Abstract
Complex periodic patterns can self-organize through dynamic interactions between diffusible activators and inhibitors. In the biological context, self-organized patterning is challenged by spatial heterogeneities (‘noise’) inherent to biological systems. How spatial variability impacts the periodic patterning mechanism and how it can be buffered to ensure precise patterning is not well understood. We examine the effect of spatial heterogeneity on the periodic patterning of the fruit fly eye, an organ composed of ∼800 miniature eye units (ommatidia) whose periodic arrangement along a hexagonal lattice self-organizes during early stages of fly development. The patterning follows a two-step process, with an initial formation of evenly spaced clusters of ∼10 cells followed by a subsequent refinement of each cluster into a single selected cell. Using a probabilistic approach, we calculate the rate of patterning errors resulting from spatial heterogeneities in cell size, position and biosynthetic capacity. Notably, error rates were largely independent of the desired cluster size but followed the distributions of signaling speeds. Pre-formation of large clusters therefore greatly increases the reproducibility of the overall periodic arrangement, suggesting that the two-stage patterning process functions to guard the pattern against errors caused by spatial heterogeneities. Our results emphasize the constraints imposed on self-organized patterning mechanisms by the need to buffer stochastic effects.
Author summary
Complex periodic patterns are common in nature and are observed in physical, chemical and biological systems. Understanding how these patterns are generated in a precise manner is a key challenge. Biological patterns are especially intriguing, as they are generated in a noisy environment; cell position and cell size, for example, are subject to stochastic variations, as are the strengths of the chemical signals mediating cell-to-cell communication. The need to generate a precise and robust pattern in this ‘noisy’ environment restricts the space of patterning mechanisms that can function in the biological setting. Mathematical modeling is useful in comparing the sensitivity of different mechanisms to such variations, thereby highlighting key aspects of their design.
We use mathematical modeling to study the periodic patterning of the fruit fly eye. In this system, a highly ordered lattice of differentiated cells is generated in a two-dimensional cell epithelium. The pattern is first observed by the appearance of evenly spaced clusters of ∼10 cells that express specific genes. Each cluster is subsequently refined into a single cell, which initiates the formation and differentiation of a miniature eye unit, the ommatidium. We formulate a mathematical model based on the known molecular properties of the patterning mechanism, and use a probabilistic approach to calculate the errors in cluster formation and refinement resulting from stochastic cell-to-cell variations (‘noise’) in different quantitative parameters. This enables us to define the parameters most influencing noise sensitivity. Notably, we find that this error is roughly independent of the desired cluster size, suggesting that large clusters are beneficial for ensuring the overall reproducibility of the periodic cluster arrangement. For the stage of cluster refinement, we find that rapid communication between cells is critical for reducing error. Our work provides new insights into the constraints imposed on mechanisms generating periodic patterning in a realistic, noisy environment, and in particular, discusses the different considerations in achieving optimal design of the patterning network.
Electronic supplementary material The online version of this article (doi: 10.1007/s10867-016-9409-4) contains supplementary material, which is available to authorized users.
Keywords: Drosophila eye, Robust periodic patterning, Mathematical modeling, Noise, Spatial heterogeneity, Lateral inhibition
Introduction
The body plan of multicellular organisms is defined during embryonic development, when uniform fields of cells are patterned into distinct tissues and organs positioned in spatially distinct domains. This patterning is defined by biochemical networks of interacting genes and proteins, some of which are secreted to the extracellular environment and mediate communication between cells. The precision by which spatial patterns are defined and their reproducibility between individuals is particularly striking when considering the inherent stochastic variability of the underlying biological processes [1–3]: external conditions such as temperature or nutrients vary, genetic polymorphisms distinguish between individuals, and random fluctuations arise due to the discrete nature of the underlying molecular processes. Understanding how such variations are buffered is a key theoretical and experimental challenge [4]. Previous studies have demonstrated, for example, how this need for robustness has shaped the molecular mechanisms that establish a spatial gradient of morphogens, molecules that can induce distinct cell fates in a concentration-dependent manner. Therefore, a robust establishment of a morphogen gradient ensures a reproducible splitting of the field into distinct spatial domains [5–7].
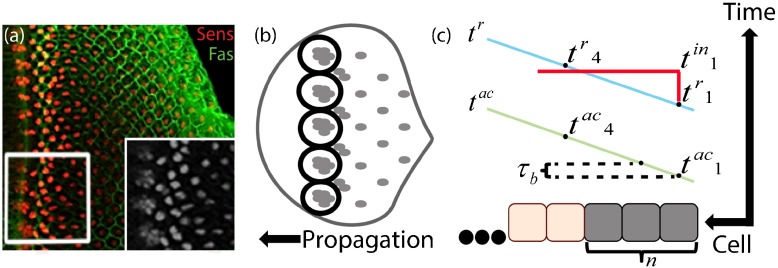
While patterning by morphogen gradients has been extensively studied, less is understood about the robustness of mechanisms that self-organize periodic patterns. Perhaps the best-studied example for a precise periodic pattern that self-organizes during development is the eye of the fruit fly Drosophila melanogaster. The complex eye of the adult fly is built of a hexagonal array of ∼800 units, the ommatidia, each of which is composed of 20 photoreceptors and supporting cells [8, 9]. This precise periodic pattern of the eye is defined during the larvae stage, when the immature adult eye is present as a two-dimensional epithelial tissue (eye disc). Patterning begins when the disc includes ∼2000 cells, all of which have an identical capacity to be selected as the pre-neuronal cells defining the future eye unit [10]. A dynamic network then selects the pre-neuronal cells through a two-stage process. First, an activating wave sweeps across the disc and leaves behind an ordered array of clusters composed of ∼10 cells, each of which expresses specific markers (the pre-neuronal clusters) [11, 12]. Subsequently, each cluster is refined into a single selected founder cell surrounded by ∼20 cells that will form an ommatidium [Fig. 1a and b] [13]. As a result of this process, the selected cells are positioned in a hexagonal lattice-like arrangement.
Fig. 1.
Pattern formation of founder photoreceptors in the fruit fly eye. Posterior is to the right. a Confocal image of a portion of a developing disc. Membrane outlines are visualized with FasIII (green) and differentiating cells with the nuclear differentiation marker Senseless (red). The inset represents a magnified view of the boxed region including and immediately after the formation of clusters. b Scheme of the developmental process. The black arrow indicates long-range activation propagation. Clusters secrete inhibitory signals (indicated by black circles) that direct the formation of the clusters ahead to discrete, evenly spaced positions. Each cluster is later refined into one cell through contact lateral inhibition. c Cluster formation in one dimension. Time is on the vertical axis and cell position on the horizontal axis. Cluster size is defined by the number of cells that become refractory (selected) before the first cell in the cluster produces inhibition (red horizontal line). t ac,t r and t in are the times when each cell is activated, becomes refractory and produces inhibition, respectively. τ b is the time gap between activation of two adjacent cells and n is the size of the cluster
Cluster selection proceeds sequentially from column to column. Each selected cell secretes an inhibitory signal that prevents selection of nearby cells. In effect, the inhibitory signals define an inhibitory circle around each selected cluster, where selection is not possible. These selection-resistant-regions are defined rapidly following cluster selection, reaching in front of the progressing activation wave. Consequently, the evenly spaced clusters in each column define the position of the clusters in the next column by the points of minimum inhibition [14]. In principle, this mechanism could also result in the immediate selection of clusters of just one cell, obviating the need for a two-stage process. For example, previous quantitative models have shown that a single-cell pattern can propagate to define the observed pattern. In this formulation, clusters appeared as a transient stage of the dynamics, but did not contribute to the inhibitory-signal pattern propagation, which was defined by the single selected cell [15]; alternatively, we reasoned that clusters could participate in the actual pattern formation if all cluster cells would contribute to the lateral inhibition process, and that this may promote patterning robustness. Here, we use a rigorous mathematical approach to show that this two-stage process, whereby the full clusters contribute to pattern propagation, indeed increases the patterning robustness.
Spatial heterogeneity is likely to be substantial within the epithelial tissue where eye patterning takes place. First, random variations in cell size, cell position and cell shape are common, and impact the effective inhibition radii defined by the secreted diffusing inhibitors. Second, stochastic fluctuations in the cell biosynthetic capacity introduce spatial variations in the rate at which the inhibitors are produced.
To examine how this spatial variability influences the precision of patterning, we formulated a mathematical model describing the patterning process, and used a probabilistic approach to calculate the probability of patterning errors. First, we examined the probability that a cluster will be larger (or smaller) than a desired cluster size, n. Second, we estimated the probability that cluster refinement will fail. Notably, the rates of errors in the cluster formation were independent of cluster size. Rather, they were largely dependent on the distributions of signaling velocities. Pre-formation of large clusters therefore greatly increases the reproducibility of the overall periodic arrangement, suggesting that the two-stages patterning process functions to guard the pattern against errors caused by spatial heterogeneities. Our results emphasize the constraints imposed on self-organized patterning mechanisms by the need to buffer stochastic effects.
Results
A model for generating a periodic pattern through sequential activation
We formulated a general model of cluster formation through sequential activation. Consider an ordered one-dimensional array of cells Ci that are activated sequentially at times being the velocity of the activation wave, which we assume to be uniform in time and space. Once activated, each cell initiates an internal process that, after some time delay, results in its selection at time . After a second time delay, each selected cell will start secreting inhibition at time . We assume that the inhibitory signal secreted by the first selected cell diffuses rapidly [10, 16] and extends beyond the area that will become the future cluster. This signal will prevent selection of all cells that have not been selected by that time, but will not influence cells that have already been selected, as these cells are refractory to inhibition. In the absence of noise, the number of selected cells n is given by the number of cells that have been selected before the first cell began producing the inhibition:
| 1 |
where is the time when the first cell in the cluster began secreting the inhibitory signal, δt is the time gap between the times the first cell was selected and the time it began to produce the inhibitor signal, and is the time lag between activation of two adjacent cells [Fig. 1c]. Thus, if 0<δt<τb, only the first cell to be activated will be selected, as this first cell will inhibit all other cells prior to their selection, leading to a cluster of size n=1. Similarly, if τb<δt<2τb both the first and second cells to be activated will become selected before the first cell begins secreting the inhibition signal, leading to a cluster of size n=2.
In the sections below, we compare error probabilities in formations of clusters of different sizes, starting with n=1. n=1 is the simple case of ‘one-step patterning’, since a pattern of single cells is formed a priori and no further step of cluster refinement is required. n>1, on the other hand, is the case of ‘two-step patterning’, as each cluster is further refined to a single cell.
Cluster formation in the presence of noise: the error probability for a cluster of size n=1
To introduce noise into this model, we choose tr out of some distribution P(tr). In the biological context, this distribution summarizes the variability in the molecular constituents of the patterning network interpreting the activation signal. For simplicity, we assume that P(tr) is uniformly distributed in a region of width 2τσ centered around the mean tr ¯. τσ captures the strength of the molecular noise, namely the cell-to-cell variability in the time it takes an activated cell to reach the selection threshold. The time at which the cell i becomes refractory is given by , where tris selected from the distribution P(tr).
Assume now that the mean (desired) number of selected cells in the cluster is n. Our goal is to calculate the probability of errors, namely the probability that a larger or smaller number of cells will be selected. For the case n=1, precise selection requires that the times by which cells Ci, i=2,3,4… are selected ( are all larger than the time the first cell begins secreting the inhibitory signal (at the time . Cells in position i, for which tr ¯+(i−1)τb−τσ>tr ¯ + δt + τσ, will comply with this demand regardless of the noise realization, and will therefore be necessarily inhibited before becoming refractory. Errors will therefore be due only to cells i for which . Note that in principle, although we assume , for all i>1 (the requirement for cluster size n=1) a noise τσ that is significantly larger than the selection bias τb would still allow multiple cells to be selected before becoming refractory.
For simplicity, we first examine the simplest case, in which only the cell positioned at the border of the cluster (i=2) can contribute to the error. In this case, the error is given by:
| 2 |
where δt = tin ¯−tr ¯ captures the time delay from the mean time of selection to the mean time of beginning producing the inhibitor. Note again that for the present case of n=1,δt<τb. The first integral in (2) sums over all possible times tin at which the first cell can produce inhibition. For each possible tin, the second integral sums over the possible times the second cell is selected aftertin (making this an irrelevant selection as the cell is already inhibited).
While the solution of the two integrals is straightforward in this case, it is useful to consider their geometrical interpretation, as this interpretation is iterated for the more complicated case. In the non-trivial case δt+2τσ>τb, Pn=1 is simply the sum of a rectangle and a trapezoid [Fig. 2a and b]. The rectangle is obtained from the inhibiting times tin that precede all possible times for the second cell selection, which is max{tin,tr ¯ + τb−τσ} = tr ¯ + τb−τσ. For these values, the second integral equals 1, as there are no further restrictions on the times in which the second cell can be selected. The area of this rectangle is simply the fraction of tin for which this constraint is obeyed, multiplied by the probability density [first term in the non-trivial solution to Pn=1 in (2)]. The trapezoid is obtained from the region where max{tin,tr ¯ + τb−τσ} = tin, in which case too early selection of the second cell will result in an error. The area of this trapezoid is given by the second term in the non-trivial solution to Pn=1 in (2). The trapezoid’s large base is given by the maximal possible range of that will give minimal selection (when tin is minimal) and its small base is defined by the minimal possible range of tr that will give minimal selection (when tin is maximal). Its height is the size of the region in which the error can occur (too early selection of the second cell), namely δt+2τσ−τb. Again, its area should be normalized by the probability density. Therefore, the rectangle represents regions of no constraint, where times can be chosen freely, whereas the non-rectangular shape (here a trapezoid) represents regions of constraint, where in order for no error to occur the times have to be chosen in some order.
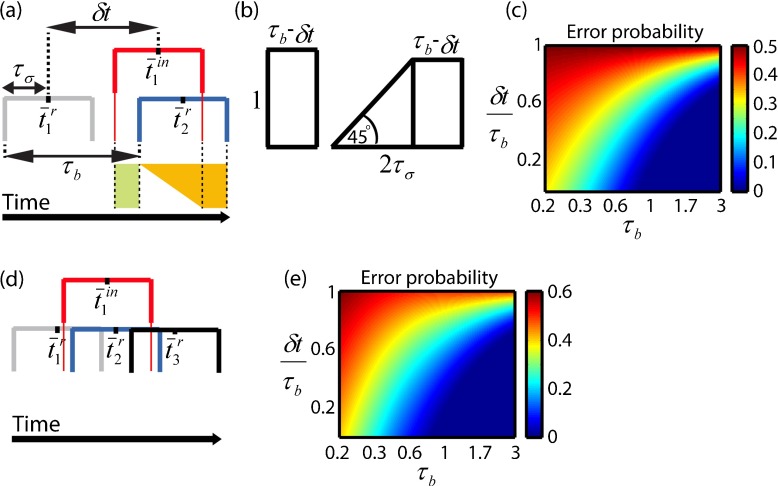
Fig. 2.
Cluster formation in the presence of noise for the case n = 1. a–b Capturing noise in time distributions. The refractory time is chosen out of a distribution P(t r) of width 2τ σ, centered at t r ¯.The red distribution represents P(t in) and is the time gap between selection and inhibition production of the first cell. The rectangle (in green) and trapezoid (in yellow) correspond to the shapes in (b) and represent the probability P n=1 in different regions for choosing t in. To obtain the terms calculated in (2), the area of the rectangle and trapezoid should be normalized by and , respectively. The trapezoid’s angle is 45∘, because for each point in the overlapping domains of and P(t in) we require that , the number of these requirements giving the trapezoid’s height. c The error probability ε is a function of τ b and δ t in the simplest case. Error was calculated accounting only for cell i=2 even when τ b was small enough to allow selection of more cells to potentially lead to error. Error probability increases sharply when τ b≲1 (for . d–e General case of n=1. d More cells can potentially be selected before t in (here also cell i=3), leading to an error. e The error probability ε for when accounting for all cells that can lead to error. This error becomes higher than the approximated error shown in (c) as τ b decreases and contributions to error from cells i≥3 become more significant
Figure 2c shows the solution for ε(τb,δt,n=1) in (2), plotted as a function of the two normalized variables and . Thus, the activation delay between two adjacent cells τb and the delay from selection to production of the inhibitor δt both refer to a noise distribution 2τσ of width 1. As shown, the error ε increases sharply with δt (in regions δt+1>τb in which ε>0) and when τb<1 (for . Erroneous instances can occur even when τb>1, depending on δt. Notably, we assumed that cell i=1 could not inhibit its own selection when δt<1, as it became refractory before producing the inhibitor, and therefore erroneous formation of a cluster of size n=0 is not possible.
A similar derivation can be used to calculate the error probability in the general case, where additional cells (not only the cell i=2) can contribute to the error by being selected before tin [Fig. 2d]. Focusing again on cluster size n=1,the error in this more general scenario is given by:
| 3 |
Cells Ci, i=2,3..K can potentially be chosen before tin, and therefore contribute to the error. Cells Ci, i>K cannot contribute to the error as tr ¯ + Kτb−τσ>tr ¯ + δt + τσ. Note that the possibility that a cell will be chosen before being inhibited and therefore contribute to the error decays with the distance of the cell from the first cell i=1,as such cells will be activated later. Therefore, for a large i the corresponding integral in the product in (3) approaches 1. In this view, the error can be presented as a series whose leading term is obtained by calculating the error probability with respect to cell i=2 alone, and whose remaining terms stem from the remaining neighbors which we consider as perturbations whose contribution to error decreases the further they are from the first cell:
| 4 |
where is the probability for no error to occur after accounting only for cell i=2 which we calculated in (2). are corrections of higher orders. As discussed above, in the geometrical approach, the leading term represents a single requirement ( and therefore contains only two-dimensional geometrical shapes. Higher order terms represent multiple constraints and will therefore be represented by shapes of higher dimension; for example, if two cells contribute significantly to the error (i=2,3), then the correct selection would require that both and . This can be represented by shapes of three dimensions. In general, when accounting for j≤K potentially selected cells before tin, the error is represented by shapes with (j+1) dimensions.
Figure 2e shows the general solution for ε(τb,δt,n=1) in (3), in which erroneous selection of all potential cells i=2,3..K was accounted for ( and . The extent to which (2) is a valid approximation of (3) depends on the value of τb; the larger is τb, the better the approximation; it is accurate when τb≥1 and differs more from the exact error given by (3) as τb decreases and the contributions of cells i≥3 become more significant.
Error probability for a cluster of size n=2
The error probability in the case n=2 can be calculated following a similar line of reasoning. Now, a proper selection of only two cells is obtained when the second cell is selected before the time at which the first cell begins secreting the inhibitory signal, whereas cells Ci, i=3,4,5… are selected after that time. Again we first start with the simplest case in which an error can only stem from cells at the cluster borders; that is if either cell i=2 is selected after tin (leading to n=1) or if cell i=3 is selected before tin (leading to n=3), but cells i>3 do not have this potential of being selected before tin [Fig. 3a]. Note that if both errors occur together, the cluster will be of the correct size n=2, but will not be continuous; hence, we consider this as an erroneous selection. The error is therefore given by:
| 5 |
As in (2), the first integral captures the possible times for the first cell to produce inhibition. For each possible tin, the second integral captures the possible times for selecting the second cell, while demanding this time to be before tin and the third integral captures the times for selecting the third cell after tin.
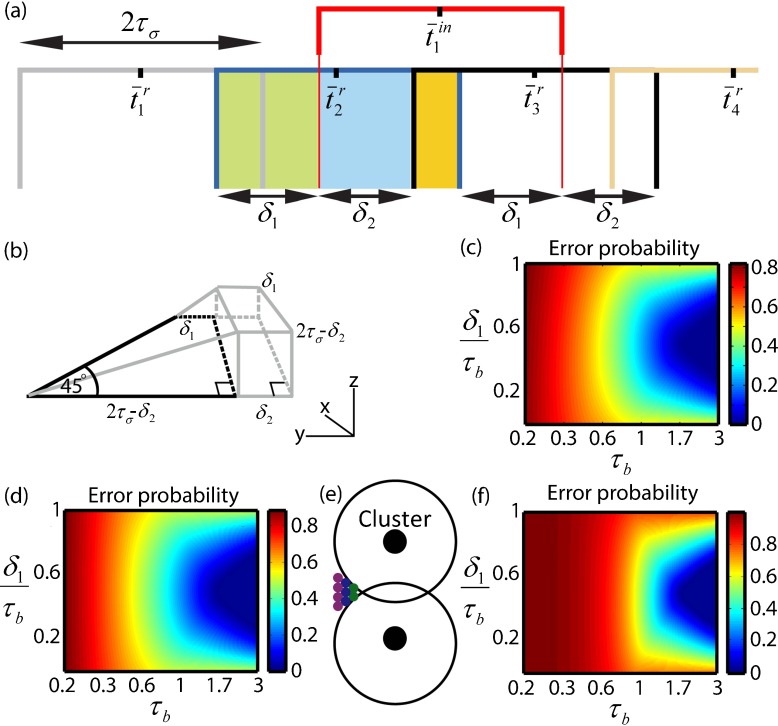
Fig. 3.
Error probability in cluster formation for the cases n≥2. a Error probability for n=2 is calculated by dividing the distribution of into different domains (in green, blue and yellow). b A three-dimensional shape whose volume represents the probability for , when is chosen from the yellow rectangle in (a). The trapezoid in black on the xy plane represents the probability . For each possible domain for choosing t in, varying between the large and small bases of the black trapezoid, a trapezoid on the yz plane can be drawn (in gray), representing . The angles of all gray trapezoids are 45∘ (not drawn). To obtain the probability calculated in (6), the volume should be normalized by . c Error probability ε as a function of τ b and δ 1 for the case n=2. ε increases sharply when τ b<1. d Error probability ε as a function of τ b and δ 1 for the case n=8. The error differs more from the case n=2 in (c) as τ b decreases and additional cells can potentially be selected after t in, compared to the case n=2 in which only cell i=2 can potentially do so. e Cluster formation in two dimensions. Different columns, represented by different colors, contain cells activated at the same time. The number of cells in each column is determined by the column’s position with respect to the inhibition circles. f Error probability for the case n=3 columns shown in (e). Since erroneous selection of many more cells (particularly at the cluster border) can now lead to error, ε increases sharply much earlier, at τ b=1.5
The major difference from the case n=1 discussed above is that for the simple case of n=1 we sought a single requirement—that is —which could be represented by two-dimensional shapes. Now, also the simplest case incorporates the multiple requirements , for correct selection of cells on both sides of the cluster border. In this aspect, all cases of n≥2 are the same, sharing the same demand for correct selection, which also gives the first-order term in the error if not obeyed. Compared to n=1, three-dimensional shapes represent the multiple requirements in the simplest cases for n≥2, as shown below. It is therefore helpful to first solve explicitly the error for n=2 and then generalize for larger clusters.
As we did for (2), we can divide (5) into different terms compatible with geometrical shapes. Let us do so by examining three possible scenarios, in which was selected from the green, blue or yellow rectangular domains in Fig. 3a and terming the probabilities for non-erroneous cluster formation in each of the cases as , and respectively. For simplicity we set tr ¯ + τb−τσ≡0 and change the notations by denoting δ1 = δt−τb and δ2 = τb−δ1, where δ1 is bounded by 0<δ1< τb [Fig. 3a]. Adjusting (5) accordingly, is simply given by setting the upper limit in the second integral to fit with the green rectangular borders in Fig. 3a:
The second integral in gives the height of the two following shapes: a cuboid obtained for when the third integral is 1 (when max{tin,τb} = τb is the lower limit of the third integral and the upper limit of the first integral) whose base is the rectangle and volume is . A prism obtained for (maxtinτb) = tin whose base is the trapezoid and volume is . Thus, .
Obtaining is similar. The only change is that now is chosen from the blue rectangular domain, and therefore:
The two possible shapes are now prisms, depending on the lower limit of the third integral (or the upper limit of the second integral): a prism for when the third integral is 1 (the prism’s height), whose base is a triangle originating from the first two integrals and whose volume is . A prism obtained when max{tin,τb} = tin, whose base is the same trapezoid as of , whose height is and volume is therefore . Thus, .
To obtain , we need to refer to the yellow rectangular domain out of which is now chosen where is required simultaneously. In Fig. 3b, the black trapezoid on the xy plane represents the chances for . As for all previous trapezoids, the trapezoid’s large base denotes the maximal possible domain for choosing tin (when is minimal) and its small base denotes the minimal possible domain for choosing tin (when is maximal). For each of the allowed domains of tin, varying between 2τσ−δ2 and δ1 (lower and upper bases of the black trapezoid), a trapezoid on the yz plane (in gray) can be drawn, representing the respective probabilities for . For instance, in the case when is minimal (lower base in black), the large base of the trapezoid on the yz plane corresponds to when is maximal (tin is minimal) and the small base corresponds to when is minimal (tin is maximal). This shapes volume can be obtained from (5), after resetting the limits of the second integral, or be calculated directly from Fig. 3b as . The error in the simplest scenario for n=2 is therefore:
| 6 |
Note that even for τσ<τb<2τσ as in Fig. 3a, cell i=4 can potentially be chosen before tin if δ1+2τσ>2τb.
In the general case for n=2, when more cells (not only cell i=3) can potentially be selected before tin, the error is:
| 7 |
As we did for n=1 we can write (7) as a series whose leading term which we calculated in (6) and contains three-dimensional shapes, captures error stemming from cells at the cluster border (cells i=2,3) where chances for mistake are the highest. The remaining terms of the series contain shapes of four and higher dimensions (i.e., requires that and and will therefore be represented by four-dimensional shapes).
Figure 3c shows the solution for ε(τb,δ1,n=2) in (7), using the normalized terms and . As shown, the error probability increases sharply when the delay in activation between adjacent cells is small relative to the distribution width of the noise (τb<1). Error is also dependent on the parameter capturing the delay between selection to inhibition production (δ1 = δt−τb); when, the error increases with δ1 as a result of erroneous selection with respect to cells i≥3 and when , the error decreases with δ1 as a result of erroneous selection with respect to cells i=2. Note that for the case n=1 [Fig. 2c and e], the error only increases with δt, since erroneous selection can only occur with respect to cells i>1. Supplementary Fig. ??1 shows the similarity between the approximated error [given by (6)] and (7) for different values of τb.
Error probability for a general n in one and two dimensions
With the same approach, we obtain the error for any given n = n′ as:
| 8 |
Cells Cj, j = n′(n′−1),…,M (M≥2) can potentially be chosen after tin and therefore contribute to the error. Cells Ci, i = n′+1,…,K can potentially be chosen before tin and therefore contribute to the error. For a given τb, which determines the number of cells that can contribute to the error, K−1≤M≤K+1, depending on δ1.
As for the case of n=2, (8) can be written as a series, whose first-order term stems from cells n′ and n′+1 at the cluster border, represented by three-dimensional shapes, whereas shapes of higher dimensions represent terms of higher order. Thus, all cases in which n≥2 are not essentially different with respect to the leading term in the series describing their error probability, but might differ in the number of the remaining terms for a given τb (in cases of a larger n, more cells might be required to be selected before tin). Figure 3d demonstrates that the solution for ε(τb,δ1,n=8) in (8) is very similar to the solution for ε(τb,δ1,n=2) in Fig. 3c, having a higher error probability of only ∼0.05 when τb is minimal (τb=0.2) and the largest number of additional cells can potentially be selected after tin . This means that when n≥2, the error is roughly independent of n, implying that in order to minimize relative fluctuations in cluster size, n should be large. These fluctuations (errors) are bound to occur in the realm of biologically measured parameters, as shown in the next section. Optimal cluster size is ultimately obtained by considering other constraints in the system impinging upon a large n, such as cluster size not exceeding the limit at which all cells in the cluster contact directly for the proper latter process of cluster refinement.
Equations (1–8) above can readily be generalized for two-dimensional clusters [Fig. 3e]. Here, n represents the number of columns in the cluster, rather than the number of cells. The number of cells in each column depends on the column’s placement between the inhibition circles. The error for any given n = n′ in two dimensions is:
| 9 |
where l(j) and q(i) are the number of cells in the columns that can potentially be selected before or after tin respectively, and all other notations are otherwise the same as in (8).
Figure 3f shows the solution for ε(τb,δ1,n=3) in (9), corresponding to the error in the formation of the two-dimensional cluster of ten cells shown in Fig. 3e. Here, the error has a high probability of occurring even when τb≲1.5, as many more cells at the cluster’s border contribute to the error’s first term (four cells in column i=3 and five cells in column i=4), compared to a maximum of two cells at the cluster border in one dimension.
Assessing error probability in cluster formation by estimation of the activation delay τb
Following the formulations above (6)–(9), we investigated whether, based on biological measurements, errors in cluster formation have a high probability of occurrence. For this purpose, we sought to evaluate the normalized , since the realm in which the activation-delay is located is a key predictor for error. In a one-dimensional system, error in cluster formation can practically be avoided if τb>1 (given , whereas if τb<1 errors are highly probable [Fig. 3c and d]. In a two-dimensional system, errors might be avoided if τb>1.5, but are practically inevitable if τb<1.5 [Fig. 3f].
For obtaining a rough estimation of , the time lag between activation of adjacent cells (or columns of cells in two dimensions) τb needs to be assessed and then divided by the distribution width capturing the variability in the molecular constituents of the patterning network interpreting the activation signal 2τσ. The long-range activation wave lays down a new column of cells roughly every two hours [17], distanced ∼6 cells from the column behind [18] and compatible with a bias of ∼20 minutes between activation of adjacent cells. The distribution width, 2τσ, can be estimated by the time lapse between activation (initial elevation of the pro-neural gene atonal) and selection (expression of the gene senseless as part of a cluster), which is roughly 15 cells [19]; namely 15 times the bias above multiplied by the noise level in the system. Altogether, the normalized bias in the process of cluster formation can be approximated as:
| 10 |
Thus, even when assuming relatively low biological noise of less than 10%, the normalized τb is less than 1.
The error in cluster formation is also dependent on δ1, which is more difficult to estimate, as there is no clear indication of when the first cell in the cluster started producing inhibition. Notably, for a given τb the error is minimal when . In the biological realm of τb≈0.67 calculated by (10) when accounting for 10% noise, the error probability is ∼0.5 in one dimension [(7) and Fig. 3c] and ∼0.8 in two dimensions [(9) and Fig. 3f]. Error probability rises even more for other values of δ1 or when accounting for more noise. The formation of relatively large clusters is therefore highly beneficial, as they are less sensitive to absolute fluctuations in cluster size that are bound to occur.
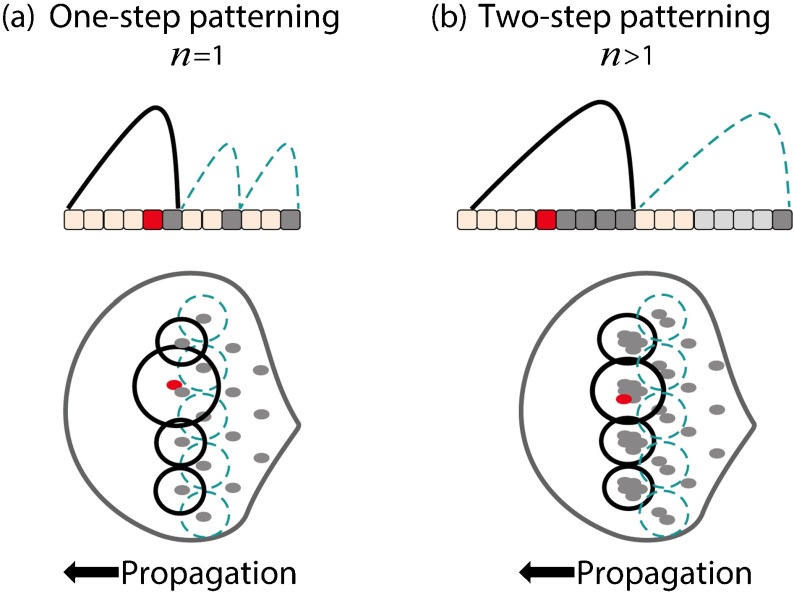
Figure 4 illustrates the benefit of two-step patterning (n>1) compared to one-step patterning (n=1). The effective inhibitor range, which determines the placement of newly forming clusters, is dependent on cluster size for given values of the inhibitor diffusion coefficient, the threshold above which a cell is inhibited and the inhibitor production and degradation rates (Appendix A). The relative difference between inhibition ranges surrounding clusters of different sizes (when all other parameters are fixed) therefore depends on the relative difference between their sizes; when the relative difference in sizes is large, the relative difference between inhibition ranges is large, and vice versa.
Fig. 4.
Comparison between one-step and two-step patterning. Posterior is to the right. a One-step patterning in one and two dimensions. A hexagonal pattern of single cells is generated a priori, separated by inhibition secreted by each cell. In one dimension (upper scheme), each cell secretes inhibition that determines the position of the cell ahead (inhibition secreted by posterior cells is in broken turquoise). If as a result of an error, a cluster of n=2 is formed (cell in red was erroneously selected), the relative error in size is , δ n being the deviation in cluster size from the desired n=1. Since the relative error is large, inhibition secreted by this cluster will reach a significantly larger effective range (black line), thus ruining periodicity. Similarly, in two dimensions (lower scheme) a new column of cells is placed in the intersection between the inhibitory circles surrounding posterior cells (broken turquoise circles). If because of an error an excess cell is selected (red cell), the inhibition surrounding this cluster will largely differ from its neighbors, severely affecting the placement of the anterior column ahead. b Two-step patterning in one and two dimensions. The relative error for n=5 is illustrated, when an excess cell was selected (in red), that is . Thus, the relative difference between the inhibition range surrounding the erroneous cluster to that of its neighbors is lower compared to that in (a), and the hexagonal periodicity is less affected. Broken turquoise line (upper scheme) and circles (lower scheme) illustrate the inhibition surrounding the original clusters of n=5
In one-step patterning, single cells produce the inhibitor [Fig. 4a]. Any deviation from n=1 [red cells in Fig. 4a] will largely affect the inhibition range surrounding the erroneous cluster, since the relative error in cluster size is large. This will lead to a distortion in the hexagonal periodicity, which may rapidly amplify in the layout of later columns. In two-step patterning, when multiple cells produce the inhibitor, the relative error in cluster size can be largely reduced [Fig. 4b]. Thus, an error in cluster formation [red cells in Fig. 4b] is of less consequence. Errors in clusters of size n≈10, like in the Drosophila eye, might hardly affect hexagonal periodicity.
A model for error probability in cluster refinement
Patterning in the eye is a sequential process in which each of the clusters, after directing the placement of the clusters ahead, is refined into a single founder cell that, together with the surrounding ∼20 cells later to differentiate, constitute a single eye unit (an ommatidium). Successful refinement of a cluster to a single founder cell is important; failure to refine, where two (or more) founder cells differentiate adjacently (‘twinning’), will lead to a distortion in the eye of the adult fly (eye roughening) [12].
Thus, we next sought to determine the probability of the failure to refine a cluster of n cells, whose formulation was described in the sections above, into a single cell. To this end, we extended a previous model by Barad and coworkers for minimizing error in lateral-inhibitory circuits [20]. Cluster refinement is achieved via transmission of inhibitory signals through direct contact of cells in the cluster [21]. The first cell to inhibit its neighboring cells will be selected as the founder cell. For simplicity and without loss of generality, we assume that contact-induced transmitted inhibition is a direct late consequence of activation by the propagating wave. Therefore, the first cell in the cluster to be activated is the one favored to become a founder cell. Since the system incorporates noise, not all cells respond the same to the activating signal, and it is therefore possible for some other cell, which was not the first to be activated, to be the first to inhibit its neighbors and become selected. The noise in the system also enables selection from a group of seemingly equivalent cells, which were activated at the same time (like cells in the same column in two dimensions), as one of these cells might be the first to interpret the activation signal and thereby inhibit its neighbors and become selected. We refer to successful refinement as the selection of a single cell, regardless of whether this cell was favorable. Refinement is considered erroneous if none of the cells in the cluster succeeded in inhibiting the remaining cells, and therefore more than one cell was selected.
We introduce noise to the process of refinement following the same approach we used in the process of cluster formation. However, since all the cells in the cluster now compete by trying to inhibit their neighboring cells, the model for cluster refinement is somewhat different than the model we formulated for cluster formation, in which only the first cell (or cells in two dimensions) produces the inhibitor. We term the time when each cell starts to induce contact-dependent inhibition of its neighbors tD (termed after the ligand mediating inhibition – Delta). To introduce noise, we choose tD out of some distribution P(tD), which we assume is uniformly distributed in a region of width 2τσ centered around the mean tD ¯. Since the molecular constituents governing the process of refinement are not the same as those governing cluster formation, τσ, which captures the strength of the molecular noise, is generally different in both processes. In two dimensions, each cell in column i is selected from P(tD) at time , depending on the column’s position [Fig. 5a].
Fig. 5.
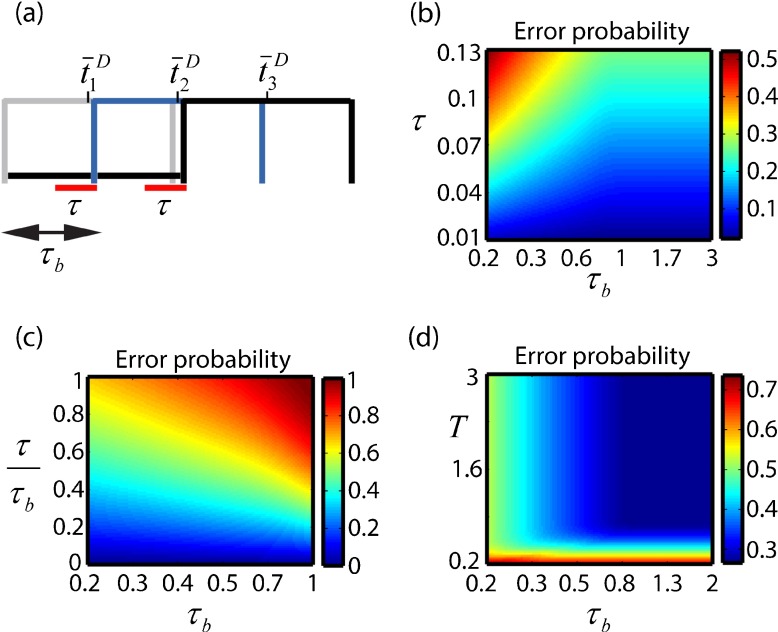
Cluster refinement in the presence of noise. a A probabilistic approach for choice of the times when contact lateral inhibition is induced. τ is the inhibition time-delay indicated in red. In this example, there is a chance for the cluster to refine into a single cell from columns 1 or 2, if all other cells in columns 1, 2 and 3 are chosen τ time units later. Though there is no chance for a cell in column 3 to inhibit cells in column 1, it is possible for a cell in column 3 not to be inhibited. b Error probability ε as a function of τ b and τ. ε increases with τ and decreases with τ b. In the region where τ b>1 + τ, the error merely stems from impaired refinement within the first column, which we assume has two cells. c Same as (b) for when is kept constant. Increasing τ b and τ at the same rate leads to an increase in ε. d ε as a function of τ b and of the developmental time window T. ε is insensitive to T down to some limit beneath which the probability error increases sharply
Based on a previous study [20], we introduce two more terms that play a role in the process of refinement: 1). The inhibition-time delay, termed τ, which is the time gap from when a cell first initiates contact-inhibitory signaling to when its neighbors start responding to that signal, i.e., are actually repressed. Note that this parameter was also incorporated indirectly in the previous process through the time gap between selection and inhibition δt (or δ1). 2) The developmental time window, termed T, which is some intrinsic time limit following activation after which, if not repressed beforehand, cells will become selected by default. Such selection by default has been proven true in other developmental processes [20].
Assume that the first cell starts inhibiting its neighbors at ; successful refinement is obtained if all the remaining cells in the cluster are chosen at some later times . Explicitly, if the second cell will be selected at , this cell will become selected before responding to inhibition from the first cell, leading to selection of two cells. In addition, if , the remaining cells are selected by default and therefore are not responsive to inhibition from the first cell. For obtaining a general term for the error ε(τb,nτ,T), namely the probability for more than one cell to be chosen from a two-dimensional cluster with n columns, it is useful to first define the probability for a cell in column i to be successfully inhibited by the first cell as . For simplicity we set the beginning of the first cell’s distribution tD ¯−τσ, to be 0. We term Pn→1 as the chance for a cluster of n columns to refine itself successfully into a single cell. The general term for the error ε(τb,nτ,T) is given by:
| 11 |
The integral in (11) sums all possibilities for inhibition production by the first cell (, which is one out of l(i) cells in any column i out of n. This cell must inhibit all other cells in its column i, reflected by the term in the integral, and further inhibit all cells in the remaining columns j≠i, reflected by the term , where q(j) is the number of cells in column j. Note that the upper limit of the integral must reach (i−1)τb+2τσ−τ only if column i has l(i)≥2 cells, for the first cell to successfully inhibit its neighbors in the column after incorporating the inhibition-delay (τ). If l(i)=1, the first cell has no neighboring cells in the column, and therefore can run until (i−1)τb+2τσ, unless the distribution for selecting a cell in an adjacent column starts beforehand. Thus, if l(i)=1, the upper limit of the integral should be min{(i−1)τb+2τσ,iτb−τ,T}.
A geometrical approach for probability calculation can be implemented as before and (11) can be represented as a series. The series leading term represents successful refinement solely in the first column i=1, out of which a cell has the highest probability to be chosen, and which will be represented by two-dimensional shapes if l(1)=2, or by higher dimensional shapes if l(1)>2. Terms of higher order successively stem from accounting for further columns i>1, namely for their inhibition or potential refinement (i.e., the second term is obtained when also accounting for successful inhibition or refinement of cells in column i=2).
The interplay between the parameters governing cluster refinement
We next wished to examine the relative contribution of each of the parameters influencing refinement to the error given by (11). To examine error dependence on the model parameters we assume the number of cells in each column to increase by one cell relative to the column behind, starting with a column of two cells [as in Fig. 3d]. We further assume at first that the number of columns in the cluster, n, is not a limiting factor in the refinement (as would be the case where, for example τb and τ require reference to three columns for refinement such as in Fig. 5a, but the cluster has only n=2 columns. See the ?? Supplementary Information for when this is not true). Normalizing by the distribution width as before ( ; ; , Fig. 5b illustrates ε(τb,τ) for when T>1 + τ. Here too, T does not play a role in cluster refinement since the first cell will surely reach within the time window T. As observed by Barad and coworkers [20], the error probability decreases with τb and increases with τ. The larger the time-gap in activation (τb), the lesser the chances for the first cell not to succeed in inhibiting the cells in further columns, and therefore the error decreases. The larger τ is, the higher the chances for inhibition by the first cell not to be interpreted before selection of other cells, and therefore the error increases. Note that when τb>1 + τ the probability for error (when τ is large enough) merely stems from failure to select one out of the two cells in the first column, as all cells in the other columns will surely be inhibited
Interestingly, when is kept constant [Fig. 5c], the relative influence of the two parameters (the activation-delay and the inhibition-delay) on the error probability is revealed, where ε(τb,τ) increases when increasing τb and τ at the same rate. These findings, concerning the limited capability of the bias (τb) to compensate for the inhibition-delay (τ), strongly suggest mechanisms that minimize τ for obtaining minimal error probability, rather than merely increasing τb.
Figure 5d illustrates ε(τb,T) when τ is a constant (τ=0.07), and shows that the error probability becomes sharply sensitive to T beneath a certain limit. Occurrences that result in an effective decrease in the developmental time window are thus unfavorable.
Assessing the key parameters controlling the error in cluster refinement
Erroneous refinement of clusters is very rare in wild type [22]. This imposes constraints on the key parameters controlling refinement. As shown above, a rapid inhibition-delay (τ) and a broad time-window (T) are the main contributors to successful refinement [Fig. 5]. However, a large activation-delay ( moderates the restraints on these parameters whereas a small one ( reinforces them; when the normalized activation-delay is large, can reach up to ∼0.04 and can reach down to ∼0.5 for error to rarely occur [Fig. 5b and d]. When is small, the requirements for a rapid inhibition-delay (τ) and a broad time-window (T) become more stringent. Thus, to assess the constraint stringency on the key parameters governing refinement, we newly estimated the normalized in the process, based on biological measurements.
We assume that initiation of the refinement process is directly dependent on long-range activation and therefore that the non-normalized bias τb is the same as in the process of cluster formation. However, the noise in the process of refinement, which we capture in a distribution of width 2τσ, is different from the noise in the process of cluster formation, as the molecular constituents of the patterning networks in both processes are different. Thus, a rough approximation of can be obtained by newly estimating the distribution width in the process of refinement, the bias itself being 20 min such as in the cluster formation. We now estimate 2τσ as the time lags between selection (first expressing the gene senseless as part of a cluster) and the time when a cell is singled out of the cluster, which is compatible with 12 cells (two columns of clusters) [19] multiplied by the bias. Altogether, the normalized bias in the process of cluster refinement can be approximated as:
| 12 |
Thus, also in the process of refinement, even when assuming relatively low biological noise of less than 10%, the normalized τb is less than 1. This implies stringent restraints on the key parameters governing refinement; namely the inhibition-delay to be sufficiently low ( and the developmental time-window to be sufficiently large () [Fig. 5b and d].
Discussion and conclusions
Biological systems are challenged by the need to perform effective computational tasks in a highly stochastic environment. The need to buffer stochastic variability largely restricts the space of possible mechanisms that can function in the biological setting. Noise buffering is particularly critical during embryonic development, where precise tissue patterning is needed to ensure the reproducibility of the adult body plan. Previously, we as well as others have shown how robustness is achieved in mechanisms splitting the tissue into distinct domains of gene expression. Here, we extended the analysis to the less studied paradigm of periodic patterning, where a lattice-like arrangement of selected cells self-organizes, as exemplified in the eye of the fruit fly Drosophila.
Periodic patterning of the fly eye is executed in two steps. First, evenly spaced clusters of ∼10 cells are defined. Second, these clusters are refined into a single selected cell. The first stage of cluster selection is critical not only for sequential refinement, but also for ensuring large-scale hexagonal periodicity. This is because clusters in each column define the position of clusters in the next column, allowing errors to propagate and in fact become rapidly amplified. Previous mathematical models that tested different aspects of robust patterning in the Drosophila eye did not highlight the significance of cluster formation and its importance for robust periodical refinement [15, 23]. We were therefore particularly interested in the parameters that control the sensitivity of cluster formation to spatial heterogeneities in cell size, shape, position or biosynthetic capacity.
For defining this sensitivity, we formulated a probabilistic model that allowed us to calculate the probability of error in cluster formation. Analytically, we approximated this error as a series, whose leading terms correspond to the error coming from misclassifying cells at the cluster border. We further showed that each term in this series can be represented as a geometrical shape, providing simple interpretation of the different error terms.
We found that cluster size has little influence on the error probability. Therefore, when attempting to reduce the relative fluctuations in cluster size it is beneficial to work with large clusters. This may explain why in the biological setting large clusters are formed prior to their refinement into a single selected cell, rather than selecting single cells already in the initial step. Related systems, which require precise ordering of solitary cells in the presence of spatial heterogeneity, might also adopt a similar strategy to buffer noise, temporarily expanding the initial selection, to be refined at a subsequent stage.
Efficient refinement of the selected clusters depends on all cluster cells being in close proximity to enable contact inhibition. This requirement limits the maximal cluster size. In the case of fly eye patterning, the effective cell proximity between cells is enhanced by the action of cytonemes [24, 25], specialized cellular extensions through which cells can communicate. Cluster size, which in the fly eye patterning contains ∼10 cells, is likely to present a compromise between maintaining of a robust cluster size (which favors large clusters) and achieving efficient refinement (which favors smaller and well-connected clusters).
The advantage of rapid signaling at the stage of cluster refinement was described in a previous study [20], which focused on refinement of clusters of fully equivalent cells. In the present context, the cluster cells are not fully equivalent, as they are selected sequentially and begin producing lateral inhibition signals at different times. This introduces an effective bias towards the selection of a particular cell—the one that was the first to be selected. We also find that in this case, rapid response to the lateral inhibition is essential and presents the main parameter controlling the error in cluster refinement. Surprisingly, perhaps, we find that biologically relevant biases (τb) cannot compensate for too slow signaling. Mechanisms that decrease τ, such as cis-mediated lateral inhibition [20], are therefore needed to eliminate erroneous refinement, as instances of twinning very rarely occur in wild type.
Another key parameter in optimizing cluster refinement is the developmental time window (T); in other words, the time over which the lateral inhibition signals can prevent selection before cells are selected by default. This time window should be sufficiently long to allow proper refinement. T can be effectively increased by local signaling that enhances the accumulation of inhibitors. In a recent work, we argued that a short-range activator must be assumed to enable robust cluster selection, and identified the secreted protein Scabrous as the predicted activator [26]. Lack of Scabrous results in a ‘twinning’ phenotype (selection of two adjacent cells) [12]. We propose that the twinning phenomenon results from an effective decrease in T when the activator Scabrous is absent.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank I. Averbukh and M. Chapal from the Barkai laboratory for useful discussion. This work was funded by grants from the ERC and the Minerva Foundation to N. B. who is an incumbent of the Lorna Greenberg Scherzer Professorial Chair.
Appendix: A: Mathematical model for cluster formation
The formulation of a time-based approach model for understanding the process of cluster formation is described in detail in a separate work (Ref. [15] and [26] in the main text), and is mentioned here briefly for completeness. This model considers three variables: a long-range, non-autonomous activator h, a non-autonomous short-range inhibitor u and a self-inducing activator a. A selected cell is a cell that stably expresses a. The equations that describe the interplay between these variables in one dimension are:
| 13 |
where τa,τh,and τu denote the typical time scales of a, h and u that we used to normalize all other parameters; Pa,Ph,and Pu are the respective production constants; λa,λh,and λu are the respective degradation rates; Dh,and Du are the respective diffusion constants and 𝜃(a) is the Heaviside step function, which is equal to 1 for positive values and 0 otherwise.
Assuming a uniform propagation of h with a speed v, we replace the precise term for h with:
| 14 |
which has the solution:
| 15 |
We can now calculate the time when a cell i at position x is activated (h>h1), assuming zero initial level of a:
| 16 |
Similarly, the times when each cell becomes selected (a>aa) and when the first cell in each cluster reaches the threshold for inhibition production are given by:
| 17 |
| 18 |
The number of cells that become refractory to inhibitory signals before the first cell reaches the threshold for inhibition production determines cluster size n. Accordingly,
| 19 |
Substituting (17), (18) and (19), we obtain:
| 20 |
Appendix: B: Analytical term for error probability in cluster formation for the case n=2
The error in the general case for n=2, given by (7) can be written analytically as:
| 21 |
where j =2,3,4… and the operators η(tin),γ(tin) and φ(tin) are given by:
| 22 |
and where .
Here, q = j if P(tin) overlaps with , corresponding with the possible overlap of P(tin) with in Fig. 3a in the main text.
The error probability ε(τb,δ1) above stands for n=2, in which tin must be chosen prior to any number of overlapping cells chosen from the distribution for and the distributions to follow, but succeeds only . Obtaining ε for a general n (8) requires demanding tin to succeed a larger number of cells determined by τb, the net effect being a higher error probability.
Appendix: C: Analytical term for error probability in cluster refinement
As described in the main text, it is convenient to first define the probability for a cell i to be successfully inhibited by the first cell producing inhibition as:
| 23 |
The probability for error refinement (after normalizing all terms by, given by (11), can be written analytically as (see Table 1 for integral limits):
| 24 |
Table 1.
Limits of the integrals in (24)
| l i m 1 | n 1=1 | |
| n 1>1 | ||
| l i m 2 | n 1=1 | |
| n 1>1 | ||
| l i m 3 | ||
| l i m 4 | ||
| l i m 5 | ||
| l i m 6 |
These limits hold for when n≥j, so all overlapping probabilities must be considered when calculating ε. If n<j, namely only part of the overlapping distributions must be considered, j should be replaced by n in (24), and to terminate integration before referring to cells that do not exist, all terms of the forms and should be replaced with and , respectively
where n1…nj are the number of cells in the columns 1…j. We assumed the inhibition delay to be smaller than the distribution width and the bias (τ< min{τb,1}). in the upper limit for τb>1 + τ is the step function that equals 1 if n1=1 (in which case we demand ε=0 regardless of T), or otherwise equals 0.
References
- 1.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 2.Bar-Even A, Paulsson J, Maheshri N, Carmi M, O’Shea E, Pilpel Y, Barkai N. Noise in protein expression scales with natural protein abundance. Nat. Genet. 2006;38:636–43. doi: 10.1038/ng1807. [DOI] [PubMed] [Google Scholar]
- 3.Newman JRS, Weissman JS. Systems biology: many things from one. Nature. 2006;444:561–562. doi: 10.1038/nature05407. [DOI] [PubMed] [Google Scholar]
- 4.Barkai N, Shilo B-Z. Variability and robustness in biomolecular systems. Mol. Cell. 2007;28:755–60. doi: 10.1016/j.molcel.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 5.Stanojević D, Hoey T, Levine M. Sequence-specific DNA-binding activities of the gap proteins encoded by hunchback and Krüppel in Drosophila. Nature. 1989;341:331–335. doi: 10.1038/341331a0. [DOI] [PubMed] [Google Scholar]
- 6.Eldar A, Rosin D, Shilo B-Z, Barkai N. Self-enhanced ligand degradation underlies robustness of morphogen gradients. Dev. Cell. 2003;5:635–646. doi: 10.1016/S1534-5807(03)00292-2. [DOI] [PubMed] [Google Scholar]
- 7.Eldar A, Shilo B-Z, Barkai N. Elucidating mechanisms underlying robustness of morphogen gradients. Curr. Opin. Genet. Dev. 2004;14:435–439. doi: 10.1016/j.gde.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 8.Voas MG, Rebay I. Signal integration during development: insights from the Drosophila eye. Dev. Dyn. 2004;229:162–175. doi: 10.1002/dvdy.10449. [DOI] [PubMed] [Google Scholar]
- 9.Kumar JP. Building an ommatidium one cell at a time. Dev. Dyn. 2012;241:136–149. doi: 10.1002/dvdy.23707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar JP. My what big eyes you have: how the Drosophila retina grows. Dev. Neurobiol. 2011;71:1133–1152. doi: 10.1002/dneu.20921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frankfort BJ, Mardon G. R8 development in the Drosophila eye: a paradigm for neural selection and differentiation. Development. 2002;129:1295–1306. doi: 10.1242/dev.129.6.1295. [DOI] [PubMed] [Google Scholar]
- 12.Baker NE, Mlodzik M, Rubin GM. Spacing differentiation in the developing Drosophila eye: a fibrinogen-related lateral inhibitor encoded by scabrous. Science. 1990;250:1370–1377. doi: 10.1126/science.2175046. [DOI] [PubMed] [Google Scholar]
- 13.Jarman AP, Grell EH, Ackerman L, Jan LY, Jan YN. Atonal is the proneural gene for Drosophila photoreceptors. Nature. 1994;369:398–400. doi: 10.1038/369398a0. [DOI] [PubMed] [Google Scholar]
- 14.Baonza A, Casci T, Freeman M. A primary role for the epidermal growth factor receptor in ommatidial spacing in the Drosophila eye. Curr. Biol. 2001;11:396–404. doi: 10.1016/S0960-9822(01)00125-7. [DOI] [PubMed] [Google Scholar]
- 15.Lubensky DK, Pennington MW, Shraiman BI, Baker NE. A dynamical model of ommatidial crystal formation. Proc. Natl. Acad. Sci. U.S.A. 2011;108:11145–11150. doi: 10.1073/pnas.1015302108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heberlein U, Wolff T, Rubin GM. The TGFβ homolog dpp and the segment polarity gene hedgehog are required for propagation of a morphogenetic wave in the Drosophila retina. Cell. 1993;75:913–926. doi: 10.1016/0092-8674(93)90535-X. [DOI] [PubMed] [Google Scholar]
- 17.Roignant J-Y, Treisman JE. Pattern formation in the Drosophila eye disc. Int. J. Dev. Biol. 2009;53:795–804. doi: 10.1387/ijdb.072483jr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee EC, Hu X, Yu SY, Baker NE. The scabrous gene encodes a secreted glycoprotein dimer and regulates proneural development in Drosophila eyes. Mol. Cell. Biol. 1996;16:1179–1188. doi: 10.1128/MCB.16.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baonza A, Freeman M. Notch signalling and the initiation of neural development in the Drosophila eye. Development. 2001;128:3889–3898. doi: 10.1242/dev.128.20.3889. [DOI] [PubMed] [Google Scholar]
- 20.Barad O, Rosin D, Hornstein E, Barkai N. Error minimization in lateral inhibition circuits. Sci. Signal. 2010;3(129):ra51. doi: 10.1126/scisignal.2000857. [DOI] [PubMed] [Google Scholar]
- 21.Treisman JE. Retinal differentiation in Drosophila. Wiley Interdiscip. Rev. Dev. Biol. 2013;2:545–557. doi: 10.1002/wdev.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ready DF, Hanson TE, Benzer S. Development of the Drosophila retina, a neurocrystalline lattice. Dev. Biol. 1976;53:217–240. doi: 10.1016/0012-1606(76)90225-6. [DOI] [PubMed] [Google Scholar]
- 23.Pennington MW, Lubensky DK. Switch and template pattern formation in a discrete reaction-diffusion system inspired by the Drosophila eye. Eur. Phys. J. E Soft. Matter. 2010;33:129–148. doi: 10.1140/epje/i2010-10647-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roy S, Hsiung F, Kornberg TB. Specificity of Drosophila cytonemes for distinct signaling pathways. Science. 2011;332:354–358. doi: 10.1126/science.1198949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chou Y-H, Chien C-T. Scabrous controls ommatidial rotation in the Drosophila compound eye. Dev. Cell. 2002;3:839–850. doi: 10.1016/S1534-5807(02)00362-3. [DOI] [PubMed] [Google Scholar]
- 26.Gavish, A., Shwartz, A., Weizman, A., Schejter, E., Shilo, B.Z., Barkai, N.: Periodic patterning of the Drosophila eye is stabilized by the diffusible activator Scabrous. Nat. Commun. 7, 10461. doi:10.1038/ncomms10461 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.