Summary
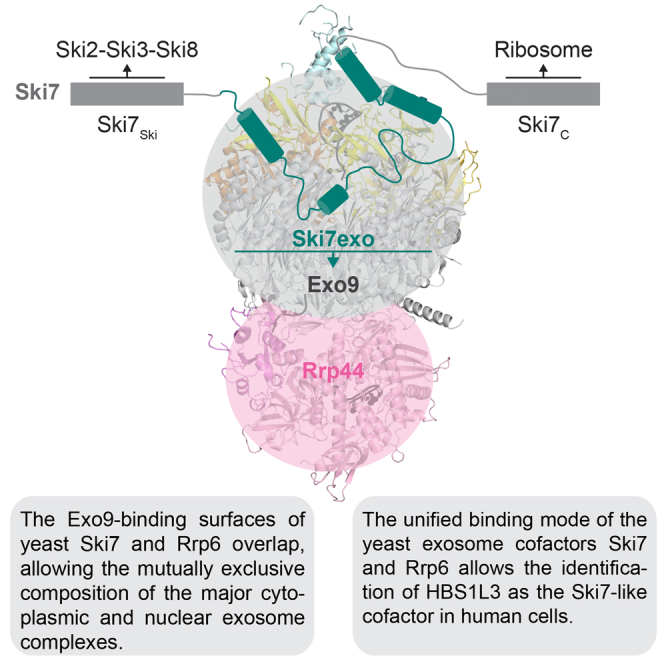
The RNA exosome complex associates with nuclear and cytoplasmic cofactors to mediate the decay, surveillance, or processing of a wide variety of transcripts. In the cytoplasm, the conserved core of the exosome (Exo10) functions together with the conserved Ski complex. The interaction of S. cerevisiae Exo10 and Ski is not direct but requires a bridging cofactor, Ski7. Here, we report the 2.65 Å resolution structure of S. cerevisiae Exo10 bound to the interacting domain of Ski7. Extensive hydrophobic interactions rationalize the high affinity and stability of this complex, pointing to Ski7 as a constitutive component of the cytosolic exosome. Despite the absence of sequence homology, cytoplasmic Ski7 and nuclear Rrp6 bind Exo10 using similar surfaces and recognition motifs. Knowledge of the interacting residues in the yeast complexes allowed us to identify a splice variant of human HBS1-Like as a Ski7-like exosome-binding protein, revealing the evolutionary conservation of this cytoplasmic cofactor.
Graphical Abstract
Highlights
-
•
The yeast exosome binds Ski7 with low nanomolar affinity and extensive interactions
-
•
The Ski7 exosome-binding domain folds upon recognizing Csl4, Mtr3, and Rrp43 subunits
-
•
Ski7 and Rrp6 lack sequence homology but form a similar interface with the exosome
-
•
The exosome interface residues of yeast Ski7 are conserved in human Hbs1L isoform 3
Kowalinski et al. (2016) show that the yeast exosome core complex recognizes the cytoplasmic cofactor Ski7 and the nuclear cofactor Rrp6 similarly. Through structural analyses, they identify a splice variant of HSB1-Like as the long-sought Ski7-like exosome binding cofactor in humans.
Introduction
The eukaryotic RNA exosome is a conserved ribonuclease complex that controls the quantity and quality of a large number of RNAs. Exosome-mediated RNA degradation leads to the elimination of nuclear and cytoplasmic transcripts in turnover and quality control pathways or to partial trimming of RNA precursors in processing pathways (reviewed in Chlebowski et al., 2013, Houseley et al., 2006, Lebreton and Séraphin, 2008, Lebreton et al., 2008, Lykke-Andersen et al., 2009, Schaeffer et al., 2011). The core complex of the RNA exosome was originally discovered from genetic and biochemical analyses in budding yeast (Mitchell et al., 1997) and has since been characterized at the molecular and structural level (reviewed in Januszyk and Lima, 2014, Makino et al., 2013). Orthologs have also been identified in other eukaryotes and have been linked to Mendelian diseases in the human population (Allmang et al., 1999, Fabre and Badens, 2014).
The yeast exosome core complex is formed by ten different proteins. Only a single subunit (Rrp44, also known as Dis3) is catalytically active (Dziembowski et al., 2007, Liu et al., 2006). The other nine core subunits (Exo9) form a cylindrical structure that threads RNA substrates to the Rrp44 exoribonuclease site (Bonneau et al., 2009, Liu et al., 2006, Makino et al., 2013, Wasmuth et al., 2014). Yeast Exo10 is present in the nucleus as well as in the cytoplasm but binds compartment-specific cofactors. In the nucleus, the exosome associates with an additional ribonuclease complex (Rrp6-Rrp47), an RNA helicase (Mtr4), and a small protein (Mpp6) to form a 14-subunit assembly (Butler and Mitchell, 2010, Schuch et al., 2014). In the cytoplasm, the exosome functions together with the Ski2-Ski3-Ski8-Ski8 (Ski) complex, a tetrameric assembly centered at an Mtr4-like RNA helicase (Ski2) (Anderson and Parker, 1998, Brown et al., 2000, Halbach et al., 2013). Orthologs of these exosome cofactors are well conserved in eukaryotes (Schilders et al., 2007, Butler and Mitchell, 2010, Schaeffer et al., 2011), and several are mutated in human diseases (reviewed in Fabre and Badens, 2014, Staals and Pruijn, 2010).
An additional cofactor, Ski7, bridges the interaction between the exosome and Ski complexes in S. cerevisiae (Araki et al., 2001, Halbach et al., 2013, van Hoof et al., 2000, Wang et al., 2005). The N-terminal exosome-binding and Ski-binding domains of Ski7 are required for all known exosome functions in the cytoplasm, including mRNA turnover and quality-control pathways (Araki et al., 2001, Schaeffer et al., 2011, van Hoof et al., 2002. The C-terminal GTPase-like domain of Ski7 has instead a specific role in nonstop decay (NSD) (van Hoof et al., 2002). NSD is one of the quality-control pathways that monitors the process of mRNA translation: it eliminates defective transcripts where the absence of in-frame termination codons causes ribosomes to stall upon translating the 3′ poly(A) tail (reviewed in Inada, 2013, Klauer and van Hoof, 2012, Lykke-Andersen and Bennett, 2014, Shoemaker and Green, 2012). S. cerevisiae Ski7 is a paralogue of the ribosome recycling factor Hbs1. Hbs1 functions in no-go decay (NGD), another translational quality-control pathway that targets and degrades transcripts with ribosomes stalled in the coding region or in the 3′ untranslated region (Doma and Parker, 2006, Guydosh and Green, 2014; and reviewed in Inada, 2013, Lykke-Andersen and Bennett, 2014, Shoemaker and Green, 2012). Yeast Hbs1 and Dom34 have also been recently implicated in NSD (Tsuboi et al., 2012).
The SKI7 and HBS1 paralogous genes originated from an ancestral genome duplication event in budding yeast (Marshall et al., 2013). The Ski7 and Hbs1 proteins comprise a similar translational GTPase-like domain (Kowalinski et al., 2015) and are expected to share similarities in recognizing stalled ribosomes (van Hoof et al., 2002). However, they diverge in their activities (GTP binding versus GTP hydrolysis), mRNA targets (nonstop versus no-go mRNAs), and interacting proteins (exosome-Ski versus Dom34). Another conspicuous difference is that Hbs1 is conserved across eukaryotes while Ski7 orthologs have only been identified in a subset of fungal species (Marshall et al., 2013). Although Ski7 is currently assumed to be a specialized protein in yeasts, several observations argue against such evolutionary restriction. First, given that the exosome and Ski complexes are conserved from yeast to human, it is reasonable to expect that a Ski7-like cofactor would bridge their interaction also in higher eukaryotes. Second, NSD is not limited to yeast but also exists in mammalian cells, where it depends on HBS1-DOM34 as well as exosome and SKI proteins (Saito et al., 2013). In this work, we set out to visualize how the yeast exosome interacts with Ski7, and based on the structural information, we identified isoform 3 of HBS1L as the human counterpart of Ski7.
Results and Discussion
Identification and Structure Determination of a Minimal Exosome-Ski7 Complex
Ski7 mediates the interaction between the RNA degrading exosome and the Ski complex in vivo (Araki et al., 2001) and in vitro (see Figure S1A available online). Coimmunoprecipitation studies from yeast strains expressing different tagged versions of Ski7 have identified two separate domains in the N-terminal region as responsible for binding the Ski and exosome complexes (Araki et al., 2001). We recapitulated the interactions in vitro with purified recombinant proteins and more precisely defined the domain boundaries within Ski7. Using a combination of secondary structure prediction, limited proteolysis, and pull-down assays (Figure S1B), we mapped the Ski2-Ski3-Ski8-interacting domain of Ski7 at residues 1–105 and the Exo9-interacting domain at residues 116–235 (Figure 1A). Both domains are predicted to contain sparse secondary structure elements.
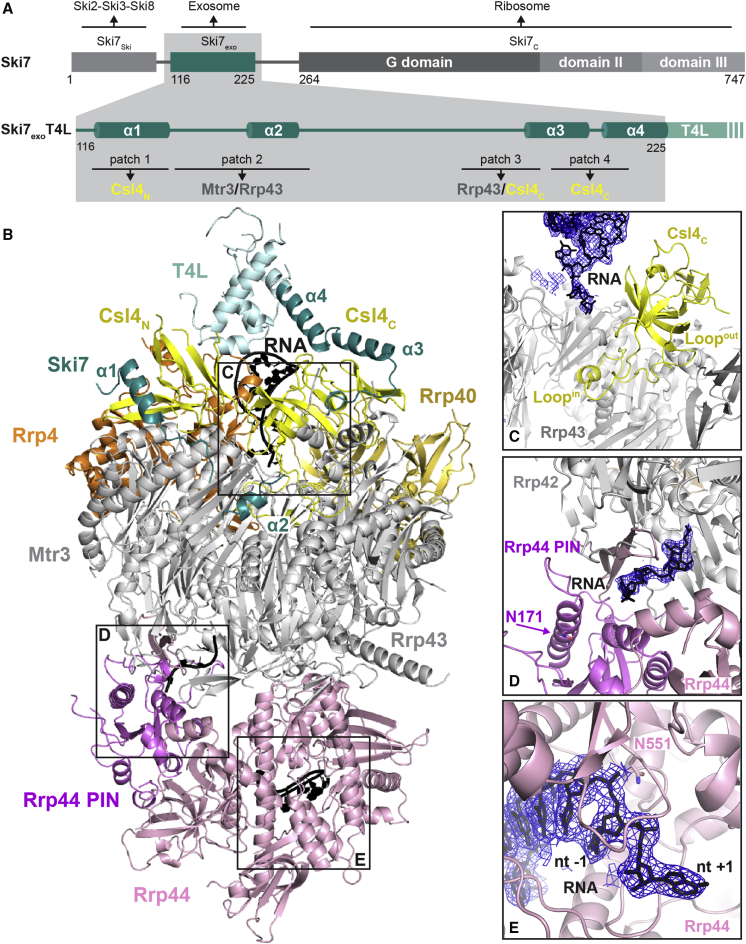
Figure 1.
The Atomic Structure of a S. cerevisiae Exo10–Ski7exo–RNA Complex
(A) The overall domain structure of Ski7 is shown on top, together with its binding partners. The region of the molecule in the present structure (Ski7exo) is shown in teal. At the bottom is a zoom-in view with the secondary structure elements of Ski7exo followed by the C-terminal T4 lysozyme (T4L). The main exosome-binding patches of Ski7exo are annotated together with the Exo9 subunits they bind to.
(B) Overview of the structure of Exo10–Ski7exo–RNA. Cartoon representation of the exosome with Rrp44 in light pink (Rrp44 PIN domain in violet), Csl4, Rrp4, and Rrp40 in yellow, orange, and champagne, respectively; the RNase PH ring in gray; and RNA in black. Ski7exoT4L is shown with Ski7exo in teal and T4L in light teal.
(C–E) Zoom-in views of three portions of the structure that are indicated in boxes in (B). Representation and colors are as in Figure 1B. RNA is in stick representation. Electron density 2Fo–Fc map is shown in blue mesh around the RNA (contoured at 0.9 σ). See also Figure S2.
We proceeded to crystallize a yeast exosome core bound to the interacting domain of Ski7. To this end, we used a catalytically inactive 10-subunit exosome complex where the Rrp44 subunit had been mutated to impair the exonuclease and endonuclease active sites (D551N, D171N substitutions), hereafter referred to as Exo10 (Bonneau et al., 2009, Liu et al., 2006, Makino et al., 2013). Based on the approach we have previously used for crystallizing an exosome core bound to the nuclear cofactor Rrp6 (Makino et al., 2013), we added an RNA substrate featuring a tetra-loop at the 5′ end and a single-stranded extension of 28 ribonucleotides at the 3′ end (Uloop-28). The Exo10-Uloop-28 complex bound to Ski7 residues 116–235 yielded crystals, but the hair-like needles were unsuitable for diffraction studies. To promote additional crystal packing interactions, we followed a protein engineering strategy (Chun et al., 2012) and fused T4 lysozyme (T4L) to the C terminus of the construct. Ski7116-235-T4L yielded thicker needles of the complex that diffracted to 4.2 Å resolution. Structure determination by molecular replacement and model building suggested that truncating the C terminus of Ski7 and thus shortening the linker to T4L could improve crystal quality (Figure S1C). Indeed, the corresponding exosome complex with a truncated Ski7 domain (residues 116–225, Ski7exo) fused to T4L showed improved diffraction properties (Figure 1A). The crystal structure of S. cerevisiae Exo10-Ski7exoT4L-Uloop-28 (hereafter referred to as Exo10-Ski7-RNA) was determined and refined to 2.65 Å resolution with Rfree of 25.9% Rfactor of 21.5% and good stereochemistry (Figure 1B and Table 1).
Table 1.
X-Ray Data Collection and Refinement Statistics
| Dataset | Exo10-Ski7-RNA |
|---|---|
| Wavelength (Å) | 0.97852 |
| Resolution range (Å) | 66.69–2.65 (2.75–2.65) |
| Space group | P 212121 |
| a, b, c (Å) | 106.08, 182.53, 250.55 |
| α, β, γ (°) | 90, 90, 90 |
| Total reflections | 2,802,181 (233,628) |
| Unique reflections | 272,923 (28,614) |
| Multiplicity | 10.26 (8.16) |
| Completeness (%) | 100.0 (99.9) |
| Mean I/sigma (I) | 7.33 (1.14) |
| Rmerge (%) | 21.1 (176.2) |
| CC(1/2) | 99.0 (36.2) |
| Refinement | |
| R work (%) | 21.52 (33.39) |
| R free (%) | 25.86 (36.24) |
| Protein residues | 3,446 |
| RNA residues | 27 |
| Ligands | 7 × MPD, 1 × Zn2+, 4 × Na+ |
| Water | 134 |
| Stereochemistry | |
| Rms (bonds) | 0.004 |
| Rms (angles) | 0.77 |
| Ramachandran favored (%) | 97 |
| Ramachandran outliers (%) | 0.27 |
Values for the highest-resolution shell are given in parentheses. MPD, 2-Methyl-2,4-pentanediol. See also Figure S1.
Exo10–Ski7–RNA Complex: The Exosome Core
The 10-subunit exosome core in the Ski7-bound complex is overall similar to that observed in Rrp6-bound complexes (Makino et al., 2013, Makino et al., 2015, Wasmuth et al., 2014) (Figures 1B and S2A). In particular, the ring formed by the six RNase PH-like proteins (Rrp41, Rrp45, Rrp46, Rrp43, Mtr3, and Rrp42) superposes well with that observed in other exosome structures (root-mean-square deviation [rmsd] of less than 1.22 Å over all α-carbon atoms) (Makino et al., 2015, Makino et al., 2013, Wasmuth et al., 2014). At the top of the RNase PH-like subunits, the three so-called cap proteins (Rrp4, Rrp40, and Csl4) form an upper ring that closes around part of the RNA 5′ tetra-loop (Makino et al., 2013). Near the bottom of the RNase PH-like subunits, the Rrp44 nuclease is in a closed conformation reminiscent of that found in an exosome complex crystallized before in the presence of a long RNA in the central channel (Makino et al., 2013) (Figure S2A).
Despite the overall similarity with previous structures, there are several differences. Starting from the top of the complex, a loop of Csl4 (residues 158–200) undergoes a large conformational change when compared to Rrp6-bound structures (Makino et al., 2013, Makino et al., 2015, Wasmuth et al., 2014): instead of protruding from the outer surface of the exosome, it lines the inner cavity of the complex, where it approaches the RNA in the central channel (Figures 1C and S2B). The outward facing conformation of this loop observed in Rrp6-bound complexes would not be compatible with the Ski7-bound complex because of steric clashes with the N terminus of Rrp43 (Figure S2C). However, whether this loop influences the specific activity of the nuclear and cytoplasmic exosomes in S. cerevisiae is currently unclear, as we could not detect obvious changes with the assays and substrates we have tested.
Another significant difference in comparison to earlier structures is that one of the three OB-fold domains of Rrp44 (CSD1) is shifted away from the RNase PH-like ring by about 10 Å. In turn, the opposite loops from Rrp42 and Rrp43 are disordered (residues 159–169 and 250–270, respectively). The alternative conformation of CSD1 changes the shape of the internal RNA-binding channel in this part of the complex, and in parallel the ribonucleotide chain also deviates in its path (Figures 1D and S2D). Although the movement of CSD1 appears to be due to crystal packing rather than to Ski7 binding, it highlights the conformational plasticity of the Rrp44 subunit and of how RNA substrates can thread through it. Interestingly, such a movement might also open an alternative RNA path from the central channel to the endonuclease site of the PIN domain (Figures 1D and S2D). This potential RNA path would be consistent with previous biochemical data supporting the presence of a channel-dependent path to the exosome endonuclease site (Drazkowska et al., 2013, Wasmuth and Lima, 2012).
Finally, a difference with previous structures is observed at the RNA 3′ end, where density for one additional ribonucleotide is present (Figures 1E and S2E). It appears that the RNA substrate continues with one nucleotide beyond the inactive exonuclease site (D551N), toward the solvent. This position likely reflects the exit route taken by the cleaved 3′ end nucleotide that would be produced and expelled by a wild-type active complex.
Exo10–Ski7–RNA Complex: The Cytoplasmic Cofactor Ski7
The N-terminal region of Ski7 is natively unfolded in solution (Figure S3A) but adopts well-defined secondary structure elements when bound to Exo10 (Figure 1B). Ski7exo folds into four helices (α1–α4) connected by extended linkers. Most of the polypeptide chain has well-ordered electron density (Figure S3B), with the exception of residues 142–147 and residues 159–179 in the linkers connecting Ski7exo helices α1-α2 and α2-α3, respectively. Ski7exo binds at the upper half of the exosome core, where it interacts with the cap protein Csl4 and with the RNase PH-like proteins Mtr3 and Rrp43 (Figure 2A). In the structure, Ski7exo helix α4 directly continues into the T4L molecule that was artificially fused to it for crystallization purposes. T4L is partially ordered at the top of the exosome and interacts with the cap protein Rrp4. With hindsight, the T4L engineering strategy not only contributed an additional crystal contact but also likely stabilized the cap proteins via a contact to Rrp4 (Figure S1C). The position of T4L in the structure is far from Ski7-exosome interaction sites (Figures 1B and 2A), and as such it will not be discussed further.
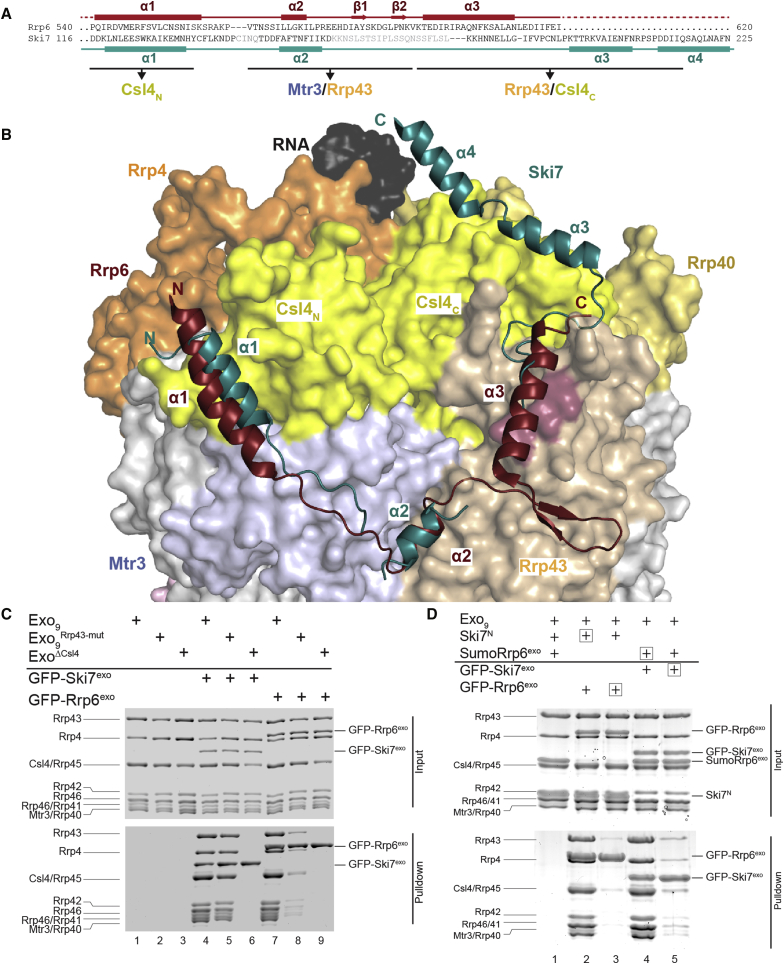
Figure 2.
Ski7 Binds the Exosome with an Extensive Interaction Network
(A) The exosome is shown in a surface representation and Ski7exoT4L in cartoon representation. Colors are as in Figure 1B, except for Mtr3 (pale blue) and Rrp43 (pale orange).
(B–D). Zoom-in views of the interaction patches 1–4 in cartoon representation, with colors as in Figure 2A. Interacting residues are in stick representation as indicated.
(F) Protein coprecipitations in GFP pull-down assays. The exosome coprecipitates with double or triple mutants of GFP-Ski7 in a single binding patch (lanes 8–11). Only when combining mutants of two patches (Ski7exo,exomut-23, lane 12) the interaction lost. See also Figure S3D.
(G) Growth assay of wild-type and mutant yeast strains. Endogenous SKI7 was replaced by wild-type or mutant SKI7-EGFP fusions in a strain where endogenous XRN1 was present (left panel) or deleted (right panel). All strains (except wild-type) also carried an YCplac33[XRN1,URA3] rescue vector. For the growth assay, cells were grown to early exponential phase and spotted in serial dilutions onto 5-fluoroorotic acid (5-FOA) medium or control plates. Medium containing 5-FOA selects for cells that have lost the YCplac33[XRN1,URA3] rescue vector. Deletions of either Ski7exo (row 7) or Ski7N (row 8) as well as the Ski7exo,exomut-23 mutant (row 6) had a lethal effect. Controls: SC w/o URA, presence of rescue shuffle vector carrying wt XRN1; YPAD, general growth control on nonselective/permissive media; YPAD/G418, maker associated with Ski7 genomic integration; YPAD/Nat marker associated with XRN1 knockout. For expression controls and yeast strains, see Figure S3E and Table S1. SC, synthetic complete medium; URA, uracil. See also Figures S3 and S4.
Ski7 Wraps around Csl4, Rrp43, and Mtr3 with Extensive Interactions
Ski7exo wraps around the exosome covering about 3,150 Å2 of its accessible surface area (1,625 Å2 of Csl4, 870 Å2 of Rrp43, and 660 Å2 of Mtr3), as calculated with the PISA server (Krissinel and Henrick, 2007). Starting from the N terminus, the α1 helix of Ski7exo docks into a conserved hydrophobic pocket present on the side of the Csl4 N-terminal domain (patch 1) (Figure 2B). Here, hydrophobic side chains of Ski7 (Leu121Ski7, Trp125Ski7, Ile128Ski7, Met131Ski7) engage in van der Waals interactions with Csl4 (Figure 2B). Following the first helix, an extended segment with hydrophobic residues (e.g., Tyr134Ski7) stretches over the surface of Mtr3, connecting to the second helix (Figure 2B). The short α2 helix of Ski7exo wedges into a hydrophobic cleft formed at the junction between Mtr3 and Rrp43 (patch 2) (Figure 2C). Here, the interaction is mediated by a cluster of apolar side chains (including Phe149Ski7, Phe151Ski7, Phe154Ski7, Ile155Ski7, Ile156Ski7) (Figure 2C). After a disordered segment, the linker connecting to the third helix coils around Rrp43 and projects hydrophobic residues (Leu186Ski7, Leu187Ski7, Ile189Ski7, Phe190Ski7, Pro192Ski7) into a cleft formed at the junction with the Csl4 C-terminal domain (patch 3) (Figure 2D). Finally, Ski7exo helix α3 and the tip of helix α4 bind onto a conserved hydrophobic surface at the side of the Csl4 C-terminal domain (via Leu195Ski7, Ala203Ski7, Phe207Ski7, Pro212Ski7) (patch 4) (Figure 2E).
The interaction between Ski7 and the exosome is overall dominated by hydrophobic contacts, but there are additional polar interactions (e.g., Asn132Ski7 at patch 1, Thr152Ski7 at patch 2, Glu185Ski7 at patch 3 and Asp213 Ski7, Asp214Ski7, Gln217Ski7 at patch 4). With hindsight, the recently reported crosslinking-mass spectrometry data on the Ski7-exosome interaction support well-ordered parts of the complex structure (crosslinks between Lys180Ski7 and Lys45Rrp43, and Lys181Ski7 and Lys45Rrp43) (Figure S3C) (Shi et al., 2015). Multiple crosslinks observed beyond the boundaries of our crystallization construct might have been caused by the collapse of flexible regions of Ski7 upon crosslinking treatment, but are consistent with the overall directionality of the Ski7 polypeptide on the exosome.
Effect of Specific Structure-Based Mutations In Vitro and In Vivo
Based on the structural analysis, we engineered a series of substitutions at the largest interaction hotspots of Ski7exo and tested the ability of the mutants to bind the exosome in pull-down assays with purified proteins. Mutations of Ski7 residues at patch 1 (Trp125Asp, Ile128Asp, or Ski7exomut-1), patch 2 (Phe149Asp, Phe151Asp, Phe154Asp, or Ski7exomut-2), or patch 3 (Ile189Asp, Phe190Asp or Ski7exomut-3) were not sufficient to individually impair the interaction with Exo9 (Figure 2F, lanes 8–11). Exo9 binding was disrupted in vitro only when impairing two interaction hotspots simultaneously (Ski7exomut-23, with 5 combined mutations at patches 2 and 3) (Figure 2F, lane 12). Similarly mutating a binding surface in patch 4 on Csl4 (Trp272Glu, Phe292Glu, Asn250Ala or Csl4exomut-4) did only disrupt interaction in combination with one of the Ski7 mutations (Figure S3D). Albeit qualitative, these assays suggested the presence of a high-affinity interaction between Ski7 and the exosome. Indeed, quantitative measurements by microscale thermophoresis (MST) revealed a dissociation constant in the low nanomolar range (Kd ∼5 nM, Figure S4A). When impairing a single patch in Ski7exomut-1, the binding affinity decreased 10-fold but remained relatively high (Kd 50 nM, Figure S4A).
Next, we tested the effect of Ski7 mutants in complementation assays in yeast. We integrated wild-type SKI7, ski7ΔN, ski7Δexo (lacking the entire exosome-binding domain), or ski7mut-23 (with the five substitutions at patches 2 and 3) as C-terminal eGFP fusions at the endogenous locus in a W303 diploid yeast strain in which one of the chromosomal copies of SKI7 had been deleted. To assess growth defects in the absence of XRN1, we used an XRN1/xrn1Δ diploid strain that we had previously generated (Halbach et al., 2013). After sporulation and tetrad dissection, haploids were mated accordingly to generate ski7Δ/xrn1Δ, ski7Δexo-eGFP/xrn1Δ, ski7ΔN/xrn1Δ, ski7exomut-23-eGFP/xrn1Δ, and SKI7-eGFP/xrn1Δ strains (Figure 2G; Table S1). Consistent with previous reports, disruption of SKI7 and XRN1 was synthetically lethal (ski7Δ/xrn1Δ, Figure 2G) (van Hoof et al., 2000). In the ski7Δexo-eGFP/xrn1Δ, ski7ΔN/xrn1Δ and ski7mut-23-eGFP/xrn1Δ strain, the mutant proteins were expressed at levels comparable to wild-type Ski7-eGFP as judged by western blot (Figure S4B), but cells showed synthetic growth defects (Figure 2G). These data are consistent with the notion that the high-affinity interactions between yeast Ski7 and the exosome are important for function in vivo.
Ski7 and Rrp6 Share a Common Interaction Surface on the Exosome Core
We analyzed the Ski7-bound and Rrp6-bound structures to compare how the cytoplasmic and nuclear cofactors recognize the core complex. Ski7exo and the C-terminal exosome-binding domain of Rrp6 (Rrp6C or Rrp6exo) do not share apparent similarity at the sequence level (Figure 3A). However, they adopt a similar topology of secondary structure elements and engage analogous surfaces of Csl4, Mtr3, and Rrp43 (Figure 3A). In particular, Ski7exo and Rrp6exo bind at the patch 1 and patch 2 surfaces of the exosome with remarkably similar interactions (Figure 3B). For example, Phe549Rrp6 and Trp125Ski7 occupy the equivalent position at patch 1, and Leu572Rrp6 and Phe151Ski7 occupy the same position at patch 2 (Figures S5A and S5B). After patch 2, the mode of interaction diverges. In the case of Rrp6exo, the polypeptide chain folds into a β-hairpin that extends toward the outer surface of Rrp43, as it does not have structural elements equivalent to Ski7exo helices α3 and α4 (Figure 3A).
Figure 3.
Ski7 and Rrp6 Share Similar Exosome-Binding Surfaces
(A) Topology-based sequence alignment of Rrp6exo and Ski7exo. Structural elements of Rrp6 are in red and of Ski7 in teal. Residues not visible in the Ski7 structure are in gray. The corresponding exosome-interacting proteins are annotated.
(B) Comparison of Ski7 and Rrp6 binding to the exosome. Top view on the superposition of the nuclear (PDB: 5c0w) and cytosolic exosome (superposition based on Mtr3, yielding an optimal fit for Ski7exo and Rrp6exo). The cytosolic exosome is shown in surface representation with colors as in Figure 2A. The Rrp43 surface residues mutated in Exo9,Rrp43-mut are in pale red. Ski7 and Rrp6 are displayed in cartoon representation. Rrp6 is in red. See also Figure S5.
(C) Pull-down of mutant exosomes on GFP-Ski7 or GFP-Rrp6, respectively. SDS PAGE of a GFP-binder pull-down experiment. Mutation of a hydrophobic patch on Rrp43 does not alter the interaction with Ski7 but weakens binding of Rrp6 (lanes 5 and 8, respectively). Neither Ski7 nor Rrp6 coprecipitates with an exosome lacking Csl4 (ExoΔCsl4) (lanes 6 and 9).
(D) Competition of Ski7 and Rrp6 binding to the exosome. SDS PAGE of a GFP-binder pull-down experiment. Tagged (1×) and nontagged (4× molar ratio) Ski7 (GFP-Ski7exo and Ski7N) and Rrp6exo (GFP-Sumo-Rrp6C and Sumo-Rrp6C) constructs were incubated with 1.2× molar excess of exosome complex. Reaction components that are annotated with a box were added 20 min after nonboxed components and before incubation with GFP-binder resin. Rrp6 does not displace bound Ski7 (lane 3), while Ski7 can partially displace Rrp6 (lane 5).
We evaluated the similarities and differences of Ski7 and Rrp6 binding on the exosome surface. Csl4 provides an important anchoring point for both cofactors (Figure 3B). Indeed, in vitro pull-down assays with GFP-tagged Ski7exo or GFP-tagged Rrp6exo showed that neither the cytoplasmic nor the nuclear cofactor coprecipitated with an exosome complex lacking Csl4 (ExoΔCsl4) (Figure 3C, lanes 6 and 9, respectively). The C-terminal domain of Csl4 is involved more specifically in the interaction with Ski7 (Figure 3B). Consistently, in the ski4-1 allele, a single-point mutation (Gly253Glu) in the C-terminal domain of Csl4 has been shown to severely reduce the association with Ski7 in vivo (van Hoof et al., 2002) and to affect mRNA turnover (van Hoof et al., 2000). Conversely, the structural analysis indicates that a surface of Rrp43 is involved in the interaction with Rrp6, but not with Ski7 (Figures 3B and S5C). We engineered an Rrp43 mutant with three substitutions at this surface (Leu37AspRrp43, Ile39AspRrp43, Leu43AspRrp43, or Rrp43mut mutant) (Figures 3B and S5C), reconstituted the corresponding exosome complex (Exo9,Rrp43-mut), and tested it in pull-down assays with GFP-tagged Rrp6exo or GFP-tagged Ski7exo. In these assays, the interaction of the mutant exosome with the nuclear cofactor was strongly reduced, while the cytoplasmic cofactor was not significantly affected (Figure 3C, lanes 5 and 8, respectively).
Given the overlapping binding sites of Ski7 and Rrp6, we assessed their competition for exosome binding. In devising the pull-down assays, we took into account the high affinity with which Exo9 binds not only Ski7exo (Kd ∼5 nM, Figure S4) but also Rrp6exo (Kd ∼10 nM, Figure S4). We preincubated Exo9 with either Ski7exo or Rrp6exo and then added the corresponding GFP-tagged competitor (GFP-Rrp6exo or GFP-Ski7exo, respectively). In the GFP pull-down assays, GFP-Rrp6exo was not able to displace bound Ski7exo from the complex (Figure 3D, lane 3), while GFP-Ski7exo could partially displace Rrp6exo (Figure 3D, lane 5), in line with the slightly higher exosome-binding affinity of Ski7exo than Rrp6exo (Figure S4A). We then reversed the order of events by incubating Exo9 with the GFP-tagged component first, before adding the untagged competitor. In the GFP pull-down assays, none of the untagged components could displace the tagged competitor (Figure 3D, lanes 2 and 4). We concluded that Ski7exo and Rrp6exo have similar and mutually exclusive binding surfaces on the exosome and that detailed differences in these interfaces can be exploited to selectively dissociate the cofactors from the complex.
HBS1-like Isoform 3 Is a Human Ski7-like Protein
The identification of the exosome-binding residues in Ski7 prompted us to search for the long sought-after ortholog in human cells. In L. kluyveri and other fungi that split off from the S. cerevisiae branch before genome duplication, a single gene (HBS1/SKI7) gives rise to separate Hbs1-like and Ski7-like proteins by alternative splicing (Marshall et al., 2013). Interestingly, human HBS1 has been shown to coimmunoprecipitate with human DIS3L and SKI2W in HeLa cells (Saito et al., 2013), but the caveat from these experiments is that the interactions could be indirect.
From the analysis of the NCBI RefSeq sequences, three splice variants are generated from a single HBS1 gene in human cells (Figure 4A). All three HBS1-Like (HBS1L) isoforms share a similar N-terminal region, encoded by exons 1, 2, and 4. While isoforms 1 and 3 also share the region encoded by exon 3 (residues 37–78), this exon is skipped in isoform 2. The major difference, however, lies in the C-terminal domain. In isoforms 1 and 2 the C-terminal domain is a GTPase-like fold that binds the protein DOM34 (Saito et al., 2013) and that is encoded by skipping exon 5 and starting from exon 6 (to 19). In contrast, isoform 3 contains an unrelated C-terminal domain that is encoded by exon 5 (residues 144–632). Bioinformatic analyses revealed a similarity between the yeast Ski7exo domain and a C-terminal segment of HBS1-like isoform 3 (HBS1L3) (residues 540–632) (Figure 4B). In particular, several residues that mediate the interaction of Ski7 with the patch 1, patch 2, and patch 4 surfaces of the exosome appeared to be conserved in HBS1L3. This analysis suggested the HBS1L3 might bind the human exosome analogously to how Ski7 binds the yeast exosome. We tested this hypothesis using in vitro pull-down assays with purified proteins. We reconstituted human Exo9 and found that indeed it coprecipitated with the GFP-tagged C-terminal segment of human HBS1L3 (HBS1L3exo) (Figure 4C, lane 4) and not with GFP-tagged Mtr4ΔN80ΔSK (Falk et al., 2014), which was used as negative control. We concluded that HBS1L3 is a human Ski7-like protein in human that is capable of interacting with the human exosome.
Figure 4.
HBS1-Like Isoform 3 Is a Human Ski7-like Protein
(A) Exon-intron structure of human HBS1L. The three verified isoforms are shown. The exosome-binding region (exon 5) and the translational GTPase-like domain (exons 7–13) are indicated in teal and blue, respectively.
(B) Multiple sequence alignment of the exosome-binding region of Ski7 from S. cerevisiae and the exosome-binding isoform HBS1L3 from different vertebrates. Structural elements of Sc. Ski7 are indicated in teal; conserved residues between Ski7 and HBS1L family members are highlighted in yellow. Abbreviations and accession numbers are as follows: Hs, Homo sapiens, NP_001138679; Mm, Mus musculus, NP_001138681; Xt, Xenopus tropicalis, XP_012818244; Tr, Takifugu rubripes, XP_011610307; Sc, Saccharomyces cerevisiae, NP_014719; Sk, Saccharomyces kudriavzevii, EJT42305; Sa, Saccharomyces arboricola, EJS41691; and Se, Saccharomyces eubayanus, KOG96583.
(B) Pull-down assay of human exosome with GFP-tagged C terminus of HBS1L3. SDS PAGE of a GFP-binder pull-down experiment. GFP-tagged Mtr4ΔN80ΔSK (Falk et al., 2014) was used as negative control.
Conclusions
The “Superkiller“ SKI genes were all originally identified from mutations resulting in the overexpression of a killer toxin in virus-infected yeast strains (Toh-E et al., 1978). Two decades later, it has become clear that Ski2, Ski3, and Ski8 form an obligate protein complex in vivo and that Ski7 is not stably associated with it (Brown et al., 2000). However, Ski7 consistently coimmunoprecipitates and copurifies with endogenous exosome subunits (Dziembowski et al., 2007, van Hoof et al., 2002). Indeed, the interaction of Ski7 with exosome subunits is even more salt resistant than that of Rrp44, an intrinsic subunit of the S. cerevisiae core complex (Dziembowski et al., 2007). Our data support the notion that Ski7 is a constitutively bound component of the cytoplasmic yeast exosome: its N-terminal domain binds with low nanomolar affinity in vitro and engages the outer surfaces of the exosome rings with strong interactions. Indeed, impairing a single interaction hotspot is not sufficient to abrogate binding: the concomitant mutation of two interaction hotspots is required to impair binding in vitro and in vivo. Interestingly, the nature of the interactions of the cytoplasmic exosome-Ski7 complex is very similar to that of the nuclear exosome-Rrp6 complex. The similarities in the structures were unexpected, as Ski7 and Rrp6 do not share noticeable homology at the sequence level. Thus, Ski7 and Rrp6 appear to have evolved to exploit the same surface features of the exosome core. We speculate that the mutually exclusive interaction of Ski7 and Rrp6 with the exosome physically separates the binding to the two cofactors and potentially prevents Ski7 from being transported into to the nucleus with Rrp6-bound exosome complexes.
The N-terminal domain of yeast Ski7 does not share noticeable sequence homology with human proteins. Yet, the interacting surfaces on the yeast exosome are conserved in higher eukaryotes. Combining structural information with bioinformatic analysis, we identified the C-terminal domain of human HBS1L3 as a Ski7-like exosome-binding region. HBS1L3 also appears to contain regions with sparse sequence similarity to the Ski-complex-binding domain of yeast Ski7, but future structural studies will be required to understand the molecular mechanisms with which Ski7-like cofactors recognize and recruit the Ski complex. In contrast to the general importance of the exosome-binding and Ski-binding regions of Ski7, the C-terminal translational GTPase-like domain is required solely for the yeast NSD pathway (Araki et al., 2001, van Hoof et al., 2002). HBS1L3 clearly lacks an analogous GTPase-like domain. This domain is instead present in human HBS1L1 and HBS1L2 as well as in yeast Hbs1, whereby it functions in the release of stalled ribosomes in the NGD pathway (Becker et al., 2011, Guydosh and Green, 2014, Matsuda et al., 2014; reviewed in Shoemaker et al., 2010). Our results thus raise a set of questions on whether the different human HBS1L isoforms might have acquired specific roles in mRNA turnover and surveillance. Understanding their detailed functions in the decay of different mRNAs in human cells is an important quest for future studies, not least because of the involvement of cytoplasmic exosome cofactors in human diseases and in the modulation of the immune system (reviewed in Fabre and Badens, 2014, Rigby and Rehwinkel, 2015).
Experimental Procedures
Expression and Purification
S. cerevisiae Exo9 subunits, Rrp44 (1–1,001, D171N, D551N) and Rrp6exo (518–693) were expressed in E. coli and purified with similar protocols as described before (Bonneau et al., 2009, Makino et al., 2013). All Ski7 constructs were expressed in E. coli as a fusion protein with an N-terminal His tag and either a thioredoxin polypeptide cleavable with prescission protease (for structural studies) or GFP (for biochemical studies). Ski7 was purified by affinity chromatography followed by ion exchange (heparin column) and size-exclusion chromatography (SEC). The S. cerevisiae Exo10 complex was reconstituted as described before (Bonneau et al., 2009, Makino et al., 2013), and after addition of 1.2× molar excess of Ski7 another size-exclusion chromatography was performed on a Superose 6 column (GE Healthcare). H. sapiens Exo9 proteins were expressed and purified similarly to the protocols previously described (Greimann and Lima, 2008). C-terminally GFP-tagged human HBS1L3exo (comprising residues 540–632) was purified via Ni-affinity and after prescission protease aided cleavage of a his-thioredoxin tag, reversely passed through an Ni-affinity column, and subjected to SEC. Detailed protocols are in the Supplemental Information.
Crystallization and Structure Determination
Exo10–Ski7 crystallized at a concentration of 12.5 mg/ml upon the addition of 1.3× molar excess of a stem-loop RNA with sequence CCCCCGAGAGGGGG(U)27A (Uloop-28). The complex (in 20 mM HEPES [pH 7.5], 100 mM NaCl, 2 mM MgCl2, and 5 mM TCEP) crystallized in 27% 2-Methyl-2,4-pentanediol (MPD), 0.1 M 2-(N-morpholino)ethanesulfonic acid (MES) (pH 6), and 10 mM CaCl2. Crystals grew typically within 10 days and were cryo-protected (in 32% MPD, 0.1 M MES [pH 6]) and flash-cooled in liquid nitrogen before X-ray exposure. All X-ray diffraction data were collected at 100 K at the Swiss Light Source (SLS) synchrotron in Villigen (Switzerland). The data were processed and scaled with XDS (Kabsch, 2010). The crystals belong to the space group P212121 and contain one complex in the asymmetric unit. The data processing statistics are summarized in Table 1. The atomic model of Exo9 (4ifd) was used as a molecular replacement model in PHASER (McCoy et al., 2007). The single domains of Rrp44 and the visible T4L moiety were placed partly manually (as rigid bodies) and partly automatically (by PHASER runs with the fixed refined model). Manual model building with COOT (Emsley and Cowtan, 2004) was aided by models resulting from automated model building with BUCCANEER (Cowtan, 2006). The refinement was performed with Phenix Refine (Afonine et al., 2012), and the stereochemistry was assessed by MolProbity (Davis et al., 2007). Interface area calculations were done with the PISA server (Krissinel and Henrick, 2007). PyMOL was used for producing the figures (Schrödinger, 2011).
Pull-Down Experiments
A total of 5 μg of tagged bait was incubated with 1.2× molar excess of untagged prey to pull down in a volume of 50 μl of pull-down buffer (20 mM Tris [pH 7.5], 100 mM NaCl, 0.01% NP40, 2 mM DTT). After incubation with GSH Sepharose (GE Healthcare) or GFP-binder resin, respectively, and three washing steps with pull-down buffer the resin was dried and taken up in SDS buffer. Input and pull-down fractions were analyzed on denaturing 12% SDS-PAGE.
Microscale Thermophoresis
For affinity measurements, Exo9 (no Rrp44 subunit) was buffered in 20 mM HEPES (pH 7.5), 100 mM NaCl, 2.5 mM MgCl2. GFP-tagged protein constructs were diluted in MST (microscale thermophoresis) buffer (20 mM HEPES [pH 7.5], 100 mM NaCl, 2.5 mM MgCl2, 0.2% Tween 20, 1 mg/ml BSA). A dilution series of Exo9 was produced and supplemented with even amounts of GFP-tagged protein (4–5 nM for WT Ski7 and Rrp6 constructs, 25 nM for the mutant Ski7). Different constructs were chosen due to difficult behavior of the highly hydrophobic Ski7 peptide. Thermophoresis was measured at 87%–90% LED power for Rrp6 and Ski7 WT and 20% LED power for the Ski7 mutant with standard parameters in a NanoTemper Monolith NT.115 machine. The data were analyzed with the MO software (NanoTemper Technologies), choosing a cold region between −1 and 0 and a hot region including the time range of 5 to 7 s as indicated in Figure S3.
Yeast Experiments
Author Contributions
E.K. and E.C. started the project and wrote the paper. Exosome subunits were purified by E.S. and J.E. The complex was crystallized by J.E. and E.K. The structure was determined by E.K.; A.K. assembled and tested human exosome and HBS1L. P.R. performed in vivo experiments in yeast. B.H. did the bioinformatical analysis.
Acknowledgments
We thank the Max Planck Institute of Biochemistry (MPIB) Crystallization and Core Facilities; the beam line scientists at PXII and PXIII at SLS for assistance with data collection and members of our lab for useful discussions and critical reading of the manuscript. We are grateful to Ajla Hrle (NanoTemper Technologies) for advice with the MST data and to Marc Baumgärtner and Max Fastner for assistance in protein purification. This study was supported by the Max Planck Gesellschaft, the European Research Council (ERC Advanced Investigator Grant 294371), and the Deutsche Forschungsgemeinschaft (DFG SFB1035, GRK1721 and CIPSM) to E.C.; and by an EMBO long-term fellowship, Marie Curie Actions Intra-European Fellowship (IEF) and a Daimler-Benz Postdoctoral stipend to E.K.
Published: June 23, 2016
Footnotes
Supplemental Information includes five figures and Supplemental Experimental Procedures and can be found with this article at http://dx.doi.org/10.1016/j.molcel.2016.05.028.
Accession Numbers
The coordinates and structure factors of Exo10-Ski7-RNA have been deposited in the Protein Data Bank with the accession code 5JEA.
Supplemental Information
References
- Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., Terwilliger T.C., Urzhumtsev A., Zwart P.H., Adams P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allmang C., Kufel J., Chanfreau G., Mitchell P., Petfalski E., Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 1999;18:5399–5410. doi: 10.1093/emboj/18.19.5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J.S., Parker R.P. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki Y., Takahashi S., Kobayashi T., Kajiho H., Hoshino S., Katada T. Ski7p G protein interacts with the exosome and the Ski complex for 3′-to-5′ mRNA decay in yeast. EMBO J. 2001;20:4684–4693. doi: 10.1093/emboj/20.17.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T., Armache J.-P., Jarasch A., Anger A.M., Villa E., Sieber H., Motaal B.A., Mielke T., Berninghausen O., Beckmann R. Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome. Nat. Struct. Mol. Biol. 2011;18:715–720. doi: 10.1038/nsmb.2057. [DOI] [PubMed] [Google Scholar]
- Bonneau F., Basquin J., Ebert J., Lorentzen E., Conti E. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell. 2009;139:547–559. doi: 10.1016/j.cell.2009.08.042. [DOI] [PubMed] [Google Scholar]
- Brown J.T., Bai X., Johnson A.W. The yeast antiviral proteins Ski2p, Ski3p, and Ski8p exist as a complex in vivo. RNA. 2000;6:449–457. doi: 10.1017/s1355838200991787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler J.S., Mitchell P. Rrp6, Rrp47 and cofactors of the nuclear exosome. Adv. Exp. Med. Biol. 2010;702:91–104. [PubMed] [Google Scholar]
- Chlebowski A., Lubas M., Jensen T.H., Dziembowski A. RNA decay machines: the exosome. Biochim. Biophys. Acta. 2013;1829:552–560. doi: 10.1016/j.bbagrm.2013.01.006. [DOI] [PubMed] [Google Scholar]
- Chun E., Thompson A.A., Liu W., Roth C.B., Griffith M.T., Katritch V., Kunken J., Xu F., Cherezov V., Hanson M.A., Stevens R.C. Fusion partner toolchest for the stabilization and crystallization of G protein-coupled receptors. Structure. 2012;20:967–976. doi: 10.1016/j.str.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- Davis I.W., Leaver-Fay A., Chen V.B., Block J.N., Kapral G.J., Wang X., Murray L.W., Arendall W.B., 3rd, Snoeyink J., Richardson J.S., Richardson D.C. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma M.K., Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drazkowska K., Tomecki R., Stoduś K., Kowalska K., Czarnocki-Cieciura M., Dziembowski A. The RNA exosome complex central channel controls both exonuclease and endonuclease Dis3 activities in vivo and in vitro. Nucleic Acids Res. 2013;41:3845–3858. doi: 10.1093/nar/gkt060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziembowski A., Lorentzen E., Conti E., Séraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. [DOI] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fabre A., Badens C. Human Mendelian diseases related to abnormalities of the RNA exosome or its cofactors. Intractable Rare Dis. Res. 2014;3:8–11. doi: 10.5582/irdr.3.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk S., Weir J.R., Hentschel J., Reichelt P., Bonneau F., Conti E. The molecular architecture of the TRAMP complex reveals the organization and interplay of its two catalytic activities. Mol. Cell. 2014;55:856–867. doi: 10.1016/j.molcel.2014.07.020. [DOI] [PubMed] [Google Scholar]
- Greimann J.C., Lima C.D. Reconstitution of RNA exosomes from human and Saccharomyces cerevisiae cloning, expression, purification, and activity assays. Methods Enzymol. 2008;448:185–210. doi: 10.1016/S0076-6879(08)02610-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guydosh N.R., Green R. Dom34 rescues ribosomes in 3′ untranslated regions. Cell. 2014;156:950–962. doi: 10.1016/j.cell.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbach F., Reichelt P., Rode M., Conti E. The yeast ski complex: crystal structure and RNA channeling to the exosome complex. Cell. 2013;154:814–826. doi: 10.1016/j.cell.2013.07.017. [DOI] [PubMed] [Google Scholar]
- Houseley J., LaCava J., Tollervey D. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Inada T. Quality control systems for aberrant mRNAs induced by aberrant translation elongation and termination. Biochim. Biophys. Acta. 2013;1829:634–642. doi: 10.1016/j.bbagrm.2013.02.004. [DOI] [PubMed] [Google Scholar]
- Januszyk K., Lima C.D. The eukaryotic RNA exosome. Curr. Opin. Struct. Biol. 2014;24:132–140. doi: 10.1016/j.sbi.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauer A.A., van Hoof A. Degradation of mRNAs that lack a stop codon: a decade of nonstop progress. Wiley Interdiscip. Rev. RNA. 2012;3:649–660. doi: 10.1002/wrna.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalinski E., Schuller A., Green R., Conti E. Saccharomyces cerevisiae Ski7 is a GTP-binding protein adopting the characteristic conformation of active translational GTPases. Structure. 2015;23:1336–1343. doi: 10.1016/j.str.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Lebreton A., Séraphin B. Exosome-mediated quality control: substrate recruitment and molecular activity. Biochim. Biophys. Acta. 2008;1779:558–565. doi: 10.1016/j.bbagrm.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Lebreton A., Tomecki R., Dziembowski A., Séraphin B. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature. 2008;456:993–996. doi: 10.1038/nature07480. [DOI] [PubMed] [Google Scholar]
- Liu Q., Greimann J.C., Lima C.D. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J., Bennett E.J. Protecting the proteome: Eukaryotic cotranslational quality control pathways. J. Cell Biol. 2014;204:467–476. doi: 10.1083/jcb.201311103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen S., Brodersen D.E., Jensen T.H. Origins and activities of the eukaryotic exosome. J. Cell Sci. 2009;122:1487–1494. doi: 10.1242/jcs.047399. [DOI] [PubMed] [Google Scholar]
- Makino D.L., Baumgärtner M., Conti E. Crystal structure of an RNA-bound 11-subunit eukaryotic exosome complex. Nature. 2013;495:70–75. doi: 10.1038/nature11870. [DOI] [PubMed] [Google Scholar]
- Makino D.L., Schuch B., Stegmann E., Baumgärtner M., Basquin C., Conti E. RNA degradation paths in a 12-subunit nuclear exosome complex. Nature. 2015;524:54–58. doi: 10.1038/nature14865. [DOI] [PubMed] [Google Scholar]
- Marshall A.N., Montealegre M.C., Jiménez-López C., Lorenz M.C., van Hoof A. Alternative splicing and subfunctionalization generates functional diversity in fungal proteomes. PLoS Genet. 2013;9:e1003376. doi: 10.1371/journal.pgen.1003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda R., Ikeuchi K., Nomura S., Inada T. Protein quality control systems associated with no-go and nonstop mRNA surveillance in yeast. Genes Cells. 2014;19:1–12. doi: 10.1111/gtc.12106. [DOI] [PubMed] [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P., Petfalski E., Shevchenko A., Mann M., Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′-->5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- Rigby R.E., Rehwinkel J. RNA degradation in antiviral immunity and autoimmunity. Trends Immunol. 2015;36:179–188. doi: 10.1016/j.it.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S., Hosoda N., Hoshino S. The Hbs1-Dom34 protein complex functions in non-stop mRNA decay in mammalian cells. J. Biol. Chem. 2013;288:17832–17843. doi: 10.1074/jbc.M112.448977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer D., Clark A., Klauer A.A., Tsanova B., van Hoof A. Functions of the cytoplasmic exosome. Adv. Exp. Med. Biol. 2011;702:79–90. doi: 10.1007/978-1-4419-7841-7_7. [DOI] [PubMed] [Google Scholar]
- Schilders G., van Dijk E., Pruijn G.J.M. C1D and hMtr4p associate with the human exosome subunit PM/Scl-100 and are involved in pre-rRNA processing. Nucleic Acids Res. 2007;35:2564–2572. doi: 10.1093/nar/gkm082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger, L. (2011). The PyMOL Graphics System. Version. v1.5.0.4.
- Schuch B., Feigenbutz M., Makino D.L., Falk S., Basquin C., Mitchell P., Conti E. The exosome-binding factors Rrp6 and Rrp47 form a composite surface for recruiting the Mtr4 helicase. EMBO J. 2014;33:2829–2846. doi: 10.15252/embj.201488757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Pellarin R., Fridy P.C., Fernandez-Martinez J., Thompson M.K., Li Y., Wang Q.J., Sali A., Rout M.P., Chait B.T. A strategy for dissecting the architectures of native macromolecular assemblies. Nat. Methods. 2015;12:1135–1138. doi: 10.1038/nmeth.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker C.J., Green R. Translation drives mRNA quality control. Nat. Struct. Mol. Biol. 2012;19:594–601. doi: 10.1038/nsmb.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker C.J., Eyler D.E., Green R. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science. 2010;330:369–372. doi: 10.1126/science.1192430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staals R.H.J., Pruijn G.J.M. The human exosome and disease. Adv. Exp. Med. Biol. 2010;702:132–142. [PubMed] [Google Scholar]
- Toh-E A., Guerry P., Wickner R.B. Chromosomal superkiller mutants of Saccharomyces cerevisiae. J. Bacteriol. 1978;136:1002–1007. doi: 10.1128/jb.136.3.1002-1007.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi T., Kuroha K., Kudo K., Makino S., Inoue E., Kashima I., Inada T. Dom34:hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3′ end of aberrant mRNA. Mol. Cell. 2012;46:518–529. doi: 10.1016/j.molcel.2012.03.013. [DOI] [PubMed] [Google Scholar]
- van Hoof A., Staples R.R., Baker R.E., Parker R. Function of the ski4p (Csl4p) and Ski7p proteins in 3′-to-5′ degradation of mRNA. Mol. Cell. Biol. 2000;20:8230–8243. doi: 10.1128/mcb.20.21.8230-8243.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A., Frischmeyer P.A., Dietz H.C., Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- Wang L., Lewis M.S., Johnson A.W. Domain interactions within the Ski2/3/8 complex and between the Ski complex and Ski7p. RNA. 2005;11:1291–1302. doi: 10.1261/rna.2060405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmuth E.V., Lima C.D. Exo- and endoribonucleolytic activities of yeast cytoplasmic and nuclear RNA exosomes are dependent on the noncatalytic core and central channel. Mol. Cell. 2012;48:133–144. doi: 10.1016/j.molcel.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmuth E.V., Januszyk K., Lima C.D. Structure of an Rrp6-RNA exosome complex bound to poly(A) RNA. Nature. 2014;511:435–439. doi: 10.1038/nature13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.