Summary
Basement membranes (BMs) are planar protein networks that support epithelial function. Regulated changes to BM architecture can also contribute to tissue morphogenesis, but how epithelia dynamically remodel their BMs is unknown. In Drosophila, elongation of the initially spherical egg chamber correlates with the generation of a polarized network of fibrils in its surrounding BM. Here, we use live imaging and genetic manipulations to determine how these fibrils form. BM fibrils are assembled from newly synthesized proteins in the pericellular spaces between the egg chamber’s epithelial cells, and undergo oriented insertion into the BM by directed epithelial migration. We find that a Rab10-based secretion pathway promotes pericellular BM protein accumulation and fibril formation. Finally, by manipulating this pathway, we show that BM fibrillar structure influences egg chamber morphogenesis. This work highlights how regulated protein secretion can synergize with tissue movement to build a polarized BM architecture that controls tissue shape.
Graphical abstract
eTOC Blurb
Isabella and Horne-Badovinac examine how basement membrane (BM) structure contributes to morphogenesis in the context of Drosophila egg chamber development. They describe how the follicular epithelium coordinates Rab10-based BM protein secretion and directed tissue migration to build a polarized BM structure. Selectively altering this BM structure affects organ morphogenesis.
Introduction
BMs are specialized extracellular matrices (ECMs) at the basal sides of epithelia. They are composed of a conserved set of core proteins, including Type IV Collagen (Col IV), Laminin, and the heparin sulfate proteoglycan Perlecan, that self-assemble into a planar network (Yurchenco, 2011). Beyond these shared features, BMs show compositional and structural diversity necessary to support the homeostatic needs of their associated tissues (Hynes and Naba, 2012). They can also be remodeled during development to facilitate tissue morphogenesis (Daley and Yamada, 2013; Morrissey and Sherwood, 2015). However, the mechanisms by which tissues modify BM structure, and how BM structure in turn modulates tissue dynamics, are poorly understood.
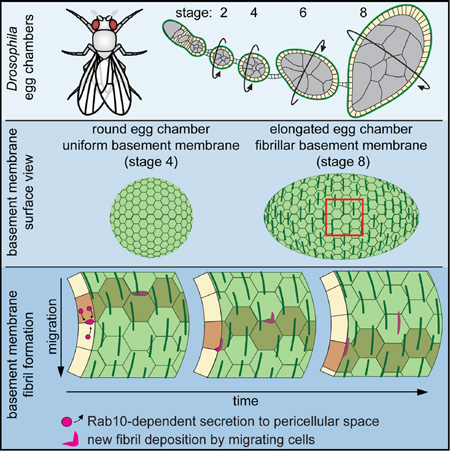
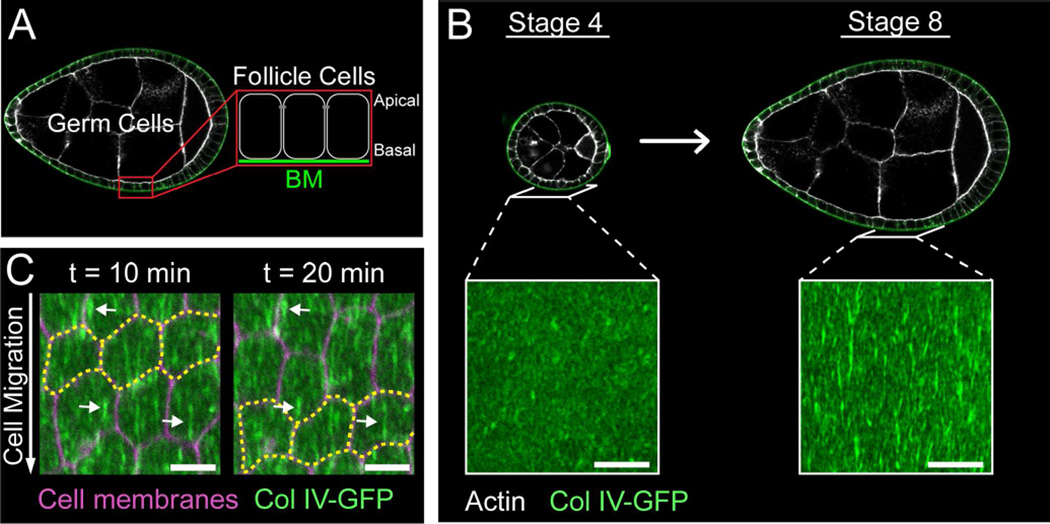
The Drosophila egg chamber offers a powerful system to study the interplay between BM structure and morphogenesis (Isabella and Horne-Badovinac, 2015a). Egg chambers are organ-like structures in the ovary that each generates one egg. They contain an interior germ cell cluster and an enveloping layer of somatic epithelial cells, called follicle cells (Figure 1A). The follicle cells are polarized with their apical membranes contacting the germ cells and their basal membranes facing outward. This epithelium produces a BM that surrounds the egg chamber. Egg chamber development is categorized into 14 morphological stages. Between stages 5 and 10, egg chambers elongate along their anterior-posterior (A-P) axes to produce the ellipsoidal shape of the egg (Figure S1A).
Figure 1. Introduction to BM fibrils.
(A) Egg chamber structure.
(B) Stage 4 egg chambers are round and have BMs that are largely uniform in structure; stage 8 egg chambers are elongated and their BMs contain polarized fibrils. Scale bars = 10 µm.
(C) Time-lapse of follicle cell migration. Follicle cells migrate along the stationary BM in the direction of fibril polarity. Yellow outline marks three cells moving over time; arrows mark three stationary fibrils over time. Stage 7. Scale bars = 5 µm.
See also Figure S1.
Egg chamber elongation coincides with a dramatic change in BM structure (Gutzeit et al., 1991; Haigo and Bilder, 2011). Before elongation, the BM is largely uniform; between stages 5 and 8, however, linear, fibril-like structures are added to the pre-existing planar matrix (Figure 1B). All major BM proteins thus far examined – Col IV, Laminin, and Perlecan – exhibit this structure (Gutzeit et al., 1991; Haigo and Bilder, 2011; Schneider et al., 2006). BM fibrils align perpendicular to the A-P axis, polarizing the matrix. The construction of this new matrix architecture was reported to depend on collective migration of the follicle cells along the BM (Figure 1C) (Haigo and Bilder, 2011); this movement causes the entire egg chamber to rotate within the BM in the same direction that it becomes polarized. Although rotation appears to be necessary for BM fibril formation, it is not sufficient, as the follicle cells begin migrating ~20 hours before fibrils appear (Cetera et al., 2014). Thus, the mechanisms underlying BM fibril formation are still unclear.
The polarized BM is thought to function as part of a molecular corset that anisotropically constrains egg chamber growth to promote elongation (Figure S1B) (Gutzeit et al., 1991). The main evidence supporting this idea comes from experiments in which collective follicle cell migration is blocked (Haigo and Bilder, 2011). Inhibiting this migration prevents BM fibril formation and blocks elongation; however, migration feeds into other cellular process that may contribute to the elongation program independently of BM structure (Cetera et al., 2014; Viktorinová and Dahmann, 2013). Thus, to show that BM fibrils play a role in elongation, we must identify a more specific way to affect their assembly.
Here, we use live imaging and genetic manipulations to determine how BM fibrils form. We find that BM fibrils are generated de novo from newly synthesized proteins. Our data suggest a model in which new BM proteins are secreted through a basal region of the lateral plasma membrane and aggregate into cohesive structures within the pericellular space between follicle cells. They are then inserted into the BM in the correct orientation by the directed migration of the tissue. We further show that the small GTPase Rab10 and its effector EH domain binding protein 1 (Ehbp1) promote BM fibril formation via this pericellular secretion pathway. Finally, we use Rab10/Ehbp1 mis-expression to selectively alter BM structure and show that increasing BM fibril formation influences egg chamber elongation. This work shows how coordinated cellular behaviors can regulate BM structure during development, and how the matrix remodeling can play an instructive role in shaping the tissue.
Results
BM fibrils form from newly synthesized proteins
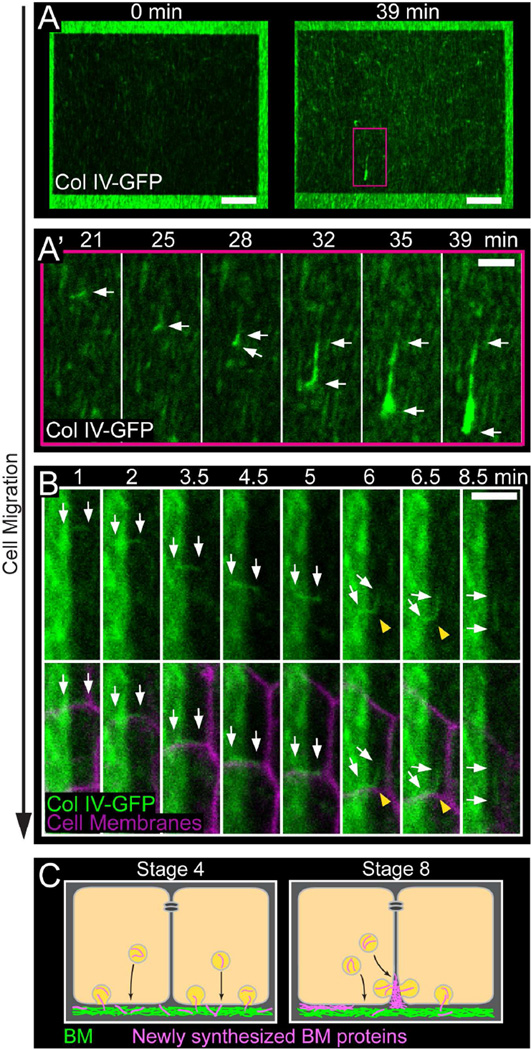
The fibrillar structures that form in the follicular BM are best visualized with a GFP protein trap in the Col IV α2 chain Viking (Col IV-GFP) (Haigo and Bilder, 2011). To visualize BM fibril formation, we performed live imaging of stage 7 BMs labeled with Col IV-GFP. This process coincides with a period of increased BM protein production (Haigo and Bilder, 2011); thus, we reasoned that fibrils might arise from newly synthesized proteins. To test this idea, we photo-bleached a large rectangular region of the BM, which allowed us to assess the contribution of newly synthesized Col IV-GFP to fibril formation (Figures 2A, 2B and S2A; Movies S1 and S2). Over the course of a 39-minute movie, two populations of Col IV-GFP can be seen moving relative to the static BM (Figures 2A and S2A; Movie S1). The first population appears as punctae that move steadily throughout the movie. This signal likely represents Col IV within the secretory pathway of the migrating cells. The second population includes structures that often appear linear - these initially move with the follicle cells but eventually integrate into the BM as new fibrils. We will refer to these structures as “nascent fibrils”. Newly incorporated fibrils always exhibit bright GFP fluorescence relative to the photo-bleached BM, indicating that they form from newly synthesized proteins (Figures 2A and S2A; Movie S1). Additionally, although the shapes of nascent fibrils are highly variable, from globules that appear to unfurl during insertion (Figure 2A’; Movie S1) to wispy, linear structures that insert with little shape change (Figures 2B and S2A’; Movies S1 and S2), their cohesiveness suggests that they are assembled prior to deposition into the BM.
Figure 2. Live imaging of BM fibril formation.
(A) Still images from Movie S1 showing fibril incorporation into the BM. (A) First and last frames for Movie S1. The dark rectangle is the photobleached region. The pink box corresponds to the region shown in (A’). (A’) Montage showing an individual nascent fibril with full GFP fluorescence moving in the direction of cell migration and then incorporating into the BM. Arrows mark both ends of the nascent fibril. Scale bars = 10 µm (A), 3 µm (A’).
(B) Montage from Movie S2 showing fibril incorporation into the BM. The dark portion on the right of each panel is the photobleached region. A nascent fibril travels with the migrating cell-cell interface until it is drawn away from this location and inserted into the BM. BM insertion causes the fibril to bend (yellow arrowheads) and then become properly aligned in the BM. Arrows mark both ends of the nascent fibril. Scale bar = 3 µm.
(C) Model for BM fibril formation. Prior to fibril formation (represented by stage 4), we envision that new BM proteins (pink) exit through the basal surface and directly incorporate into the planar BM (green). During fibril formation (represented by stage 8), a portion of the BM traffic may be redirected to a basal region of the lateral surface. BM proteins would then aggregate in the pericellular space before being deposited in the BM as fibrils.
Experiments performed at stage 7. See also Figure S2 and Movies S1 and S2.
Performing the same experiment with follicle cell plasma membranes marked suggested that BM fibrils form at cell-cell interfaces (Figure 2B; Movie S2). At the beginning of the movie shown in Figure 2B, a linear nascent fibril sits at the interface between two follicle cells, where it is oriented perpendicular to the mature fibrils within the BM. As the nascent fibril moves with the migrating tissue, it maintains association with the cell-cell interface. After 8 minutes, one side of the nascent fibril stops moving, likely due to adhesion to the BM, while the other side stays associated with the cell-cell interface. Continued cell migration draws the nascent fibril away from the interface, first inducing a 90° bend and then rotating it into the proper orientation. The fibril ultimately loses contact with the moving interface and takes up its final position in the BM (Figure 2B; Movie S2). We made 19 such movies, and observed 88 fibril formation events. In all cases, new fibrils were incorporated into the BM as a cohesive structure from a cell-cell interface.
We observed that only a fraction of newly synthesized Col IV is incorporated into the BM as easily visualized fibrils at stage 7, while the rest continues to undergo constitutive deposition into the planar BM. Live total internal reflection fluorescence (TIRF) microscopy of a photobleached BM revealed Col IV-GFP incorporation into regions containing new fibrils and into regions where no obvious fibrils appear (Figure S2B; Movie S3). Thus, new proteins appear to be apportioned between these two BM populations. BM protein secretion appears to largely cease between stages 8 and 9, concurrent with the end of rotational migration, as we observe a sharp drop in intracellular Col IV and Laminin levels at this time (Figures S2C–S2F).
These live imaging experiments led us to generate a model for how BM fibrils form (Figure 2C). Prior to stage 5, we envision that newly synthesized BM proteins are trafficked to the basal cell surface. Immediate contact with the BM upon secretion would promote even incorporation across the BM and expansion of the isotropic planar matrix. In contrast, during fibril formation, a portion of the BM traffic may be redirected to a basal region of the lateral cell surface. This would cause BM proteins to accumulate in the pericellular spaces between follicle cells and promote their aggregation into nascent fibrils. The migration of the epithelium would then provide the directional information to orient fibrils perpendicular to the A-P axis as they incorporate into the BM.
BM fibrils form in the pericellular space between follicle cells
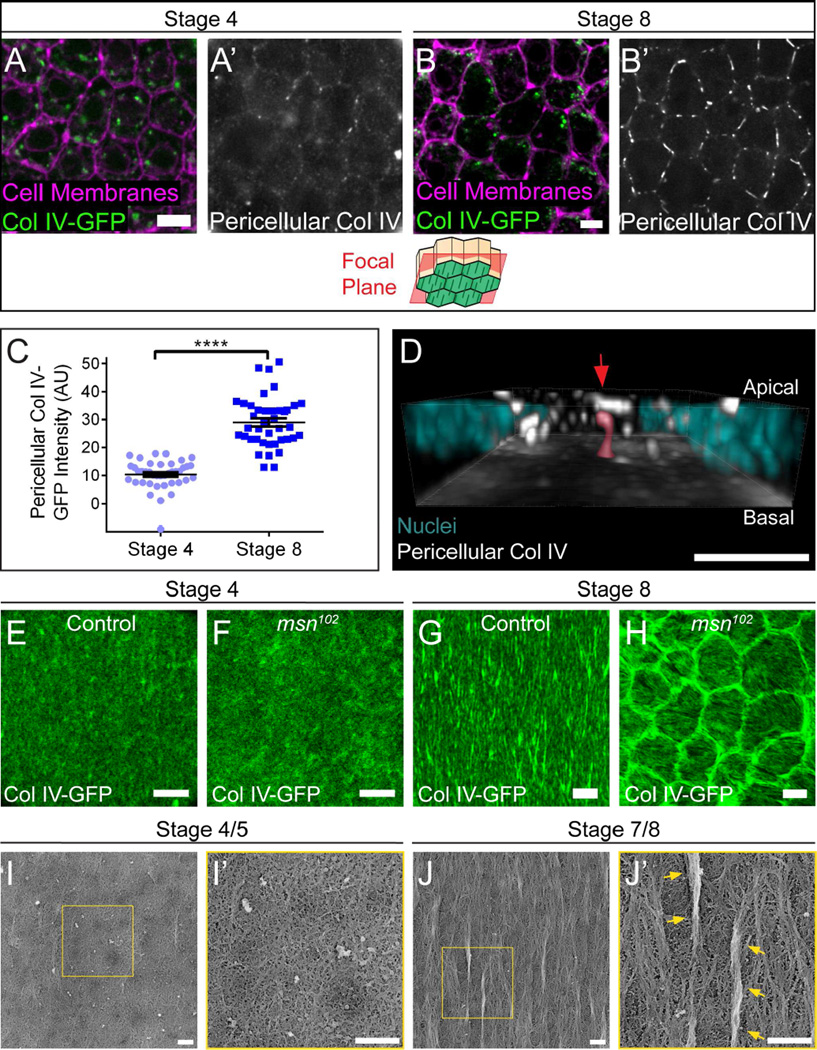
To test our model, we first asked whether Col IV accumulates in the pericellular space between follicle cells during fibril formation. We stained non-permeabilized tissue expressing Col IV-GFP with an anti-GFP antibody to selectively label extracellular Col IV. Because pericellular Col IV-GFP is dim relative to intracellular Col IV-GFP, it is difficult to see without enhancing the signal with a non-permeabilized stain. Images were obtained at a focal plane ~1–1.5 µm apical to the BM. Very little Col IV is present in the pericellular space at stage 4, before fibril formation begins; however, we detected significant pericellular Col IV at stage 8 when fibrils are forming (Figures 3A–3C).
Figure 3. BM fibrils form in the pericellular space between follicle cells.
(A and B) Representative images of Col IV in the pericellular space. Staining non-permeabilized tissue expressing Col IV-GFP with a GFP antibody reveals pericellular Col IV (white) and does not label intracellular Col IV-GFP (green). The illustration shows the rough distance from the BM (green) at which the images were taken. Pericellular Col IV is low at stage 4 (A) but high at stage 8 (B). Scale bars = 5 µm.
(C) Quantification of pericellular Col IV. Data represent mean ± s.e.m. t-test: **** = P<0.0001.
(D) 3D reconstruction of the basal half of a stage 8 follicular epithelium, showing pericellular Col IV aggregates. The image is oriented with the BM down; most BM fluorescence has been removed to allow visualization of nascent fibrils. The highlighted nascent fibril (red highlight and arrow) contacts the BM and is likely in the process of BM incorporation. For animation of this 3D reconstruction, see Movie S4. Scale bar = 5 µm.
(E–H) At stage 4, the BMs of non-migrating msn102 epithelia (F) show little difference from controls (E). However, at stage 8, the BMs of non-migrating msn102 epithelia (H) show ring-like aggregates around cells, which likely represent nascent fibrils that could not exit the pericellular space. Scale bars = 5 µm.
(I–J) Platinum replica electron micrographs of the inner surface of de-cellularized follicular BMs. (I) Stage 4/5 BMs are primarily composed of an isotropic planar matrix. (J) Stage 7/8 BMs contain large linear aggregates that lie atop the planar matrix (arrows), as well as small polarized regions that appear to be integrated within the planar matrix. (I’ and J’) Blow-ups of the boxed regions in (I and J). Scale bars = 500 nm.
Although linear structures can often be seen in individual focal planes at stage 8 (Figure 3B), 3D reconstruction of pericellular Col IV-GFP across the tissue reveals that nascent fibrils have diverse shapes and orientations (Figure 3D; Movie S4). Many nascent fibrils, including the one highlighted in Figure 3D, make contact with the BM at one end, suggesting that they are in the early stages of BM deposition.
To ensure that the pericellular Col IV is newly synthesized protein, we examined stage 8 epithelia in which some cells express Col IV-GFP and some express unmarked Col IV. Due to follicle cell migration, the BM typically becomes uniformly labelled with GFP under these conditions. We envisioned two possible outcomes from this experiment. (1) Col IV-GFP could be present in the pericellular space throughout the epithelium, which would suggest that at least some of this population is derived from the BM, via diffusion and/or endocytic recycling. (2) Col IV-GFP could be restricted to the pericellular space around Col IV-GFP-expressing cells, a scenario that can only be achieved by direct secretion of new protein into this location. We observed the latter, indicating that pericellular Col IV-GFP is new material that has not yet reached the BM (Figure S3A).
Further support for our model comes from changes in BM structure that occur when follicle cell migration is blocked. We previously showed that loss of the Ste20-family kinase Misshapen (Msn) blocks follicle cell migration (Lewellyn et al., 2013). Under these conditions, Col IV-GFP forms ring-like aggregates around the edges of non-motile cells. These rings show the same stage-specificity as pericellular Col IV accumulation – they are absent at stage 4, but strongly apparent at stage 8 (Figures 3E–3H). Because loss of Msn increases integrin levels at the basal cell surface, we initially proposed that this phenotype might be due to heightened adhesion to the BM (Lewellyn et al., 2013). However, we have now found that two conditions that block collective follicle cell migration by other means – loss of the Fat2 cadherin (Viktorinová and Dahmann, 2013) and RNAi knockdown of the SCAR complex component Abelson interacting protein (Abi) (Cetera et al., 2014) – cause the same change in BM structure (Figures S3B–S3D). We also observed that the rings extend up to 2 µm into the pericellular space, consistent with the idea that BM proteins exit the cell at the lateral surface during fibril formation (Figure S3D). Thus, the rings likely represent the subset of BM proteins that would have formed fibrils, but were unable to exit the pericellular space in the absence of follicle cell migration. Collectively, these data support a model in which nascent fibrils are assembled in the pericellular space between follicle cells and are then drawn into the BM by directed epithelial motility.
Notably, in our non-migratory conditions small, polarized ridges can be seen in the planar BM overlaying individual follicle cells (Figures 3H, S3B and S3C). This suggests that the epithelium has an alternate method to induce BM polarity, perhaps by reorganizing the planar matrix. To more closely examine BM structure, we generated platinum replicas of decellularized BMs for electron microscopic (EM) analysis. As expected, stage 4/5 BMs exist as a largely isotropic planar meshwork (Figure 3I), while large fibrils - generally 1–3 µm in length - are distributed throughout the BM at stage 7/8 (Figures 3J and S3E). These fibrils appear as distinct aggregates of aligned proteins that sit atop the planar matrix, and can only be seen on the inner, cell-facing surface of the BM (Figure S3F). In addition to fibrils, there are also smaller regions of local alignment that appear to be integrated within, rather than on top of, the planar matrix (Figures 3J and S3E). While we cannot rule out that these regions simply represent very small fibrils, these data open the possibility that there may be two levels of polarization in the follicular BM - large fibrils that are deposited on top of the pre-existing planar matrix and smaller regions of local alignment within the planar matrix itself.
Rab10 promotes BM fibril formation
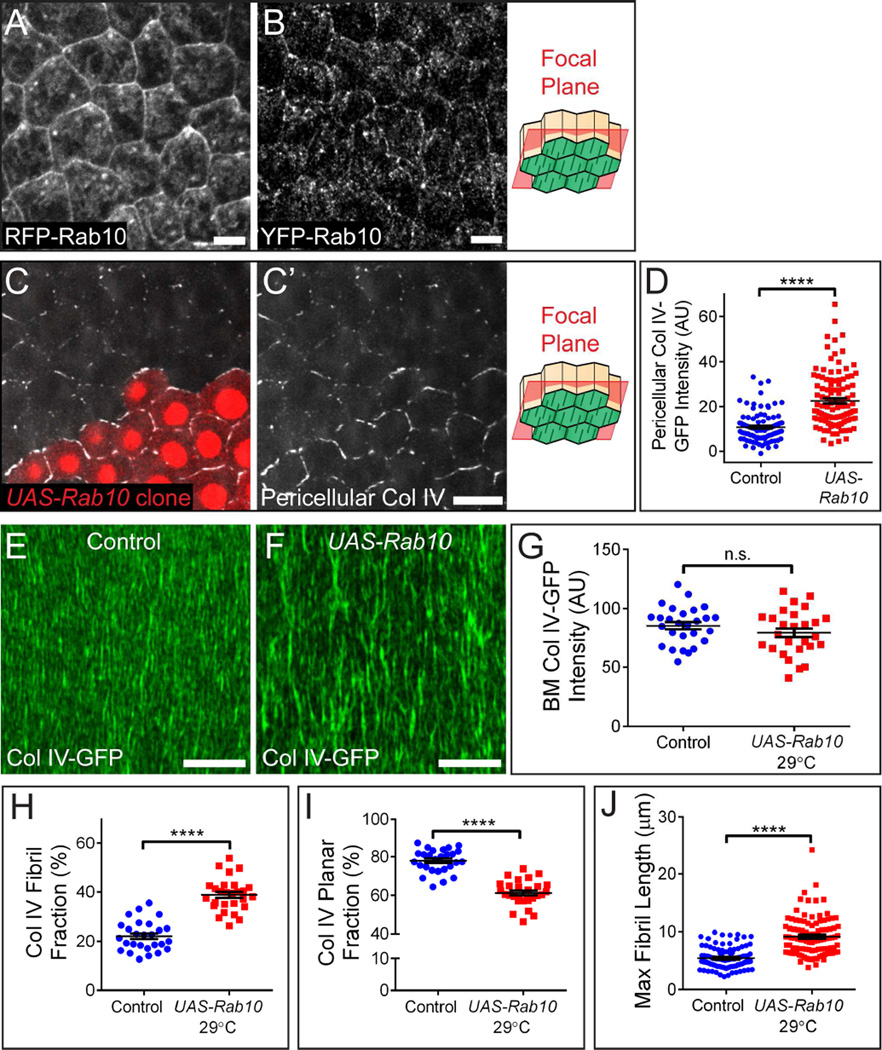
Given that BM fibrils form during a specific developmental time period from newly secreted proteins, we reasoned that there must be a regulated change in the BM secretion machinery that promotes their formation. We previously identified the small GTPase Rab10 as a central regulator of polarized BM protein transport and showed that this protein labels membrane-bound compartments in the basal region of the cell (Lerner et al., 2013). This Rab10 population could represent endosomal sorting compartments and/or exocytic carriers, but the location of these structures is not necessarily indicative of where BM proteins exit the cell. Further examination showed that Rab10 also accumulates on the follicle cells’ lateral surfaces. We observed this pattern with both a UAS-RFP-Rab10 transgene (Figure 4A) and a construct in which YFP was inserted into the endogenous Rab10 locus (Figure 4B). This localization could reflect fusion of Rab10 vesicles with lateral cell membranes, as Rab10 can function with the exocyst to control late stages of secretion (Babbey et al., 2010; Sano et al., 2015; Taylor et al., 2015; Zou et al., 2015). Endogenous Rab10 levels also increase significantly between stages 4 and 6 (Figures S4A–S4B). Thus, Rab10 might direct BM protein traffic to the lateral plasma membranes, and thereby promote BM fibril formation.
Figure 4. Rab10 promotes BM fibril formation.
(A and B) RFP-Rab10 produced from a UAS transgene (A) and endogenous YFP-Rab10 (B) both localize to lateral membranes. Antibody staining was used to enhance YFP-Rab10 signal. Scale bars = 5 µm.
(C) Representative image showing that clonal UAS-Rab10 expression (red cells) increases pericellular Col IV relative to wild-type cells. Scale bars = 10 µm.
(A–C) The illustrations show the rough distance from the BM (green) at which the images were taken.
(D) Quantification of the condition shown in (C).
(E and F) Representative images showing that UAS-Rab10 expression in all follicle cells at 29°C enhances the incorporation of Col IV into fibrils. Scale bars = 10 µm.
(G) UAS-Rab10 does not alter Col IV-GFP levels in the BM.
(H) UAS-Rab10 increases the fraction of BM Col IV-GFP contained within fibrils.
(I) UAS-Rab10 decreases the fraction of BM Col IV-GFP in the planar matrix.
(J) UAS-Rab10 increases maximum BM fibril length.
(D and G–J) Data represent mean ± s.e.m. t-test: n.s. = P>0.05, **** = P<0.0001.
Experiments performed at stage 8. See also Figure S4.
Consistent with this idea, Rab10 over-expression increases both the amount of Col IV in the pericellular space (Figures 4C–4D) and the fibrillar nature of the BM (Figures 4E–4F) at stage 8. We considered two possible explanations for this effect. Increased Rab10 activity might cause more Col IV to be secreted. Alternatively, it might cause a shift in protein distribution, such that more Col IV is directed into fibrils at the expense of the planar matrix. To distinguish between these ideas, we measured Col IV-GFP levels in the BM, and then determined the percentage of that Col IV that is in fibrils (fibril fraction) vs. the planar matrix (planar fraction) (Figure S4C). Given the resolution limits of light microscopy and our image processing algorithm, this method likely underestimates the fibril fraction of the matrix, as very small fibrils will elude detection and be assigned to the planar fraction. This method does, however, provide a sensitive metric to compare relative changes to BM structure between conditions. For example, it allowed us to confirm that the fibrillar structure of the BM is not altered by the GFP tag on Col IV (Figures S4D–S4F).
Using this strategy, we found that Rab10 over-expression does not change the amount of Col IV in the BM at stage 8 (Figure 4G); instead, the fibril fraction increases at the expense of the planar fraction (Figures 4H–4I). This condition also produces longer BM fibrils (Figure 4J). The fibril fraction is further increased by co-over-expressing Rab10’s GDP/GTP exchange factor (GEF) Crag (Denef et al., 2008; Lerner et al., 2013); thus, the active, GTP-bound form of Rab10 is responsible for this effect (Figures S4G–S4J). We were unable to determine whether the opposite occurs under Rab10 loss of function, as BM proteins are mis-trafficked to the apical surface under these conditions (Lerner et al., 2013). However, our data strongly suggest that Rab10 helps to guide newly synthesized Col IV into a laterally-directed, fibril-forming pathway.
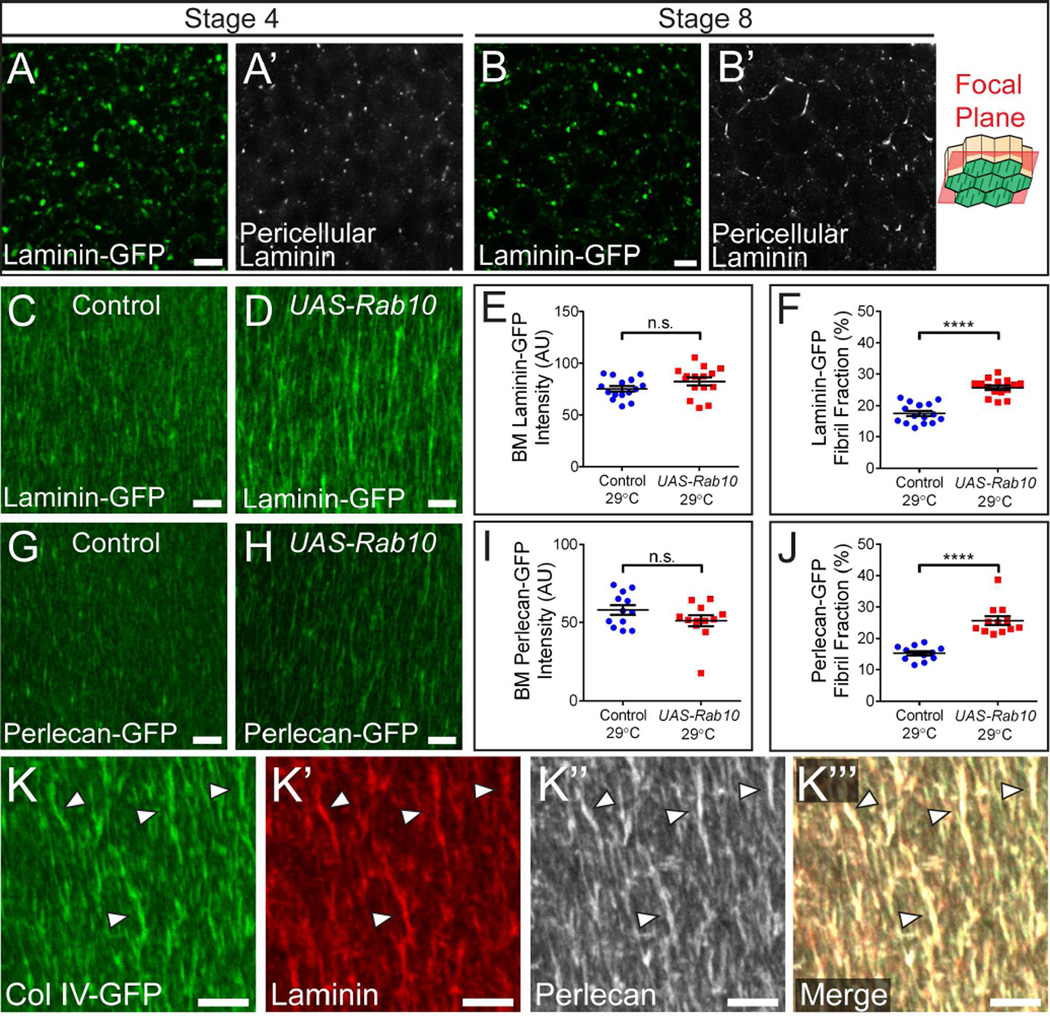
Thus far, we have primarily used Col IV-GFP to study BM fibril formation. However, Laminin and Perlecan also form fibrils (Gutzeit et al., 1991; Schneider et al., 2006). We therefore examined whether these proteins do so via the same mechanism as Col IV. We first examined GFP-tagged versions of the Perlecan homolog Terribly reduced optic lobes (Perlecan-GFP) and the Laminin β subunit LanB1 (Laminin-GFP). Although it is difficult to observe Perlecan-GFP before it reaches the BM, Laminin-GFP accumulates in the pericellular space weakly at stage 4 and strongly at stage 8 (Figures 5A–5B). Moreover, Rab10 over-expression increases the BM fibril fraction of Laminin-GFP and Perlecan-GFP at stage 8 without altering the amount of these proteins in the BM, similarly to Col IV (Figures 5C–5J). Finally, we simultaneously visualized Col IV-GFP, Laminin, and Perlecan in the BM and found that all three co-localize in individual fibrils (Figure 5K). These data show that fibrils are compositionally similar to the planar BM, and that a common mechanism governs the secretion and fibrillogenesis of Col IV, Laminin, and Perlecan.
Figure 5. Rab10 also targets Laminin and Perlecan into BM fibrils.
(A and B) Laminin is low in the pericellular space at stage 4 (A) but high at stage 8 (B). Scale bars = 5 µm. The illustration shows the rough distance from the BM (green) at which the images were taken.
(C–D) Representative images showing that UAS-Rab10 expression in all follicle cells at 29°C enhances the incorporation of Laminin into fibrils. Scale bars = 5 µm.
(E) UAS-Rab10 does not alter Laminin-GFP levels in the BM.
(F) UAS-Rab10 increases the fibril fraction of Laminin-GFP.
(G–H) Representative images showing that UAS-Rab10 expression in all follicle cells at 29°C enhances the incorporation of Perlecan into fibrils. Scale bars = 5 µm.
(I) UAS-Rab10 does not alter Perlecan-GFP levels in the BM.
(J) UAS-Rab10 increases the fibril fraction of Perlecan-GFP.
(E,F, I and J) Data represent mean ± s.e.m. t-test: n.s. = P>0.05, **** = P<0.0001.
(K) Col IV, Laminin, and Perlecan co-localize in individual fibrils (arrowhead). Scale bars = 5 µm.
Experiments performed at stage 8 unless otherwise noted in figure.
Modulating Rab10 activity provides a tunable mechanism to control BM structure
Our discovery that Rab10 over-expression increases pericellular BM protein accumulation and fibril formation without affecting bulk BM protein composition suggests that the Rab10 pathway may exist in competitive balance with another pathway that directs secretion to the planar matrix. If true, the fibril fraction should scale with Rab10 activity. We utilized the fact that UAS transgene expression increases with temperature to examine the fibril fraction across a range of Rab10 expression levels. Although temperature-independent in controls, the stage 8 fibril fraction rises with temperature in the UAS-Rab10 condition (Figure S5A). The amount of Rab10 in the follicle cells, therefore, provides a tunable mechanism to control the relative distribution of BM proteins into the fibrillar vs. planar BM populations.
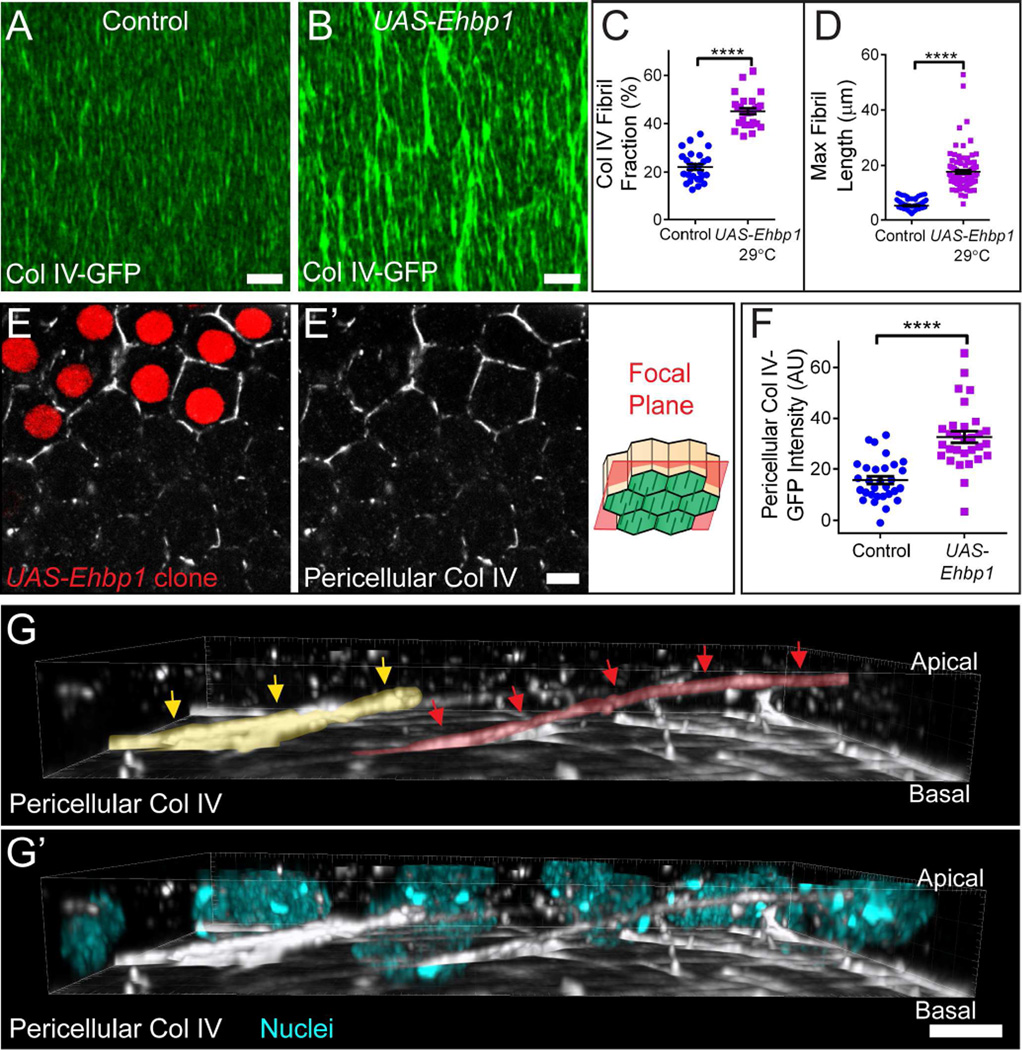
As an alternative approach to enhancing Rab10-based secretion, we over-expressed Ehbp1, which is a Rab10 effector in C. elegans (Shi et al., 2010). This role appears to be conserved, as Ehbp1 depletion phenocopies loss of Rab10 (Figures S5B–S5D), and Ehbp1 over-expression tunably increases the stage 8 BM fibril fraction and maximum BM fibril length (Figures 6A–6D and S5A) without altering BM Col IV levels (Figure S5E). In fact, over-expressing Ehbp1 increases fibril formation to an even greater extent than Rab10 over-expression (Figure S5A). Ehbp1 over-expression also increases pericellular Col IV (Figures 6E–6F) at stage 8. 3D reconstruction of this pericellular pool revealed extremely long aggregates that span multiple cell lengths, consistent with the extremely long BM fibrils seen in this condition (Figure 6G; Movie S5). These observations strengthen the link between pericellular Col IV and BM fibril formation. Further, although it is unclear why manipulating Ehbp1 has a stronger effect on BM protein trafficking than Rab10, it provides us with a practical means to increase the fibril fraction beyond what can be accomplished by over-expressing Rab10.
Figure 6. Ehbp1 promotes BM fibril formation.
(A and B) Representative images showing that UAS-Ehbp1 expression at 29°C in all follicle cells enhances BM fibril formation. Scale bars = 5 µm.
(C and D) UAS-Ehbp1 expression increases BM fibril fraction (C) and maximum BM fibril length (D). Graphs use same control data as Figures 4H and J.
(E) Representative image showing that clonal UAS-Ehbp1 expression (red cells) increases pericellular Col IV relative to neighboring wild-type cells. The illustration shows the rough distance from the BM (green) at which the images were taken. Scale bar = 5 µm.
(F) Quantification of the condition shown in (E).
(C, D, and F) Data represent mean ± s.e.m. t-test: **** = P<0.0001.
(G–G’) 3D reconstruction of the basal 3/4 of the follicular epithelium, showing pericellular Col IV in the UAS-Ehbp1 condition. Extremely long pericellular aggregates can be seen, consistent with the long BM fibrils seen in (B and D). Two nascent fibrils are indicated by red and yellow arrows and highlights. Image is oriented with BM down; most BM fluorescence has been removed to allow visualization of nascent fibrils. For animation of this 3D reconstruction, see Movie S5. Scale bars = 5 µm.
Experiments performed at stage 8. See also Figure S5 and Movie S5.
BM fibrils play an instructive role in egg chamber elongation
We next asked how altering BM architecture affects egg chamber elongation. Having identified conditions that allow us to control the extent of BM fibril formation over a large range, we examined the effects of both a modest (UAS-Rab10 23°C) and a strong (UAS-Ehbp1 29°C) increase in fibril fraction. Modestly increasing the fibril fraction leads to an increase in the egg’s aspect ratio (length/width) (Figures 7A–7C). This enhanced elongation is first seen at stage 7 (Figure 7A), indicating that increasing the fibrillar nature of the BM is sufficient to augment the egg chamber’s morphogenetic program. Conversely, strongly increasing the fibril fraction decreases the egg’s aspect ratio (Figures 7D–7F). In this case, the egg chamber elongates normally, but fails to maintain its shape after stage 10 (Figure 7F). These changes are not due to an indirect effect on egg chamber rotation, as follicle cell migration rates are unaffected by changes in the BM’s fibril fraction (Figures S6A–S6E; Movie S6). These data show that differing BM architectures can influence both the establishment and maintenance of the egg chamber’s elongated shape.
Figure 7. BM fibrils play an instructive role in egg chamber elongation.
(A) UAS-Rab10 expression at 23°C increases the egg chamber’s aspect ratio. This effect is first seen at stage 7, suggesting that this BM structure augments elongation morphogenesis. n = 25–32 egg chambers/data point.
(B and C) Representative images showing that 23°C UAS-Rab10 expression results in eggs that are longer and narrower than controls.
(D and E) Representative images showing that 29°C UAS-Ehbp1 expression results in eggs that are shorter and wider than controls.
(B–E) For each pair of eggs, the length and width of the control egg (orange lines) is mapped onto the experimental egg for reference. Scale bars = 50 µm.
(F) UAS-Ehbp1 expression at 29°C reduces the egg’s aspect ratio. This effect is not seen until stage 12, suggesting that this BM structure is defective in maintaining the elongated state. n = 23–27 egg chambers/data point.
(A and F) Data represent mean ± s.e.m. t-test: * = P<0.05, ** = P<0.01, *** = P<0.001, **** = P<0.0001.
(G) Graph showing how egg aspect ratio changes as a function of Col IV fibril fraction. Fibril fractions of 30–33% increase egg aspect ratio compared to controls, whereas fibril fractions of 38% and above reduce it. Grey bars show control ranges for both measurements. X axis: n = 14–27 stage 8 egg chambers/condition. Y axis: n = 40–60 stage 14 egg chambers/condition. Both axes: data represent mean ± s.e.m. t-test values are in Figure S6F. Fibril fraction values represent same data shown in Figures 4H, 6C and S5A.
(H) Proposed model for BM fibril formation. The images are stills from the animation shown in Movie S7. During fibril formation, Rab10 directs a portion of newly synthesized BM proteins to a basal region of the lateral plasma membrane for secretion. It may do so in competition with an unidentified pathway that directs BM protein secretion to the basal surface for incorporation into the planar matrix. Secretion to the lateral surface causes BM proteins to aggregate in the pericellular space between follicle cells. Directed follicle cell migration then inserts the nascent fibrils into the BM in the correct orientation. Cell migration direction is to the right.
The interplay between BM architecture and elongation can best be seen by plotting the aspect ratios of eggs across the full range of fibril fractions that we can generate (Figures 7G and S6F). Under control conditions, the fibril fraction ranges from 22–25%. Increasing the fibril fraction to 30–33%, via moderate Rab10 over-expression (23°C and 25°C), maximally increases the egg’s aspect ratio compared to controls, whereas increases above 37% via Epbh1 or 29°C Rab10 over-expression ultimately decrease the egg’s aspect ratio compared to controls. These observations demonstrate how fine regulation of BM structure, in this case by tunable regulation of Rab10 pathway activity, can precisely influence morphogenesis. They also reveal the importance of tightly controlling BM structure during development.
Discussion
BMs are often viewed as largely static structures that provide mechanical support for tissues. However, the changes in tissue size and shape that occur during development necessitate that BMs be similarly malleable. Regulated changes in BM structure may also actively promote tissue deformation. Thus, understanding morphogenesis will require that we know how tissues dynamically remodel their BMs, and how differing BM architectures affect tissue shape. Focusing on the fibrillar BM that surrounds the Drosophila egg chamber, we used live imaging to watch this matrix being remodeled in real time. The resulting information allowed us to determine how BM fibrils form, and to establish a direct role for these structures in egg chamber elongation.
We found that fibril formation begins when newly synthesized BM proteins aggregate in the pericellular space between follicle cells. These nascent fibrils then incorporate into the BM by first making a single point of contact with the planar matrix, and then being pulled out of the pericellular space as the follicle cells migrate away from this site. In this way, the motion of the migrating epithelium also provides the directional information needed to align BM fibrils perpendicular to the egg chamber’s A-P axis and polarize the matrix (Figure 7H; Movie S7).
3D reconstructions of nascent fibrils in the pericellular space showed that they often appear globular, despite the fact that most mature fibrils are linear. In rare cases, we have seen globules deposited into the BM; however, most eventually resolve into linear structures. The globules could be aggregation intermediates that will fuse into linear structures in the pericellular space. Alternatively, tension placed on a globule during the deposition process by the migrating cells may cause its elongation, similar to pulling two sides of a cotton ball in opposite directions. In either case, the model that emerges is that BM fibril formation occurs through a series of seemingly disordered events that, through bulk action, produce a robustly polarized matrix.
Although nascent fibril deposition appears to be the main way that the follicular BM becomes polarized, other mechanisms may contribute to this process. Although impeding collective follicle cell migration blocks the fibril formation, the resulting BMs still exhibit small, aligned ridges that tend to be oriented in the same direction as the actin bundles at the basal surface of each adjacent follicle cell. A seemingly similar phenomenon was also recently reported by another group (Aurich and Dahmann, 2016). Because the actin bundles are contractile elements that interact with the BM through integrin-based adhesions (Bateman et al., 2001; Cetera et al., 2014; Delon and Brown, 2009), these ridges could form via physical deformation of the planar matrix. Whether such a mechanism contributes to BM polarity under wild-type conditions is currently unclear, although it could produce the small-scale alignment we observed by EM.
We found that Rab10 and its putative effector Ehbp1 promote BM fibril formation. We previously showed that Rab10 prevents improper sorting of BM proteins into an apically-directed secretory pathway (Lerner et al., 2013). Our data now suggest that Rab10 may perform this function, in part, by targeting BM proteins to a basal region of the lateral plasma membrane for secretion. In support of this notion, Rab10 accumulates on lateral membranes and its over-expression increases the amount of BM proteins in the pericellular space. Work from other systems has shown that Rab10 localizes to exocytic vesicles bound for a basal region of the lateral plasma membrane (Cao et al., 2008), and that this protein functions with the exocyst to control the docking of exocytic vesicles at the cell surface (Babbey et al., 2010; Sano et al., 2015; Taylor et al., 2015; Zou et al., 2015). Altogether, these data suggest a model wherein Rab10 promotes BM fibril formation by determining the location where new BM proteins exit the cell.
Our data further suggest that Rab10 functions in competitive balance with a second BM secretion pathway. New BM material is deposited into the planar matrix at the same time that fibrils are generated. That Rab10 (or Ehbp1) over-expression can increase pericellular BM protein accumulation and fibril formation without altering bulk protein levels in the BM indicates that a portion of BM proteins – likely those bound for the planar matrix – are normally secreted via a different pathway. We propose that there may be two distinct pathways for BM deposition – one directed to the basal surface for planar matrix assembly and one directed to the lateral surface for fibril formation (Figure 7H; Movie S7). This model offers a simple mechanism to control the allocation of BM proteins into distinct populations by tuning activity levels of the two pathways.
What sets the developmental timing of BM fibril formation remains an open question. Although this process coincides with increased Col IV incorporation into the BM (Haigo and Bilder, 2011), this does not appear to be the initiating factor, as we have previously shown that abrogating this increase does not inhibit BM fibril formation (Isabella and Horne-Badovinac, 2015b). Instead, because this phenomenon coincides with an increase in Rab10 expression and is sensitive to Rab10 levels, we propose that a regulated increase in Rab10 pathway activity may specify both the timing and extent of fibril formation; however, future work is required to test this hypothesis.
It has been speculated that polarization of the follicular BM allows it to function as a molecular corset for egg chamber elongation (Gutzeit et al., 1991). In this model, the aligned fibrils provide an anisotropic constraining force that channels egg chamber growth along the A-P axis. Our finding that a modest increase in fibril formation enhances elongation supports this idea. It is becoming increasingly clear, however, that fibril formation is only one of many structural changes that must occur in the BM for it to exert this corset function. For example, we recently showed that a temporally regulated increase in Col IV levels and concomitant decrease in Perlecan levels are also necessary for proper egg chamber elongation (Isabella and Horne-Badovinac, 2015b). Moreover, we have found that BM architecture influences egg chamber shape beyond the active elongation phase. When the fibril fraction in the BM exceeds 37%, egg chambers elongate normally, but this matrix can no longer maintain the elongated state. We envision that this failure is due not to increased fibril formation per se, but to a global weakening of the BM caused by the accompanying depletion of the planar matrix. This idea is consistent with previous work showing that collagenase-mediated digestion of the BM causes the egg chamber to round up during this maintenance phase (Haigo and Bilder, 2011). Altogether, these observations suggest that the ways in which the follicular BM influences the shape of the egg are complex, and that there is still much to be learned about this process.
To our knowledge, a polarized BM structure akin to that of the follicular BM has not been seen outside of insects, although there is currently very little data describing in vivo BM architecture with sufficient detail to reveal such structure. However, the fibrillar BM is one instance of what appears to be a common strategy of using a surrounding ECM to provide a stabilizing/constraining force during morphogenesis. For instance, the developing Xenopus notochord is ensheathed by an oriented network of Collagen fibrils that channel its elongation along the A-P axis (Adams et al., 1990). Additionally, thickening of the BM around epithelial ducts in mouse mammary and salivary glands is believed to stabilize these structures (Fata et al., 2004; Harunaga et al., 2014); whether these BMs also exhibit circumferential polarity has not been examined. Further understanding the roles of BMs in morphogenesis will require detailed examination of BM structure in other tissues.
In conclusion, this work highlights how coordinated tissue behaviors – in this case regulated protein secretion and tissue movement – can synergize to remodel BM architecture during development, and how matrix remodeling can play an active role in tissue morphogenesis.
Experimental Procedures
Drosophila genetics
Experimental genotypes are in supplemental experimental procedures. Experimental crosses were raised at 25°C and females aged on yeast for 3 days at 29°C with exceptions listed in supplemental experimental procedures. UAS transgenes were driven with traffic jam-Gal4 or traffic jam-Gal4, Mef2.mb247-Gal80 for follicle cell expression or hs-Flp;Act5c>>Gal4 for FLP-out. FLP-out was induced by 37°C heat shock for 1 hour, twice daily for 3 days on yeast with intermittent periods at 25°C. Mitotic clones were generated using FRT80B or FRT40A and e22c-Gal4 or T155-Gal4, respectively, to drive UAS-Flp expression. Most lines were obtained from the Bloomington Drosophila stock center except: nls-mRFP, vkg, FRT40 is from (Haigo and Bilder, 2011). vkg-GFP and Trol-GFP are from Flytrap (Buszczak et al., 2007). abi RNAi and traffic jam-Gal4 are from the Drosophila Genetic Resource Center (Kyoto Institute of Technology, Kyoto, Japan). fat2N103-2 is from (Horne-Badovinac et al., 2012). UAS-flag-Rab10 and UAS-RFP-Rab10 are from this study. Mef2.mb247-Gal80 was a gift from Martin Heisenberg. Endogenous YFP-Rab10 is from (Dunst et al., 2015). UAS-flag-Ehbp1 is from (Giagtzoglou et al., 2012). LanB1-GFP (Sarov et al., 2016) and UAS-Ehbp1 RNAi are from Vienna Drosophila Resource center (Vienna, Austria). UAS-Crag is from (Denef et al., 2008).
Staining and microscopy
Ovaries were dissected in S2 medium and fixed for 15 minutes in PBS + 0.1% Triton (PBT) + 4% EM-grade formaldehyde (Polysciences), then separated from the muscle sheath by gentle pipetting. Antibody stains were performed in PBT and detected with Alexa Fluor-conjugated secondary antibodies (1:200, Invitrogen). Actin was labeled with TRITC-Phalloidin (1:200, Sigma) or Alexa-647 Phalloidin (1:50, Invitrogen), and nuclei labeled with DAPI (1:1000, Sigma). For non-permeabilized stains, ovarioles were dissected from the muscle sheath in S2 medium and fixed for 6 minutes in PBS + 4% EM-grade formaldehyde. GFP antibody stains were performed as above using PBS instead of PBT. Antibodies used: guinea pig α-Laminin (1:400) (Harpaz and Volk, 2012), rabbit α-Trol (1:1000) (Friedrich et al., 2000), rabbit α-GFP (1:200, Molecular Probes A21311 and A31852). Fluorescent images were obtained using Zeiss LSM 510 or LSM 880 confocal microscopes. Images for stage 14 aspect ratios were obtained using a Leica DM550B microscope with a Leica DFC425C camera. Image processing and custom image analysis were performed using ImageJ and Python. For 3D reconstructions, confocal Z-stacks were deconvolved (Huygens) and 3D images made in Imaris (Bitplane). Graphing and statistical analysis were performed in Prism (GraphPad). For all graphs, error is presented as mean ± s.e.m. and statistical differences between conditions were determined with 2-tailed, unpaired t-tests.
Live imaging
Live imaging was performed as described (Prasad et al., 2007), with the following modifications. Dissected egg chambers were placed on a pad of 0.4% NuSieve GTG low melt agarose (Lonza) in live imaging medium, and follicle cell membranes marked with CellMask (1:1000, Molecular Probes) or UAS-mCD8-RFP. The coverslip was cushioned with vacuum grease at each corner. For confocal imaging of BM fibril formation, a region of the Col IV-GFP in the BM was photobleached with a 488nm laser at 100% power for 10 iterations, 10 minutes prior to imaging. TIRF movies were taken on an Olympus IX-50 microscope equipped with an iXon EMCCD camera (Andor) and a 100× objective fitted with through-the-objective TIRF illumination. TIRF photobleaching was achieved by exposing the BM to the 488nm TIRF laser at 50% power for ~2 minutes, 10–15 minutes before imaging. Movies were processed using ImageJ.
Platinum replica electron microscopy
To decellularize the follicular BM, egg chambers were dissected as for live imaging in HL3.1 (70mM NaCl, 5mM KCl, 1.5mM CaCl, 4mM MgCl2, 10mM NaHCO3, 5mM trehalose, 115mM sucrose, 5mM HEPES), adhered to poly-lysine (Sigma) coated slides, incubated 10–20 minutes in PBT, sonicated, and washed in PBS. BMs were then prepared for EM using the protocol in (Svitkina, 2009). Briefly, BMs were fixed with 2% Glutaraldehyde (Electron Microscopy Sciences), tannic acid, and uranyl acetate; critical point dried; coated with platinum and carbon; and transferred onto EM grids for observation. Samples were imaged using a FEI Tecnai Spirit G2 transmission electron microscope (FEI Company, Hillsboro, OR) operated at 80 kV. Images were captured by Eagle 4k HR 200kV CCD camera and presented in inverted contrast.
Production of UAS-RFP-Rab10 and UAS-Flag-Rab10 transgenic flies
The Rab10 coding sequence was PCR-amplified from genomic DNA isolated from; UAS-YFP-Rab10; flies (Zhang et al., 2006). The PCR product was gel extracted, digested with BamHI and Xhol and cloned into the Gateway pENTR3C Dual Selection Entry Vector (Invitrogen). It was then recombined (LR clonase reaction, Invitrogen) into pTRW (uasT promoter, N-terminal mRFP tag) or pTFW (uasT promoter, N-terminal 3xFLAG tag) (Carnegie Drosophila Gateway Vector Collection). Transgenic flies were generated via P-element-mediated transformation (Best Gene).
Measurement of Pericellular Col IV-GFP intensity
Confocal sections were acquired 1–1.5 µm apical of the BM. For wild-type measurements, 5 cells/egg chamber (n = 8 egg chambers/stage) were randomly selected for analysis. When measuring the effect of transgene expression on pericellular signal, experiments were performed in mosaic epithelia via FLP-out, allowing us to collect data for the control and experimental condition from the same tissues. In this case, 1–10 cells of each genotype/egg chamber (n = 20 egg chambers (Ehbp1), 27 egg chambers (Rab10)) were randomly selected for analysis. The edges of each cell were manually outlined with 3 pixel-thick lines; mean intensity of all pixels contained within the line was measured. For each cell, background fluorescence values were measured over cell centers and subtracted from outline intensity. All images were acquired with the same settings.
Measurement of fibril fraction and BM fluorescence intensity
A confocal section capturing the entire thickness of the BM was acquired. All images were obtained at the same settings. Because the BM generally did not take up the entire imaging frame, the largest possible representative rectangle was cropped to remove background pixels. Mean intensity of all pixels in the cropped image was measured to determine overall GFP intensity. Pixels contained within fibrils were isolated by successive intensity and size thresholds. To determine appropriate thresholding parameters, 10 Col IV-GFP BMs from each of the control, UAS-Rab10, and UAS-Ehbp1 conditions were manually thresholded by eye to maximally include fibrils and exclude the planar matrix. A conservative threshold was used to ensure exclusion of all non-fibrillar pixels, meaning our analysis likely underestimates the fibril fraction. By analyzing our by-eye thresholds, we found that we consistently applied an intensity cutoff corresponding to 1.35× the median image intensity independently for all conditions, and used this value for subsequent analysis. We then applied an object size threshold of 20 pixels to remove image noise. Using custom Python and ImageJ scripts, we applied these thresholds to all experimental images to isolate pixels within fibrils and, inversely, in the planar matrix. Fibril fraction was measured by dividing the sum intensity of all fibrillar pixels by the sum intensity of all pixels in the image. Planar fraction was measured by dividing the sum intensity of all non-fibrillar pixels by the sum intensity of all pixels in the image. By definition, fibril fraction + planar fraction = 100%. This method was used for analysis of Col IV-GFP, Laminin-GFP, and Perlecan-GFP egg chambers.
Measurement of maximum fibril length
In each of 10 cropped images/condition from our fibril fraction analysis, length of the 10 longest fibrils was manually measured in ImageJ.
Measurement of Rab10 protein levels
In central transverse sections of stage 3–8 egg chambers with endogenously tagged YFP-Rab10, the entire epithelium of each egg chamber was manually outlined and its mean fluorescence intensity measured in ImageJ. All images were acquired with the same settings.
Measurement of egg chamber aspect ratios
In central transverse sections, egg chamber length (anterior to posterior tip) and width (widest region perpendicular to anterior-posterior axis) were measured, and ratio of length:width was calculated. Dorsal appendages were excluded from measurements.
Measurement of follicle cell migration rates
20 minute time-lapse movies were acquired from egg chambers labeled with CellMask or UAS-mCD8-RFP. The leading edge of a single follicle cell was marked at the start and end of the movie and distance traveled was measured and divided by movie length (minutes). Three distant cells were measured and their rates averaged for each egg chamber.
Supplementary Material
Highlights.
Basement membrane (BM) fibrils form in the epithelial pericellular space
Directed epithelial migration orients new fibrils as they are inserted into the BM
Rab10 promotes BM protein secretion to the pericellular space for fibril formation
Increasing fibril formation influences elongation morphogenesis in the egg chamber
Acknowledgments
We thank David Lerner for helpful discussions and insights, Alice Readick and Allison Zajac for experimental assistance, Muriel Grammont and members of her lab for experimental advice, Farida Korobova for EM assistance, and Matt Vanderzalm for the fly illustration in the graphical abstract. We thank Stefan Baumgartner, Hugo Bellen, David Bilder, Suzanne Eaton, Martin Heisenberg, Trudi Schüpbach, Brian Stramer, and Talila Volk for providing reagents, and Ed Munro, Chip Ferguson, and members of the Horne-Badovinac lab for helpful discussions and manuscript comments. This work was supported by NIH T32 HD055164 and a NSF Graduate Research Fellowship to A.J.I., and grants from the NIH (R01-GM094276) and American Cancer Society (RSG-14-176) to S.H-B. This project was also supported by the National Center for Advancing Translational Sciences of the NIH through Grant Number UL1 TR000430. EM was performed at the Northwestern University Center for Advanced Microscopy supported by NCI CCSG P30 CA060553.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
A.J.I. and S.H-B. designed experiments. A.J.I. performed experiments, analyzed data and prepared figures. A.J.I. and S.H-B. wrote the manuscript.
References
- Adams DS, Keller R, Koehl MA. The mechanics of notochord elongation, straightening and stiffening in the embryo of Xenopus laevis. Development. 1990;110:115–130. doi: 10.1242/dev.110.1.115. [DOI] [PubMed] [Google Scholar]
- Aurich F, Dahmann C. A Mutation in fat2 Uncouples Tissue Elongation from Global Tissue Rotation. Cell Rep. 2016;14:2503–2510. doi: 10.1016/j.celrep.2016.02.044. [DOI] [PubMed] [Google Scholar]
- Babbey CM, Bacallao RL, Dunn KW. Rab10 associates with primary cilia and the exocyst complex in renal epithelial cells. Am. J. Physiol. Renal Physiol. 2010;299:F495–F506. doi: 10.1152/ajprenal.00198.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman J, Reddy RS, Saito H, Van Vactor D. The receptor tyrosine phosphatase Dlar and integrins organize actin filaments in the Drosophila follicular epithelium. Curr. Biol. 2001;11:1317–1327. doi: 10.1016/s0960-9822(01)00420-1. [DOI] [PubMed] [Google Scholar]
- Buszczak M, Paterno S, Lighthouse D, Bachman J, Planck J, Owen S, Skora AD, Nystul TG, Ohlstein B, Allen A, Wilhelm JE, Murphy TD, Levis RW, Matunis E, Srivali N, Hoskins RA, Spradling AC. The Carnegie Protein Trap Library: A Versatile Tool for Drosophila Developmental Studies. Genetics. 2007;175:1505–1531. doi: 10.1534/genetics.106.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Li C, Higginbotham JN, Franklin JL, Tabb DL, Graves-Deal R, Hill S, Cheek K, Jerome WG, Lapierre La, Goldenring JR, Ham A-JL, Coffey RJ. Use of fluorescence-activated vesicle sorting for isolation of Naked2-associated, basolaterally targeted exocytic vesicles for proteomics analysis. Mol. Cell. proteomics. 2008;7:1651–1667. doi: 10.1074/mcp.M700155-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cetera M, Ramirez-San Juan GR, Oakes PW, Lewellyn L, Fairchild MJ, Tanentzapf G, Gardel ML, Horne-Badovinac S. Epithelial rotation promotes the global alignment of contractile actin bundles during Drosophila egg chamber elongation. Nat. Commun. 2014;5:5511. doi: 10.1038/ncomms6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley WP, Yamada KM. ECM-modulated cellular dynamics as a driving force for tissue morphogenesis. Curr. Opin. Genet. Dev. 2013;23:408–414. doi: 10.1016/j.gde.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delon I, Brown NH. The integrin adhesion complex changes its composition and function during morphogenesis of an epithelium. J. Cell Sci. 2009;122:4363–4374. doi: 10.1242/jcs.055996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denef N, Chen Y, Weeks SD, Barcelo G, Schüpbach T. Crag regulates epithelial architecture and polarized deposition of basement membrane proteins in Drosophila. Dev. Cell. 2008;14:354–364. doi: 10.1016/j.devcel.2007.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunst S, Kazimiers T, von Zadow F, Jambor H, Sagner A, Brankatschk B, Mahmoud A, Spannl S, Tomancak P, Eaton S, Brankatschk M. Endogenously Tagged Rab Proteins: A Resource to Study Membrane Trafficking in Drosophila. Dev. Cell. 2015;33:351–365. doi: 10.1016/j.devcel.2015.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fata JE, Werb Z, Bissell MJ. Regulation of mammary gland branching morphogenesis by the extracellular matrix and its remodeling enzymes. Breast cancer Res. 2004;6:1–11. doi: 10.1186/bcr634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich MV, Schneider M, Timpl R, Baumgartner S. Perlecan domain V of Drosophila melanogaster. Eur. J. Biochem. 2000;267:3149–3159. doi: 10.1046/j.1432-1327.2000.01337.x. [DOI] [PubMed] [Google Scholar]
- Giagtzoglou N, Yamamoto S, Zitserman D, Graves HK, Schulze KL, Wang H, Klein H, Roegiers F, Bellen HJ. dEHBP1 controls exocytosis and recycling of Delta during asymmetric divisions. J. Cell Biol. 2012;196:65–83. doi: 10.1083/jcb.201106088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutzeit H, Eberhardt W, Gratwohl E. Laminin and basement membrane-associated microfilaments in wild-type and mutant Drosophila ovarian follicles. J. Cell Sci. 1991;100:781–788. doi: 10.1242/jcs.100.4.781. [DOI] [PubMed] [Google Scholar]
- Haigo SL, Bilder D. Global tissue revolutions in a morphogenetic movement controlling elongation. Science. 2011;331:1071–1074. doi: 10.1126/science.1199424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpaz N, Volk T. A novel method for obtaining semi-thin cross sections of the Drosophila heart and their labeling with multiple antibodies. Methods. 2012;56:63–68. doi: 10.1016/j.ymeth.2011.09.017. [DOI] [PubMed] [Google Scholar]
- Harunaga JS, Doyle AD, Yamada KM. Local and global dynamics of the basement membrane during branching morphogenesis require protease activity and actomyosin contractility. Dev. Biol. 2014;394:197–205. doi: 10.1016/j.ydbio.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horne-Badovinac S, Hill J, Gerlach G, Menegas W, Bilder D. A screen for round egg mutants in Drosophila identifies tricornered, furry, and misshapen as regulators of egg chamber elongation. 2012;G3 2:371–378. doi: 10.1534/g3.111.001677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes RO, Naba A. Overview of the matrisome--an inventory of extracellular matrix constituents and functions. Cold Spring Harb. Perspect. Biol. 2012;4:a004903. doi: 10.1101/cshperspect.a004903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabella AJ, Horne-Badovinac S. Building from the Ground up: Basement Membranes in Drosophila Development. In: Miner J, editor. Current Topics in Membranes. Elsevier Ltd.; 2015a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isabella AJ, Horne-Badovinac S. Dynamic regulation of basement membrane protein levels promotes egg chamber elongation in Drosophila. Dev. Biol. 2015b;406:212–221. doi: 10.1016/j.ydbio.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner DW, McCoy D, Isabella AJ, Mahowald AP, Gerlach GF, Chaudhry Ta, Horne-Badovinac S. A Rab10-dependent mechanism for polarized basement membrane secretion during organ morphogenesis. Dev. Cell. 2013;24:159–168. doi: 10.1016/j.devcel.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewellyn L, Cetera M, Horne-Badovinac S. Misshapen decreases integrin levels to promote epithelial motility and planar polarity in Drosophila. J. Cell Biol. 2013;200:721–729. doi: 10.1083/jcb.201209129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey MA, Sherwood DR. An active role for basement membrane assembly and modification in tissue sculpting. J. Cell Sci. 2015;128:1–8. doi: 10.1242/jcs.168021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad M, Jang AC-C, Starz-Gaiano M, Melani M, Montell DJ. A protocol for culturing Drosophila melanogaster stage 9 egg chambers for live imaging. Nat. Protoc. 2007;2:2467–2473. doi: 10.1038/nprot.2007.363. [DOI] [PubMed] [Google Scholar]
- Sano H, Peck GR, Blachon S, Lienhard GE. A potential link between insulin signaling and GLUT4 translocation: Association of Rab10-GTP with the exocyst subunit Exoc6/6b. Biochem. Biophys. Res. Commun. 2015;465:6–10. doi: 10.1016/j.bbrc.2015.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarov M, Barz C, Jambor H, Hein MY, Schmied C, Suchold D, Stender B, Janosch S, KJ VV, Krishnan R, Krishnamoorthy A, Ferreira IR, Ejsmont RK, Finkl K, Hasse S, Kämpfer P, Plewka N, Vinis E, Schloissnig S, Knust E, Hartenstein V, Mann M, Ramaswami M, VijayRaghavan K, Tomancak P, Schnorrer F. A genome-wide resource for the analysis of protein localisation in Drosophila. Elife. 2016;5:1689–1699. doi: 10.7554/eLife.12068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M, Khalil Aa, Poulton J, Castillejo-Lopez C, Egger-Adam D, Wodarz A, Deng W-M, Baumgartner S. Perlecan and Dystroglycan act at the basal side of the Drosophila follicular epithelium to maintain epithelial organization. Development. 2006;133:3805–3815. doi: 10.1242/dev.02549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi A, Chen CC, Banerjee R, Glodowski D, Audhya A, Rongo C, Grant BD. EHBP-1 functions with RAB-10 during endocytic recycling in Caenorhabditis elegans. Mol. Biol. Cell. 2010;21:2930–2943. doi: 10.1091/mbc.E10-02-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina T. Imaging Cytoskeleton Components by Electron Microscopy. Methods Mol Biol. 2009;586:187–206. doi: 10.1007/978-1-60761-376-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CA, Yan J, Howell AS, Dong X, Shen K. RAB-10 Regulates Dendritic Branching by Balancing Dendritic Transport. PLOS Genet. 2015;11:e1005695. doi: 10.1371/journal.pgen.1005695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viktorinová I, Dahmann C. Microtubule Polarity Predicts Direction of Egg Chamber Rotation in Drosophila. Curr. Biol. 2013;23:1472–1477. doi: 10.1016/j.cub.2013.06.014. [DOI] [PubMed] [Google Scholar]
- Yurchenco PD. Basement membranes: cell scaffoldings and signaling platforms. Cold Spring Harb. Perspect. Biol. 2011;3:a004911. doi: 10.1101/cshperspect.a004911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Schulze KL, Hiesinger PR, Suyama K, Wang S, Fish M, Acar M, Hoskins RA, Bellen HJ, Scott MP. Thirty-One Flavors of Drosophila Rab Proteins. Genetics. 2006;176:1307–1322. doi: 10.1534/genetics.106.066761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou W, Yadav S, DeVault L, Nung Jan Y, Sherwood DR. RAB-10-Dependent Membrane Transport Is Required for Dendrite Arborization. PLOS Genet. 2015;11:e1005484. doi: 10.1371/journal.pgen.1005484. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.