signals from deep tissues than firefly luciferase and other bioluminescent proteins.
INTRODUCTION
Fluorescent proteins (FPs) are useful in living cells to report gene expression, cell localization, protein levels and locations, and biochemical activities1. When using FPs, researchers often need to simultaneously image multiple channels, either to assay multiple biological events or to use one fluorophore as a reference in quantitative studies2. In one-photon imaging, this is routinely accomplished by exciting multiple FPs with multiple excitation wavelengths. However, in the case of multi-photon microscopy, due to the expense of adding a second Ti-Sapphire laser or an optical parametric oscillator (OPO) and the complexity of controlling them together, simultaneous imaging of multiple FPs is uncommon. An easier way to visualize multiple events in multi-photon microscopy may be to use fluorophores with large Stokes shifts, so that one excitation wavelength can be used to excite two FPs that emit in different wavelength regions3. Given the large number of existing GFP-based reporters, an orange-red FP that is bright and well excited at GFP excitation wavelengths would be especially useful. However, existing large-Stokes-shift (LSS) orange-red FPs all show low brightness with quantum yields below 50%4.
A bright LSS orange-red FP would also facilitate the application of recently developed microscopy methods to simultaneously visualize two biological processes. These methods include structured illumination for super-resolution imaging in thin specimens5,6, adaptive optics for enhancing resolution through scattering tissue7–9, random-access two-photon microscopy for increasing speed10, and light sheets generated by Bessel beams11–13, optical lattices14, or incoherent extended focusing15,16 for increasing speed and reducing phototoxicity and photobleaching. Using two excitation wavelengths with these microscopy methods is possible, but present technical challenges. Besides being expensive to implement, different excitation wavelengths produce differently sized excitation volumes for each fluorophore in two-photon microscopy and different illumination patterns in structured illumination, optical lattice, or Bessel-beam microscopy. Thus, for visualization of two events using newly developed advanced microscopy methods, an orange-red FP that can be excited simultaneously with but detected separately from GFP would be highly desirable.
A different optical imaging modality, bioluminescence imaging (BLI), could also benefit from the development of a bright LSS orange-red FP. In animal preclinical studies, BLI with non-secreted luciferases offers a relatively inexpensive and simple method for in vivo tracking of cells, for example to assess stem cell survival or tumor growth17. For in vivo imaging, luciferases with emission above 600 nm are preferred, as red photons escape absorbance by hemoglobin and exhibit longer mean free paths than bluer photons, thereby enabling deeper imaging in tissue18. Firefly luciferase (FLuc), the first BLI reporter19 and still the most commonly used reporter for intravital imaging in rodents, has peak emission of 612 nm at 37 ºC and a bioluminescence quantum yield of 0.41, but very low catalysis rates of 1.6 reactions per second18,20,21. FLuc has been mutated for even redder emission and for higher specific activity in vitro, but comparisons of these new mutants to wild- type FLuc have found them to perform worse in cells and animals22–24. Variants of the related click beetle luciferase also produce fewer photons in vivo than codon-optimized FLuc22. Among other non-secreted luciferases, Renilla luciferase (RLuc) has higher catalytic activity, but emits natively at wavelengths below 500 nm and with lower bioluminescent quantum yield25,26. RLuc has been engineered to emit at up to 550 nm27, but these wavelengths are still efficiently absorbed by hemoglobin. NanoLuc, a recently engineered version of a luciferase from the shrimp Oplophorus, has ~100x faster catalysis than FLuc28, but its output is even bluer, peaking at 460 nm, and it has not been demonstrated to improve detectability in vivo versus FLuc29. Thus, no existing luciferase combines high enzymatic turnover, high brightness and a red-shifted emission spectrum to generate high red output.
An alternative to luciferase engineering for improving BLI performance is bioluminescence resonance energy transfer (BRET). In BRET, the energy of the excited electron in the initial reaction product of the luciferase reaction is transferred to an acceptor chromophore. Like fluorescence resonance energy transfer, the efficiency of this process depends on the relative distance and orientation of donor and acceptor dipoles and energetic overlap between donor and acceptor excited states. If the acceptor is a fluorescent protein with a more red-shifted emission and higher quantum yield (QY), then BRET results in the generation of redder photons at higher efficiency. Indeed, this phenomena is used in nature; RLuc naturally binds to Renilla GFP, enabling BRET and thereby increasing emission red-shift and intensity30. Fusion to yellow or red FPs also intensifies or red-shifts the emission of RLuc31,32. However, BRET systems have not yet been shown to improve the sensitivity of BLI in vivo beyond FLuc. We considered the possibility of building a BRET system using the blue-emitting NanoLuc luciferase. A high-QY LSS orange-red FP would make a desirable BRET acceptor for NanoLuc, as the combined system could feature the high enzymatic turnover of NanoLuc with the high QY and red-shifted emission of the FP.
Here we present a bright cyan-excitable orange-red FP, discover an unique basis for its large Stokes shift, demonstrate its usefulness in two-photon and light-sheet microscopy, and use it to develop a bioluminescent enzyme that allows imaging from deep tissues in living mice with significantly higher sensitivity than firefly luciferase.
RESULTS
Development of a cyan-excitable orange fluorescent protein
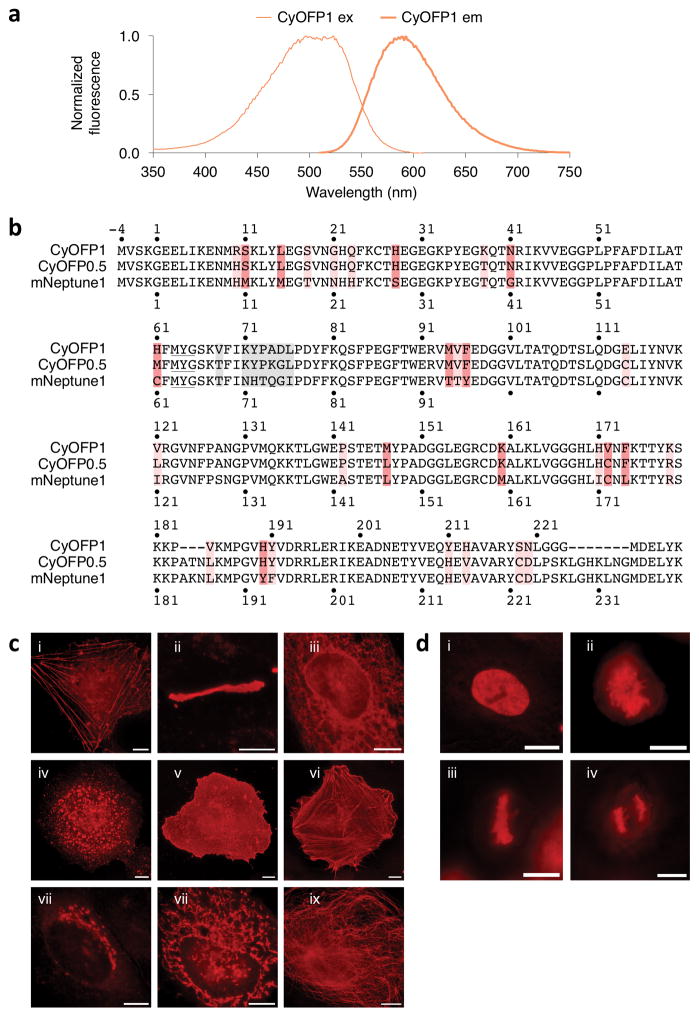
Through semi-rational mutagenesis of the far-red FP mNeptune2 and library screening in bacterial colonies33, we obtained an extremely bright cyan-excitable orange-red fluorescent protein which we named CyOFP1 (Fig. 1a). CyOFP1 differs from mNeptune2 by 33 mutations and 2 deletions (Fig. 1b, Supplementary Table 1). CyOFP1 has a broad excitation spectrum with wavelengths from 488 to 526 nm exciting with > 95% of peak efficiency. This is distinct from standard orange-red FPs, which are maximally excited in the 550–610 nm range. CyOFP1 has peak emission of 589 nm and an average emission wavelength of 595 nm. As 620 nm is considered the orange/red boundary34, it is best described as being orange35, although other FPs with similar or bluer emissions, such as tdTomato and DsRed, are often called red FPs (Supplementary Fig. 1a).
Figure 1.
Development of CyOFP1, a cyan-excitable red fluorescent protein. (a) Normalized excitation and emission spectra of CyOFP1. (b) Sequence alignment of CyOFP1 and mNeptune1. Amino acids forming the chromophore are underlined. Interior mutations are in red and outer barrel mutations are colored pink. Mutations in the upper loop (when the barrel is oriented with termini pointing upwards) are colored gray. Amino acid numbering begins at −4 so that homologous sequences are numbered as in PDB file 3IP2 for the structure of Neptune. (c) Fluorescence images of HeLa CCL2 cells expressing CyOFP1 fused to various domains. For each fusion, the linker amino acid (aa) length is indicated in between the two domains, and the origin of the fusion partner and its normal subcellular location are indicated in parentheses. i, CyOFP1-18aa-actin (β-actin, actin cytoskeleton); ii, Cx43-7aa-CyOFP1 (rat α-1 connexin 43, gap junctions); iii, CytERM-17aa-CyOFP1 (rabbit cytochrome p450 aa1-29, endoplasmic reticulum); iv, CyOFP1-14aa-RhoB (human RhoB, endosomes); v, CyOFP1-5aa-CAAX (human c-Ha-Ras 20- aa farnesylation signal, plasma membrane); vi, Tractin-11aa-CyOFP1 (rat F-Tractin; actin cytoskeleton); vii, SiT-7aa-CyOFP1, human sialyltransferase aa1-45, Golgi apparatus); viii, COX8A-7aa-CyOFP1 (human cytochrome C oxidase subunit VIIIA; mitochondria); ix, CyOFP1-18aa-tubulin (human α-tubulin, microtubules). (d) Fluorescence images of CyOFP1-10aa-H2B (human histone 2B) in HeLa S3 cells; i, interphase; ii, prophase; iii, metaphase; iv, anaphase. Scale bars, 10 Mm.
CyOFP1 has several unique and desirable characteristics. In being excited by cyan light, CyOFP1 differs from previous LSS orange-red FPs, which are excited by more phototoxic blue light around 440 nm. CyOFP1 has a peak extinction coefficient of 40 mM−1cm−1 and a QY of 0.76. The QY of CyOFP1 is the highest characterized for an orange-red FP, even surpassing the 0.70 QY of DsRed36,37 (Table 1). CyOFP1 fluorescence lifetime was well fit to a single exponential with τ = 3.663 ± 0.002 ns (mean of 59 cells ± standard error of the mean, Supplementary Fig. 1b), also the longest reported lifetime for an orange-red FP. CyOFP1 exhibits high photostability under arc-lamp illumination in vitro (120 s compared to 150 s for mEGFP, Supplementary Fig. 1c). CyOFP1 matures with a maturation half-time of 15 min (Supplementary Fig. 1d), making it one of the fastest-maturing orange or red fluorescent proteins38, and its fluorescence is stable across a range of pH values, with a pKa of 5.5 (Supplementary Fig. 1e). CyOFP1 is monomeric at 0.1 μM (Supplementary Fig. 1f), shows excellent performance in various fusion proteins (Fig. 1c), and does not block mitotic progression when fused to histones (Fig. 1d).
Table 1.
Properties of CyOFP1 and other large-Stokes-shift orange-red fluorescent proteins
| CyOFP1 | mBeRFP | LSSmOrange | LSSmKate1 | LSSmKate2 | mKeima | |
|---|---|---|---|---|---|---|
| Excitation peak (nm) | 497, 523a | 446 | 437 | 463 | 460 | 440 |
| Emission peak (nm) | 589 | 611 | 572 | 624 | 605 | 620 |
| EC at peak (mM−1cm−1) | 40 | 65 | 52 | 31 | 26 | 13 |
| QY | 0.76 | 0.27 | 0.45 | 0.08 | 0.17 | 0.24 |
| Brightnessb | 31 | 18 | 19 | 2.5 | 4.4 | 3.2 |
| Lifetime (ns) | 3.6 | 2.0 | ND | ND | 1.4e | 1.8e |
| Photostability (s)c | 111 | ND | ND | ND | ND | ND |
| pKa | 5.5 | 5.6 | 5.7 | 3.2 | 2.7 | 6.5 |
| Maturation (min)d | 15 | 60 | 140 | 100 | 150 | 160e |
| Reference | this work | 4 | 47 | 39 | 39 | 48 |
EC, extinction coefficient; QY, quantum yield; ND, not determined.
Peaks at 497 and 523 nm differ in excitation efficiency by less than 0.3%.
Calculated as the product of EC at peak and QY.
Predicted time for fluorescence to photobleach by 50% under arc-lamp illumination from initial emission rate of 1000 photons per molecule per second. For EGFP, this time is 170 s.
Half-time to full maturation after exposure to normoxic conditions.
Values from ref. 4.
Structural basis of the large Stokes shift in CyOFP1
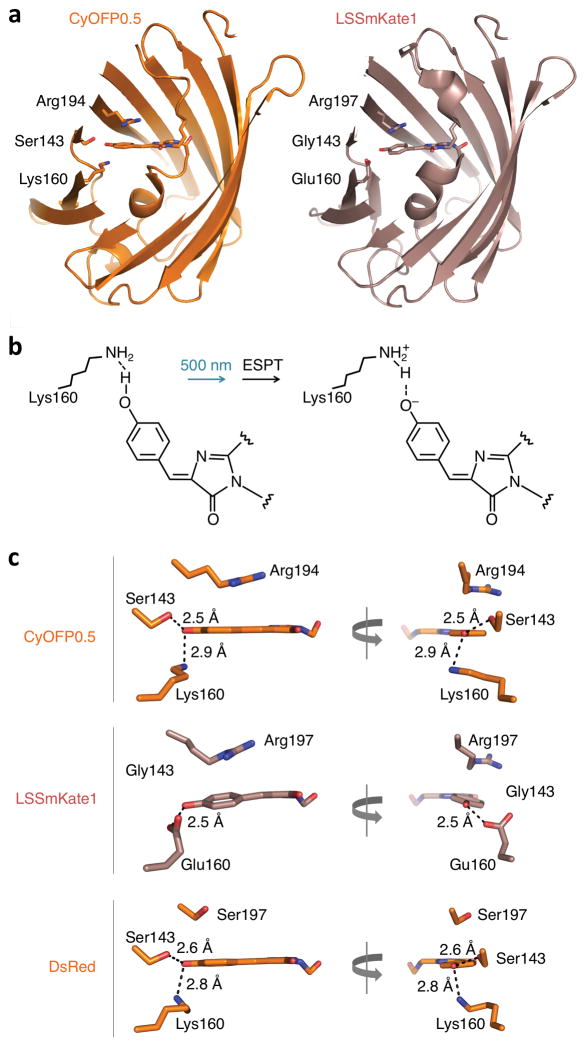
To understand the origin of the large Stokes shift and high QY of CyOFP1, we determined the atomic structure of CyOFP0.5 (Fig. 1b), an earlier variant with similar spectral characteristics. We obtained an X-ray crystal structure at 2.39 Å resolution with four chains per crystallographic unit (see Supplementary Table 2 for crystallographic data). As expected, the chromophore is similar to that of DsRed and tdTomato, and, unlike the mKO and mOrange family of orange FPs39, does not contain a ring in place of the acylimine group. One residue, Lys160, accounts for the large Stokes shift in CyOFP0.5 (Supplementary Fig. 2a). The structure revealed that Lys160 is located beneath the chromophore (when the barrel is oriented with termini pointing upwards), and its amino group is positioned to engage in a hydrogen bond interaction with the phenolic hydroxyl group of the chromophore (Fig. 2a–c). In chains A–C of the crystal, the Lys160 side chain is in an extended conformation with the amino group beneath the phenolic hydroxyl, with donor-acceptor distances of 2.7–3.0 Å, whereas in chain D, turns at the Cγ and Cδ atoms cause the amino group of Lys160 to face away from the phenolic hydroxyl group.
Figure 2.
Structural characterization of CyOFP0.5. (a) Cutaway view of CyOFP0.5 (left) and LSSmKate (right) with main chain in cartoon format and chromophores and their hydrogen-bonding partners in stick format. Chromophore and side-chain atoms are colored as carbon orange (CyOFP0.5) or pink (LSSmKate1), nitrogen blue, and oxygen red. In both proteins, the side chain of aa160 serves as a hydrogen bond donor and ESPT acceptor. (b) Proposed ESPT in CyOFP0.5. The proton from the phenolic hydroxyl group of the chromophore is transferred to the Lys160 after the chromophore excited by cyan light. Hydrogen bonds are represented as dashed black lines. (c) The CyOFP0.5 chromophore (top) is more coplanar than the LSSmKate1 chromophore (middle), and is engaged in two hydrogen bonds compared to one in LSSmKate1. The hydrogen bond partners of the CyOFP0.5 chromophore are similar to those of the DsRed chromphore (bottom).
The presence of a hydrogen bond between Lys160 and the chromophore phenol group suggests a mechanism for the large Stokes shift that, to our knowledge, has not previously been described. As the absorbance peak of CyOFP0.5 and CyOFP1.0 is blue-shifted by ~100 nm relative to its predecessor, and chromophore protonation causes similarly large absorbance blue-shifts in other FPs, the phenolic hydroxyl group is likely to exist during the ground state in the protonated neutral form. The protonated chromophore phenol group would then serve as a hydrogen bond donor to a neutral Lys160 side chain (Fig. 2b). Consistent with this, PROPKA calculated the pKa of Lys160 as 6.06–6.73 in chains A–C of the crystal, implying deprotonation of the Lys160 amino groups at neutral pH. Chromophore protonation at neutral pH is also supported by the appearance at high pH of a species with a red-shifted and sharper absorbance spectrum similar to mKate (Supplementary Fig. 2b). Starting from the doubly neutral state, excited state proton transfer (ESPT) from the chromophore to the Lys160 amino group (Fig. 2b) would explain the large Stokes shift of CyOFP0.5. ESPT also underlies large Stokes shifts in wild-type GFP and other LSS proteins (Supplementary Fig. 2c), but the use of lysine as an ESPT acceptor appears unique to the CyOFP family.
CyOFP0.5 shows substantial similarity to DsRed in the arrangement of hydrogen bond partners near the chromophore phenol group. Specifically, positions 143 and 160 are also Ser and Lys respectively in DsRed and mApple (Fig. 2c, Supplementary Fig. 2c). Indeed, following EPST, the chromophore of CyOFP0.5 would be predicted to be similar to those of DsRed and mApple in being anionic and hydrogen-bonded to Ser143 and Lys160 (Fig. 2b, Supplementary Fig. 2c). Thus, differences elsewhere in CyOFP0.5 compared to DsRed and mApple apparently modulate the pKa of the chromophore or Lys160 to favor the protonation of the chromophore over Lys160 at neutral pH specifically in CyOFP0.5.
The CyOFP0.5 structure also suggests several possible explanations for CyRFP1’s higher QY compared to other LSS orange-red FPs. The chromophore interactions of CyOFP0.5 are most similar to those of LSSmKate1, whose structure has also been characterized40. LSSmKate1 also contains an ESPT acceptor, a Glu side chain, at position 160, but the QY of LSSmKate1 is 0.08 versus 0.76 for CyRFP0.5 and CyRFP1.0. Compared to LSSmKate1, CyOFP0.5 shows three major differences (Fig. 2c). First, the CyOFP0.5 chromophore is more planar. Second, the CyOFP0.5 chromophore is more tightly packed on its top surface by Arg194 (homologous to Arg197 in LSSmKate1) due to a more planar conformation of the Arg side chain. Finally, the chromophore phenol group in CyOFP0.5 engages in hydrogen bonds with two amino acids, Ser143 and Lys160, as opposed to only Glu160 in LSSmKate1, due to a mutation of position 143 to Gly in LSSmKate1. Each of these differences would be expected to constrain chromophore motion and thus increase QY in CyOFP0.5 compared to LSSmKate1. In particular, hydrogen bonding to Ser143 and Lys160 may be a common mechanism for high QY in CyOFP0.5, mApple, and DsRed. As the above amino acids are preserved between CyOFP0.5 and CyOFP1.0, the same interactions presumably exist in CyOFP1.0 as well.
Single-excitation dual-emission imaging with CyOFP1 and GFP
CyOFP1’s spectral attributes make it ideal for single-excitation dual-emission imaging together with a GFP. As a demonstration, we imaged CyOFP1 and EGFP simultaneously with a single excitation window in a wide field set up (Supplementary Fig. 3a,b). We next tested CyOFP1 performance in single-excitation dual-emission microscopy. Specifically, we imaged MV3 melanoma cells expressing CyOFP1 fused to the actin marker tractin and cytosolic EGFP in a three-dimensional (3D) environment using axially swept light-sheet microscopy, exciting both fluorophores with a 488-nm excitation laser and collecting green and orange emissions on two high-speed cameras15. Simultaneous 3D imaging of EGFP and CyOFP1-tractin over time allowed us to ascertain the role of actin in the formation of non-apoptotic blebs (Supplementary Fig. 3c). We observed that these blebs formed initially as actin-free structures marked exclusively by cytosolic EGFP, but were later enriched for polymerized actin (Supplementary Movie 1). In addition, we observed that CyOFP1 retained the majority of its fluorescence for over 40 stacks, each consisting of 126 optical sections spaced 160 nm apart (Supplementary Fig. 3d). Thus CyOFP1 can be used in one-photon light-sheet excitation for simultaneous imaging with EGFP over long time courses.
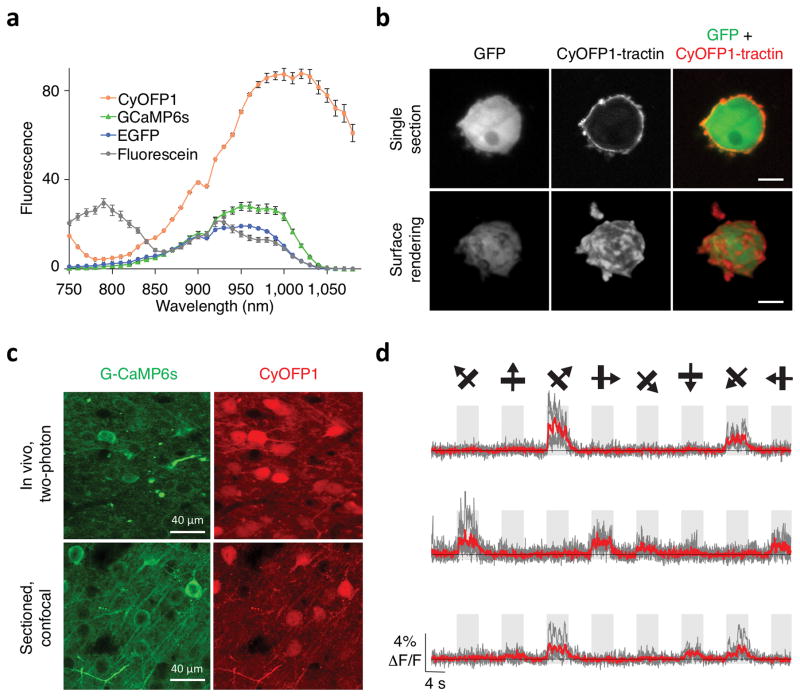
CyOFP1 may be especially useful together with GFP-based sensors for multi-photon imaging, where using multiple Ti-Sapphire lasers or adding an OPO to excite green and red fluorophores simultaneously is uncommon due to expense and complexity. CyOFP1 is well excited in two-photon mode with a broad peak between 900 and 1060 nm (Fig. 3a). At 920–940 nm, wavelengths commonly used to excite GFP-based probes41–43, CyOFP1 has higher brightness than EGFP or the GFP- based calcium sensor GCaMP6s (Fig. 3a). CyOFP1 and GFP-based probes can thus be simultaneously excited in two-photon mode using a single excitation wavelength and their emissions separated with appropriate filters.
Figure 3.
Simultaneous dual-emission two-photon imaging of CyOFP1 with GFP-based reporters. (a) Two- photon excitation spectra of CyOFP1, GCaMP6s, EGFP, and fluorescein (as reference standard). Intensity is presented as thousands of counts per s per μM protein at 1 mW excitation. Error bars are standard deviation. n = 3 for CyOFP1, 4 for GCaMP6s, 2 for EGFP, and 4 for fluorescein. (b) Single optical section (upper row) and surface rendering (lower row) of tractin-CyOFP1 and cytosolic EGFP in MV3 melanoma cells acquired by two-photon Bessel-beam light-sheet microscopy. CyOFP1 is localized to the cortex of the cell and small membrane protrusions. (c) Fluorescence images of CyOFP1 and GCaMP6s in layer-2/3 pyramidal neurons in mouse brain V1 cortex in a single optical section acquired by two-photon excitation at 940 nm. (d) GCaMP6s responses of three mouse neurons co- expressing CyOFP1 in response to drifting gratings. Single sweeps (grey) and averages of 5 sweeps (red) are overlaid. Directions of grating motion (8 directions) are shown above traces (arrows).
Two-photon excitation is advantageous for Bessel-beam light-sheet microscopy, where the nonlinearity of two-photon excitation attenuates the side lobes of the effective Bessel beam, reducing out- of-focus excitation11–13. Here, imaging of two fluorescent probes would be greatly simplified by the use of simultaneously excitable but distinctly emitting fluorophores. To test this, we imaged the MV3 cells coexpressing CyOFP1-tractin and cytoplasmic EGFP with two-photon Bessel-beam light-sheet microscopy13. We observed dynamic actin protrusions with enhanced resolution compared to one-photon excitation (Fig. 3b, Supplementary Movie 2). CyOFP1 again performs well in time-lapse fluorescence imaging, retaining most of its fluorescence over 25 stacks of 126 sections each (Supplementary Fig. 3e).
In the mouse brain, where GCaMP-family sensors are typically used with two-photon excitation to visualize neuronal activity in deep layers41–43, CyOFP1 could serve as an anatomical fill or a reference channel. To test this, we used a bicistronic adeno-associated virus to co-express CyOFP1 with GCaMP6s in mouse primary visual cortex in vivo. Upon two-photon excitation at 940 nm, CyOFP1 fluorescence could be seen to fill the neuronal cell bodies and dendritic processes without noticeable subcellular aggregation after 4 weeks of expression (Fig. 3c, Supplementary Movie 3). Somatic calcium transients of orientation-tuned neurons were simultaneously recorded with GCaMP6s, similar to previously reported studies41–43 (Fig. 3d). Thus CyOFP1 and GCaMP6s enable simultaneous two-photon imaging of cellular structure and calcium transients in living mouse brains using a single excitation wavelength.
Development of a bright orange-red bioluminescent protein
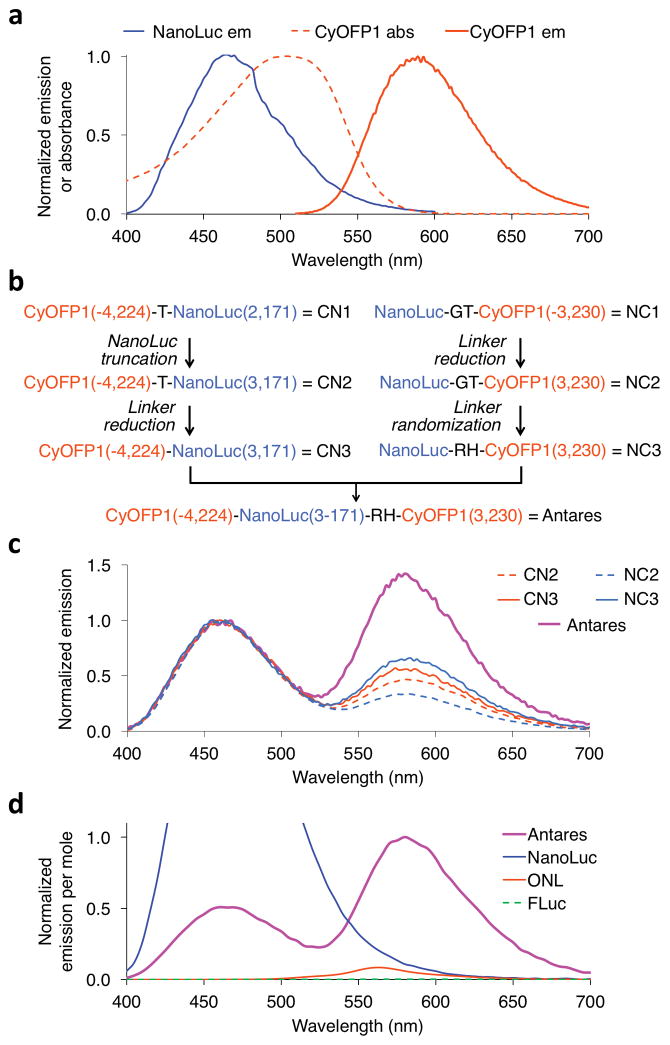
We noticed that CyOFP1 has ideal spectral characteristics to be a BRET acceptor for NanoLuc, potentially providing a way to improve in vivo BLI. The emission spectrum of NanoLuc overlaps substantially with the absorption spectrum of CyOFP1 (Fig. 4a), so if the two proteins are in close enough proximity, the excited-state energy of the oxidized reaction intermediate in the NanoLuc active site could be transferred to excite CyOFP1 by BRET, leading to emission with the spectrum and QY of CyOFP1. In addition to its unusually high QY of 0.76, nearly half of CyOFP1 emission occurs above 600 nm, which is highly desirable as absorbance and scattering of light in mammalian tissue decreases dramatically above 600 nm. We therefore hypothesized that we could create a fusion of CyOFP1 and NanoLuc that would effectively perform as a highly active red-emitting bioluminescent protein.
Figure 4.
Development of a BRET system with NanoLuc and CyOFP1. (a) NanoLuc and CyOFP1 are well matched for BRET, as the emission spectrum of NanoLuc overlaps the absorbance spectrum of CyOFP1. (b) Protein engineering flow chart leading to Antares. Numbers in parentheses refer to the first and last amino acid of the domain fragment contained in the fusion protein, with numbering as in Figure 1b. Linker sequences are indicated in single-letter amino acid code. Deletions of the C-terminus of NanoLuc yielded inactive enzyme and were not pursued further. (c) Emission spectra of the constructs depicted in b. Spectra were normalized to the NanoLuc emission peak at 460 nm. Higher 584-nm emission peaks indicate higher BRET efficiencies. (d) Comparison of emission spectra of purified Antares, NanoLuc, Orange Nano-lantern (ONL), and FLuc. Emissions from 100 μL of 2 nM protein immediately after addition of 20 μM substrate were measured, then normalized to peak emission of Antares.
To created and testesd a series of fusions proteins to optimize BRET between CyOFP1 and NanoLuc (Fig. 4b). We first optimized linkages for high BRET in CyOFP1-NanoLuc and NanoLuc-CyOFP1 fusions (Supplementary Fig. 4a–d). We then combined the fusions and found that BRET was further enhanced with CyOFP1 at both ends of NanoLuc (Fig. 4c). We named this final fusion protein Antares, after the binary star system that consists of a dim blue star and an exceptionally bright orange-red star.
We compared Antares in vitro to other bioluminescent reporters. First, Antares shows less blue and more red emission per enzyme molecule than NanoLuc, as expected with BRET (Fig. 4d, Table 2). Specifically, emissions of red photons > 600 nm were increased 20-fold over NanoLuc. Antares also produced > 500-fold more photons per molecule than FLuc across the spectrum, and > 430-fold more above 600 nm (Fig. 4d, Table 2). During the course of this work, blue and orange color variants of Nano- lantern, composed of RLuc fused with a fluorescent protein, were developed. Among the Nano-lanterns, Orange Nano-lantern (ONL) is the most similar to CyOFP1 in emission spectra. We hypothesized that the greater catalytic rate of NanoLuc with FRZ compared to RLuc with coelenterazine would allow Antares to produce light at a higher rate than ONL. Indeed, Antares produced 20-fold more photons across all wavelengths than ONL at equal protein and substrate concentrations, and 28-fold more above 600 nm (Fig. 4d, Table 2).
Table 2.
Relative in vitro photon outputs of Antares, NanoLuc, FLuc, and ONL
| Reporter | Total emission | Emission > 600 nm |
|---|---|---|
| Antares | 1 | 0.28 |
| NanoLuc | 1.7 | 0.014 |
| FLuc | 0.0018 | 0.00065 |
| ONL | 0.051 | 0.010 |
Values are normalized to Antares total emission.
Improving bioluminescence imaging in vivo with Antares
We systematically tested Antares in non-invasive deep-tissue BLI of gene expression using both cell-based and live-animal models. First, to identify the best-performing BLI reporters to compare with Antares, we performed a small screen of reporters in cells in a mouse tissue phantom with optical transmission and scattering properties similar to a living animal. As substrates are applied directly to cells in a tube before placement in the phantom, this method allowed us to test luciferases at equal concentrations of substrate and not be confounded by the uncertainty of substrate uptake in vivo. We included in this screen FLuc (expressed from the codon-optimized luc2 gene) as the most commonly used luciferase for in vivo mammalian imaging, yellow Nano-lantern as a BRET system with higher turnover and higher QY than FLuc, a fusion of RLuc8.6–535 and TurboFP635 (BRET6) as a BRET system with higher turnover and redder emission than FLuc31, and NanoLuc alone. For yellow Nano-lantern and BRET6, we tested a soluble formulation of coelenterazine (sCTZ) and ViviRen, a coelenterazine derivative with less auto-oxidation, both of which have been observed to yield larger BLI signals than standard coelenterazine formulations44,45. For NanoLuc and Antares, we tested sCTZ and furimazine (FRZ). Finally, we also tested ONL with sCTZ.
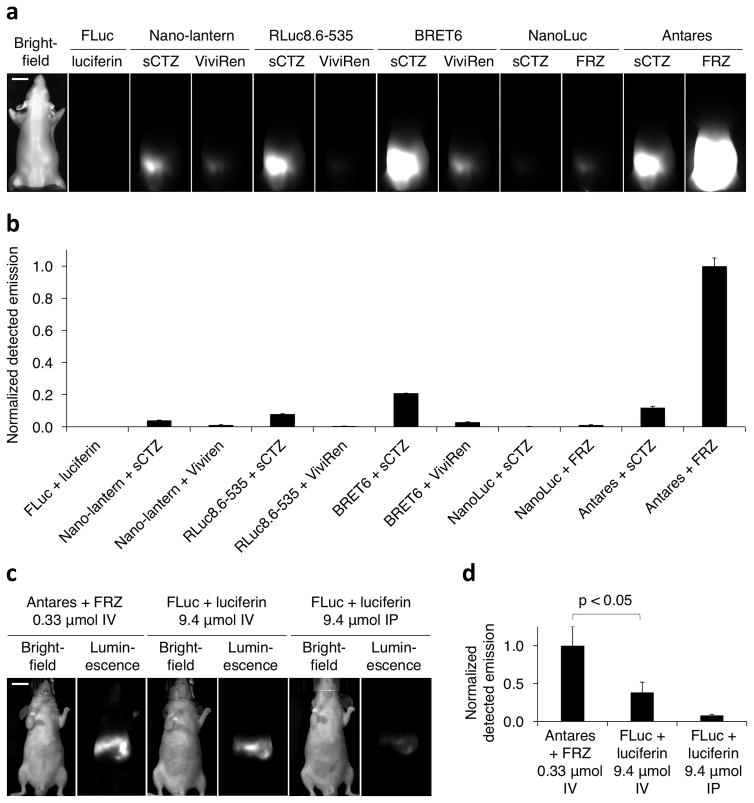
We observed that Antares with FRZ produced the most detectable emission of all luciferase-enzyme combinations tested in the mouse tissue phantom (Fig. 5a,b). Of note, Antares with FRZ was 4.8-fold brighter than the second-brightest reporter-substrate combination, BRET6 with sCTZ, and > 2000-fold brighter than FLuc with luciferin (Fig. 5a,b). Antares with sCTZ, which is utilized at 31-fold lower rates than FRZ by NanoLuc, produced fewer detectable photons than BRET6. However, NanoLuc catalyzes sCTZ oxidation as efficiently as RLuc in BRET6, so the superior performance of BRET6 could arise from longer emissions of TurboFP635 and/or different BRET efficiencies in the two constructs. Notably, NanoLuc alone with FRZ was 84-fold weaker than Antares with FRZ, and indeed was weaker than most other reporter-substrate combinations (Fig. 5a,b), demonstrating the beneficial effect on tissue penetration of shifting NanoLuc emission to redder wavelengths via BRET to CyOFP1. Lastly, Antares produced 22-fold higher detectable emissions than ONL in the mouse phantom (Supplementary Fig. 5a,b), consistent with the 28-fold higher production of red photons per enzyme molecule previously observed (Table 2).
Figure 5.
Antares is superior to other reporters for BLI in mice. (a) Cells expressing Antares produce larger detectable bioluminescence signal in mouse phantoms than cells expressing other reporters. Cells expressing the indicated reporters with the indicated substrates were injected into a mouse phantom at 0.7-cm depth, and images were acquired in an IVIS Spectrum for 1 s in bioluminescence mode. Although peak pixel intensity of Antares was 46325 counts, the displayed intensity range was set to 0–5000 to allow visual confirmation of signal from all reporter proteins. (b) Quantitation of cellular bioluminescence in phantom mice. Total counts were normalized to co-expressed CFP intensity and then normalized to mean counts from Antares with FRZ. The second-brightest reporter-substrate combination, BRET6 with sCTZ, produced 21% of the detectable emission of Antares with FRZ. Error bars represent standard error of the mean (SEM) from 3 replicate measurements. (c) In mice, Antares produces more detectable signal upon IV administration of 0.33 μmol FRZ than FLuc upon IV or IP administration of 9.4 μmol luciferin. (d) Quantification of bioluminescence in living mice. Total counts were normalized to mean counts from Antares with FRZ. Error bars represent SEM of measurements from multiple mice (n = 5 for Antares, n = 6 for FLuc with IV luciferin, n = 16 for FLuc with IP luciferin).
Although our experiments using a phantom mouse allowed comparison among BLI reporters expressed in cells, they do not account for differences in vivo in substrate availability. We thus also performed non-invasive BLI of each reporter protein expressed in living mice. We first compared emissions from Antares and FLuc expressed in the liver. To compare their highest obtainable signals, we injected maximum substrate amounts determined by solubility and injection volume limits, although these were much higher for D-luciferin than for FRZ. Nevertheless, emissions from Antares following IV administration of 0.33 μmol FRZ were 2.6-fold higher (p < 0.05 by t-test) than those from FLuc with 9.4 μmol luciferin injected IV. Of note, these Antares emissions were also 13-fold higher than those detected from FLuc in a standard BLI protocol of IP administration of 9.4 μmol luciferin (Fig. 5c,d). We also compared Antares with BRET6, as this was the second highest-performing bioluminescent reporter in the phantom mouse experiment. Antares with FRZ produced 3.8-fold and 12-fold more detectable signal than BRET6 with CTZ upon IV and IP administration of substrate, respectively (p = 0.0049 and 0.019 by t-test, Supplementary Fig. 5c,d). Taken together, these results demonstrate that Antares produces more detectable photons than FLuc or BRET6 when expressed in deep tissues of living mice.
DISCUSSION
In this study, we describe CyOFP1, an exceptionally bright orange FP with cyan excitability. CyOFP1 addresses long-standing issues in two distinct optical imaging modalities, optical sectioning microscopy and bioluminescence imaging. We found that CyOFP1 provides bright signals that can be co-excited with a GFP and its spectrally similar derivatives, but collected separately. We demonstrated that CyOFP1 and a GFP can be imaged simultaneously using advanced light-sheet and two-photon techniques without using two lasers or an OPO. We further found that CyOFP1 is a suitable acceptor for resonance energy transfer from NanoLuc, and engineered a fusion protein, Antares, that effectively couples the high substrate turnover of NanoLuc to the high quantum yield and orange-red emission of CyOFP1.
CyOFP1 is distinct from previous LSS FPs in its optical properties, with a less dramatic Stokes shift but with much higher brightness. The large Stokes shift in previously described LSS orange-red FPs is believed to involve ESPT from the protonated phenolic hydroxyl group of the chromophore to either Asp158 or Asp/Glu160, either directly or via a relay involving a serine side chain (Supplementary Fig. 3a)40. In CyOFP1, a neutral Lys side chain at position 160 appears to be the proton acceptor. As with other LSS FPs, CyOFP1 likely exhibits blue-shifted absorbance due to protonation of the chromophore in the ground state, and a large Stokes shift due to an EPST process. The redder absorbance of CyOFP1 compared to other LSS FPs and its smaller Stokes shift is indicative of differences in molecular orbital energies between the chromophores of CyOFP1 and the other LSS FPs. This could be due to differences in the positioning of the proton that undergoes ESPT; for example it could be less tightly bound to the phenol group in CyOFP1 than in other LSS orange-red FPs.
CyOFP1 meets a need for an orange-red fluorophore that can be co-excited with GFP. Recently developed microscopy methods such as random-access two-photon illumination, adaptive optics, Bessel-beam light-sheet microscopy, optical lattices, and incoherent extended focusing all use dynamic modulation of a laser to pattern illumination5–16. Three-photon excitation uses far-infrared laser illuimination to extend imaging depths46. All these techniques require custom hardware for the excitation beam, making simultaneous excitation at two different wavelengths cumbersome. Co-expressing CyOFP1 together with GFP would be an easier way to image two structures or biological events than implementing two excitation wavelengths. For example, in the brain, CyOFP1 can serve as the basis for a structural marker co-imaged with a GFP-based reporter of calcium, voltage, or neurotransmitter release. This marker can be used for motion correction, especially when using an indicator with a low fluorescence baseline like those of the GCaMP6 family, and for estimating the expression level of the reporter. In non-neuronal cells, CyOFP1 and GFP can be used to image two different structures, or two different cell populations. An additional benefit of truly simultaneous excitation of CyOFP1 and GFP markers is that temporal analysis does not need to adjust for any time shift in acquisition of different image channels.
The engineering of Antares, an orange-red bioluminescent reporter comprising a fusion of NanoLuc to CyOFP1 domains, addresses another long-standing need to improve the sensitivity of BLI in mammals beyond FLuc. FLuc has been used as a reporter gene in mammals for nearly two decades19, but no alternative natural or engineered bioluminescent protein had been found to be superior to FLuc in detectable output from mammalian tissue22–24. Our experiments in a mouse tissue phantom, the first to our knowledge to make any comparisons amongst FLuc, Nano-lantern, BRET6, and NanoLuc for red photon output, identify Antares as the reporter with the most red emission when expressed in mammalian cells. Antares also produced the highest levels of detectable emissions when expressed from deep locations in living mice. Indeed, our experiments indicate that Antares is capable of producing an order of magnitude more detectable signal than the most commonly used BLI protocol utilizing FLuc.
In summary, with its combination of cyan excitability and high brightness, CyOFP1 should be useful in a wide variety of non-linear and light-sheet microscopy applications. In addition, the red- emitting bioluminescent protein Antares created from NanoLuc and CyOFP1 substantially improves bioluminescence signals in vivo in mammals over firefly luciferase, and should be the BLI reporter of choice whenever the highest detection sensitivity is required.
ONLINE METHODS
Mutagenesis and screening of libraries
Mutations for specific residues were introduced by overlap-extension PCR. Mutants were expressed and screened in constitutively active bacterial expression vector pNCS (Allele Biotech). Plasmids were transformed into chemically competent XL-10 Gold E. coli (Agilent), and colonies were grown on LB/agar plates at 37 °C for 16–20 hours and at room temperature for an additional 20–24 hours. For each round of mutagenesis, the number of colonies screened was 10- fold the expected library diversity to ensure full coverage. Colonies expressing blue-shifted mNeptune variants were screened for transmitted color by eye and for fluorescence in a dark enclosure with a KL2500 fiber-optic light source (Leica), 610/20-nm excitation and 645/30-nm emission filters (Chroma), and a ST-8300M cooled CCD camera controlled with CCDOps software (Santa Barbara Instrument Group). Colonies expressing large-Stokes shift variants were screened for transmitted color by eye and for fluorescence under blue LED excitation with an orange acrylic filter.
Protein production and characterization
For spectral characterization, bacterial pellets were lysed in B-PER II (Pierce) and hexahistidine-tagged proteins were purified with HisPur Cobalt Resin (Pierce). Proteins were desalted into phosphate-buffered saline (PBS) pH 7.4 using Econo-Pac 10DG gravity flow chromatography columns (Bio-Rad). Absorbance, excitation spectra, and emission spectra were measured with a Lambda35 UV/VIS and LS-55 fluorescence spectrometers (Perkin Elmer). Extinction coefficients were calculated using the base-denaturation method49. Quantum yields were determined using cresyl violet in methanol as a standard (QY = 0.54). pH titrations were performed using a series of buffers (1 M HOAc, 1 M NaOAc, 5 M NaCl for pH 3.0–4.5; 1 M NaH2PO4, 1 M Na2HPO4, 5 M NaCl for pH 5.0–9.0). In vitro photobleaching measurements were performed in PBS droplets under mineral oil on an IX81 inverted microscope with a 40×/1.15-numerical aperture (NA) water-immersion objective, an X-Cite 120-W metal halide lamp (Lumen Dynamics) at 100% neutral density, a 485/22-nm excitation filter (Omega), and an Orca ER camera (Hamamatsu) controlled by Micro-Manager software. Images were acquired every 1 s under continuous illumination. Times were scaled to produce photon output rates of 1000 per molecule per s as previously described30. Maturation experiments were performed by measuring change in fluorescence following exposure of CyOFP1-expressing E. coli grown in deoxygenated media to normal atmosphere, corrected for photobleaching of a fully mature CyOFP1 sample.
Fluorescence lifetime characterization
HEK293T cells (GE Healthcare Dharmacon, HCL4517) were transfected using Lipofectamine 2000 with a plasmid encoding CyOFP1 under the CAG promoter. Cells were imaged using a custom-built two-photon fluorescence lifetime imaging microscope described previously50. Briefly, CyOFP1 was excited with a Ti:sapphire laser (Chameleon, Coherence) tuned at a wavelength of 920_nm, and collected through a photoelectron multiplier tube with low transfer time spread (H7422–40p; Hamamatsu). Fluorescence lifetime images were obtained using a time-correlated single photon counting board (SPC-150; Becker and Hickl) controlled with custom software51.
Fusion protein construction and imaging
pCyOFP1-N1 and pCyOFP1-C1 expression vectors were first constructed based on pEGFP-C1 and pEGFP-N1 (Clontech). Briefly, the CyOFP1 cDNA was amplified by PCR with a 5’ primer encoding an AgeI site and a 3’ primer encoding either a BspEI (C1) or NotI (N1) site, and the PCR products were digested, purified, and ligated into similarly treated pEGFP-C1 or pEGFP-N1 vector backbones. Then, to construct plasmids encoding CyOFP1 fusions, fragments encoding various protein domains were excised from existing plasmids encoding mEmerald fusions using available restriction sites and ligated into similarly treated pCyOFP1-N1 and pCyOFP1-C1 vector backbones. Domains for fusion proteins were derived from the following sources: human β-actin, NM_001130442.1; human RhoB, NM_004040.2; human H2B, NM_021058.3; human α-tubulin, NM_006082; rat connexin-43, NM_012326.2; rabbit cytochrome p450, XM_002718526.1; human cytochrome C oxidase subunit VIIIA, NM_004074.2; human sialyltransferase, NM_173216.2; c-Ha-Ras, NM_001130442.1; rat F-Tractin, NM_031045.2. For epifluorescence imaging, DNA was prepared using the Plasmid maxi kit (Qiagen) and transfected into HeLa CCL-2 or HeLa S3 cells (ATCC) using Effectene (Qiagen) grown on coverslips in a 50:50 mixture of Dulbecco’s modified Eagle’s medium (DMEM) and Ham’s F12 with 12.5% HyClone Cosmic Calf Serum (GE Healthcare Life Sciences). Cells were fixed after 48 h, mounted with gelvatol52, and imaged by epifluorescence with an Eclipse 80i microscope (Nikon). For light-sheet imaging, the CyOFP1-tractin and cytosolic EGFP constructs were cloned into the pLVX lentiviral vector (Clontech). Viruses were produced and MV3 cells were infected according to the manufacturer’s instructions.
Crystallization and structure solution of CyOFP0.5
Prior to crystallization, hexahistadine-CyOFP0.5 was purified using size exclusion chromatography to remove aggregation. Thereafter, CyOFP0.5 was buffer-exchanged with 50 mM Tris (hydroxymethyl)aminomethane, 25 mM NaCl, 4.0 mM TCEP at pH 7.4 and concentrated to 13 mg/mL for crystallization. CyOFP0.5 was crystallized at 12°C in a dark chamber by sitting-drop vapor diffusion against 0.16 M MgCl, 0.08 M Tris HCl at pH 8.5, 24% PEG 4000, and 20% glycerol. CyOFP0.5 crystals were cryoprotected in paratone oil and flash frozen in liquid nitrogen. X- ray diffraction data were collected at the Advanced Lightsource on SIBYLS Beamline 12.3.1. The crystal structure was solved by molecular replacement using Phaser53 as part of the CCP4 suite54. Models of CyOFP0.5 and the fluorophore were built with Coot v0.8.155. Refinement to 2.39Å was performed in REFMAC556 with model rebuilding in Coot. Water molecules were manually added by inspection throughout the refinement process. The final model is composed of 44 water molecules and all residues except residues 1–4 and 233–243 of chain A, 1–6 and 233–243 of chain B, 1–6 and 232–243 of chain C, 1–7 and 232–243 of chain D, as these did not show interpretable electron density. The quality of the model was then analyzed using the programs MolProbity57 and PROCHECK58. The coordinates and reflections are deposited in the PDB with accession code 5BQL.
Two-photon spectroscopy
Two-photon excitation spectra were obtained on an epifluorescence microscope (IX81, Olympus) combined with a Ti:Sapphire laser source (Chameleon Ultra II, Coherent). The laser beam, attenuated by a computer-controlled achromatic half-wave plate followed by a fixed polarizer to control laser power, was reflected off a dichroic mirror (675DCSPXR, Omega), and focused into a protein solution using a 60×/1.2-NA apochromatic water-immersion objective (UPLSAPO60XW, Olympus), producing an e−2 beam radius at focus of 450 nm at a laser wavelength of 940 nm. To block spontaneous emission from the laser cavity at wavelengths below 700 nm, a long-pass filter (715LP, Semrock) was placed in the beam path before the microscope. Fluorescence collected by the objective was filtered twice (720SP, Semrock) to block emission above 700 nm, and focused by the tube lens onto a fiber-coupled avalanche photodiode (SPCM-AQRH14-FC, Excelitas Technologies). The output pulse count rate was stored on computer as a function of excitation wavelength. An additional microscope port was used periodically to check fluorescence emission spectra, using a fiber coupled spectrometer. The average laser power at the sample was held at 0.5 mW across the excitation spectrum (730–1080 nm). Fluorescent proteins were measured in PBS, except for GCaMP6s which was measured in MOPS/KCl with 10 mM CaEGTA, pH 7.2 (C-3008MP, Invitrogen). Fluorophore concentrations were determined from absorption measurements and published extinction coefficients (CyOFP1 reported here is 40,400 M−1cm−1 at 505 nm). Excitation spectra were adjusted for differences (< 20%) in detector quantum efficiency for the emission spectra of green and orange fluorophores.
3D light sheet imaging
3D collagen samples were created inside a custom agarose mold in order to avoid proximity to stiff surfaces and to reduce refractive index mismatch and light scatter (Welf et al. in review). Briefly, 2% agarose was heated and placed in a rectangular mold containing a cylindrical void for sample containment. Agarose molds were incubated in complete medium prior to sample addition in order to heat the agarose and allow medium components to permeate the agarose. Collagen gels were created by mixing bovine collagen I (Advanced Biomatrix) with concentrated phosphate buffered saline (PBS) and water to create gels of 2.0 mg/mL collagen. This collagen solution was then neutralized with 1N NaOH and mixed with MV3 cells (gift of P. Friedl, University of Texas MD Anderson Cancer Center) just prior to incubation at 37°C to induce collagen polymerization. Samples were imaged in phenol red-free DMEM with 10% fetal bovine serum and antibiotic-antimycotic (Life Technologies) with an one-photon axially swept light-sheet microscope and a two-photon Bessel beam microscope. The one-photon light-sheet microscope illuminates the sample with a tightly focused light-sheet that is scanned in its propagation direction to evenly excite a large field of view in a time-averaged fashion12. Illumination and detection objectives had NA of 0.8, yielding an isotropic 3D resolution of 380 nm. The excitation wavelength was 488 nm and dual-emission imaging of GFP and CyOFP1 was accomplished using two Orca Flash 4.0 CMOS cameras. The two-photon Bessel beam microscope used in this study was conceptually similar to one previously described10, but featured a much longer Bessel beam (FWHM in propagation direction ~100 μm) and improved telecentric scan optics13. Illumination and detection objectives had NA of 0.8, yielding a near-isotropic resolution of ~350 nm. The Bessel beam was scanned five times over the field of view to generate a time-averaged light sheet. No structured illumination was applied; instead for each focal plane only one image was acquired. The excitation wavelength was 900 nm and dual-emission imaging of GFP and CyOFP1 was accomplished using two Orca Flash 4.0 CMOS cameras. For photostability measurements of CyOFP1, powers in the backpupil were 16 μW for one-photon light-sheet microscopy and 92 mW for two-photon Bessel-beam light-sheet microscopy. Images were cropped, scaled, and quantified in ImageJ (NIH), background- subtracted brightness values were calculated in Excel (Microsoft).
Two-photon functional imaging of mouse primary visual cortex in vivo
Mouse procedures were performed in accordance with protocols approved by the Janelia Research Campus Institutional Animal Care and Use Committee and Institutional Biosafety Committee. AAV1 vectors expressing CyOFP1 and GCaMP6s were used to inject the mouse visual cortex as described42. 3–4 weeks after virus injection, a cranial window implantation surgery and in vivo functional imaging were performed, following a previously published protocol42,43. Mice were anesthetized using isoflurane (2.5% for induction, 1.5–2% during surgery). A 2.0–2.5 mm-diameter circular craniotomy was placed above V1 (centered 2.7 mm left, and 0.2 mm anterior to Lambda suture). The craniotomy was covered with 1% agarose (Sigma), a 3-mm round no. 1 thickness glass coverslip (Warner Instruments) and a custom titanium head post were cemented to the brain using black dental cement (Contemporary Ortho-Jet). The animal was then placed under the imaging microscope on a blanket warmed to 37 °C and kept anesthetized using 0.5% isoflurane and sedated with 20 μL at 0.33 mg/mL chlorprothixene injected intramuscularly. Imaging was performed with a custom-built two-photon microscope with a resonant scanner. The light source was a Mai Tai HP 100 femtosecond-pulse laser (Spectra-Physics) running at 940 nm. A 16× 0.8-NA water- immersion objective (Nikon) was used. Images were acquired using ScanImage 5 (vidriotechnologies.com)59. Functional images (512 × 512 pixels, 250 × 250 μm2) of layer 2/3 cells (100–250 μm under the pia) were collected at 15 Hz. Maximal laser power was 145 mW at the front aperture of the objective, and satisfactory signal-to-noise images were typically acquired using 5–25 mW. Mice were presented with moving grating stimuli generated using the Psychophysics Toolbox60 in MATLAB (Mathworks). Each stimulus trial consisted of a 4 s blank period followed by a 4 s drifting sinusoidal grating (0.05 cycles/degree, 1 Hz temporal frequency, 8 different directions). The visual stimuli were presented using an LCD monitor (30 × 40 cm), placed 25 cm in front of the center of the right eye of the mouse. The monitor subtended an angle of ±38° horizontally and ±31° vertically around the eye of the mouse. Regions of interest corresponding to visually identifiable cell bodies were selected using a semi-automated algorithm, and changes to fluorescence signal were calculated as previously described41–43.
Bioluminescence characterization in vitro
Hexahistidine-tagged proteins were expressed in E. coli at 30° C for 30 h, lysed in B-PER II (Pierce), and purified with HisPur Cobalt Resin (Pierce). Luminescent spectra of purified proteins were measured on a Safire2 microplate reader (Tecan). Each protein was diluted to 2 nM in 30mM MOPS 100mM KCl pH 7.5 with 0.1% BSA, and a final concentration of 20 μM substrate was used.
Bioluminescence imaging in a mouse tissue phantom
HEK293A cells (Life Technologies) were transfected using Lipofectamine LTX (Life Technologies) with plasmids expressing bioluminescence probes following mTurquoise2 and a P2A sequence. Cells were harvested 48 h later, and 300000 cells in 150 μL Live Cell Imaging Solution (Life Technologies) were loaded onto a 96-well plate, and mTurquoise2 fluorescence was obtained with a Safire2 microplate reader (Tecan). Fluorometry settings were: excitation wavelength and bandpass width, 434 and 6 nm; emission wavelength and bandpass width, 474 and 10 nm. During fluorometry, 50000 cells in 10 μL Live Cell Imaging Solution were loaded into glass capillary tubes with inner diameter 0.8–1.1 mm (Kimble Chase, cat. #34507–99) and gently pelleted by centrifugation for 15 s at 500 g. 10 μL of Live Cell Imaging Solution with 20 μM D-luciferin (Research Products International), coelenterazine (Promega), ViviRen (Promega), or furimazine (Promega) was then added, tubes were inserted into the mouse tissue phantom (PerkinElmer), and bioluminescence images were acquired in an IVIS Spectrum imaging system (PerkinElmer). Imaging settings were: field of view, 13 cm; aperture, f/1; binning, 2 × 2; and exposure time, 1 s. Images were acquired every 20 s for 5 min, in which time luminescence signals were observed to plateau or drop. All fluorescence and bioluminescence measurements were performed in triplicate. Transfection-normalized bioluminescence intensity was calculated as net bioluminescence divided by net fluorescence, where net bioluminescence was obtained on the IVIS Spectrum by integrating intensity in a common region of interest encompassing all transfected cells and subtracting signal from a similar region encompassing untransfected control cells, and where net fluorescence was obtained on the Safire2 as fluorescence of transfected cells with fluorescence of untransfected cells subtracted.
Bioluminescence imaging in deep tissues of living mouse
For live mouse imaging, required sample numbers were calculated using the program G*Power based on the signal differences observed in the mouse phantom and a two-tailed significance (α) level of 0.05. Livers of male 6- to 7-week-old nude mice (strain J:NU #7850 EC, Jackson Laboratories) were hydrodynamically transfected by injecting 20 μg of plasmid DNA in 1.8 mL of 0.9% NaCl into the tail vein within 5–6 seconds. Animals were randomly assigned to receive FLuc2, BRET6, or Antares plasmids. The person performing the tail vein injections was blinded as to the plasmids being injected. 48 h after injection, mice were anesthesized with inhaled isoflurane via face mask for substrate injection and imaging. For FLuc-expressing mice, 9.42 μmol (3 mg) of D-luciferin sodium salt (Research Products International) in 100 μL water was injected intraperitoneally. For BRET6 and Antares-expressing mice, tail vein injections were performed with 333 nmol (141 μg) of coelenterazine (NanoLight Technology, water-soluble formulation) in 120 μL water, or 333 nmol (123 μg) of furimazine (Promega) in 120 μL of 8% glycerol, 12% ethanol, and 50% polyethylene glycol 400 in water. The person performing the substrate injections was blinded as to the identify of the mouse being injected and substrate being used. Images were immediately acquired in the IVIS Spectrum or IVIS 100 every 20 s for 5 min, in which time luminescence signals were observed to plateau or drop. The person performing the imaging was blinded as to the treatments the mouse received. The highest intensity image from each mouse was selected for quantitation. Image settings were: field of view, 13 cm; aperture, f/4; binning, 2 × 2; and exposure time, 2 s. Quantitation was performed as with phantom mice above. Three or four mice were analyzed per condition in each of three separate experiments, then data were pooled for each condition for statistical analysis. A normal distribution of intensities in each condition was verified by the Shapiro-Wilk test and differences between conditions were assessed by ANOVA followed by Tukey’s posthoc tests, using the program Prism (GraphPad). Mouse procedures were carried out in accordance with applicable regulations and the Stanford Administrative Panel on Laboratory Animal Care.
Supplementary Material
Acknowledgments
We thank Tim Doyle (Stanford University) for assistance with bioluminescence imaging, Jianhua Chen (Shenzhen Institutes of Advanced Technology) for analysis of light-sheet microscopy data, Scott Classen (ALS SIBYLS) for help with synchrotron X-ray data collection, Poncho Meisenheimer and Thomas Kirkland (Promega) for furimazine, and and members of the Lin laboratory for general assistance and advice. The Advanced Light Source (ALS), a national user facility operated by Lawrence Berkeley National Laboratory on behalf of the Department of Energy Office of Basic Energy Sciences, through the Integrated Diffraction Analysis Technologies (IDAT) program, is supported by DOE Office of Biological and Environmental Research and National Institute of Health project MINOS (R01GM105404). This work was supported by the University of Hawaii at Manoa Undergraduate Research Opportunities Program (M.H. and C.T.T.), a long-term fellowship from the Human Frontier Science Program (T.L.), NIH grants R01HL064274 (M.K.), R01MH080047 (R.Y.), 1U01NS090600 (M.Z.L.), and P50GM107615 (M.Z.L.), Shenzhen Basic Research Foundation grant JCYJ20150521144320987 (J.C.), the Hundred Talents Program award from the Chinese Academy of Sciences (J.C.), a Burroughs Wellcome Foundation Career Award for Medical Scientists (M.Z.L.), and a Rita Allen Foundation Scholar Award (M.Z.L.).
Footnotes
AUTHOR CONTRIBUTIONS
J.C. engineered and characterized CyOFP1 and Antares, performed bioluminescence experiments, and co-wrote the manuscript. Y.O. performed bioluminescence experiments. A.S., N.A., C.T.T., M.H., and H-L.N. obtained the crystal structure of CyOFP1. H.D. performed two-photon imaging in mice. J.J.M. and T.L. performed optical characterization of CyOFP1. F.Z. performed injections in mice. B.B.K. characterized CyOFP1 monomericity. E.S.W., K.M.D., and R.F. performed light-sheet microscopy. M.A.B. characterized CyOFP1 fusions. M.W.D., R.F., M.A.K., R.Y., D.S.K., and H.-L.N. provided supervision. M.Z.L. characterized CyOFP1 maturation, assisted in data analysis, and co-wrote the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: J.C. and M.Z.L. have filed a patent application for CyOFP and Antares.
References
- 1.Newman RH, Fosbrink MD, Zhang J. Genetically encodable fluorescent biosensors for tracking signaling dynamics in living cells. Chem Rev. 2011;111:3614–3666. doi: 10.1021/cr100002u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Depry C, Mehta S, Zhang J. Multiplexed visualization of dynamic signaling networks using genetically encoded fluorescent protein-based biosensors. Pflugers Arch. 2013;465:373–381. doi: 10.1007/s00424-012-1175-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawano H, Kogure T, Abe Y, Mizuno H, Miyawaki A. Two-photon dual-color imaging using fluorescent proteins. Nat Methods. 2008;5:373–374. doi: 10.1038/nmeth0508-373. [DOI] [PubMed] [Google Scholar]
- 4.Yang J, et al. mBeRFP, an improved large stokes shift red fluorescent protein. PLoS One. 2013;8:e64849. doi: 10.1371/journal.pone.0064849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shao L, Kner P, Rego EH, Gustafsson MG. Super-resolution 3D microscopy of live whole cells using structured illumination. Nat Methods. 2011;8:1044–1046. doi: 10.1038/nmeth.1734. [DOI] [PubMed] [Google Scholar]
- 6.Li D, et al. ADVANCED IMAGING. Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. Science. 2015;349:aab3500. doi: 10.1126/science.aab3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu Y, et al. Optical focusing deep inside dynamic scattering media with near-infrared time-reversed ultrasonically encoded (TRUE) light. Nat Commun. 2015;6:5904. doi: 10.1038/ncomms6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Si K, Fiolka R, Cui M. Fluorescence imaging beyond the ballistic regime by ultrasound pulse guided digital phase conjugation. Nat Photonics. 2012;6:657–661. doi: 10.1038/nphoton.2012.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ji N, Milkie DE, Betzig E. Adaptive optics via pupil segmentation for high-resolution imaging in biological tissues. Nat Methods. 2010;7:141–147. doi: 10.1038/nmeth.1411. [DOI] [PubMed] [Google Scholar]
- 10.Katona G, et al. Fast two-photon in vivo imaging with three-dimensional random-access scanning in large tissue volumes. Nat Methods. 2012;9:201–208. doi: 10.1038/nmeth.1851. [DOI] [PubMed] [Google Scholar]
- 11.Planchon TA, et al. Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat Methods. 2011;8:417–423. doi: 10.1038/nmeth.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao L, et al. Noninvasive imaging beyond the diffraction limit of 3D dynamics in thickly fluorescent specimens. Cell. 2012;151:1370–1385. doi: 10.1016/j.cell.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Welf ES, et al. Quantitative Multiscale Cell Imaging in Controlled 3D Microenvironments. Dev Cell. 2016;36:462–475. doi: 10.1016/j.devcel.2016.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen BC, et al. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science. 2014;346:1257998. doi: 10.1126/science.1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dean KM, Roudot P, Welf ES, Danuser G, Fiolka R. Deconvolution-free subcellular imaging with axially swept light sheet microscopy. Biophys J. 2015;108:1–9. doi: 10.1016/j.bpj.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dean KM, Fiolka R. Uniform and scalable light-sheets generated by extended focusing. Opt Express. 2014;22:26141–26152. doi: 10.1364/OE.22.026141. [DOI] [PubMed] [Google Scholar]
- 17.Close DM, Xu T, Sayler GS, Ripp S. In vivo bioluminescent imaging (BLI): noninvasive visualization and interrogation of biological processes in living animals. Sensors (Basel) 2011;11:180–206. doi: 10.3390/s110100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao H, et al. Emission spectra of bioluminescent reporters and interaction with mammalian tissue determine the sensitivity of detection in vivo. J Biomed Opt. 2005;10:41210. doi: 10.1117/1.2032388. [DOI] [PubMed] [Google Scholar]
- 19.Contag CH, et al. Visualizing gene expression in living mammals using a bioluminescent reporter. Photochem Photobiol. 1997;66:523–531. doi: 10.1111/j.1751-1097.1997.tb03184.x. [DOI] [PubMed] [Google Scholar]
- 20.Ando Y, Niwa K, Yamada N, Enomoto T, Irie… T. Firefly bioluminescence quantum yield and colour change by pH-sensitive green emission. Nature Photonics. 2007;2:44–47. [Google Scholar]
- 21.Branchini BR, Magyar RA, Murtiashaw MH, Anderson SM, Zimmer M. Site-directed mutagenesis of histidine 245 in firefly luciferase: a proposed model of the active site. Biochemistry. 1998;37:15311–15319. doi: 10.1021/bi981150d. [DOI] [PubMed] [Google Scholar]
- 22.Liang Y, Walczak P, Bulte JW. Comparison of red-shifted firefly luciferase Ppy RE9 and conventional Luc2 as bioluminescence imaging reporter genes for in vivo imaging of stem cells. J Biomed Opt. 2012;17:016004. doi: 10.1117/1.JBO.17.1.016004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazo-Vargas A, Park H, Aydin M, Buchler NE. Measuring fast gene dynamics in single cells with time-lapse luminescence microscopy. Mol Biol Cell. 2014;25:3699–3708. doi: 10.1091/mbc.E14-07-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mezzanotte L, et al. Evaluating reporter genes of different luciferases for optimized in vivo bioluminescence imaging of transplanted neural stem cells in the brain. Contrast Media Mol Imaging. 2013;8:505–513. doi: 10.1002/cmmi.1549. [DOI] [PubMed] [Google Scholar]
- 25.Matthews JC, Hori K, Cormier MJ. Purification and properties of Renilla reniformis luciferase. Biochemistry. 1977;16:85–91. doi: 10.1021/bi00620a014. [DOI] [PubMed] [Google Scholar]
- 26.Shimomura O, Johnson FH, Masugi T. Cypridina bioluminescence: light-emitting oxyluciferin-luciferase complex. Science. 1969;164:1299–1300. doi: 10.1126/science.164.3885.1299. [DOI] [PubMed] [Google Scholar]
- 27.Loening AM, Dragulescu-Andrasi A, Gambhir SS. A red-shifted Renilla luciferase for transient reporter-gene expression. Nat Methods. 2010;7:5–6. doi: 10.1038/nmeth0110-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimomura O, Masugi T, Johnson FH, Haneda Y. Properties and reaction mechanism of the bioluminescence system of the deep-sea shrimp Oplophorus gracilorostris. Biochemistry. 1978;17:994–998. doi: 10.1021/bi00599a008. [DOI] [PubMed] [Google Scholar]
- 29.Hall MP, et al. Engineered luciferase reporter from a deep sea shrimp utilizing a novel imidazopyrazinone substrate. ACS Chem Biol. 2012;7:1848–1857. doi: 10.1021/cb3002478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ward WW, Cormier MJ. Energy transfer via protein-protein interaction in renilla bioluminescence. Photochem Photobiol. 1978;27:389–396. [Google Scholar]
- 31.Dragulescu-Andrasi A, Chan CT, De A, Massoud TF, Gambhir SS. Bioluminescence resonance energy transfer (BRET) imaging of protein-protein interactions within deep tissues of living subjects. Proc Natl Acad Sci U S A. 2011;108:12060–12065. doi: 10.1073/pnas.1100923108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saito K, et al. Luminescent proteins for high-speed single-cell and whole-body imaging. Nat Commun. 2012;3:1262. doi: 10.1038/ncomms2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chu J, et al. Non-invasive intravital imaging of cellular differentiation with a bright red-excitable fluorescent protein. Nat Methods. 2014;11:572–578. doi: 10.1038/nmeth.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bruno TJ, Svoronos PDN. CRC handbook of fundamental spectroscopic correlation charts. CRC Press; Boca Raton, FL: 2006. [Google Scholar]
- 35.Chu J, Xing Y, Lin MZ. In: The Fluorescent Protein Revolution. Day R, Davidson M, editors. United States: 2014. pp. 153–167. [Google Scholar]
- 36.Baird GS, Zacharias DA, Tsien RY. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci U S A. 2000;97:11984–11989. doi: 10.1073/pnas.97.22.11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lounis B, Moerner WE. Single photons on demand from a single molecule at room temperature. Nature. 2000;407:491–493. doi: 10.1038/35035032. [DOI] [PubMed] [Google Scholar]
- 38.Lam AJ, et al. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat Methods. 2012;9:1005–1012. doi: 10.1038/nmeth.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miyawaki A, Shcherbakova DM, Verkhusha VV. Red fluorescent proteins: chromophore formation and cellular applications. Curr Opin Struct Biol. 2012;22:679–688. doi: 10.1016/j.sbi.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piatkevich KD, Malashkevich VN, Almo SC, Verkhusha VV. Engineering ESPT pathways based on structural analysis of LSSmKate red fluorescent proteins with large Stokes shift. J Am Chem Soc. 2010;132:10762–10770. doi: 10.1021/ja101974k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Akerboom J, et al. Optimization of a GCaMP calcium indicator for neural activity imaging. J Neurosci. 2012;32:13819–13840. doi: 10.1523/JNEUROSCI.2601-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen TW, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dana H, et al. Thy1-GCaMP6 transgenic mice for neuronal population imaging in vivo. PLoS One. 2014;9:e108697. doi: 10.1371/journal.pone.0108697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morse D, Tannous BA. A water-soluble coelenterazine for sensitive in vivo imaging of coelenterate luciferases. Mol Ther. 2012;20:692–693. doi: 10.1038/mt.2012.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Otto-Duessel M, et al. In vivo testing of Renilla luciferase substrate analogs in an orthotopic murine model of human glioblastoma. Mol Imaging. 2006;5:57–64. [PubMed] [Google Scholar]
- 46.Horton NG, et al. Three-photon microscopy of subcortical structures within an intact mouse brain. Nat Photonics. 2013;7:205–209. doi: 10.1038/nphoton.2012.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shcherbakova DM, Hink MA, Joosen L, Gadella TW, Verkhusha VV. An orange fluorescent protein with a large Stokes shift for single-excitation multicolor FCCS and FRET imaging. J Am Chem Soc. 2012;134:7913–7923. doi: 10.1021/ja3018972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kogure T, et al. A fluorescent variant of a protein from the stony coral Montipora facilitates dual- color single-laser fluorescence cross-correlation spectroscopy. Nat Biotechnol. 2006;24:577–581. doi: 10.1038/nbt1207. [DOI] [PubMed] [Google Scholar]
- 49.Chalfie M, Kain SR. Green Fluorescent Protein: Properties, Applications, and Protocols. Wiley-Liss; 1998. [Google Scholar]
- 50.Yasuda R. Imaging spatiotemporal dynamics of neuronal signaling using fluorescence resonance energy transfer and fluorescence lifetime imaging microscopy. Curr Opin Neurobiol. 2006;16:551–561. doi: 10.1016/j.conb.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 51.Yasuda R, et al. Supersensitive Ras activation in dendrites and spines revealed by two-photon fluorescence lifetime imaging. Nat Neurosci. 2006;9:283–291. doi: 10.1038/nn1635. [DOI] [PubMed] [Google Scholar]
- 52.Harlow E, Lane D. Mounting samples in gelvatol or mowiol. CSH Protoc. 2006;2006 doi: 10.1101/pdb.prot4461. [DOI] [PubMed] [Google Scholar]
- 53.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 56.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 57.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J Mol Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 59.Pologruto TA, Sabatini BL, Svoboda K. ScanImage: flexible software for operating laser scanning microscopes. Biomed Eng Online. 2003;2:13. doi: 10.1186/1475-925X-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brainard DH. The Psychophysics Toolbox. Spat Vis. 1997;10:433–436. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.