Abstract
Thermodynamics is an important driving factor for chemical processes and for life. Earlier work has shown that each cancer has its own molecular signaling network that supports its life cycle and that different cancers have different thermodynamic entropies characterizing their signaling networks. The respective thermodynamic entropies correlate with 5-year survival for each cancer. We now show that by overlaying mRNA transcription data from a specific tumor type onto a human protein–protein interaction network, we can derive the Gibbs free energy for the specific cancer. The Gibbs free energy correlates with 5-year survival (Pearson correlation of –0.7181, p value of 0.0294). Using an expression relating entropy and Gibbs free energy to enthalpy, we derive an empirical relation for cancer network enthalpy. Combining this with previously published results, we now show a complete set of extensive thermodynamic properties and cancer type with 5-year survival.
Keywords: Cancer, Signaling networks, Gibbs free energy, Entropy, Protein-protein interactions, 5-year survival
Introduction
Early insights into protein–protein interaction (PPI) networks suggest that the complexity of changes in PPI network topology correlates with cancer stage [1, 2] and clinical outcomes [3]. If validated prospectively, this would offer a powerful tool not only in better prognostic but also in therapeutic applications by providing a rational basis for personalized drug selection that is informed by the mRNA expression data.
Several published studies appear to corroborate the above hypothesis by linking molecular data with clinical outcomes. Paliouras et al. [2] used mass spectrometry on prostate clinical samples to show how changes in the protein–protein interaction network architecture relate to Gleason score and prostate specific antigen (PSA). Similarly, Freije et al. [3] showed that gene expression profiling of gliomas correlated with patient survival. We expect any dimensionality reduction of the expression vector to correlate with cancer stage, although the correlation may be poor because of inherent noise in the data and/or from assumptions inherent in the dimensionality reduction algorithm. In order to reduce the combined uncertainty inherent to a PPI network or the expression datasets and to better reconcile disparate PPI networks, one can combine PPI networks, transcriptome, and survival data. The consolidation of these data sets into a coherent abstract model is not only likely to improve the quality of the information in each of these previously unrelated data types, but may improve the data quality sufficiently to use the information for personalized therapies.
There are several ways of measuring complexity of protein–protein interaction networks. Chung et al. [4] observe that loops of three to six proteins are highly prevalent in PPIs and that 96% of the proteins in these loops play a significant role in some biological function including mRNA metabolic processing and cell cycle regulation. Recent papers [5, 6] describe topological metrics of PPI cancer networks that correlate with 5-year cancer patient survival. Of particular interest are Breitkreutz et al. [7] and Takemoto and Kaori [8] who introduce a thermodynamic measure based on degree distribution. A degree distribution is essentially a Boltzmann probability distribution [9, 10], which allows us to consider real-world statistical thermodynamics as a conceptual framework within which to view cancer initiation and progression. This is easy to visualize at a molecular level because Boltzmann’s entropy is a function of the natural-log of the number of equivalent ways, or the number of energy states, for a molecule (a protein). If a protein interacts with two neighbors, it has two different energy configurations for that molecule. If a protein interacts with 20 neighbors, it has a greater number of energy configurations and hence higher entropy. Boltzmann entropy is directly related to network degree entropy in PPIs and represents a quantitative measure of the network’s complexity. A simple and ordered network will have a low value of entropy associated with it. A complex and less-ordered network will be characterized by a higher value of entropy. However, in thermodynamics, entropy reflects only one aspect of a statistical system consisting of many units, its arrangement among possible microstates. In addition, the constituent units may physically interact, which brings another aspect into the picture, the system’s enthalpy. Together, enthalpy and entropy define a function of state called the Gibbs free energy that contains both aspects of the system’s behavior.
As motivated by the statements above, the main focus of this paper is on thermodynamics of protein–protein interaction networks in cancer, with an emphasis on entropy and Gibbs free energy as two key measures describing the complexity and chemical energetics of interactions in these networks. We therefore do not discuss Shannon, Kolmogorov–Sinai, or other information-based entropies, which may be relevant to the problem in general but not particularly useful in the present context. In the present manuscript, we review some thermodynamic entropy measures of PPI networks and then describe how to compute Gibbs free energy for cancer networks and show its correlation with 5-year survival, which provides retrospective validation of these concepts. In a more general context, we suggest that these energy views of the PPI are close analogies to the Waddington epigenetic landscape.
Brief review of entropy measures to PPI networks
Without attempting an exhaustive review of entropy of PPI networks, we discuss a few key papers. Rashevsky [11] was the first to suggest degree entropy as a complexity measure for graphs. His “graphs” were aliphatic molecular structures, so by modern standards, they were small graphs. The extension of information theory to thermodynamics in networks was made by Dehmer and Mowshowitz [12], in their review of the application of various entropy measures to network analysis. One of the first papers to discuss an information-based entropy was by Demetrius and Manke [13] who studied evolution of networks as a means to understand biological fitness. Their model assumes directed links in the network and they utilize Kolmogorov–Sinai entropy along with Markov processes to describe the evolution (and thus the robustness) of the networks. They extended that work to include cellular robustness [14].
As we pointed out in the introduction, being able to combine PPI and transcriptome data could enable more accurate use of these inherently noisy data, and with appropriate analysis may lead to actionable insight for clinical applications.
West et al. [15] describe fixed PPI architecture and mRNA expression data to derive unique weighted networks for each cancer studied. They start with a PPI from www.pathwaycommons.org and transcription data for different cancers. They modified the PPI to contain weighted connections by incorporating the transcriptome data. The weights are Pearson correlation coefficients of gene expression between genes i and j, across multiple samples of the same cancer type. Then computing entropy, they suggest that the best drug targets are those protein nodes with the highest robustness. Their suggested targets are strongly based on the mRNA expression levels, across a population of samples. If some protein has a very high up-regulated mRNA expression, one could have deduced the importance of that node in the network without actually computing the entropy. This is how many targets are “discovered.” Benzekry et al. [6] suggests a different approach to target discovery based on the unique architecture of each cancer PPI.
Other recent attempts are being made to combine PPI network data and RNA expression data. The quest to find correlations between the PPI networks/transcription data and survival/prognosis has continued. In 2012, Liu et al. [16] defined a measure called state-transition-based local network entropy (SNE). It is a Shannon information measure that is probabilistically, or conditionally, dependent on the previous state of a local dynamical network—a Markov process. They used mRNA expression data at different stages of tumor development, overlaid it on PPI network data, and showed that SNE changes significantly with cancer progression. Others have used Shannon entropy measure to show that gene expression patterns of melanoma and prostate cancers group according to cancer stage [17]. Shannon entropy, unlike degree entropy, is not a thermodynamic measure.
Banerji et al. [18] use a slightly similar method to West et al. [15] to devise a different network entropy. They also use the PathwayCommons network and gene expression data. Using a mass action principle they assume a higher interaction probability if two genes are highly expressed and their protein products interact. Their main point is to show a difference in entropy between stem cells and differentiated cells. They also show entropy differences, with linear correlation, between normal tissue, cancer tissue, and cancer cell lines. This is very similar to the work of Rietman et al. [1], and the work we describe here.
The work we describe here is an extension of the results by [7, 8] who used unique KEGG (www.genome.jp/kegg) pathway networks for each cancer. They then computed the degree-entropy, or as we argued above, Boltzmann entropy, for the nodes in the network and the overall network. They showed a linear correlation between this entropy measure and overall 5-year cancer survival rate. Here, we describe how to calculate Gibbs free energy for the nodes in the PPI, and we show a linear correlation between Gibbs free energy and 5-year survival rate. We also derive an empirical relation between the observed entropy and the observed Gibbs free energy.
We now introduce Gibbs free energy, a thermodynamic measure encompassing both network complexity and cell thermodynamics (as represented by transcriptome), and show that it can be correlated with cancer survival. As we will see, Gibbs free energy is correlated with network complexity because it is thermodynamically a function of entropy and the network entropy is correlated with network complexity by degree distribution (Boltzmann distribution).
Theoretical background
The homeostasis of cells is maintained by a complex, dynamic network of interacting molecules ranging in size from a few dozen Daltons to hundreds of thousands of Daltons. Any change in concentration of one or more of these molecular species alters the chemical balance, or in terms of thermodynamics, chemical potential. These changes then percolate through the network, affecting the chemical potential of other species. The end result represents perturbations in the network manifesting as concentration changes, giving rise to changes in the energetic landscape of the cell. These energetic changes can be described as chemical potential on an energetic landscape only different in kind from the Waddington epigenetic landscape.
Mutational events invariably alter the chemical potential of one or more proteins and/or other molecular species within a single cell. Yet, two neighboring cancer cells in the same microenvironment may exhibit a different energetic landscape because the chemical potential is different within the two cells. Naturally, when a bundle of cells is harvested, for example in a biopsy, and the cells are digested to extract RNA for transcription analysis, the transcriptome is essentially an average of that bundle of cells. Since any given gene is typically transcribed into multiple copies of its corresponding mRNA molecule, the transcriptome can act as a surrogate for the concentration of the proteins. To support this conjecture, several research groups have described correlations of mRNA with protein concentrations [19, 20] and found Pearson correlation, r, to range from 0.4 to 0.8, in a large number of experiments across five different species. More recently studies of the human proteome across multiple tissue types included in the relevant transcriptomic analysis, and found an average correlation between transcription signal and mass spectrometry proteomic information to be 83% [21, 22].
In this connection, Huang et al. [23] proposed that RNA expression data are surrogate metrics for the protein state of cells and represent the concentration of specific numbers of individual proteins exposed to either dimethylsulfoxide or all-trans-retinoic acid. Thus, the authors first introduced the concept of a chemical energy landscape for cells. Following exposure to the chemical perturbation, the gene expression data were collected at different time points, cleaned to remove low-expression genes, and a self-organizing map created. A principal component analysis was then used to produce a map showing the energetic (chemical potential) trajectory of the cells. The transcriptome has been shown to correlate with protein concentrations [21, 22], and can be generally correlated to the state of the cell. Certainly there are high-throughput protein concentration techniques [24], but the transcriptome provides a higher number of measurements (probes) identified with gene label and readily mapped to protein–protein interaction networks (e.g., thebiogrid.org).
The dynamics of cells are coordinated and controlled by protein–protein interactions, and the complete set (known) PPIs gives rise to a network. The state-of-the-art database of these PPI networks is Biogrid (http://thebiogrid.org), described by Breitkreutz et al. [25]. It should be stressed that, even though state-of-the-art today, it is not complete, and does not describe the full species-specific PPI networks. There are several reasons for this including the fact that the proteome has not been fully mapped from open-reading frames to genes and proteins. Consequently, calculations of the networks’ properties such as entropy or the Gibbs free energy should be taken as estimates reflecting the present state of knowledge about these networks.
Here, we report the outcomes of merging two types of data, transcriptome and PPI networks, to compute the energetic state of cancer. We show a correlation between the Gibbs free energy and 5-year patient survival for different cancers. Below, we describe the calculation of Gibbs free energy of cells, outline the data sources, and present the results and discussion.
Proteins do not interact simultaneously with large numbers of neighbors, as would be implied by the PPI network view of some hub proteins (e.g., p53). Instead, the hub protein may be interacting with one or two neighbors at a time, forming a complex nanomachine part such as a ribosome. We make the ensemble assumption that many copies of the hub protein may be located in many places in cells and each of the copies may be interacting with a different protein partner. Therefore, we can assume an ensemble of the protein of interest, as well as that its interactions with its neighbors, are akin to an ideal gas mixture.
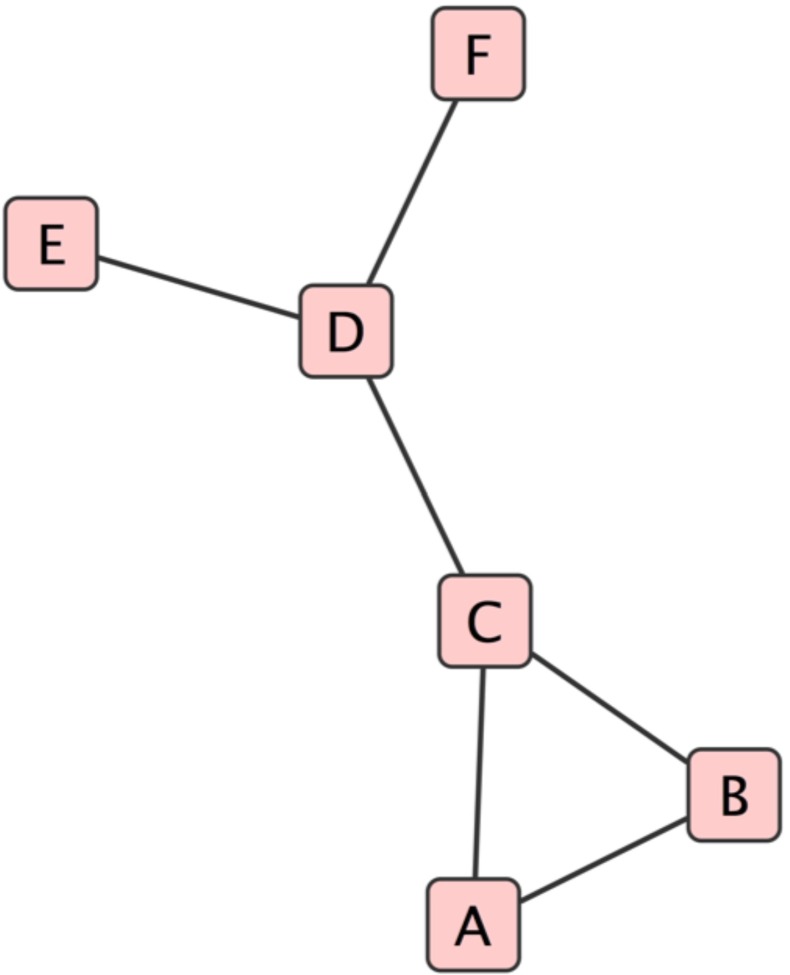
To help in the understanding of the calculation of Gibbs free energy from the transcriptome and the PPI perspective, we present a simple example shown in Fig. 1. Figure 1 shows a small network with individual nodes (proteins) within the network (labeled A, B, C, D, E, and F). For example, D represents a protein connected to E, C, and F by its edges (or links), which represent the interactions between the proteins. Because there is no directionality assigned to the links, the network is said to be an undirected graph. We compute the Gibbs free energy for protein D below. The network reveals that protein D interacts with proteins C, E, and F, and assuming an ideal mixture of these three proteins, we can assign a nominal chemical potential:
| 1 |
where ci denotes the concentration of protein i. Since (1) is written as a ratio, we can replace the concentrations with mole fractions, or even normalized expression, to give the same chemical potential. This relation is known as the entropy of mixing [26]. The nominal chemical potentials, represented with either concentration or expression, can be used to calculate a nominal Gibbs free energy for not only a single protein with its neighbors, but also for the entire network, for the cell, and the tumor as represented by the transcriptome.
Fig. 1.
An example of a small protein–protein interaction network created using Cytoscape®. The nodes (A–F) represent individual proteins, the lines, called edges, represent protein–protein interactions. No information about directionality of the interactions is implied. Protein D, for example, represents a protein connected to E, C, and F by its edges (or links). To compute the Gibbs free energy for node D in this network, we start with the normalized gene expression data as a surrogate for protein concentration of each node. Gibbs free energy for node D would be: normalized gene expression value divided by the sum of normalized expression of node D + the normalized gene expression values of the neighbors (E, F, C). This quotient becomes the argument for the natural logarithm. The coefficient of the natural logarithm is the normalized expression value for node D. All of this is summarized in (2)
The chemical potential can be used to compute the Gibbs free energy for any protein in the network as follows:
| 2 |
where the sum is taken over all neighbors and includes the concentration of the protein in question, ci. In conventional thermodynamics, Gibbs free energy scales the expression to thermal energy units, and we can drop the usual convention of including the RT coefficient. Furthermore, because we do not have information on the molar fractions, or molar concentrations, we substitute a normalized (rescaled) [0, 1] RNA transcription value in place of the concentrations, still maintaining the value of the Gibbs energy.
The rescaling of the transcriptome is performed in order to convert it to units of “concentration.” The data from TCGA are log-2 normalized and already collapsed to gene symbols. The log-2 normalization comes about from preprocessing the gene probe data and represents the transcription values. These preprocessed data are typically in the range [–10, 10]. To rescale, we find the minimum value of the transcription dataset in question (e min) and the maximum value (e max). Giving the range these data fall in, the maximum will be about 10 and the minimum will be about –10. The expression value for each gene in that transcriptome vector is then processed as: ci = (ei−emin)/(emax−emin).
This rescaling is justified from both a mathematical perspective and a chemical physics perspective. Negative values in the argument of the natural logarithm are undefined. The argument from a chemical physics perspective is based on concentrations. If a gene is very down regulated, it is not producing much protein. We assign the protein concentration to 0 for the gene that is the most down regulated. Whereas a gene that is highly up regulated will be producing a great deal of protein, the rescaling assigns the gene product that is most up-regulated, the highest concentration, to a value of 1.
Results
Using (1) and (2), we computed the Gibbs free energy for each node in the network as well as the sum of all nodes, i.e., total Gibbs free energy. The analysis is limited to cancers for which transcription data existed in the TCGA database. All of the data sets had used the Agilent®; platform, providing a very good gene ID match across all cancers listed in Table 1. The data, which were already log-2 transformed and collapsed into gene IDs, were averaged across samples for each gene to create a single expression vector representing the entire set for each cancer. Table 1 also shows the number of samples, the types of cancers, and the respective survival rates.
Table 1.
Summary table of the number of subjects in TCGA data sets and respective 5-year survival of individual cancer types from SEER
| TCGA | Cancer type | N | Percent | Gibbs |
|---|---|---|---|---|
| Name | 5-year survival | |||
| KIRC | Kidney renal clear cell | 72 | 68 | –5687 |
| KRIP | Kidney renal papillary cell | 16 | 68 | –4944 |
| LGG | Low-grade glioma | 27 | 50 | –6411 |
| GBM | Glioblastoma multiforme | 483 | 2 | –5668 |
| BRCA | Breast invasive carcinoma | 590 | 88 | –6674 |
| UCEC | Uterine corpus endometrial | 54 | 84 | –6310 |
| OV | Serous cystadenocarcinoma | 562 | 45 | –6233 |
| COAD | Colon adenocarcinoma | 174 | 65 | –6099 |
| READ | Rectum adenocarcinoma | 72 | 64 | –5861 |
| LUAD | Lung adenocarcinoma | 32 | 17 | –5916 |
| LUSC | Lung squamous cell | 155 | 40 | –6212 |
We collected a total of 11 cancers: KIRC (kidney renal clear cell, TCGA 2013b) (1); KIRP (kidney renal papillary cell); LGG (low-grade glioma); GBM (glioblastoma multiforme, TCGA, 2008); COAD (colon adenocarcinoma, TCGA 2012a); BRCA (breast invasive carcinoma, TCGA 2012c) (2); LUAD (lung adenocarcinoma); LUSC (lung squamous cell, TCGA 2012b) (3); UCEC (uterine corpus endometrial, TCGA, 2013a) (4); OV (ovarian serous cystadenocarcinoma); READ (rectum adenocarcinoma). Gibbs free energy included in this table is the average of the respective number for each individual cancer and was computed using Eq. (2)
Before actually overlaying the expression data on the PPI network, the average expression vector is rescaled to be in the range [0, 1], effectively setting highly up-regulated gene expressions to 1 and highly down-regulated gene expressions to 0. A base assumption was made that previously established correlation that highly up-regulated genes result in a high protein concentration and highly down-regulated genes result in a very low protein concentration [22, 23]. This prevented any negative argument in the natural logarithm of Eq. (3), and provided consistency from a chemical physics perspective. The calculated Gibbs values are shown in Table 1.
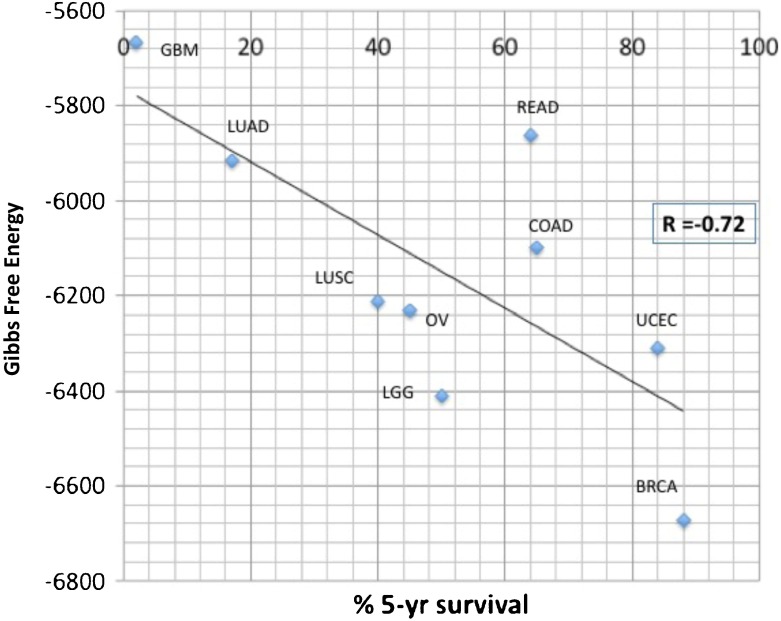
A plot of Gibbs free energy values versus percent 5-year survival for these cancers is shown in Fig. 2. There are nine cancers shown in the graph (GBM, LUAD, LUSC, READ, COAD, OV, LGG, UCEC, BRCA) with Pearson r correlation of –0.7181, and p value of 0.0294. The Spearman correlation is –0.633 with a p value of 0.0671. The Kendall tau test correlation is –0.555 with a p value of 0.0371. These statistics do not include KRIC (“kidney renal clear cell”) and KRIP (“kidney renal papillary cell”) abnormal tissue growths, which, even though highly proliferative and destructive, are of questionable malignant potential. If one were to include these two abnormal growths (KRIC and KIRP) in the analysis, the correlation would drop to –0.016.
Fig. 2.
Gibbs free energy and the probability of 5-year survival. Data from the TCGA gene list were overlaid on BioGRID® in order to merge protein–protein interaction network data with transcription data using Eq. (3). As evident, Gibbs free energy can be correlated with 5-year survival with an r coefficient of –0.72. We have excluded KIRC and KIRP because the biology of neuroectodermal and epithelial cancers differ from KIRC and KIRP. The inclusion of KIRC and KRIP in the calculation decreased correlation to –0.21
For comparisons, we used another measure of the expression data versus survival. We calculated singular values using numpy.lanalg.svd(X) in Python and compared them to survival. The first three singular values versus survival gave r correlations of –0.070, + 0.115, + 0.176, respectively (leaving out KIRC, KRIP). These are very poor correlations, and it is reasonable to conclude that Gibbs free energy is more effective in evaluating a real effect on survival, because it is associated with significant changes in energy of a signaling protein network in a cancer cell. An important implication of the correlation between Gibbs free energy and survival is that the higher the Gibbs free energy absolute value of a given cancer type, the more robust it is against external perturbations and the lower the probability of patient survival over a 5-year period. This is consistent with other concepts in physics where Gibbs free energy is a measure of stability of a thermodynamic system. Gibbs free energy and entropy are both thermodynamic measures, and because the observations are similar, we can compare the two thermodynamic measures. Physical systems in equilibrium have a statistical tendency to reach states of maximum entropy (when thermally isolated) or minimum Gibbs free energy (when kept at a constant ambient temperature). Although biological systems are open and far from thermodynamic equilibrium, we expect some aspects of their behavior to be driven by tendencies dictated by thermodynamics or thermodynamic-like considerations. In this paper, we show that reaching a Gibbs free energy minimum for the PPI aspect of cancer cell dynamical interactions is akin to a principle of maximum entropy (second law of thermodynamics).
As noted in the Introduction, the degree distribution used by Breitkreutz et al. [7] is essentially a Boltzmann distribution. This allows us to compare entropy with Gibbs free energy. The empirical equation for the linear fit of the Gibbs free energy with survival without kidney cancer is G = 8.112σ+5753.9 (Fig. 2). Using the data from Breitkreutz et al. [7], we can write the empirical equation for the liner fit of entropy as S = −0.0087σ + 2.2731. Solving both of these equations for 5-year survival probability, σ, and equating, we find an empirical relationship between the PPI entropy and Gibbs free energy for cancer cells, namely G = 7873 − 932S. Note that in order to relate G and S, we used the absolute value of the Gibbs free energy. This is consistent with the fundamental thermodynamic relationship linking Gibbs free energy and entropy: G = H − TS. What remains to be analyzed in the future as more data sets become available is the nature of the proportionality constant playing the role of the absolute temperature, the character of which may either be a biological constant of fundamental importance or simply a fitting parameter. It is tantalizing to speculate that a fundamental biological constant similar to temperature exists. This has already been postulated in the context of metabolism in physiology where a formal analogy was made by Demetrius between temperature and cycle time for the turnover of metabolic biochemical reactions [27].
Discussion
Among other features, cancers can be viewed as severely mutated cells. The PPI networks we used do not consider mutations. In future analysis, we would expect to be able to include PPI networks that incorporate gene fusion protein products—the result of mutations. This would enhance the analysis considerably.
As information about cancer-related genomic alterations emerge and more and more data becomes available, we can begin to establish the relationships between protein–protein interaction network complexity and cancer progression. We provide Gibbs free energy, a thermodynamic measure encompassing both network complexity and protein concentration (transcriptome), and show that thermodynamics can be correlated with cancer survival. This allows us to potentially differentiate between normal and cancer cells using thermodynamic measures.
We have shown that there is no correlation between the singular values of the expression and survival, and pointed out that the first three singular values (leaving out kidney) versus survival gave r correlations of –0.070, + 0.115, + 0.176. This suggests that the expression data is not the most significant component for the analysis and that the PPI network must be playing a significant part. To establish that the network architecture itself does not account for the correlation of Gibbs free energy and survival either, we tested a random network. One can view the mathematical steps in (1, 2) as follows:
The symbol qG represent a quasi-Gibbs free energy, the symbol ξ represent the expression vector and the little network symbol represents the PPI network. This is analogous to a vector, vector-like product producing a scalar (vector dot product). In these calculations, the network architecture is fixed for all expression vectors, for all cancers. To evaluate whether the architecture of the network itself may play a role, we used random networks, more specifically, random perceptrons [28], and found the dot product for each expression vector with this perceptron network. We computed the indicated dot product, and found that these random networks did not correlate with survival (r = 0.094). Thus, the expression data and the PPI network are both needed for a meaningful Gibbs free energy. In effect, the PPI network provides a structure to the expression data.
It is worth mentioning that our approach to describe and quantify cancer cell networks in terms of statistical thermodynamics is deeply rooted in the methodology relevant to the cascades of biochemical reactions linking it to bioenergetics [29]. In fact, we may be representing only some aspects of the cancer cell’s complexity, namely the topology of the signaling networks, protein expression levels, and protein–protein affinities. Cellular metabolism may well be an additional aspect that needs future integration [30], provided sufficient empirical data can be obtained. Moreover, as shown in [31], a complete picture may require the incorporation of time-dependence. Interestingly, the time scales of biochemical reaction rates also differ between cancer and normal cells [31].
Data sources and methods
Data for several cancers from The Cancer Genome Atlas (TCGA) hosted by the National Institute of Health (http://cancergnome.nih.gov) were collected. The Cancer Genome Atlas is described by the TCGA-Research Network [32]. More specifically, we collected a set of data that used the Agilent platform G4502A and was pre-collapsed on gene symbols. We collected a total of 11 cancers: KIRC (kidney renal clear cell, TCGA 2013b) [33]; KIRP (kidney renal papillary cell); LGG (low-grade glioma); GBM (glioblastoma multiforme, TCGA), [34]; COAD (colon adenocarcinoma, TCGA 2012a) [35]; BRCA (breast invasive carcinoma, TCGA 2012c) [36]; LUAD (lung adenocarcinoma); LUSC (lung squamous cell, TCGA 2012b) [37]; UCEC (uterine corpus endometrial, TCGA, 2013a) [38]; OV (ovarian serous cystadenocarcinoma); READ (rectum adenocarcinoma).
We used the human protein–protein interaction network (Homo sapiens, 3.3.99, March, 2013) from BioGrid, which contains 9561 nodes and 43,086 edges. BioGrid (http://thebiogrid.org) [39, 40]. The entire human PPI was loaded into Cytoscape (version 2.8.1) [41]. The list of genes obtained from TCGA (full-length expression set was 17,814 genes) for a specific cancer was “selected” using the Cytoscape functions, the “inverse selection” of Cytoscape function applied, and the nodes and their edges were removed. The resulting network, which now included only those genes found in both Biogrid and TCGA, consisted of 7951 nodes and 36,509 edges. This Cytoscape network was unloaded as an adjacency list for processing by custom Python code using Python (2.6.4) with appropriate NetworkX functions.
We used two databases for survival data: The Surveillance Epidemiology and End Results (SEER) National Cancer Institute database, which contains detailed statistical information about the 5-year survival rates of patients with cancer, and the National Brain Tumor Society database.
Acknowledgments
EAR was partly funded by the Newman Lakka Cancer Foundation, and CSTS Healthcare. JAT acknowledges funding from NSERC, Canadian Breast Cancer Foundation and the Allard Foundation. GLK was funded by NIH NIGMS RO1 GM93050, and philanthropic funds from Newman Lakka Cancer Foundation, Campanelli Foundation, Jack in the Beanstalk Foundation, and Binational Science Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Heath.
Author Contributions
EAR conceived the idea. JAT and EAR collaborated on the thermodynamics. JP contributed key chemical physics concepts. GLK contributed cancer biology concepts. All authors contributed to writing the manuscript.
Contributor Information
Jack A. Tuszynski, Email: jtus@phys.ualberta.ca
Giannoula Lakka Klement, Email: glakkaklement@tuftsmedicalcenter.org.
References
- 1.Rietman, E., Bloemendal, A., Platig, J., Tuszynski, J., Klement, G.L.: Gibbs free energy of protein–protein interactions reflects tumor stage. http://biorxiv.org/content/early/2015/07/13/022491 (2015)
- 2.Paliouras M, Zaman N, Lumbroso R, Kapogeorgakis L, Beitel LK, Wang E, Trifiro M. Dynamic rewiring of the androgen receptor protein interaction network correlates with prostate cancer clinical outcomes. Integr. Biol. (Camb.) 2011;3:1020–1032. doi: 10.1039/c1ib00038a. [DOI] [PubMed] [Google Scholar]
- 3.Freije WA, Castro-Vargas FE, Fang Z, Horvath S, Cloughesy T, Liau LM, Mischel PS, Nelson SF. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 2004;64:6503–6510. doi: 10.1158/0008-5472.CAN-04-0452. [DOI] [PubMed] [Google Scholar]
- 4.Chung SS, Pandini A, Annibale A, Coolen ACC, Thomas NSB, Fraternali F. Bridging topological and functional information in protein interaction networks by short loops profiling. Sci. Rep. 2015;5:8540. doi: 10.1038/srep08540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hinow PR, Rietman EA, Omar SI, Tuszynski JA. Algebraic and topological indices of molecular pathway networks in human cancers. Math. Biosci. Eng. 2015;12(6):1289–1302. doi: 10.3934/mbe.2015.12.1289. [DOI] [PubMed] [Google Scholar]
- 6.Benzekry ST, Tuszynski JA, Rietman EA, Klement GL. Design principles for cancer therapy guided by changes in complexity of protein–protein interaction networks. Biol. Direct. 2015;10:32. doi: 10.1186/s13062-015-0058-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breitkreutz D, Hlatky L, Rietman E, Tuszynski JA. Molecular signaling network complexity is correlated with cancer patient survivability. Proc. Natl. Acad. Sci. U.S.A. 2012;109:9209–9212. doi: 10.1073/pnas.1201416109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takemoto K, Kihara K. Modular organization of cancer signaling networks is associated with patient survivability. Biosystems. 2013;113:149–154. doi: 10.1016/j.biosystems.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 9.Gronholm T, Annila A. Natural distribution. Math. Biosci. 2007;210:659–667. doi: 10.1016/j.mbs.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 10.Richmod P, Solomon S. Power laws are disguised Boltzmann laws. Int. J. Mod. Phys. C. 2001;12:333. doi: 10.1142/S0129183101001754. [DOI] [Google Scholar]
- 11.Rashevsky N. Topology and life: in search of general mathematical principles in biology and sociology. Bull. Math. Biophys. 1954;16:317–348. doi: 10.1007/BF02484495. [DOI] [Google Scholar]
- 12.Dehmer M, Mowshowitz A. A history of graph entropy measures. Inf. Sci. 2011;181:57–78. doi: 10.1016/j.ins.2010.08.041. [DOI] [Google Scholar]
- 13.Demetrius L, Manke T. Robustness and network evolution - a entropic principle. Physica A. 2005;346:682–696. doi: 10.1016/j.physa.2004.07.011. [DOI] [Google Scholar]
- 14.Manke T, Demetrius L, Vingron M. An entropic characterization of protein interaction networks and cellular robustness. J. R. Soc. Interface. 2006;3:843–850. doi: 10.1098/rsif.2006.0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.West J, Bianconi G, Severini S, Teschendorff AE. Differential network entorpy reveasl cancer system hallmarks. Sci. Rep. 2012;2:802. doi: 10.1038/srep00802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu R, Li M, Liu ZP, Wu J, Chen L, Aihara K. Identifying critical transitions and their leading biomolecular networks in complex diseases. Sci. Rep. 2012;2:813. doi: 10.1038/srep00813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berretta R, Moscato P. Cancer biomarker discovery: the entropic hallmark. PLoS One. 2010;5:e12262. doi: 10.1371/journal.pone.0012262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banerji CRS, Miranda-Saavedra D, Severini S, Widschwendter M, Enver T, Zhou JX, Teschendroff AE. Cellular network entropy as the energy potential in Wadddingtons’s differentiation landscape. Sci. Rep. 2013;3:3039. doi: 10.1038/srep03039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003;4:117. doi: 10.1186/gb-2003-4-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maier T, Guell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583:3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 21.Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S, et al. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilhelm M, Schlegl J, Hahne H, Moghaddas Gholami A, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H, et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 23.Huang S, Eichler G, Bar-Yam Y, Ingber DE. Cell fates as high-dimensional attractor states of a complex gene regulatory network. Phys. Rev. Lett. 2005;94:128701. doi: 10.1103/PhysRevLett.94.128701. [DOI] [PubMed] [Google Scholar]
- 24.Spindel S, Sapsford K. Evaluation of optical detection platforms for multiplexed detection of proteins and the need for point-of-care biosensors for clinical use. Sensors. 2014;14:22313–22341. doi: 10.3390/s141222313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Breitkreutz BJ, Stark C, Tyers M. The GRID: the general repository for interaction datasets. Genome Biol. 2002;3:PREPRINT0013. [PubMed] [Google Scholar]
- 26.Maskill H. The Physical Basis of Organic Chemistry. New York: Oxford University Press; 1986. [Google Scholar]
- 27.Demetrius L. The origin of allometric scaling laws in biology. J. Theor. Biol. 2006;243:455–467. doi: 10.1016/j.jtbi.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 28.Anderson J. An Introduction to Neural Networks. Cambridge: MIT Press; 1995. [Google Scholar]
- 29.Demirel Y, Sandler SI. Thermodynamics and bioenergetics. Biophys. Chem. 2002;97:87–111. doi: 10.1016/S0301-4622(02)00069-8. [DOI] [PubMed] [Google Scholar]
- 30.Demirel Y. Modeling of thermodynamically coupled reaction-transport systems. Chem. Eng. J. 2008;139:106–117. doi: 10.1016/j.cej.2007.07.079. [DOI] [Google Scholar]
- 31.Lucia U. Different chemical reaction times between normal and solid cancer cells. Med. Hypotheses. 2013;81:58–61. doi: 10.1016/j.mehy.2013.04.007. [DOI] [PubMed] [Google Scholar]
- 32.Cancer Genome Atlas Research Network. Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cancer Genome Atlas Research N. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cancer Genome Atlas Research Network Comprehensive moleclar characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cancer Genome Atlas Research Network. Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, Robertson AG, Pashtan I, Shen R, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Breitkreutz BJ, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, Lackner DH, Bahler J, Wood V, et al. The Biogrid Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]