Introduction
The human gut and the rhizosphere are environments colonized by highly diverse communities of microbes, which perform complex interactions with their host and carry out important functions including enhanced disease resistance and nutrient uptake. In humans they are involved in energy harvest and storage as well as in interactions with the immune system (Clemente et al., 2012). In plants they have profound effects on seed germination, seedling vigor, nutrition, plant health, and development of the innate immune system (Mendes et al., 2013; Berg et al., 2014a; Schikora et al., 2016). The composition of the microbial communities is host-specific and related to its health status (Smalla et al., 2001; Kinross et al., 2011; Berg et al., 2014a). Imbalances caused by disturbance-induced shifts in microbial species abundances can lead to disease outbreaks in both environments (Berendsen et al., 2012; Robles Alonso and Guarner, 2013; Berg et al., 2014b) and further to probable proliferation of pathogenic species (Van Elsas et al., 2012; Van Agtmaal et al., 2015).
To restore or maintain the health of the host, bio-based solutions supporting the pathogen-suppressing ability of the hosts' native microbiome can be applied, including probiotics, synbiotics and biocontrol agents (de Vrese and Schrezenmeir, 2008). Such methods aim to increase the abundance and activity of host beneficial bacteria (HBB). However, addition of HBB does not always result in the desired pathogen suppression due to insufficient establishment, i.e., lower survival and/or poor colonization rates of the HBB (Bashan et al., 2014).
Concepts from invasion ecology suggest that survival rates of invaders are inversely related to the diversity of the native microbiome. This can be explained by higher resource uptake and consequent reduction in niche availability (Mallon et al., 2015). In addition, prevailing physical and chemical parameters in the respective environment like texture, pore size distribution, and moisture content might not favor the establishment of the introduced HBB. For a long-term establishment of the HBB in the soil these abiotic factors have to be considered. In the gut, the colonization resistance determined by the commensal microbiome is linked to its capacity to exploit the available niches and to prevent the establishment of invaders via niche occupation (reviewed in Stecher et al., 2013). The knowledge on mechanisms of microbial invasions (Mallon et al., 2015) can be used to improve the survival of HBB in both environments.
Given that similar mechanisms drive microbial colonization and establishment in the gut and rhizosphere microbiomes, we suggest that biocontrol strategies could be similar for both environments (Ramírez-Puebla et al., 2013; Berg et al., 2015; Mendes and Raaijmakers, 2015). Here we develop possible strategies to ensure long-term establishment of HBB by manipulating niche availability.
Creating microhabitats for host beneficial bacteria by introducing minor disturbances
Several studies have shown that soils harboring low microbial biomass or low microbial diversity are more susceptible to colonization by other organisms (Fließbach et al., 2009; Van Elsas et al., 2012). Certain agricultural practices can result in major disturbances of the rhizosphere microbiome. Examples include disinfestation with chemical pesticides, heat treatment (Stapleton, 2000), radiation or anaerobic disinfestation (Van Agtmaal et al., 2015). Moreover, tillage systems may have major effects on the established community by reducing certain soil microbial populations, particularly fungi (Ventorino et al., 2012). Analogous events, leading to changes in the human gut microbiome, are the application of broad spectrum antibiotics, fecal transplantations (Landy et al., 2011; de Vos, 2013) or considerable changes in diet (Turnbaugh et al., 2009). Whilst major disturbances are frequently used to eliminate pathogens, those methods possibly also disrupt beneficial functions of the indigenous microbial community (Altieri, 1999; Geiger et al., 2010).
An alternative strategy is to introduce minor disturbances to create free niches for HBB's in both the rhizosphere and the human gut microbiome. This strategy aims to selectively empty niches in the existing community.
In the rhizosphere the introduction of accessory bacterial predators such as protozoa (e.g., flagellates, ciliates) or nematodes (Jousset et al., 2006; Abada et al., 2009; Pedersen et al., 2009; Freyth et al., 2010; Neidig et al., 2011; Müller et al., 2013) could foster biocontrol strains via enhanced selective predation when the biocontrol strain protects itself through production of antibiotics. The increase in predation pressure might also stimulate biocontrol strategies by direct predation on pathogens as well as nutrient turnover and bacterial activity in soil. Likewise, specific bacteriophages could be applied to selectively eliminate target bacterial species or strains. This strategy has been effectively shown as part of disease management for Rhizobium sp., Bacillus sp., Burkholderia sp., Xanthomonas sp., Pectobacterium sp. and Dickeya sp. (Evans et al., 1979; Sharp et al., 1986; Lynch et al., 2012; Chae et al., 2014; Santamaría et al., 2014; Czajkowski et al., 2015). For this approach, elimination of pathogens and reduction of soil bacterial species that directly compete with the biocontrol agents (i.e., those sharing similar metabolic capacities) are desirable. Due to their specificity, bacteriophages have also been used to treat gastrointestinal infections of bacterial origin in humans (Sulakvelidze et al., 2001; Abedon et al., 2011). Moreover, they were successfully used together with bifidobacteria to treat antibiotic-associated dysbacteriosis in infants (Litvinova et al., 1978). Therefore, bacteriophages represent an alternative to selectively wipe out bacteria (either pathogens or strong competitors) in the gut and to form a niche for potential HBBs to thrive. In the rhizosphere the use of bacterial helper strains, an application of targeted specific antibiotics or enzymes (e.g., chitinases; Herrera-Estrella and Chet, 1999) might affect the microbiome composition sufficiently to form free niches for HBB. Another possibility is to introduce minor changes in physical properties like pH value (Rousk et al., 2010), temperature (Van Veen et al., 1997; Haas and Défago, 2005), moisture dynamics or salinity (Canfora et al., 2014; Dini-Andreote et al., 2014).
Most of the methods described here apply to the rhizosphere (e.g., substantial temperature or salinity changes), but due to ethical concerns can only be considered in a limited manner for the human gut. Thus, the direct applicability to the human gut remains to be investigated. The concept of freeing/forming a microhabitat for HBB by minor disturbances in the rhizosphere or the human gut should be developed and optimized for different situations.
Apart from making an existing niche available for the HBB by removal of at least a part of the adapted community, creation of a new niche could also be taken into consideration.
Improvement of the environment—the human gut as a paragon for concepts in biocontrol
To alleviate competition and increase the chance of establishment of HBB in an environment that harbors a highly diverse microbial community utilizing all available resources can be enabled by adding specific energy resources, for example prebiotics. Prebiotics selectively stimulate growth and/or activity of the beneficial bacteria and facilitate their establishment in the heavily colonized gut (Teitelbaum and Walker, 2002; Tuohy et al., 2003). Moreover, administration of synbiotics, a combination of a probiotic (i.e., the HBB) and a prebiotic, has recently attracted attention (Schrezenmeir and de Vrese, 2001). The prebiotic provides a selective food source for the HBB enhancing its growth and establishment (Teitelbaum and Walker, 2002; Saulnier et al., 2008). The success of synbiotics has been demonstrated in vitro as well as in vivo (Bartosch et al., 2005; Saulnier et al., 2008). We suggest that the use of synbiotics in the human gut can serve as a paragon to enhance the establishment of HBB in the soil. In the rhizosphere the addition of a selective food source e.g., rhizopins (Oger et al., 2004) could be used to stimulate specific bacteria in the rhizosphere community.
Synbiotics for the soil
Parallels with prebiotics can be seen in the application of general resources to the soil, such as composts and green manures. These strategies have shown to be effective in the control of soil-borne diseases as they combine the introduction of biocontrol microorganisms with organic matter after the thermophilic phase low in competition and free nutrients. This substrate favors the growth of beneficial microbes and suppresses the growth of saprophytic pathogens (Hoitink et al., 1997; Hoitink and Boehm, 1999). A disadvantage of using this method, however, is varying compost quality, which results in inconsistent colonization by biocontrol agents and subsequent effects on disease-suppression (Sturz and Christie, 2003). To ensure the presence of the desired HBB, composts can be fortified with specific beneficial microorganisms or amended with substrates that stimulate growth and activity of a selected group of microorganisms (Haggag and Abo-Sedera, 2005; Chae et al., 2006; Dukare et al., 2011).
In addition, specific substrates and HBB can be combined to complement each other. Several studies have shown that certain carbon sources and minerals increase the activity of biocontrol bacteria (Duffy and Défago, 1999; Shaukat and Siddiqui, 2003; Kim et al., 2008). Moreover, plants are able to select for specific bacteria by exudation of sugars, polysaccharides, amino acids, and a variety of secondary metabolites (Teplitski et al., 2000; Badri et al., 2009). These compounds are comparable to mucosal glycans in the human gut. As a soil synbiotic, these compounds could be artificially applied in combination with the respective HBB. Not only nutrient sources, but also signaling molecules and chemo-attractants should be taken into account, which are often highly specific for certain bacterial species or even strains. Ultimately, engineering beneficial microbes or genetically modified plants that are capable of synthesizing certain enzymes quenching bacterial signal particles might allow for shaping microbial communities against plant host pathogens (Dong et al., 2001; Ryan et al., 2009).
To support a long shelf life and stability of the product, these compounds can be formulated with specific carrier materials, membrane stabilizers and buffering agents in fine-tuned quantities (Paau, 1998; Bashan et al., 2014). An example for such soil inoculum carrier is biochar (charcoal used as soil amendment), known to have positive effects on soil properties such as pH (Saxena et al., 2013; Hale et al., 2014) and potentially be amended with extra HBB-specific resources.
Utilizing the specificity of host-bacterium interactions
Selection of the appropriate crop plant or a particular bacterial genotype can significantly influence the growth and establishment of HBB in soil (Mazzola, 2004) as interactions between plant and bacterial genotypes are assumed to be highly specific. This specificity could also be used in the human gut hosting defined beneficial strains (Tap et al., 2009). This selection could counteract down-sides of synbiotics, in which the presence of the HBB usually decreases dramatically once the consumption of the prebiotic stops (Bezkorovainy, 2001). For the same reasons, enhancing indigenous soil bacteria should be considered as an alternative to introducing new strains as they are likely to be better adapted to the respective environment (Chaparro et al., 2012).
Outlook and conclusions
In our opinion, the future of the HBB application lies in milder treatment of soils by using case-specific nutrient-microbe combinations as well as individualized treatments of patchy field sites after field structure analysis. As summarized in Figure 1, we suggest considering approaches such as the use of minor disturbance combined with timely application of HBB to improve their establishment in the soil. Soil treatments could be selected in analogy to therapies chosen for human guts. A new term “synbiotics for the rhizosphere” could reflect such intention.
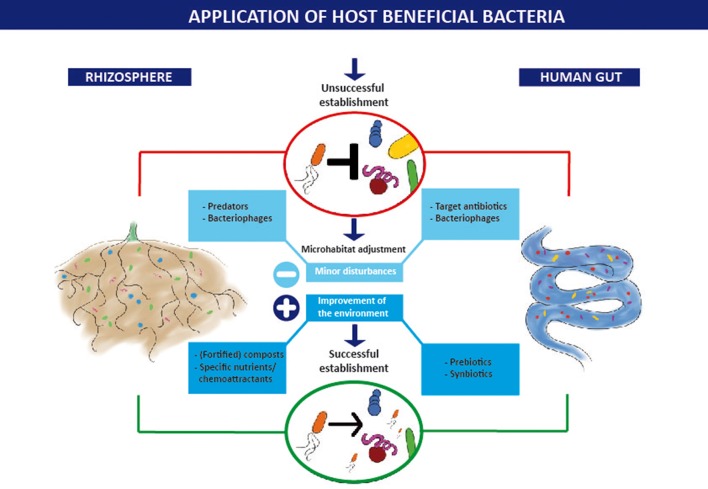
Figure 1.
Process of establishment of host beneficial bacteria (HBB) in the rhizosphere soil and the human gut. Application of HBB without additional measures frequently results in an unsuccessful establishment. To achieve a successful establishment of the HBB introduction of minor disturbances that empty host-associated niches in combination with improvements of the environment that create a new niche are suggested. Examples for the rhizosphere and the human gut.
It is assumed, that modern crop plants lost beneficial traits due to breeding programs conducted under conditions with high nutrient supply and the use of chemical pesticides. Consequently, breeding plants for beneficial plant-microbe interactions is an emerging research topic that might give birth to cultivars, which interact more efficiently with beneficial indigenous strains or with the applied HBB.
We see a sustainable future for agriculture by comparing methods for restoring or retaining the human gut microbiome and those altering the rhizosphere microbiome. Therefore, we suggest a paradigm shift in agricultural practices toward specialized treatment of the rhizosphere microbiomes as described in this work. We invite researchers of agricultural and human health related research areas to compare the methods of both fields and take into consideration findings of the other for their own future work.
Author contributions
The authors EA, AG, VK, MR, RS, OT, and SW contributed equally to this opinion paper. GB, WD, and JF shared senior contribution.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the graduate schools PE&RC, Ecology & Evolution, SENSE, and the organizers of the post graduate course “Microbial ecology.” This course gave the authors the platform for creating the outline of this article. MR participation in the course was possible through the Mobi4Health programme, within the European Union Seventh Framework Programme FP7/2007-2013 under grant agreement no. 316094. SW contribution was funded by the Netherlands Organisation for Scientific Research NWO (project no. 870.15.050). EA participation through fundings of the Austrian Research Promotion Agency FFG (project no. 836466). AG participation through fundings of NOW-WOTRO. The authors thank Monique Beijaert (NIOOKNAW) who contributed to the design of the figure. This is publication 6097 of the NIOO-KNAW.
References
- Abada E. A., Sung H., Dwivedi M., Park B. J., Lee S. K., Ahn J. (2009). C. elegans behavior of preference choice on bacterial food. Mol. Cells 28, 209–213. 10.1007/s10059-009-0124-x [DOI] [PubMed] [Google Scholar]
- Abedon S. T., Kuhl S. J., Blasdel B. G., Kutter E. M. (2011). Phage treatment of human infections. Bacteriophage 1, 66–85. 10.4161/bact.1.2.15845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altieri M. A. (1999). The ecological role of biodiversity in agroecosystems. Agric. Ecosyst. Environ. 74, 19–31. 10.1016/S0167-8809(99)00028-6 [DOI] [Google Scholar]
- Badri D. V., Weir T. L., van der Lelie D., Vivanco J. M. (2009). Rhizosphere chemical dialogues: plant–microbe interactions. Curr. Opin. Biotechnol. 20, 642–650. 10.1016/j.copbio.2009.09.014 [DOI] [PubMed] [Google Scholar]
- Bartosch S., Woodmansey E. J., Paterson J. C. M., McMurdo M. E. T., Macfarlane G. T. (2005). Microbiological effects of consuming a synbiotic containing Bifidobacterium bifidum, Bifidobacterium lactis, and oligofructose in elderly persons, determined by real-time polymerase chain reaction and counting of viable bacteria. Clin. Infect. Dis. 40, 28–37. 10.1086/426027 [DOI] [PubMed] [Google Scholar]
- Bashan Y., de-Bashan L., Prabhu S. R., Hernandez J.-P. (2014). Advances in plant growth-promoting bacterial inoculant technology: formulations and practical perspectives (1998–2013). Plant Soil 378, 1–33. 10.1007/s11104-013-1956-x [DOI] [Google Scholar]
- Berendsen R. L., Pieterse C. M. J., Bakker P. A. H. M. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. 10.1016/j.tplants.2012.04.001 [DOI] [PubMed] [Google Scholar]
- Berg G., Grube M., Schloter M., Smalla K. (2014a). Unraveling the plant microbiome: looking back and future perspectives. Front. Microbiol. 5:148. 10.3389/fmicb.2014.00148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg G., Grube M., Schloter M., Smalla K. (2014b). The plant microbiome and its importance for plant and human health. Front. Microbiol. 5:491. 10.3389/fmicb.2014.00491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg G., Krause R., Mendes R. (2015). Cross-kingdom similarities in microbiome ecology and biocontrol of pathogens. Front. Microbiol. 6:1311. 10.3389/fmicb.2015.01311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezkorovainy A. (2001). Probiotics: determinants of survival and growth in the gut. Am. J. Clin. Nutr. 73(2 suppl.), 399S–405S. [DOI] [PubMed] [Google Scholar]
- Canfora L., Bacci G., Pinzari F., Lo Papa G., Dazzi C., Benedetti A. (2014). Salinity and bacterial diversity: to what extent does the concentration of salt affect the bacterial community in a saline soil? PLoS ONE 9:e106662. 10.1371/journal.pone.0106662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae D. H., De Jin R., Hwangbo H., Kim Y. W., Kim Y. C., Park R. D., et al. (2006). Control of late blight (Phytophthora capsici) in pepper plant with a compost containing multitude of chitinase-producing bacteria. BioControl 51, 339–351. 10.1007/s10526-005-2934-x [DOI] [Google Scholar]
- Chae J.-C., Hung N. B., Yu S.-M., Lee H. K., Lee Y. H. (2014). Diversity of bacteriophages infecting Xanthomonas oryzae pv. oryzae in paddy fields and its potential to control bacterial leaf blight of rice. J. Microbiol. Biotechnol. 24, 740–747. 10.4014/jmb.1402.02013 [DOI] [PubMed] [Google Scholar]
- Chaparro J. M., Sheflin A. M., Manter D. K., Vivanco J. M. (2012). Manipulating the soil microbiome to increase soil health and plant fertility. Biol. Fertil. Soils 48, 489–499. 10.1007/s00374-012-0691-4 [DOI] [Google Scholar]
- Clemente J. C., Ursell L. K., Parfrey L. W., Knight R. (2012). The impact of the gut microbiota on human health: an integrative view. Cell 148, 1258–1270. 10.1016/j.cell.2012.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czajkowski R., Ozymko Z., de Jager V., Siwinska J., Smolarska A., Ossowicki A., et al. (2015). Genomic, proteomic and morphological characterization of two novel broad host lytic bacteriophages ΦPD10.3 and ΦPD23.1 infecting pectinolytic Pectobacterium spp. and Dickeya spp. PLoS ONE 10:e0119812. 10.1371/journal.pone.0119812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vos W. M. (2013). Fame and future of faecal transplantations – developing next-generation therapies with synthetic microbiomes. Microb. Biotechnol. 6, 316–325. 10.1111/1751-7915.12047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vrese M., Schrezenmeir J. (2008). Probiotics, prebiotics, and synbiotics. Adv. Biochem. Eng. Biotechnol. 111, 1–66. 10.1007/10_2008_097 [DOI] [PubMed] [Google Scholar]
- Dini-Andreote F., de Cassia Pereira e Silva M., Triado-Margarit X., Casamayor E. O., van Elsas J. D., Falcao Salles J. (2014). Dynamics of bacterial community succession in a salt marsh chronosequence: evidences for temporal niche partitioning. ISME J. 8, 1989–2001. 10.1038/ismej.2014.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y. H., Wang L. H., Xu J. L., Zhang H. B., Zhang X. F., Zhang L. H. (2001). Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 411, 813–817. 10.1038/35081101 [DOI] [PubMed] [Google Scholar]
- Duffy B. K., Défago G. (1999). Environmental factors modulating antibiotic and siderophore biosynthesis by Pseudomonas fluorescens biocontrol strains. Appl. Environ. Microbiol. 65, 2429–2438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dukare A. S., Prasanna R., Chandra Dubey S., Nain L., Chaudhary V., Singh R., et al. (2011). Evaluating novel microbe amended composts as biocontrol agents in tomato. Crop Prot. 30, 436–442. 10.1016/j.cropro.2010.12.017 [DOI] [Google Scholar]
- Evans J., Barnet Y. M., Vincent J. M. (1979). Effect of a bacteriophage on colonization and nodulation of clover roots by paired strains of Rhizobium trifolii. Can. J. Microbiol. 25, 974–978. 10.1139/m79-149 [DOI] [PubMed] [Google Scholar]
- Fließbach A., Winkler M., Lutz M. P., Oberholzer H.-R., Mäder P. (2009). Soil amendment with Pseudomonas fluorescens CHA0: lasting effects on soil biological properties in soils low in microbial biomass and activity. Microb. Ecol. 57, 611–623. 10.1007/s00248-009-9489-9 [DOI] [PubMed] [Google Scholar]
- Freyth K., Janowitz T., Nunes F., Voss M., Heinick A., Bertaux J., et al. (2010). Reproductive fitness and dietary choice behavior of the genetic model organism Caenorhabditis elegans under semi-natural conditions. Mol. Cells 30, 347–353. 10.1007/s10059-010-0125-9 [DOI] [PubMed] [Google Scholar]
- Geiger F., Bengtsson J., Berendse F., Weisser W. W., Emmerson M., Morales M. B., et al. (2010). Persistent negative effects of pesticides on biodiversity and biological control potential on European farmland. Basic Appl. Ecol. 11, 97–105. 10.1016/j.baae.2009.12.001 [DOI] [Google Scholar]
- Haas D., Défago G. (2005). Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 3, 307–319. 10.1038/nrmicro1129 [DOI] [PubMed] [Google Scholar]
- Haggag W. M., Abo-Sedera S. A. (2005). Characteristics of three Trichoderma species in peanut haulmscompost involved in biocontrol of cumin wilt disease. Int. J. Agric. Biol. 7, 222–229. [Google Scholar]
- Hale L., Luth M., Crowley D. (2014). Biochar characteristics relate to its utility as an alternative soil inoculum carrier to peat and vermiculite. Soil Biol. Biochem. 81, 228–235. 10.1016/j.soilbio.2014.11.023 [DOI] [Google Scholar]
- Herrera-Estrella A., Chet I. (1999). Chitinases in biological control, in Chitin and Chitinases, eds Jolles P., Muzzarelli R. A. A. (Basel: Birkhäuser Publishing; ), 171–184. [DOI] [PubMed] [Google Scholar]
- Hoitink H. A. J., Boehm M. J. (1999). Biocontrol within the context of soil microbial communities: a substrate-dependent phenomenon. Annu. Rev. Phytopathol. 37, 427–446. 10.1146/annurev.phyto.37.1.427 [DOI] [PubMed] [Google Scholar]
- Hoitink H. A. J., Stone A. G., Han D. Y. (1997). Suppression of plant diseases by composts. HortScience 32, 184–187 [Google Scholar]
- Jousset A., Lara E., Wall L. G., Valverde C. (2006). Secondary metabolites help biocontrol strains Pseudomonas fluorescens CHA0 to escape protozoan grazing. Appl. Environ. Microbiol. 72, 7083–7090. 10.1128/AEM.00557-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Jung H., Kim K., Park S. (2008). An effective biocontrol bioformulation against Phytophthora blight of pepper using growth mixtures of combined chitinolytic bacteria under different field conditions. Eur. J. Plant Pathol. 120, 373–382. 10.1007/s10658-007-9227-4 [DOI] [Google Scholar]
- Kinross J. M., Darzi A. W., Nicholson J. K. (2011). Gut microbiome-host interactions in health and disease. Genome Med. 3, 1–12. 10.1186/gm228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landy J., Al-Hassi H. O., McLaughlin S. D., Walker A. W., Ciclitira P. J., Nicholls R. J., et al. (2011). Review article: faecal transplantation therapy for gastrointestinal disease. Aliment. Pharmacol. Ther. 34, 409–415. 10.1111/j.1365-2036.2011.04737.x [DOI] [PubMed] [Google Scholar]
- Litvinova A. M., Chtetsova V. M., Kavtreva I. G. (1978). Evaluation of efficacy of the use of coli-Proteus bacteriophage in intestinal dysbacteriosis in premature infants. Vopr. Okhr. Materin. Det. 23, 42–44 [PubMed] [Google Scholar]
- Lynch K. H., Stothard P., Dennis J. J. (2012). Comparative analysis of two phenotypically-similar but genomically-distinct Burkholderia cenocepacia-specific bacteriophages. BMC Genomics 13:223. 10.1186/1471-2164-13-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallon C. A., van Elsas J. D., Salles J. F. (2015). Microbial invasions: the process, patterns, and mechanisms. Trends Microbiol. 23, 719–729. 10.1016/j.tim.2015.07.013 [DOI] [PubMed] [Google Scholar]
- Mazzola M. (2004). Assessment and management of soil microbial community structure for disease suppression. Annu. Rev. Phytopathol. 42, 35–59. 10.1146/annurev.phyto.42.040803.140408 [DOI] [PubMed] [Google Scholar]
- Mendes R., Garbeva P., Raaijmakers J. M. (2013). The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37, 634–663. 10.1111/1574-6976.12028 [DOI] [PubMed] [Google Scholar]
- Mendes R., Raaijmakers J. M. (2015). Cross-kingdom similarities in microbiome functions. ISME J. 9, 1905–1907. 10.1038/ismej.2015.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M. S., Scheu S., Jousset A. (2013). Protozoa drive the dynamics of culturable biocontrol bacterial communities. PLoS ONE 8:e66200. 10.1371/journal.pone.0066200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidig N., Paul R. J., Scheu S., Jousset A. (2011). Secondary metabolites of Pseudomonas fluorescens CHA0 drive complex non-trophic interactions with bacterivorous nematodes. Microb. Ecol. 61, 853–859. 10.1007/s00248-011-9821-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oger P. M., Mansoure H., Nese X., Dessaux Y. (2004). Engineering root exudation of Lotus toward the production of two novel carbon compounds leads to the selection of distinct microbial populations in the rhizosphere. Microb. Ecol. 47, 96–103. 10.1007/s00248-003-2012-9 [DOI] [PubMed] [Google Scholar]
- Paau A. (1998). Formulation of beneficial organisms applied to soil, in Formulation of Microbial Biopesticides, ed Burges H. D. (Dordrecht: Kluwer Academic Publishers; ), 235–254. [Google Scholar]
- Pedersen A. L., Nybroe O., Winding A., Ekelund F., Bjornlund L. (2009). Bacterial feeders, the nematodes Caenorhabditis elegans and the flagellate Cercomonas longicauda, have different effects on outcome of competition among the Pseudomonas biocontrol strains CHA0 and DSS73. Microb. Ecol. 57, 501–509. 10.1007/s00248-008-9455-y [DOI] [PubMed] [Google Scholar]
- Ramírez-Puebla S. T., Servín-Garcidueñas L. E., Jiménez-Marín B., Bolaños L. M., Rosenblueth M., Martínez J., et al. (2013). Gut and root microbiota commonalities. Appl. Environ. Microbiol. 79, 2–9. 10.1128/AEM.02553-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robles Alonso V., Guarner F. (2013). Linking the gut microbiota to human health. Br. J. Nutr. 109, 21–26. 10.1017/S0007114512005235 [DOI] [PubMed] [Google Scholar]
- Rousk J., Bååth E., Brookes P. C., Lauber C. L., Lozupone C., Caporaso J. G., et al. (2010). Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 4, 1340–1351. 10.1038/ismej.2010.58 [DOI] [PubMed] [Google Scholar]
- Ryan P. R., Dessaux Y., Thomashow L. S., Weller D. M. (2009). Rhizosphere engineering and management for sustainable agriculture. Plant Soil 321, 363–383. 10.1007/s11104-009-0001-6 [DOI] [Google Scholar]
- Santamaría R. I., Bustos P., Sepúlveda-Robles O., Lozano L., Rodríguez C., Fernández J. L., et al. (2014). Narrow-host-range bacteriophages that infect Rhizobium etli associate with distinct genomic types. Appl. Environ. Microbiol. 80, 446–454. 10.1128/AEM.02256-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saulnier D. M. A., Gibson G. R., Kolida S. (2008). In vitro effects of selected synbiotics on the human faecal microbiota composition. FEMS Microbiol. Ecol. 66, 516–527. 10.1111/j.1574-6941.2008.00561.x [DOI] [PubMed] [Google Scholar]
- Saxena J., Rana G., Pandey M. (2013). Impact of addition of biochar along with Bacillus sp. on growth and yield of French beans. Sci. Hortic. 162, 351–356. 10.1016/j.scienta.2013.08.002 [DOI] [Google Scholar]
- Schikora A., Schenk S. T., Hartmann A. (2016). Beneficial effects of bacteria-plant communication based on quorum sensing molecules of the N–acyl homoserine lactone group. Plant Mol. Biol. 90, 605–612. 10.1007/s11103-016-0457-8 [DOI] [PubMed] [Google Scholar]
- Schrezenmeir J., de Vrese M. (2001). Probiotics, prebiotics, and synbiotics—approaching a definition. Am. J. Clin. Nutr. 73(suppl.), 361S–364S. [DOI] [PubMed] [Google Scholar]
- Sharp R. J., Ahmad S. I., Munster A., Dowsett B., Atkinson T. (1986). The isolation and characterization of bacteriophages infecting obligately thermophilic strains of Bacillus. J. Gen. Microbiol. 132, 1709–1722 10.1099/00221287-132-6-1709 [DOI] [PubMed] [Google Scholar]
- Shaukat S. S., Siddiqui I. A. (2003). The influence of mineral and carbon sources on biological control of charcoal rot fungus, Macrophomina phaseolina by fluorescent pseudomonads in tomato. Lett. Appl. Microbiol. 36, 392–398. 10.1046/j.1472-765X.2003.01325.x [DOI] [PubMed] [Google Scholar]
- Smalla K., Wieland G., Buchner A., Zock A., Parzy J., Kaiser S., et al. (2001). Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl. Environ. Microbiol. 67, 4742–4751. 10.1128/AEM.67.10.4742-4751.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton J. J. (2000). Soil solarization in various agricultural production systems. Crop Prot. 19, 837–841. 10.1016/S0261-2194(00)00111-3 [DOI] [Google Scholar]
- Stecher B., Berry D., Loy A. (2013). Colonization resistance and microbial ecophysiology: using gnotobiotic mouse models and single-cell technology to explore the intestinal jungle. FEMS Microbiol. Rev. 37, 793–829. 10.1111/1574-6976.12024 [DOI] [PubMed] [Google Scholar]
- Sturz A. V., Christie B. R. (2003). Beneficial microbial allelopathies in the root zone: the management of soil quality and plant disease with rhizobacteria. Soil Tillage Res. 72, 107–123. 10.1016/S0167-1987(03)00082-5 [DOI] [Google Scholar]
- Sulakvelidze A., Alavidze Z., Morris J. G., Jr. (2001). Bacteriophage therapy. Antimicrob. Agents Chemother. 45, 649–659. 10.1128/AAC.45.3.649-659.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tap J., Mondot S., Levenez F., Pelletier E., Caron C., Furet J.-P., et al. (2009). Towards the human intestinal microbiota phylogenetic core. Environ. Microbiol. 11, 2574–2584. 10.1111/j.1462-2920.2009.01982.x [DOI] [PubMed] [Google Scholar]
- Teitelbaum J. E., Walker W. A. (2002). Nutritional impact of pre- and probiotics as protective gastrointestinal organisms. Annu. Rev. Nutr. 22, 107–138. 10.1146/annurev.nutr.22.110901.145412 [DOI] [PubMed] [Google Scholar]
- Teplitski M., Robinson J. B., Bauer W. D. (2000). Plants secrete substances that mimic bacterial N-acyl homoserine lactone signal activities and affect population density-dependent behaviors in associated bacteria. Mol. Plant Microbe. Interact. 13, 637–648. 10.1094/MPMI.2000.13.6.637 [DOI] [PubMed] [Google Scholar]
- Tuohy K. M., Probert H. M., Smejkal C. W., Gibson G. R. (2003). Using probiotics and prebiotics to improve gut health. Drug Discov. Today 8, 692–700. 10.1016/S1359-6446(03)02746-6 [DOI] [PubMed] [Google Scholar]
- Turnbaugh P. J., Ridaura V. K., Faith J. J., Rey F. E., Knight R., Gordon J. I. (2009). The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 1, 6ra14. 10.1126/scitranslmed.3000322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Agtmaal M., van Os G., Hol G., Hundscheid M., Runia W., Hordijk C., et al. (2015). Legacy effects of anaerobic soil disinfestation on soil bacterial community composition and production of pathogen-suppressing volatiles. Front. Microbiol. 6:701. 10.3389/fmicb.2015.00701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Elsas J. D., Chiurazzi M., Mallon C. A., Elhottovā D., Krištůfek V., Salles J. F. (2012). Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc. Natl. Acad. Sci. 109, 1159–1164. 10.1073/pnas.1109326109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Veen J. A., van Overbeek L. S., van Elsas J. D. (1997). Fate and activity of microorganisms introduced into soil. Microbiol. Mol. Biol. Rev. 61, 121–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventorino V., De Marco A., Pepe O., De Santo A. V., Moschetti G. (2012). Impact of innovative agricultural practices of carbon sequestration on soil microbial community, in Carbon Sequestration in Agricultural Soils: a multidisciplinary approach to innovative methods, ed Piccolo A. (Heidelberg: Springer-Verlag; ), 145–177. [Google Scholar]