Summary
The Golgi apparatus undergoes a ubiquitin-dependent disassembly and reassembly process during each cycle of cell division. Here we report the identification of the Golgi t-SNARE syntaxin 5 (Syn5) as the ubiquitinated substrate. Syn5 is monoubiquitinated by the ubiquitin ligase HACE1 in early mitosis and deubiquitinated by the deubiquitinase VCIP135 in late mitosis. Syn5 ubiquitination on Lysine 270 (K270) in the SNARE domain impairs the interaction between Syn5 and the cognate v-SNARE Bet1, but increases its binding to p47, the adaptor protein of p97. Expression of the Syn5 K270R mutant in cells impairs post-mitotic Golgi reassembly. Therefore, monoubiquitination of Syn5 in early mitosis disrupts SNARE complex formation. Subsequently, ubiquitinated Syn5 recruits p97/p47 to the mitotic Golgi fragments and promotes post-mitotic Golgi reassembly upon ubiquitin removal by VCIP135. Overall, this study reveals both the substrate and the mechanism of ubiquitin-mediated regulation of Golgi membrane dynamics during the cell cycle.
Graphical abstract
eTOC blurb
The Golgi undergoes ubiquitin-dependent disassembly and reassembly during the cell cycle. Huang et al. identify Syntaxin 5 (Syn5) as the ubiquitinated substrate on the Golgi. They show that Syn5 ubiquitination by HACE1, and deubiquitination by VCIP135, regulate Syn5 interactions with, and hence recruitment of, proteins involved in Golgi assembly.
Introduction
In mammalian cells, the Golgi apparatus undergoes extensive fragmentation in mitosis and is reassembled into intact Golgi in newly divided cells (Tang and Wang, 2013; Wang and Seemann, 2011). Fusion of mitotic Golgi fragments is catalyzed by two independent and non-additive pathways mediated by two AAA (ATPases Associated with diverse cellular Activities) ATPases: NSF (N-ethylmaleimide [NEM] sensitive factor) with its cofactors α/γ-SNAP, and p97/VCP (valosin-containing protein) with its adaptor proteins p47 and p37 (Rabouille et al., 1995; Tang et al., 2008). Both pathways require the Golgi t-SNARE (soluble NSF attachment protein receptor) syntaxin (Syn5), but only the NSF pathway requires its cognate SNARE GS28 (also called Gos28) (Rabouille et al., 1998; Tai et al., 2004). SNARE proteins are the key components for membrane fusion. Four α-helices of the SNARE domains, one from the v-SNARE on the vesicle and three from the t-SNAREs on the target membrane, zipper up to form a trans-SNARE complex that mediates membrane fusion (Weber et al., 1998). NSF disassembles the SNARE complex for the next round of fusion (Jahn and Scheller, 2006); however, the mechanism for p97-mediated membrane fusion is unknown (Rabouille et al., 1998).
A major difference between the two pathways is the involvement of ubiquitin in the p97 pathway (Wang et al., 2004). Monoubiquitination occurs during mitosis and is required for subsequent p97-mediated post-mitotic Golgi membrane fusion (Meyer et al., 2002). Ubiquitination is mediated by a Golgi-localized ubiquitin E3 ligase HACE1 (Tang et al., 2011). Monoubiquitin on a Golgi substrate does not result in protein degradation by proteasomes (Wang et al., 2004); rather it binds the UBA domain of p47, the adaptor protein of p97, and recruits the p97/p47 membrane fusion machinery to the mitotic Golgi fragments (Meyer et al., 2002). Prior to membrane fusion in telophase, ubiquitin is removed by the deubiquitinating (DUB) enzyme VCIP135 (valosin-containing protein [VCP][p97]/p47 complex-interacting protein, p135) (Wang et al., 2004). VCIP135 activity is regulated by phosphorylation in the cell cycle; it is inactivated by phosphorylation in metaphase and reactivated upon dephosphorylation in telophase (Zhang and Wang, 2015; Zhang et al., 2014). In this study, we report the identification of Syn5 as the ubiquitinated substrate on the Golgi membrane and the mechanism of ubiquitination in Golgi disassembly and reassembly during the cell cycle.
Results
Syn5 is monoubiquitinated in mitosis on the Golgi
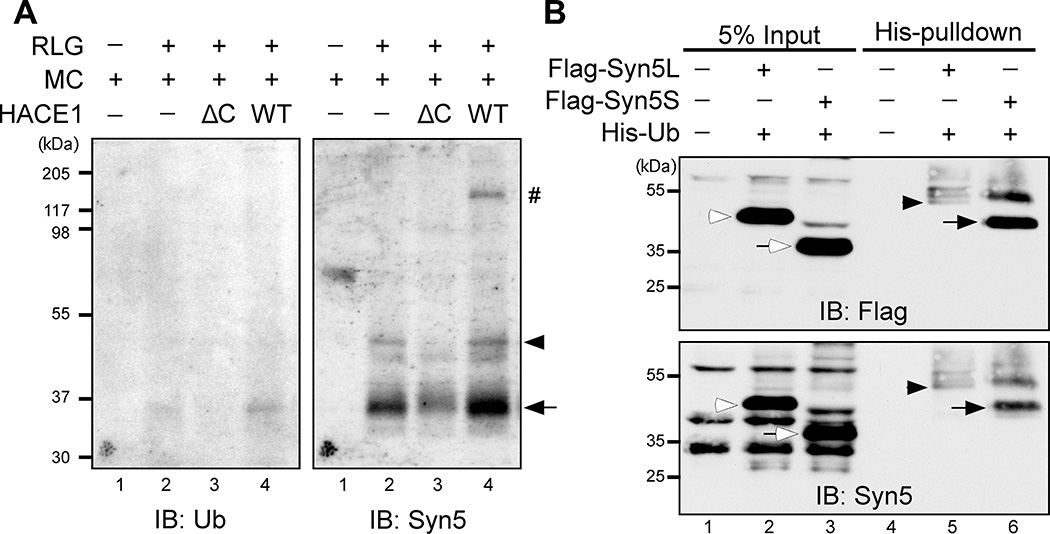
To identify the ubiquitinated substrate on the Golgi membranes, we performed an in vitro ubiquitination assay utilizing the ubiquitin ligase HACE1 (Fig. 1A). Purified rat liver Golgi membranes (RLG) (Tang et al., 2010) were incubated with recombinant wild type (WT) HACE1 or its inactive ΔC mutant, which lacks the HECT (Homologous to the E6-AP Carboxyl Terminus) domain (Tang et al., 2011). A minimal amount of mitotic cytosol was titrated into the reaction to provide ubiquitin E1 and E2 enzymes, and His-tagged ubiquitin (His-Ub) was added along with an ATP regeneration system. After incubation, HACE1 was inactivated by NEM, membranes were solubilized in detergent and 8 M urea, and ubiquitinated proteins were pulled down by a Ni-NTA affinity column. Western blot of the elute using an anti-ubiquitin antibody revealed one clear band of 35 kD (Fig. 1A), similar to the molecular weight of syntaxin 5 (Syn5), an ideal candidate in this pathway. Indeed, this band was recognized by Syn5 antibodies and correlated with the short form of Syn5 (Syn5S). Blotting with Syn5 antibodies also displayed a weak band of 42 kD equivalent to the long form of Syn5 (Syn5L). Notably, both bands were highly detectable when Golgi membranes were incubated with mitotic cytosol alone that contains endogenous HACE1 (Fig. 1A, lane 2); but the intensity was significantly increased upon the addition of WT HACE1, and decreased by HACE1 ΔC mutant (lane 3 & 4 vs. 2). Ubiquitinated Syn5 exists as distinct bands rather than smears, strongly suggesting that it is monoubiquitinated by HACE1 during mitosis.
Figure 1. Syn5 is monoubiquitinated in mitosis.
(A) Identification of Syn5 as a substrate of HACE1. Purified rat liver Golgi (RLG) membranes were mixed with mitotic cytosol (MC), recombinant WT HACE1 or ΔC mutant, and His-ubiquitin. After incubation, re-isolated membranes were solubilized in detergent, ubiquitinated proteins were isolated by Ni-NTA affinity purification and analyzed by Western blot with antibodies to ubiquitin (left panel) and Syn5 (right panel). Shown are representative results from four independent experiments. Note that the signals for both short (Syn5S, arrow) and long (Syn5L, arrowhead) forms of Syn5 were highly detectable when Golgi membranes were incubated with mitotic cytosol alone that contains endogenous HACE1 (lane 2), but the intensity was significantly reduced by HACE1 ΔC (lane 3) and enhanced by WT HACE1 (lane 4). The Syn5 antibody was generated by a GST fusion protein, and thus recognizes copurified GST-HACE1 (indicated by #).
(B) HeLa cells co-transfected with Syn5 (Flag-Syn5L or Flag-Syn5S) and His-Ubiquitin (His-Ub) were synchronized to mitosis followed by Ni-NTA affinity purification and Western blot for Flag and Syn5. Note that both the long and short forms of Syn5 are ubiquitinated, displaying an 8 kD shift: Syn5S (empty arrow) in lane 3 vs. Syn5S-ub (solid arrow) in lane 6; and Syn5L (empty arrowhead) in lane 2 vs. Syn5L-ub (solid arrowhead) in lane 5.
See also Figure S1.
In this experiment (Fig. 1A), ubiquitinated Syn5 did not display a strong mobility shift on the SDS-PAGE compared to the theoretical molecular weight of non-ubiquitinated Syn5, but since non-ubiquitinated Syn5 was not pulled down by the nickel beads, a direct comparison of ubiquitinated and non-ubiquitinated Syn5 was not possible. In addition, the use of 8 M urea may also affect the migration of the protein. To confirm that Syn5 is monoubiquitinated, Flag-tagged short or long form Syn5 was co-expressed with His-Ub in HeLa cells and cells were arrested in mitosis by nocodazole treatment. His-Ub linked proteins were pulled down by Ni-NTA affinity resin followed by Western blotting for Flag. Both short and long forms of Syn5 were detected, with an 8 kD shift from the original size of Syn5 (Fig. 1B), indicating that both forms are monoubiquitinated. This result was confirmed by reciprocal immunoprecipitation of Syn5 with a Flag antibody and Western blotting using an HA antibody to detect ubiquitin (Fig. S1). In Fig. 1B, 3.28 ± 0.01% of Syn5 short form was ubiquitinated in mitosis based on the quantitation result of ubiquitinated Syn5 in the pulldown and the total Syn5 in the input. Given that both cell synchronization and the protein pulldown efficiency were less than 100%, actual ubiquitination should be higher than this level. Consistent with this idea, in Fig. S1, 7.11% ± 0.01% of Syn5 short form was ubiquitinated based on the quantitation of the shifted ubiquitinated Syn5 band and the total Syn5 immunoprecipitated from mitotic HeLa cells. Taken together, a significant amount of Syn5 is monoubiquitinated in mitosis.
Syn5 is monoubiquitinated by HACE1 in mitosis
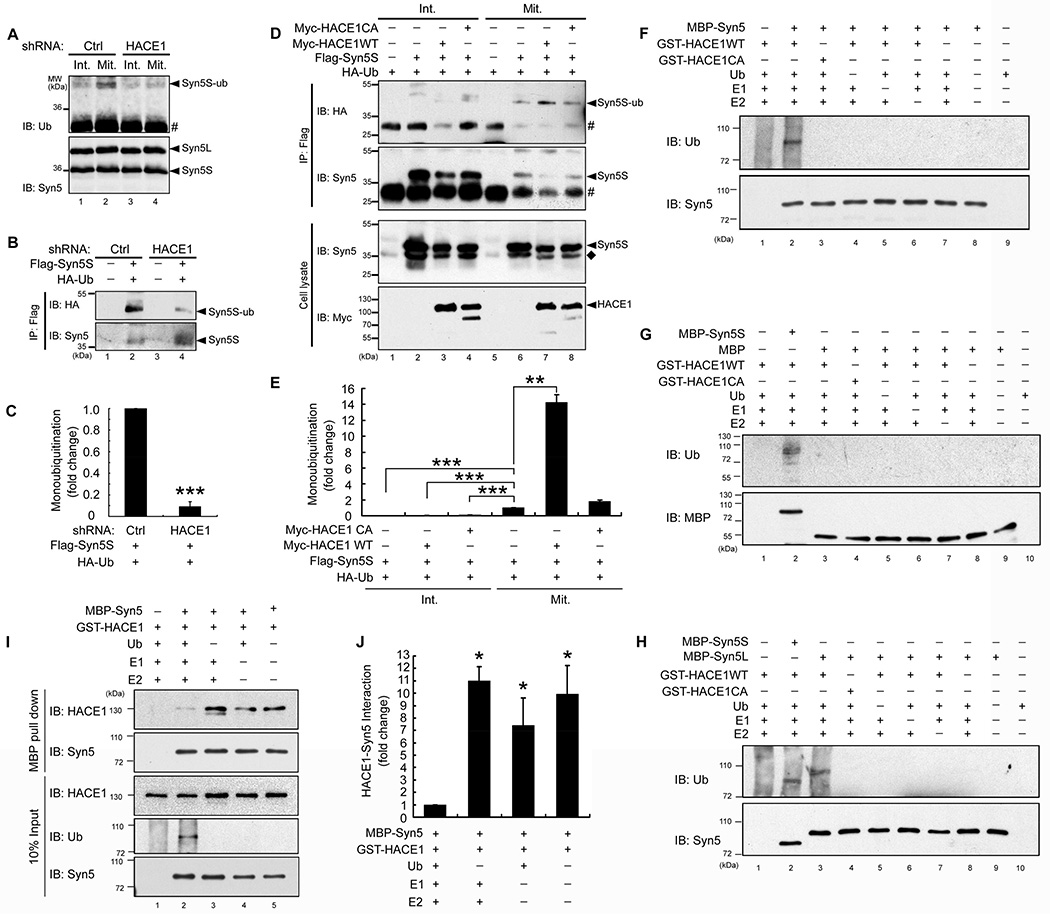
Both long and short forms of Syn5 are generated from the same mRNA by alternative translation initiation (Hui et al., 1997). The long form has an N-terminal cytoplasmic extension (1–54 aa) that contains an endoplasmic reticulum (ER) retrieval motif, and thus is more abundant in the ER and has been related to ER organization (Miyazaki et al., 2012). The short form lacks the ER retrieval motif and mainly localizes and functions in the Golgi (Hui et al., 1997). Given that the short form is ubiquitinated to a much higher level than the long form (Fig. 1A–B), we focused on the short form of Syn5 in the following experiments and hereafter we refer to the short form as “Syn5” unless specified. To assess the ubiquitination of endogenous Syn5 at different cell cycle states, Syn5 was immunoprecipitated from nonsynchronized interphase cells and nocodazole-arrested mitotic cells followed by Western blotting for ubiquitin. As shown in Fig. 2A, Syn5 is highly ubiquitinated in mitosis compared to interphase (lanes 2 vs. 1). To determine whether Syn5 is specifically ubiquitinated by HACE1, similar experiments were performed using cells in which HACE1 was depleted (Fig. S2A) by stably expressing HACE1 shRNA (Tang et al., 2011). HACE1 depletion strongly reduced Syn5 ubiquitination in mitotic cells (Fig. 2A, lane 4).
Figure 2. Monoubiquitination of Syn5 depends on HACE1.
(A) Endogenous Syn5 is ubiquitinated by HACE1 in mitotic cells. HeLa cells were infected by control (Ctrl) or HACE1 shRNA lentivirus to establish stable cell lines (Tang et al., 2011). Untreated interphase cells (Int.) or nocodazole-arrested mitotic cells (Mit.) were solubilized and immunoprecipitated with Syn5 antibodies followed by Western blotting for Syn5 and ubiquitin. Note that Syn5 was ubiquitinated in mitotic but not interphase cells (lanes 2 vs. 1), and depletion of HACE1 reduced Syn5 ubiquitination in mitosis (lanes 4 vs. 2). #, IgG light chain.
(B) HACE1 depletion decreases monoubiquitination of exogenously expressed Syn5 (Flag-Syn5S). Mitotic cells stably expressing Ctrl shRNA or HACE1 shRNA were transfected with indicated constructs and immunoprecipitated with a Flag antibody followed by Western blotting.
(C) Quantitation of (B) from three independent experiments.
(D) Co-expression of WT HACE1, but not the C876A mutant, increases mitotic Syn5 monoubiquitination. HeLa cells were transfected with indicated constructs. Interphase and mitotic cells were immunoprecipitated with a Flag antibody and analyzed by Western blotting. ◆, endogenous Syn5S; #, IgG light chain.
(E) Quantitation of (D) from three independent experiments.
(F–G) HACE1 directly ubiquitinates Syn5 in vitro. Indicated proteins were mixed and incubated, followed by Western blotting for ubiquitin and Syn5. Note the ubiquitin signal on MBP-Syn5 but not MBP.
(H) Both short and long forms of Syn5 are ubiquitinated in vitro.
(I) Ubiquitination reduces Syn5 interaction with HACE1. MBP-Syn5 was re-isolated after the in vitro ubiquitination assay and bound proteins were assessed by Western blot. Note the increased ubiquitin signal and reduced HACE1 in lane 2.
(J) Quantitation of (I) from three independent experiments.
Data are represented as mean ± s.e.m. Statistical significance was assessed by student’s t-test. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
See also Figure S2.
To further confirm that Syn5 ubiquitination is HACE1 dependent, HeLa cells were co-transfected with Flag-Syn5 and HA-Ub, synchronized into mitosis with nocodazole, and Syn5 was immunoprecipitated using a Flag antibody. Syn5 immunoprecipitated from control cells was recognized by an HA antibody for ubiquitin, but the signal was significantly reduced by HACE1 depletion (Fig. 2B, lane 4 vs. 2; Fig. 2C). Conversely, cotransfection of Flag-syn5S with WT HACE1, but not the C876A (CA) inactive mutant (Tang et al., 2011), enhanced Syn5 ubiquitination in mitosis (Fig. 2D, lanes 7 and 8 vs. 6; Fig. 2E). We also found that Syn5 interacted with HACE1 in mitosis by co-immunoprecipitation (Fig. S2B). Therefore, Syn5 is monoubiquitinated by HACE1 in mitosis in vivo.
To determine whether Syn5 is a direct substrate of HACE1, an in vitro ubiquitination assay was performed by treating recombinant MBP-tagged Syn5 with purified HACE1 (WT or the C876A mutant) in the presence of the E1 Ub-activating enzyme (UBE1), the E2 Ub conjugating enzyme (UbcH7), ubiquitin, and ATP (Tang et al., 2011; Torrino et al., 2011). Syn5 was ubiquitinated by WT HACE1 but not the CA mutant (Fig. 2F, lane 2 vs. 3). In this experiment, auto-ubiquitination of HACE1 gave a background smear of ubiquitin signal only when E1, E2, HACE1, and ubiquitin were all present at the same time, but not when one of these components was omitted, as previously reported (Anglesio et al., 2004). Both Syn5S and Syn5L were ubiquitinated to a similar extent in vitro (Fig. 2H, lane 2 and 3), while MBP alone was not ubiquitinated (Fig. 2G, lane 3). The ubiquitin signal of Syn5 existed as distinct bands rather than a smear (Fig. 2F–G, lane 2), supporting the notion that Syn5 is monoubiquitinated by HACE1. To further determine whether HACE1-mediated Syn5 ubiquitination results in Syn5 degradation, we assessed Syn5 level in interphase and mitotic cells. As shown in Fig. S2C, Syn5 protein level does not change during the cell cycle progression or upon HACE1 depletion. In addition, treatment of cells with the proteasome inhibitor MG132 did not affect Syn5 level, although MG132 treatment increased cyclin B1 level in mitosis (Fig. S2D).
To determine whether HACE1 directly interacts with Syn5 and whether the interaction is affected by Syn5 ubiquitination, Syn5 was pulled down after in vitro ubiquitination and probed for HACE1. As shown in Fig. 2I–J, the Syn5-ubiquitin signal negatively correlated with the HACE1 signal in the pulldown. These results suggest that Syn5-HACE1 interaction is transient and Syn5 is released by HACE1 upon Syn5 ubiquitination. Taken together, our results indicate that HACE1 directly interacts with and monoubiquitinates Syn5.
Post-mitotic deubiquitination of Syn5 depends on VCIP135
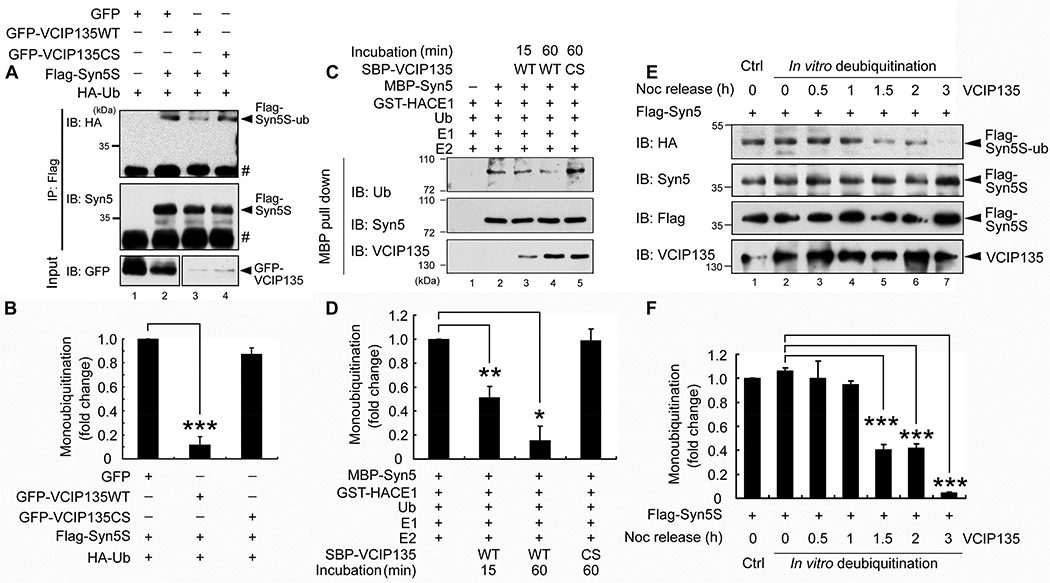
The DUB activity of VCIP135 is required for p97/p47-mediated post-mitotic Golgi membrane fusion (Uchiyama et al., 2006; Zhang and Wang, 2015). Therefore, we determined whether Syn5 is deubiquitinated by VCIP135. HeLa cells were cotransfected with Flag-Syn5 and HA-Ub with either WT VCIP135 or its inactive C218S mutant (Wang et al., 2004). Syn5 was immunoprecipitated from nocodazole-arrested mitotic cells with a Flag antibody and probed for ubiquitin with an HA antibody. As shown in Fig. 3A (lanes 3 and 4 vs. 2), co-expression of WT VCIP135, but not the C218S mutant, significantly reduced the ubiquitin signal (Fig. 3B). To investigate whether ubiquitinated Syn5 is a direct substrate of VCIP135, in vitro ubiquitinated Syn5 was treated with recombinant VCIP135 (Zhang and Wang, 2015; Zhang et al., 2014). As shown in Fig. 3C–D, incubation with WT VCIP135, but not the CS mutant, significantly reduced the ubiquitin signal on Syn5 over time. These results suggest that Syn5 is deubiquitinated by VCIP135.
Figure 3. Deubiquitination of Syn5 by VCIP135.
(A) Co-expression of WT VCIP135, but not the C218S mutant, reduces Syn5 ubiquitination. HeLa cells were transfected with indicated constructs. Nocodazole-arrested mitotic cells were immunoprecipitated with a Flag antibody and analyzed by Western blotting. #, IgG light chain.
(B) Quantitation of (A) from three independent experiments.
(C) VCIP135 deubiquitinates Syn5 in vitro. In vitro ubiquitinated MBP-Syn5 was re-isolated with Amylose beads and further incubated with recombinant SBP-VCIP135 for 15 or 60 min as indicated. Beads were washed and analyzed by Western blot. Note the reduced ubiquitin signal upon incubation with WT but not the C218S (CS) mutant of VCIP135.
(D) Quantitation of (C) from three independent experiments.
(E) Monoubiquitinated Syn5 is deubiquitinated by VCIP135 in late mitosis. HeLa cells were first blocked to mitosis and then released. At each indicated time point, endogenous VCIP135 was immunoprecipitated and used to treat Flag-Syn5S immunoprecipitated from mitotic cells expressing Flag-Syn5S and HA-Ub. After treatment, samples were analyzed by Western blot for indicated proteins.
(F) Quantitation of (E) from three independent experiments.
Data are represented as mean ± s.e.m. *, p < 0.05**; p < 0.01; ***, p < 0.001.
It has been shown that the DUB activity of VCIP 135 is inactivated by phosphorylation in mitosis and reactivated by dephosphorylation in late mitosis (Zhang and Wang, 2015; Zhang et al., 2014). To determine the temporal dynamics of VCIP135-mediated Syn5 deubiquitination during cell cycle progression, HeLa cells were arrested in early mitosis by nocodazole treatment, released by washing out the drug to allow cells progress to later mitosis and G1 phase (Zhang and Wang, 2015), and followed by immunoprecipitation of endogenous VCIP135 at different time points. Subsequently, the immunoprecipitated VCIP135 was used to treat Flag-Syn5 immunoprecipitated from mitotic cells expressing Flag-Syn5 and HA-Ub. As shown in Fig. 3E, treatment of Flag-Syn5 with VCIP135 immunoprecipitated from mitotic cells, either on ice (Fig. 3 E, lane 1) or at 30°C (lane 2), did not reduce the ubiquitin signal as indicated by the HA (HA-Ub) signal, consistent with the previous report that VCIP135 is inactivated in early mitosis (Zhang and Wang, 2015). The ubiquitin signal of Syn5 was significantly reduced when incubated with VCIP135 precipitated from cells 1.5 and 2 h after nocodazole washout when the cells were in late mitosis (Fig. 3E, lanes 5 and 6; Fig. 3F), and was almost eliminated when the washout was extended to 3 h when the cells entered G1 phase (Fig. 3E, lane 7; Fig. 3F) (Zhang and Wang, 2015). Taken together, these results demonstrate that Syn5 is deubiquitinated by VCIP135 in late mitosis.
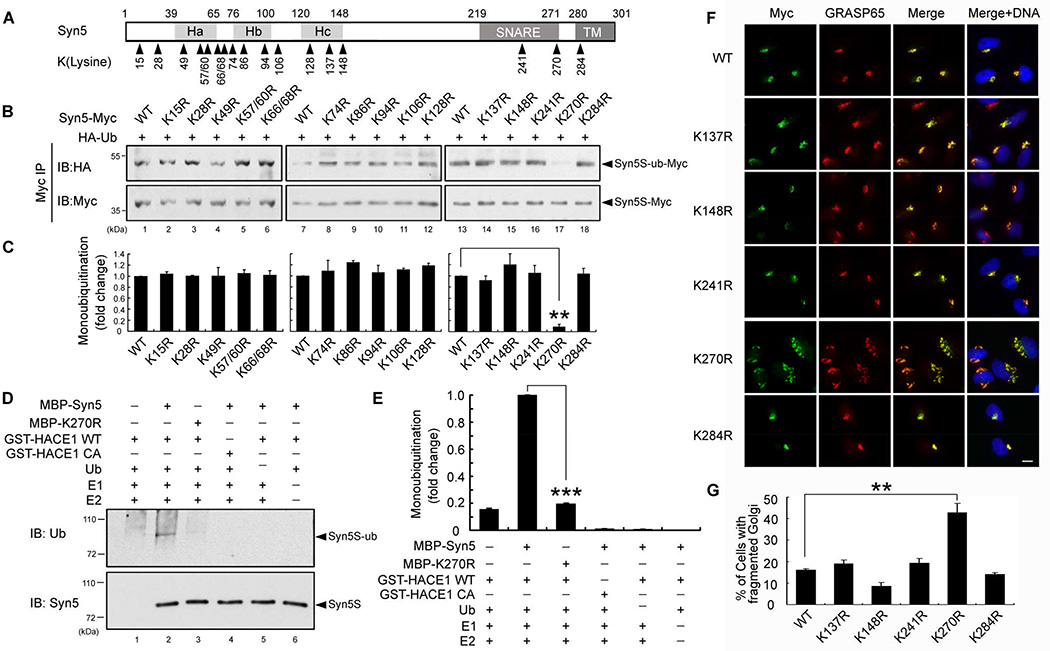
Syn5 is monoubiquitinated on lysine 270
Syn5 contains 17 lysines that are conserved between rat and human (Fig. 4A). To identify the ubiquitinated residue, all 17 lysine residues were mutated to arginines, individually or in combination, to generate 15 mutants: K15R, K28R, K49R, K57/60R, K66/68R, K74R, K86R, K94R, K106R, K128R, K137R, K148R, K241R, K270R, and K284R (Fig. 4A), all with a C-terminal Myc tag. HeLa cells were cotransfected with Syn5 KR mutants and HA-Ub followed by immunoprecipitation of Syn5 from mitotic cells with a Myc antibody. The ubiquitination signal from all KR mutants was detected by Western blotting with an HA antibody, with the exception of K270R, in which the ubiquitin signal was almost completely abolished (Fig. 4B, lane 17; Fig. 4C). This result reveals that Syn5 is ubiquitinated on K270 in mitotic cells. K270 was then confirmed as the major ubiquitination site by in vitro ubiquitination. WT Syn5 is readily ubiquitinated by HACE1; the ubiquitin signal was reduced to background level upon K270R mutation (Fig. 4D, lane 3 vs. 2; Fig. 4E). These results demonstrate that Syn5 is monoubiquitinated by HACE1 on K270.
Figure 4. Syn5 is ubiquitinated on lysine 270.
(A) Schematic representation of Syn5 short form. Syn5 possesses a three helical bundle (consisting of Ha, Hb and Hc regions), a SNARE motif and a transmembrane (TM) domain. The 17 lysines conserved between rat and human are indicated.
(B–C) Mutation of K270 reduces Syn5 monoubiquitination. (B) HeLa cells were co-transfected with Syn5-Myc or the indicated KR mutant and HA-Ub, synchronized to mitosis, immunoprecipitated by anti-Myc and blotted for HA-Ub and Syn5-Myc. (C) Quantitation of (B) from three independent experiments.
(D) K270R mutation reduces Syn5 ubiquitination in vitro. Indicated proteins were mixed and incubated followed by Western blotting for ubiquitin. Note the reduced ubiquitin signal for K270R compared to WT Syn5 (lane 3 vs. 2).
(E) Quantitation of the ubiquitin signal in (D) from three independent experiments.
(F) Expression of the K270R mutation results in Golgi fragmentation. Representative fluorescence images of HeLa cells transfected with indicated Syn5 constructs and stained for Myc (Syn5-Myc) and a Golgi marker GRASP65. Note the fragmented Golgi in K270R-expressing cells. Scale bar, 10 µm.
(G) Quantitation of (F) from three independent experiments.
Data are represented as mean ± s.e.m. *, p < 0.05**; p < 0.01; ***, p < 0.001.
See also Figure S3–S4.
Syn5 monoubiquitination is required for Golgi structure formation
Lysine 270 is located in the SNARE domain, which is important for membrane fusion. Therefore, we determined the effect of K270 mutation on Golgi morphology. Since overexpression of WT Syn5 results in Golgi fragmentation and vesicle accumulation (Hardwick and Pelham, 1992), we expressed WT Syn5 and mutants in HeLa cells at a modest expression level by reducing the transfection time. Under this condition, expression of WT Syn5 had no significant effects on the Golgi morphology. Expression of K270R, but not any other KR mutants, resulted in Golgi fragmentation (Fig. 4F–G; Fig. S3A–B). Among these lysine residues, K241 and K284 are the two lysine residues closest to K270 in the protein; K241 is also located in the SNARE domain. As both K241R and K284R mutants had no effects on the Golgi morphology, they served as excellent controls for the specificity of K270R-induced Golgi fragmentation. Consistent with the K270R mutant, expression of K270A, but not K241A or K284A mutant, also resulted in Golgi fragmentation (Fig. S4A–B). Like K270R, K270A mutation also decreased Syn5 monoubiquitination in mitosis compared to WT Syn5 and the K241A and K284A mutants (Fig. S4C).
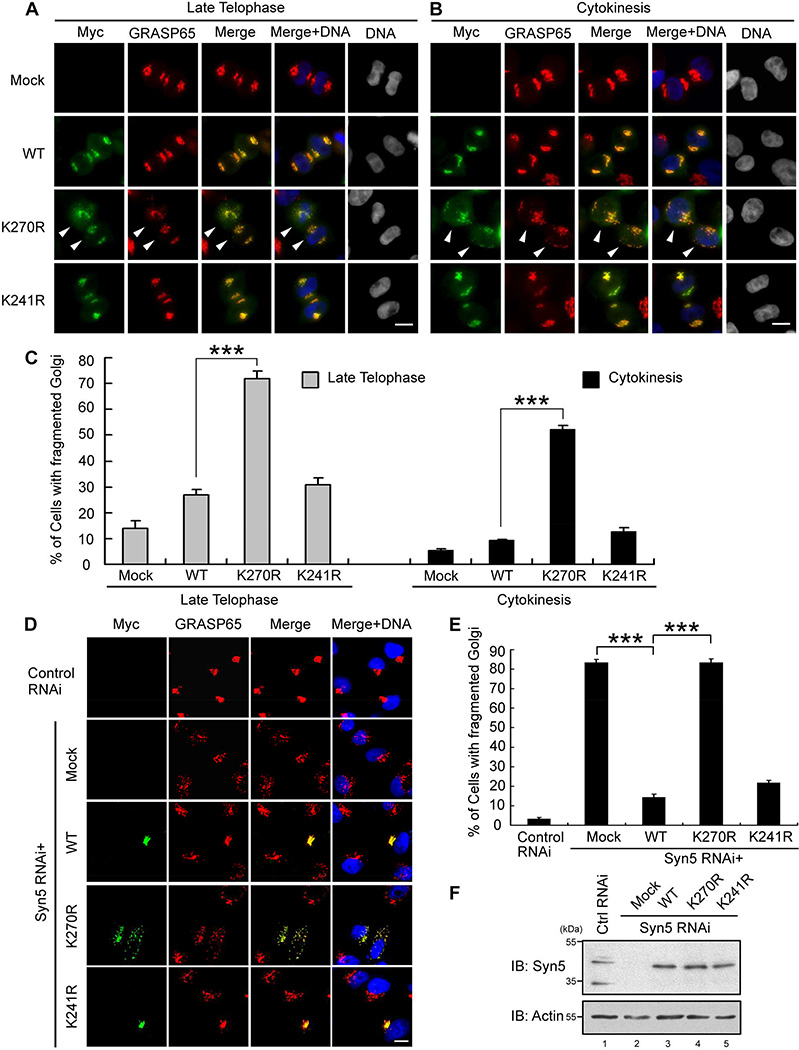
To further determine the role of Syn5 ubiquitination in mitotic Golgi disassembly and post-mitotic reassembly, we expressed Syn5 WT, K270R, or K241R and determined the Golgi morphology in cells at different cell cycle stages. The disassembly process in prometaphase, metaphase, anaphase, and telophase seemed to be unaffected by the expression of Syn5 mutants (Fig. S5A–D). However, K270R expression significantly delayed Golgi reassembly in late telophase and cytokinesis (Fig. 5A–C). This result is consistent with K270R inducing Golgi fragmentation in post-mitotic cells as shown in Fig. 4F. Taken together, these results demonstrate that mutation of the ubiquitination site K270 of Syn5 results in Golgi fragmentation.
Figure 5. Syn5 K270R mutation impairs post-mitotic Golgi reassembly.
(A–B) HeLa cells were transfected with Myc-tagged Syn5 WT, K270R, or K241R mutant and stained for Myc (Syn5) and GRASP65. Shown are representative fluorescence images of late telophase (A) and cytokinesis (B) cells. Arrowheads indicate cells with fragmented Golgi. Scale bar, 10 µm.
(C) Quantitation of cells with fragmented Golgi in (A-B) from three independent experiments.
(D) Representative fluorescence images of interphase HeLa cells in which endogenous Syn5 was replaced with exogenously expressed Myc-tagged WT or mutant Syn5. Note the fragmented Golgi in K270R expressing cells. Scale bar, 10 µm.
(E) Quantitation of (D) from three independent experiments.
(F) The expression level of Syn5 in cells used in (D and E) by Western blot.
Data are represented as mean ± s.e.m. ***, p < 0.001.
See also Figure S5–S6.
To further specify the effects caused by K270R expression, endogenous Syn5 was depleted by RNA interference (RNAi), which resulted in Golgi fragmentation (Fig. S6A–G), and then RNAi-resistant WT or mutant Syn5 was expressed to rescue the phenotype. As shown in Fig. 5D–E, WT Syn5 and K241R, but not K270R, rescued the fragmented Golgi phenotype. Depletion of endogenous Syn5 resulted in Golgi fragmentation in over 80% of the cells; expression of RNAi-resistant WT or K241R Syn5 reduced this number to about 20%. In contrast, expression of the K270R mutant had no effect on the fragmented Golgi phenotype in Syn5-depleted cells (Fig. 5D–E). In this experiment, all proteins were expressed at a similar level (Fig. 5F). These results demonstrate that monoubiquitination of Syn5 on lysine 270 is required for assembly of the Golgi structure.
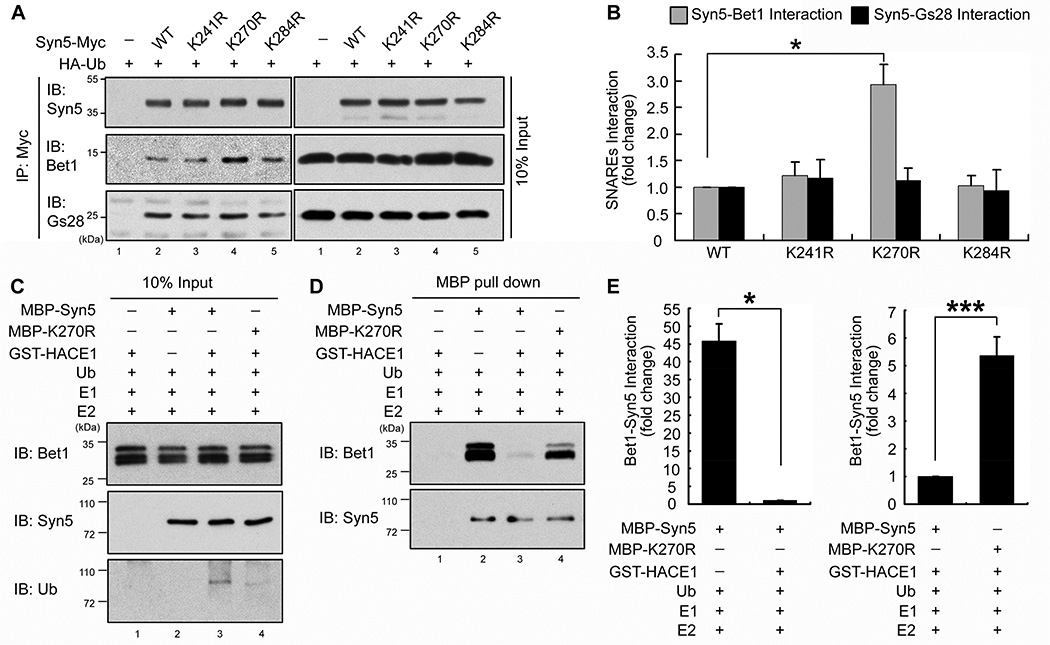
Syn5 K270R mutation increases its interaction with the v-SNARE Bet1 in mitosis
Syn5 is a master SNARE in the Golgi and is present in at least three distinct SNARE complexes: the Syn5/Gs27/Sec22b (t-SNAREs) - Bet1 (v-SNARE) complex is responsible for ER to ERGIC (ER-Golgi intermediate compartment) trafficking, the Syn5/GS28/Ykt6 - Bet1 complex for late stage of transport from the ER to cis-Golgi, and the Syn5/GS28/Ykt6 - GS15 complex mediates intra-Golgi transport and also endosome to TGN (trans-Golgi network) trafficking (Hay et al., 1997; Hong and Lev, 2014; Xu et al., 2002; Zhang and Hong, 2001). Post-mitotic Golgi membrane fusion by p97 requires Syn5 (Rabouille et al., 1998), but it is not known which other SNAREs are involved. To determine whether SNARE pairing in mitosis is affected by Syn5 ubiquitination, Myc-tagged WT or mutant Syn5 was expressed in nocodazole-arrested HeLa cells followed by immunoprecipitation of Syn5 and Western blotting for GS28 and Bet1. As shown in Fig. 6A–B, K270R mutation increased Syn5 interaction with Bet1 compared to WT Syn5, K241R, and K284R. However, none of the mutations affected the interaction between Syn5 and GS28, consistent with the report that GS28 is not required for post-mitotic Golgi cisternae regrowth (Rabouille et al., 1998). These data demonstrate that Syn5 ubiquitination regulates Syn5-Bet1 interaction in mitosis, but not Syn5-GS28 interaction.
Figure 6. K270 ubiquitination reduces Syn5 interaction with Bet1 in mitosis.
(A) Syn5-Bet1 interaction is enhanced upon K270R mutation. HeLa cells expressing indicated constructs were synchronized in mitosis and immunoprecipitated with an anti-Myc antibody and blotted for indicated proteins.
(B) Quantitation of Bet1 or GS28 interaction with Syn5-Myc in (A) from three independent experiments.
(C–E) Syn5 ubiquitination inhibits Syn5-Bet1 interaction in vitro. MBP-Syn5 was pulled down after in vitro ubiquitination reaction and further incubated with recombinant Bet1 (C). After re-isolation of Syn5, bound Bet1 was assessed by Western blot (D). (E) Quantitation of (D) for Bet1 binding to Syn5 from three independent experiments. Left panel: lane 2 vs. 3 in (D). Right panel: lane 3 vs. 4 in (D).
Data are represented as mean ± s.e.m. *, p < 0.05; **, p < 0.01; ***, p <0.001.
We also determined the effect of Syn5 ubiquitination on Syn5-Bet1 interaction in vitro (Fig. 6C–E). MBP-Syn5 was pulled down from an in vitro ubiquitination reaction and incubated with purified GST-Bet1 (Fig. 6C). MBP-Syn5 was then re-isolated and bound Bet1 was assessed by Western blotting (Fig. 6D). Non-ubiquitinated WT Syn5 had a strong interaction with Bet1, while Syn5 ubiquitination reduced this interaction to an almost undetectable level (Fig. 6D, lane 3 vs. 2; Fig. 6E). Significantly, like non-ubiquitinated WT Syn5, the K270R mutant had a strong interaction with Bet1 even in the presence of the ubiquitination machinery (Fig. 6D, lane 4; Fig. 6E). Together, these results revealed a mechanism that monoubiquitination regulates SNARE complex formation and membrane fusion.
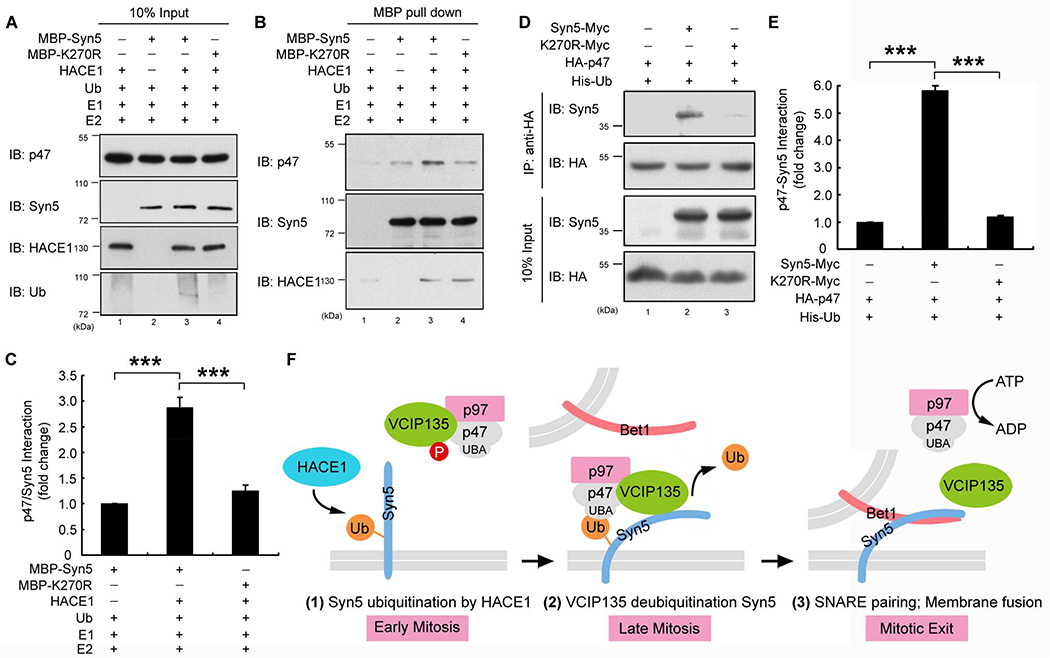
Monoubiquitination of Syn5 increases its interaction with p47
It has been reported that p47 directly binds Syn5 (Rabouille et al., 1998). In addition, p47 contains a UBA domain that preferably binds monoubiquitin (Meyer et al., 2002). These results suggest a possibility that monoubiquitination of Syn5 may control its interaction with p47 for recruitment of the p97/p47 membrane fusion machinery to the mitotic Golgi fragments. To test this possibility, MBP-Syn5 was pulled down from an in vitro ubiquitination reaction and incubated with recombinant p47 (Fig. 7A) (Zhang et al., 2015). MBP-Syn5 was then re-isolated and blotted for p47. As shown in Fig. 7B, p47 had a weak interaction with non-ubiquitinated Syn5 when HACE1 was absent (Fig. 7B, lane 2), as previously reported (Rabouille et al., 1998). However, Syn5 ubiquitination significantly increased this interaction (Fig. 7B, lane 3; Fig. 7C). In contrast to WT Syn5, the K270R mutant had a weak interaction, even in the presence of the complete ubiquitination machinery (Fig. 7B, lane 4; Fig. 7C).
Figure 7. Syn5 ubiquitination increases its interaction with p47.
(A–C) Syn5 ubiquitination enhances Syn5-p47 interaction in vitro. MBP-Syn5 was pulled down after in vitro ubiquitination reaction and further incubated with recombinant p47 (A). After re-isolation of Syn5, bound p47 was assessed by Western blot (B). (C) Quantitation of p47 bound to MBP-Syn5 in (B) from three independent experiments.
(D) Syn5 ubiquitination enhances Syn5-p47 interaction in vivo. HeLa cells expressing indicated constructs were synchronized in mitosis and immunoprecipitated with an anti-HA antibody and blotted for both Syn5-Myc and HA-p47.
(E) Quantitation of p47-Syn5 interaction in (D) from three independent experiments.
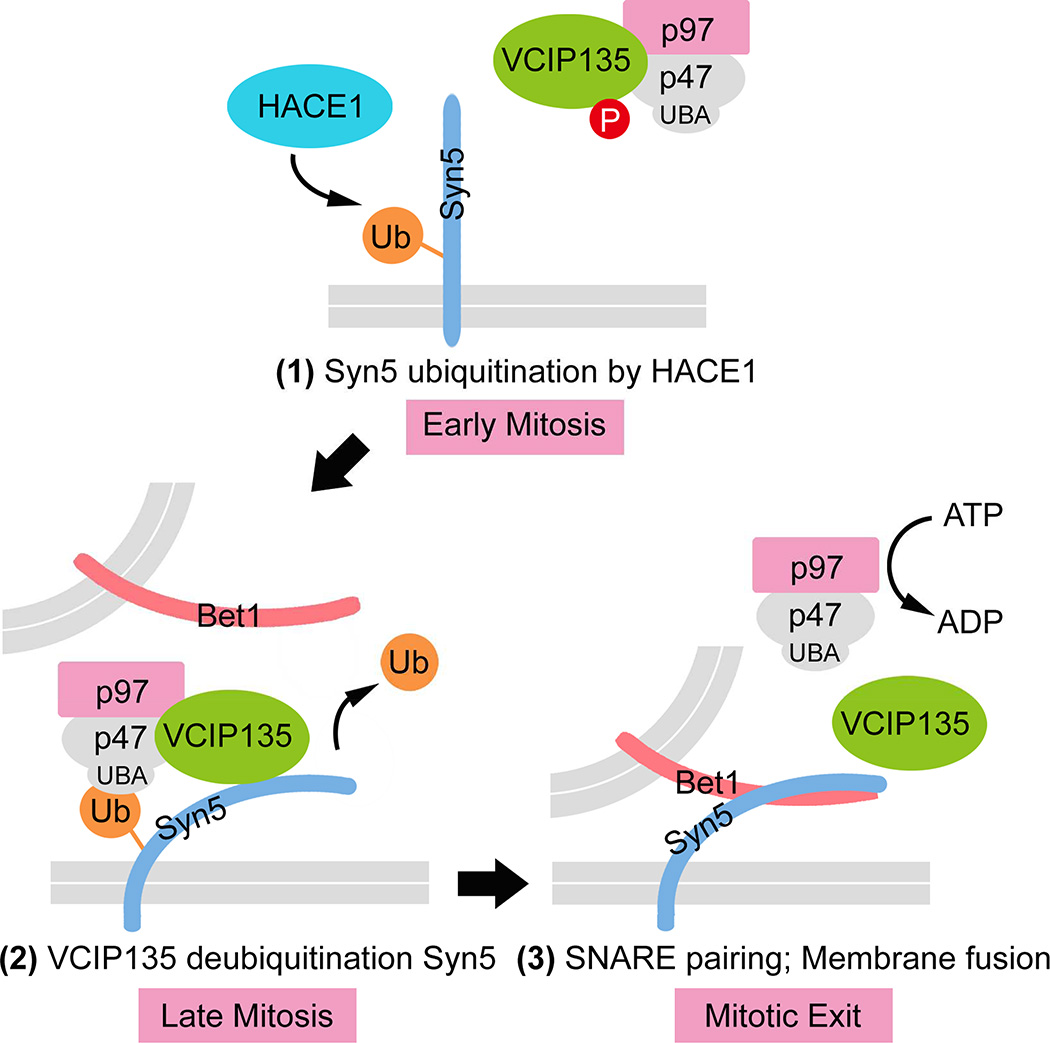
(F) A hypothetic model for the role of Syn5 ubiquitination in p97/p47-mediated post-mitotic Golgi membrane fusion. In early mitosis (left panel), Syn5 is monoubiquitinated by HACE1 on the Golgi membranes, while VCIP135 is inactivated by mitotic phosphorylation. Syn5 recruits p97/p47 complex and VCIP135 to the Golgi membranes through the interaction between the ubiquitin moiety on Syn5 and the UBA domain of p47 (middle panel). In late mitosis, ubiquitin on Syn5 is removed by VCIP135 that is reactivated by dephosphorylation, enabling Syn5-Bet1 SNARE complex formation and thus membrane fusion by p97 at mitotic exit (right panel).
Data are represented as mean ± s.e.m. ***, p < 0.001.
To test whether Syn5-p47 interaction is also regulated by Syn5 ubiquitination in cells, we co-expressed HA-tagged p47 with Myc-tagged WT Syn5 or K270R mutant and determined their interaction in mitotic cells by co-immunoprecipitation. The results showed that WT Syn5 interacted with p47 in mitotic cells, but this interaction was significantly reduced by the K270R mutation (Fig. 7D, lane 3 vs. 2; Fig. 7E). These results demonstrate that Syn5 monoubiquitination in mitosis enhances Syn5-p47 interaction to facilitate membrane association of the p97/p47 membrane fusion machinery and promote post-mitotic Golgi reassembly. K270R induced Golgi fragmentation in post-mitotic cells (Fig. 4F–G; Fig. 5; Fig. S4A–B) is likely due to its inability of binding to p47 and recruiting p97/p47 for post-mitotic Golgi membrane fusion. Overall, this study revealed the Syn5 monoubiquitination modulates Golgi membrane dynamics in the cell cycle by the control of SNARE complex formation and recruitment of the p97/p47 membrane fusion machinery to mitotic Golgi fragments.
Discussion
Several converging lines of evidence have suggested that monoubiquitination plays an essential role in the regulation of p97-mediated post-mitotic Golgi membrane fusion (Tang and Wang, 2013; Wang and Seemann, 2011). Monoubiquitination, as a regulatory signal, occurs during mitotic Golgi disassembly and is required for subsequent reassembly (Meyer et al., 2002); proteasome activity is not involved in disassembly or reassembly (Wang et al., 2004). p97 binds to monoubiquitin through the UBA domain of its adaptor protein p47; this interaction is required for p97-mediated Golgi membrane fusion (Meyer et al., 2002). Ubiquitination is mediated by the Golgi-localized ubiquitin E3 ligase HACE1 (Tang et al., 2011). Deubiquitination requires the deubiquitinase VCIP135 (Wang et al., 2004), whose activity is regulated by phosphorylation in the cell cycle; it is inactivated by phosphorylation in metaphase and re-activated upon dephosphorylation in telophase (Zhang and Wang, 2015; Zhang et al., 2014). The ubiquitination substrate, however, has remained unknown for many years.
In this study, we have identified the Golgi t-SNARE Syn5 as the ubiquitination substrate that regulates Golgi membrane dynamics in the cell cycle. This allows us to propose a working model for p97/p47-mediated Golgi membrane fusion (Fig. 7F). In early mitosis, Syn5 is monoubiquitinated on K270 by the ubiquitin ligase HACE1 on Golgi membranes (Fig. 7F, left panel). Mutation of K270 does not seem to affect mitotic Golgi disassembly (Fig. S5A–D), but it impairs post-mitotic Golgi membrane fusion (Fig. 5). Monoubiquitination of Syn5 recruits p97/p47 and VCIP135 to Golgi membranes through the interaction with the UBA domain of p47 (Fig. 7F, middle panel). At this time VCIP135 is inactivated by mitotic phosphorylation (Zhang and Wang, 2015; Zhang et al., 2014) and thus monoubiquitinated Syn5, p97, p47, and VCIP135 form a transient quaternary complex (Uchiyama et al., 2002). In late mitosis, VCIP135 is reactivated by dephosphorylation, removes ubiquitin from Syn5, and allows Syn5 interacting with its cognate SNARE Bet1 for membrane fusion at mitotic exit (Fig. 7F, right panel). The exact mechanism for p97-mediated membrane fusion remains unclear. It is possible that p97 (together with p47 and VCIP135) may change Syn5 conformation and prime it for membrane fusion, or that p97 may function in a similar way to NSF in disassembling the cis-SNARE complexes. However, Syn5 ubiquitination does not seem to cause Syn5 degradation as Syn5 protein level does not change during the cell cycle (Fig. 2A; Fig. S2C–D). Taken together, our results revealed that ubiquitination - through cycles of adding and removing ubiquitin to and from Syn5 - regulates Golgi membrane dynamics during the cell cycle.
It is interesting to see that only a relative small proportion (5–10%) of Syn5 is ubiquitinated in mitosis (Fig. 1B and Fig. S1). One possible explanation, as described in our model (Fig. 7F), is that monoubiquitination of Syn5 occurs only transiently in early mitosis and is quickly reversed by the deubiquitinase VCIP135 in late mitosis. Given that our synchronized cells are somewhat heterogeneous, ranging from relatively early to later mitotic cells and even interphase cells, the ubiquitination level is likely underestimated. On the other hand, ubiquitination of a small population of Syn5 may be able to exhibit significant effects on membrane fusion. It has been shown that a coordination of multiple SNARE complexes is required for rapid membrane fusion as in post-mitotic Golgi reassembly and neurotransmission. For example, each synaptic vesicle contains on average 6.2 molecules of syntaxin 1, several other syntaxins, and 70 active synaptobrevin molecules (Takamori et al., 2006), while each 25 nm fusion site on the plasma membrane of PC12 cells contains on average 75 clustered syntaxin molecules (Sieber et al., 2007). How these SNAREs coordinate with each other to catalyze the rapid membrane fusion has not been determined, but it is possible that a large ubiquitin moiety on one Syn5 molecule may disrupt the coordination between adjacent SNARE complexes. Consistent with this notion, our results demonstrated that elimination of this small proportion of Syn5 monoubiquitination significantly enhanced Syn5-Bet1 interaction (Fig. 6A–E) and reduced Syn5-p47 interaction (Fig. 7A–E), and is sufficient to cause Golgi defects in post-mitotic cells (Fig. 4F–G; Fig. 5; Fig. S4A–B).
The significance of our findings is three-fold. First, our findings help explain why the p97 pathway is important for post-mitotic Golgi reassembly. Syn5 is a common component of both NSF/SNAP- and p97/p47-mediated post-mitotic Golgi membrane fusion (Rabouille et al., 1998). p47 and α-SNAP compete for binding with Syn5 and thus the p97 and NSF pathways have non-overlapping functions in Golgi membrane fusion (Rabouille et al., 1998). Although ubiquitin is only involved in the p97 pathway, Syn5 is required for both pathways, and thus monoubiquitination of Syn5 may restrict the role of NSF in post-mitotic Golgi membrane fusion and control the balance between the NSF and p97 pathways. This helps to understand the relationship between the NSF and p97 pathways in post-mitotic Golgi membrane fusion.
Second, our results improve the mechanistic understanding of p97/p47-mediated membrane fusion. Unlike NSF whose role in cis-SNARE complex disassembly is well-established, the mechanism of p97-mediated membrane fusion has remained a mystery. The identification of Syn5 as the ubiquitinated substrate and our results demonstrating that Syn5 monoubiquitination regulates Syn5-Bet1 and Syn5-p47 interactions set the foundation for further exploration of the underlying mechanism. Although Syn5 is reported to be involved in three different SNARE complexes during vesicle trafficking and membrane fusion, it is the only t-SNARE known for Golgi reassembly (Rabouille et al., 1998). Our results indicate that Bet1 may be the v-SNARE in post-mitotic Golgi membrane fusion. The GS28-Syn5 interaction is not affected by monoubiquitination of Syn5, indicating that GS28 is probably not involved in Golgi reassembly in the p97 pathway, consistent with the previous report that an antibody block of GS28 does not affect p97-mediated post-mitotic Golgi membrane fusion (Rabouille et al., 1998). Future studies will determine which cognate SNAREs of Syn5 are involved in p97-mediated post-mitotic membrane fusion and how monoubiquitination regulates SNARE complex formation.
Third, Syn5 monoubiquitination may provide a possible mechanism for blocked membrane trafficking in mitosis. The disassembly of the Golgi in mitosis indicates that membrane fusion must be inhibited. This has been observed for many years (Warren et al., 1983), but the exact mechanism remained unknown. During mitosis, the Golgi is possibly disassembled by multiple mechanisms, including phosphorylation of GRASP65 and GRASP55 for Golgi unstacking (Xiang and Wang, 2010), phosphorylation of membrane tethers such as GM130 to facilitate vesicle budding (Lowe et al., 1998), and ubiquitination of Syn5 to block membrane fusion. Syn5 is the common t-SNARE in all three SNARE complexes involved in Golgi related trafficking, while Bet1 is a common v-SNARE in the two complexes for ER-to-Golgi trafficking. Syn5 ubiquitination provides a plausible explanation of how ER to Golgi and intra-Golgi trafficking is blocked during mitosis. Attaching a ubiquitin moiety to the SNARE domain of Syn5 provides an effective way to inhibit membrane fusion in mitosis, perhaps more efficient than phosphorylation of GRASP65/55 and Golgi tethering proteins (Xiang and Wang, 2010).
Taken together, our study uncovers a mechanism that controls Golgi membrane fusion. Ubiquitination of Syn5 is likely achieved by a balance between the activities of the ubiquitin ligase HACE1 and the DUB VCIP135. It is not known how HACE1 is regulated in the cell cycle, but the activity of VCIP135 is controlled by phosphorylation (Zhang and Wang, 2015). Ubiquitination of Syn5 may have two functions, one is to impair both NSF- and p97-mediated membrane fusion, which may be part of the mechanism of blocked membrane trafficking in mitosis; the other is to recruit the p97/p47 membrane fusion machinery to the Golgi fragments through interaction with the UBA domain of p47. Removal of ubiquitin from Syn5 promotes SNARE complex formation and facilitates efficient post-mitotic Golgi membrane fusion. Therefore, ubiquitin functions as a multifaceted regulator in cell cycle-regulated Golgi membrane dynamics.
Experimental Procedures
RNAi and microscopy
For knockdown experiments, HeLa cells were transfected using Lipofectamine RNAiMAX (Invitrogen) following the manufacturer’s instructions. For Syn5 depletion, cells were analyzed 72 h after Syn5 specific RNAi transfection. For the rescue of Syn5 knockdown with exogenously expressed Syn5, cells were transfected with Syn5 specific RNAi for 48 h and then by RNAi-resistant form of WT Syn5 or KR mutants for another 24 h. The control or HACE1 shRNA lentivirus infected stable cell lines were described previously (Tang et al., 2011).
To enrich mitotic cells for immunofluorescence, HeLa cells were first synchronized to G1/S by a double-thymidine block (2.5 mM thymidine for 16 h, released in fresh medium for 12 h, followed by 2.5 mM thymidine for 16 h) and then released in fresh medium for 9–10 h to mitosis before fixation. Fragmented Golgi was defined as disconnected or scattered dots. To quantify the percentage of cells with fragmented Golgi, more than 200 cells were counted in each experiment. The results are presented as the mean ± s.e.m from three independent experiments. Electron microscopy was performed as previously described (Tang et al., 2011).
In vitro ubiquitination and deubiquitination assay and protein-protein interactions
For in vitro ubiquitination of Golgi membranes, 100 µg purified rat liver Golgi (RLG) membranes (Tang and Wang, 2015) were incubated at 30°C for 60 min with 2 µM purified GST-HACE1 or GST-HACE1 ΔC, 480 µg mitotic cell cytosol (MC, to provide E1 and E2s) (Tang et al., 2010) and 50 µM purified His-tagged ubiquitin with 0.5 µM Epoxomicin, and an ATP-regenerating system in MEB buffer (50 mM Tris-HCl, pH 7.4, 0.2 M sucrose, 50 mM KCl, 20 mM β-glycerophosphate, 15 mM EGTA, 10 mM MgCl2, 2 mM ATP, 1 mM GTP, 1 mM glutathione and protease inhibitors) at a final volume of 1 ml (Tang et al., 2010). The membranes were re-isolated by centrifuge at 14000 rpm 4°C for 30 min in a benchtop centrifuge, solubilized in 1 ml lysis buffer (100 mM Tris-HCl, pH 8.0, 8 M urea, 0.5% TX-100 w/v, 150 mM NaCl, 10 mM NEM), and ubiquitinated proteins were isolated using 30 µl Ni-NTA beads (Thermo Scientific).
For in vitro ubiquitination of purified proteins, reactions were carried out with 0.3 µg MBP-tagged Syn5-ΔTM, K270R-ΔTM, long form Syn5-ΔTM, or MBP, 0.4 µg GST-HACE1, 0.35 µg human E1 ubiquitin activating enzyme (Boston Biochem), 0.4 µg UbcH7 recombinant E2 Ub conjugating enzyme (Boston Biochem), and 10 µg ubiquitin (Sigma) in 50 µl of ubiquitination buffer (50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 100 mM NaCl, 1 mM DTT, 2 mM ATP) at 30°C for 2 h. The samples were analyzed by SDS-PAGE and Western blotting.
To deubiquitinate Syn5 by immunoprecipitated VCIP135, HeLa cells were treated with 100 ng/ml nocodazole (Sigma) for 18 h, washed with fresh medium without nocodazole for five times, reseeded onto new plates, and then incubated for another 0.5, 1, 1.5, 2 and 3 hours. Immunoprecipitated VCIP135 from these cells of different time points was incubated with Flag-Syn5 immunoprecipitated from Flag-Syn5 and HA-Ub expressing mitotic cells in 20 µl buffer (150 mM KCl, 50 mM Hepes, pH 7.4, 10 mM DTT, 5% glycerol, 0.01% Triton X-100) for 1 h at 30°C as previously described (Zhang and Wang, 2015; Zhang et al., 2014). Aliquots of the reaction were analyzed by Western blot with indicated antibodies.
For in vitro deubiquitination using purified proteins, 0.3 µg MBP-Syn5-ΔTM or MBP was first incubated with 0.4 µg GST-HACE1 WT, 0.35 µg human E1 ubiquitin activating enzyme (Boston Biochem), 0.4 µg UbcH7 recombinant E2 Ub conjugating enzyme (Boston Biochem), and 10 µg ubiquitin (Sigma) in 50 µl of ubiquitination buffer (50 mM Tris-HCl pH 7.5, 5 mM MgCl2, 100 mM NaCl, 1 mM DTT, 2 mM ATP) at 30 °C for 2 h. MBP-Syn5-ΔTM or MBP were then pulled down by Amylose resin (New England Biolabs) from the 50 µl reactions diluted to 500 µl with the ubiquitination buffer. The beads were washed 3 times with deubiquitination buffer (150 mM KCl, 50 mM Hepes, pH 7.4, 10 mM DTT, 5% glycerol, 0.01% Triton X-100), MBP-Syn5 ΔTM or MBP on the Amylose beads were further incubated with 1 µg SBP-VCIP135 (WT or C218S) in 50 µl deubiquitination buffer at 30°C for 15 min or 1 h as indicated (Zhang and Wang, 2015). The beads were washed and bound proteins were analyzed by SDS-PAGE and Western blotting. To determine Syn5-Bet1 or Syn5-p47 interaction in vitro, MBP-Syn5 ΔTM, K270R-ΔTM or MBP on Amylose beads described above were further incubated with 0.63 µg GST-Bet1 or 1 µg His-p47 at 4 °C for 3 h with gentle agitation. The beads were washed and bound proteins were analyzed by SDS-PAGE and Western blotting.
Co-immunoprecipitation
HeLa cells were scraped and lysed in immunoprecipitation buffer (40 mM Hepes, pH 7.4, 200 mM KCl, 5 mM MgCl2, 1% Triton X-100, 50 mM β-glycerolphosphate, protease inhibitors) for 20 min at 4°C and cleared by centrifugation at 14,000 rpm and 4°C for 20 min in a benchtop centrifuge. Myc antibodies pre-loaded to protein G beads were added to the lysate and incubated overnight at 4°C with gentle agitation. After extensive washing by the immunoprecipitation buffer, the beads were analyzed by SDS-PAGE and Western blot. To detect ubiquitinated Syn5, HeLa cells transfected with HA-Ub and the Myc-tagged WT Syn5 or KR mutants were synchronized to mitosis by 100 ng/ml nocodazole treatment for 18 h followed by immunoprecipitation in the immunoprecipitation buffer supplemented with 50 mM NEM and Western blot. To determine the interactions between Syn5 and other SNAREs, HeLa cells were incubated in tissue culture medium containing 1 mM NEM for 15 min on ice, and quenched with 2 mM DTT in medium for 15 min on ice. Cells were finally incubated in complete medium at 37°C for 30 min before the extraction with lysis buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 5 mM EDTA, protease inhibitors) and co-immunoprecipitation (Mallard et al., 2002). To determine the interaction between Syn5 and p47, HeLa cells transfected with HA-p47, His-Ub and Myc-tagged WT Syn5 or K270R mutant were synchronized to mitosis by 100 ng/ml nocodazole treatment for 18 h followed by immunoprecipitation in the lysis buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 5 mM EDTA, protease inhibitors) supplemented with 50 mM NEM and Western blot.
Ni-NTA pulldown assay
HeLa cells transfected with His-Ub and Flag-Syn5 were synchronized to mitosis, scraped and lysed with RIPA buffer (50 mM Tris-Cl, pH 7.4, 150 mM NaCl, 1% NP-40, 0.5% deoxycholic acid, 0.1% SDS, 1 mM DTT, protease inhibitors, 1 mM PMSF, 50 mM NEM) at 4°C for 20 min. The lysate was cleared by centrifugation at 14000 rpm 4°C for 20 min, and the supernatant was added one volume of denature buffer (1% SDS, 10 mM imidazole, 4 M urea, 150 mM NaCl, 25 mM NaH2PO4, 0.25% NP40, pH 8.0) and incubated with 40 µl Ni-NTA resin overnight at 4°C with gentle agitation. After extensive washing, the beads were analyzed by SDS-PAGE and Western blot.
Supplementary Material
Highlights.
Syn5 is ubiquitinated by HACE1 and deubiquitinated by VCIP135 during the cell cycle
Syn5 monoubiquitination on Lysine 270 impairs Syn5-Bet1 interaction in mitosis
Syn5 ubiquitination recruits p97/p47 onto mitotic Golgi fragments
Expression of the Syn5 K270R mutant in cells impairs post-mitotic Golgi reassembly
Acknowledgments
We thank Drs. Tsui-Fen Chou, Jesse Hay, Wanjin Hong, Vladimir Lupashin, Hemmo Meyer, Mitsuo Tagaya, and Graham Warren for cDNA constructs, recombinant proteins and antibodies to Syn5, Bet1 and p47, Dr. Xiaoyan Zhang for ubiquitin and VCIP135 cDNAs and recombinant proteins as well as ideas and technical support, other members of the Wang Lab for suggestions and reagents, and Dr. Michael E. Bekier and Courtney Killeen for editing the manuscript. Special thanks to Dr. Honghao Zhang who provided the results shown in Figure S6. This work was supported in part by the National Institutes of Health (Grants GM087364 and GM112786), MCubed and the Fast forward Protein Folding Disease Initiative of the University of Michigan to Y. Wang.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplemental Information
Supplemental Information includes Supplemental Experimental Procedures and six figures.
Author Contributions
Y.W. conceived the research; Y.W. and S.H. designed the experiments; S.H. and D.T. performed the experiments; Y.W. and S.H. analyzed the data and wrote the paper.
References
- Anglesio MS, Evdokimova V, Melnyk N, Zhang L, Fernandez CV, Grundy PE, Leach S, Marra MA, Brooks-Wilson AR, Penninger J, et al. Differential expression of a novel ankyrin containing E3 ubiquitin-protein ligase, Hace1, in sporadic Wilms' tumor versus normal kidney. Hum Mol Genet. 2004;13:2061–2074. doi: 10.1093/hmg/ddh215. [DOI] [PubMed] [Google Scholar]
- Hardwick KG, Pelham HR. SED5 encodes a 39-kD integral membrane protein required for vesicular transport between the ER and the Golgi complex. J Cell Biol. 1992;119:513–521. doi: 10.1083/jcb.119.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay JC, Chao DS, Kuo CS, Scheller RH. Protein interactions regulating vesicle transport between the endoplasmic reticulum and Golgi apparatus in mammalian cells. Cell. 1997;89:149–158. doi: 10.1016/s0092-8674(00)80191-9. [DOI] [PubMed] [Google Scholar]
- Hong W, Lev S. Tethering the assembly of SNARE complexes. Trends in cell biology. 2014;24:35–43. doi: 10.1016/j.tcb.2013.09.006. [DOI] [PubMed] [Google Scholar]
- Hui N, Nakamura N, Sonnichsen B, Shima DT, Nilsson T, Warren G. An isoform of the Golgi t-SNARE, syntaxin 5, with an endoplasmic reticulum retrieval signal. Molecular biology of the cell. 1997;8:1777–1787. doi: 10.1091/mbc.8.9.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R, Scheller RH. SNAREs - engines for membrane fusion. Nature reviews. 2006;7:631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- Lowe M, Rabouille C, Nakamura N, Watson R, Jackman M, Jamsa E, Rahman D, Pappin DJ, Warren G. Cdc2 kinase directly phosphorylates the cis-Golgi matrix protein GM130 and is required for Golgi fragmentation in mitosis. Cell. 1998;94:783–793. doi: 10.1016/s0092-8674(00)81737-7. [DOI] [PubMed] [Google Scholar]
- Mallard F, Tang BL, Galli T, Tenza D, Saint-Pol A, Yue X, Antony C, Hong W, Goud B, Johannes L. Early/recycling endosomes-to-TGN transport involves two SNARE complexes and a Rab6 isoform. J Cell Biol. 2002;156:653–664. doi: 10.1083/jcb.200110081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HH, Wang Y, Warren G. Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1-Npl4. The EMBO journal. 2002;21:5645–5652. doi: 10.1093/emboj/cdf579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki K, Wakana Y, Noda C, Arasaki K, Furuno A, Tagaya M. Contribution of the long form of syntaxin 5 to the organization of the endoplasmic reticulum. Journal of cell science. 2012;125:5658–5666. doi: 10.1242/jcs.105304. [DOI] [PubMed] [Google Scholar]
- Rabouille C, Kondo H, Newman R, Hui N, Freemont P, Warren G. Syntaxin 5 is a common component of the NSF- and p97-mediated reassembly pathways of Golgi cisternae from mitotic Golgi fragments in vitro. Cell. 1998;92:603–610. doi: 10.1016/s0092-8674(00)81128-9. [DOI] [PubMed] [Google Scholar]
- Rabouille C, Levine TP, Peters JM, Warren G. An NSF-like ATPase, p97, and NSF mediate cisternal regrowth from mitotic Golgi fragments. Cell. 1995;82:905–914. doi: 10.1016/0092-8674(95)90270-8. [DOI] [PubMed] [Google Scholar]
- Sieber JJ, Willig KI, Kutzner C, Gerding-Reimers C, Harke B, Donnert G, Rammner B, Eggeling C, Hell SW, Grubmuller H, et al. Anatomy and dynamics of a supramolecular membrane protein cluster. Science. 2007;317:1072–1076. doi: 10.1126/science.1141727. [DOI] [PubMed] [Google Scholar]
- Tai G, Lu L, Wang TL, Tang BL, Goud B, Johannes L, Hong W. Participation of the syntaxin 5/Ykt6/GS28/GS15 SNARE complex in transport from the early/recycling endosome to the trans-Golgi network. Molecular biology of the cell. 2004;15:4011–4022. doi: 10.1091/mbc.E03-12-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamori S, Holt M, Stenius K, Lemke EA, Gronborg M, Riedel D, Urlaub H, Schenck S, Brugger B, Ringler P, et al. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–846. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- Tang D, Mar K, Warren G, Wang Y. Molecular mechanism of mitotic Golgi disassembly and reassembly revealed by a defined reconstitution assay. J Biol Chem. 2008;283:6085–6094. doi: 10.1074/jbc.M707715200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Wang Y. Cell cycle regulation of Golgi membrane dynamics. Trends in cell biology. 2013;23:296–304. doi: 10.1016/j.tcb.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Wang Y. Golgi isolation. Cold Spring Harb Protoc. 2015;2015:562–567. doi: 10.1101/pdb.prot075911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Xiang Y, De Renzis S, Rink J, Zheng G, Zerial M, Wang Y. The ubiquitin ligase HACE1 regulates Golgi membrane dynamics during the cell cycle. Nat Commun. 2011;2:501. doi: 10.1038/ncomms1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Xiang Y, Wang Y. Reconstitution of the cell cycle-regulated Golgi disassembly and reassembly in a cell-free system. Nat Protoc. 2010;5:758–772. doi: 10.1038/nprot.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrino S, Visvikis O, Doye A, Boyer L, Stefani C, Munro P, Bertoglio J, Gacon G, Mettouchi A, Lemichez E. The E3 ubiquitin-ligase HACE1 catalyzes the ubiquitylation of active Rac1. Developmental cell. 2011;21:959–965. doi: 10.1016/j.devcel.2011.08.015. [DOI] [PubMed] [Google Scholar]
- Uchiyama K, Jokitalo E, Kano F, Murata M, Zhang X, Canas B, Newman R, Rabouille C, Pappin D, Freemont P, et al. VCIP135, a novel essential factor for p97/p47-mediated membrane fusion, is required for Golgi and ER assembly in vivo. J Cell Biol. 2002;159:855–866. doi: 10.1083/jcb.200208112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama K, Totsukawa G, Puhka M, Kaneko Y, Jokitalo E, Dreveny I, Beuron F, Zhang X, Freemont P, Kondo H. p37 is a p97 adaptor required for Golgi and ER biogenesis in interphase and at the end of mitosis. Developmental cell. 2006;11:803–816. doi: 10.1016/j.devcel.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Wang Y, Satoh A, Warren G, Meyer HH. VCIP135 acts as a deubiquitinating enzyme during p97-p47-mediated reassembly of mitotic Golgi fragments. J Cell Biol. 2004;164:973–978. doi: 10.1083/jcb.200401010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Seemann J. Golgi biogenesis. Cold Spring Harb Perspect Biol. 2011;3:a005330. doi: 10.1101/cshperspect.a005330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G, Featherstone C, Griffiths G, Burke B. Newly synthesized G protein of vesicular stomatitis virus is not transported to the cell surface during mitosis. J Cell Biol. 1983;97:1623–1628. doi: 10.1083/jcb.97.5.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Sollner TH, Rothman JE. SNAREpins: minimal machinery for membrane fusion. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Wang Y. GRASP55 and GRASP65 play complementary and essential roles in Golgi cisternal stacking. J Cell Biol. 2010;188:237–251. doi: 10.1083/jcb.200907132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Martin S, James DE, Hong W. GS15 forms a SNARE complex with syntaxin 5, GS28, and Ykt6 and is implicated in traffic in the early cisternae of the Golgi apparatus. Molecular biology of the cell. 2002;13:3493–3507. doi: 10.1091/mbc.E02-01-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Hong W. Ykt6 forms a SNARE complex with syntaxin 5, GS28, and Bet1 and participates in a late stage in endoplasmic reticulum-Golgi transport. J Biol Chem. 2001;276:27480–27487. doi: 10.1074/jbc.M102786200. [DOI] [PubMed] [Google Scholar]
- Zhang X, Gui L, Zhang X, Bulfer SL, Sanghez V, Wong DE, Lee Y, Lehmann L, Lee JS, Shih PY, et al. Altered cofactor regulation with disease-associated p97/VCP mutations. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1418820112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Wang Y. Cell cycle regulation of VCIP135 deubiquitinase activity and function in p97/p47-mediated Golgi reassembly. Molecular biology of the cell. 2015 doi: 10.1091/mbc.E15-01-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhang H, Wang Y. Phosphorylation regulates VCIP135 function in Golgi membrane fusion during the cell cycle. Journal of cell science. 2014;127:172–181. doi: 10.1242/jcs.134668. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.