Abstract
Embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) have promise in regenerative medicine because of their ability to differentiate into all 3 primary germ layers. This review describes recent advances in the understanding of the link between the metabolism of ESCs/iPSCs and their maintenance/differentiation in the cell culture setting, with particular emphasis on amino acid (AA) metabolism. ESCs are endowed with unique metabolic features with regard to energy consumption, metabolite flux through particular pathways, and macromolecular synthesis. Therefore, nutrient availability has a strong influence on stem cell growth, self-renewal, and lineage specification, both in vivo and in vitro. Evidence from several laboratories has documented that self-renewal and differentiation of mouse ESCs are critically dependent on proline metabolism, with downstream metabolites possibly serving as signal molecules. Likewise, catabolism of either threonine (mouse) or methionine (human) is required for growth and differentiation of ESCs because these AAs serve as precursors for donor molecules used in histone methylation and acetylation. Epigenetic mechanisms are recognized as critical steps in differentiation, and AA metabolism in ESCs appears to modulate these epigenetic processes. Recent reports also document that, in vitro, the nutrient composition of the culture medium in which ESCs are differentiated into embryoid bodies can influence lineage specification, leading to enrichment of a specific cell type. Although research designed to direct tissue specification of differentiating embryoid bodies in culture is still in its infancy, early results indicate that manipulation of the nutrient milieu can promote or suppress the formation of specific cell lineages.
Keywords: amino acids, protein metabolism, differentiation, primary germ layers, stem cells
Introduction
The investigation of embryonic stem cell (ESC)6 maintenance and differentiation in tissue culture has revealed a wealth of information about the earliest steps of development. There are some differences in chromatin architecture and dynamics between ESCs in culture and those in the preimplantation embryo (1). However, in contrast to in vivo tissue, cultured cells provide sufficient material for molecular analysis of the basic processes associated with differentiation into the primary germ layers. Furthermore, given the interest in using cultured ESCs or in vitro differentiated somatic cells for possible clinical applications, understanding the mechanisms that regulate in vitro development becomes critical. To take full advantage of the therapeutic potential of cultured ESCs, one of the primary goals is to develop methods to reproducibly direct their differentiation into specific lineages. Therefore, discovering mechanisms by which ESC pluripotency or differentiation can be modulated represents an emerging opportunity in stem cell biology. It is becoming increasingly clear that the nutrient composition of the culture medium plays a key role in the differentiation process, and nutrient amounts can be manipulated to direct lineage specification. Given their rapid growth rate, ESCs exhibit unique metabolic features with regard to energy consumption, metabolite flux through particular pathways, and macromolecular synthesis. Therefore, nutrient availability has a strong influence on stem cell growth, self-renewal, and lineage specification, both in vivo and in vitro. This review describes recent advances in the understanding of the link between ESC metabolism and their maintenance/differentiation in the cell culture setting, particularly with regard to amino acid (AA) metabolism. Topics to be covered include the following: 1) the contrast between in vivo development and differentiation of mouse ESCs (mESCs) in cell culture, 2) how epigenetic regulation of gene expression controls the differentiation process, 3) the unique contribution that AA metabolism appears to play in development and differentiation, and 4) modulation of in vitro tissue-specific cell specification by the composition of the cell culture medium.
Cell Populations and In Vitro Differentiation of mESCs
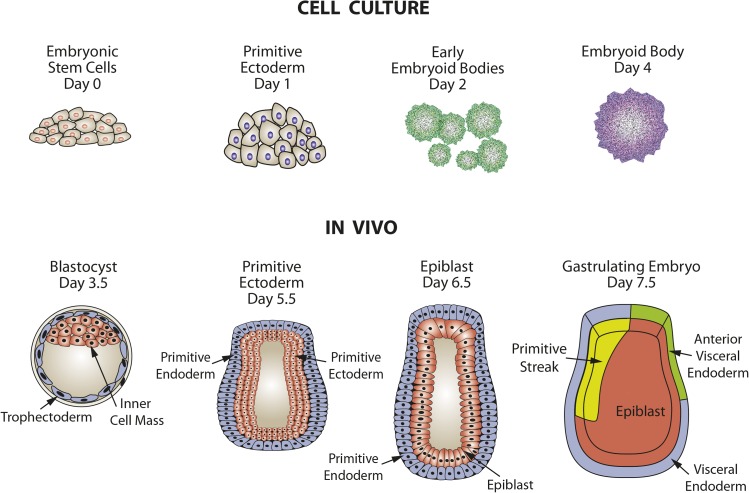
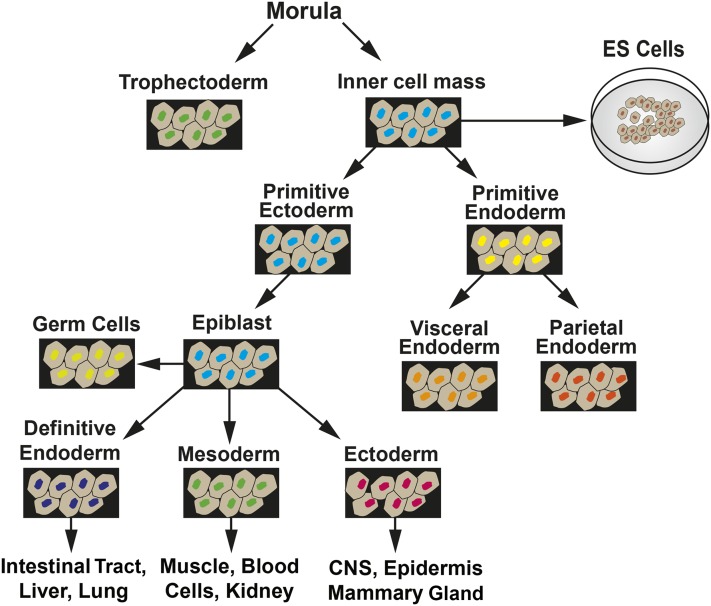
In mice, during the first 4 d after fertilization, the embryo is in a preimplantation phase in which there is cell specification into extraembryonic tissue precursors (trophectoderm and primitive endoderm) and embryonic tissue, which arises from the inner cell mass (ICM) via progression through primitive ectoderm (Figure 1). ESCs are isolated from the ICM and, when maintained in culture, can be induced to follow a similar path of differentiation to form embryoid bodies (EBs), which contain cells representative of all 3 germ layers. In the postimplantation embryo at day 5, epiblast stem cells (EpiSCs) are derived from the primitive ectoderm (Figure 2). EpiSCs give rise to the 3 primary germ layers and, eventually, all somatic cells of the embryo. Cultured ESCs and EpiSCs both exhibit self-renewal and express genes that are hallmarks of pluripotent cells, such as Nanog, sex-determining region Y-box 2 (Sox2), and octamer-binding transcription factor 4 (Oct4). However, there are also significant differences that can be detected by marker gene expression. For example, cultured mESCs display reduced expression protein 1 (Rex1)+/fibroblast growth factor 5 (Fgf5−) expression, whereas mouse EpiSCs in vivo are Rex1−/Fgf5+ in genotype. In vivo studies have documented that during the initial steps of embryogenesis, cell lineage specification is accompanied by temporal and spatial changes in gene expression, with subsets of genes either increased or decreased in specific cell types (3). Therefore, the loss of undifferentiated ESCs as well as the development of EpiSC-like cells and, subsequently, particular cell lineages can be monitored within cell cultures by measuring the expression of genes, either by mRNA or protein known to be specific for that lineage (Figure 2). There are also clear differences in morphology, with the ESCs growing as mounds of rounded colonies with a smooth border and the EpiSCs forming flat, monolayer colonies with irregular borders. Cultures of ESCs are heterogeneous and can contain EpiSC-like cells, even in the presence of differentiation inhibitor leukemia inhibitory factor (LIF). Correcting this heterogeneity is one of the ongoing goals of stem cell research. As Tan et al. (4) suggested, the heterogeneity within ESC cultures may be a result of culture medium composition influencing intracellular signaling pathways. Consequently, more homogeneous ESC cultures may be obtained through changes in culture medium composition once we understand how specific nutrients modulate the differentiation pathways.
FIGURE 1.
Comparison of embryo development in vivo with differentiation of embryonic stem cells to embryoid bodies in tissue culture. Adapted from reference 2 with permission.
FIGURE 2.
Summary of embryo development showing the transitions among tissue types that lead to extraembryonic tissues and the 3 primary germ layers. CNS, central nervous system; ES, embryonic stem. Adapted from reference 2 with permission.
ESC Differentiation and Epigenetics
During differentiation there are no changes in genomic sequence. Consequently, development must be driven by epigenetic-based changes in gene expression. DNA methylation at cytosine-phospho-guanine (CpG) sites, which is generally associated with gene repression, plays a crucial role in early development. DNA methylation–depleted ESCs undergo apoptosis when committed to differentiation (5, 6). Furthermore, there is erasure of DNA methylation in the oocyte and ICM, but re-establishment of the global pattern in a lineage-specific manner during early embryogenesis (5, 6). In contrast to the repressive function of CpG methylation in gene regulation, studies have shown that non-CpG DNA methylation is highly enriched in actively transcribed genes in ESCs (7). This non-CpG DNA methylation is diminished upon lineage commitment, suggesting that this modification may play a positive role in transcription.
Histone modification represents another level of complexity given the number of modifications and the subsequent influence on gene expression (8, 9). The chemical modifications of histone tails known to exist include acetylation, methylation, citrullination (also known as deimination), phosphorylation, ubiquitinylation, sumoylation, and biotinylation. Although the precise role in gene regulation of each modification is still not fully understood, pan acetylation of H3 and H4, as well as methylation of H3K4 and H3K36, are usually associated with gene activation, whereas methylation of H3K9, H4K20, and H3K27 are related to compacted chromatin and gene repression. Consistent with the high level of global transcription in stem cells, they contain more acetylated chromatin and smaller regions of H3K9me3 and H3K27me3 typical of heterochromatin relative to differentiated cells (9, 10). A seminal observation that illustrates the dynamic change in histone modification during ESC differentiation is the presence of bivalent regions or “domains” in a subset of developmentally related genes (7). These bivalent domains are enriched in both the transcriptional active marker H3K4Me3 and the repressive marker H3K27Me3 and, upon differentiation, resolve to either H3K4Me3 or H3K27Me3 predominantly in a lineage-specific manner (9, 11).
In addition to post-translational modification, incorporation of different histone variants can also contribute to nucleosome function and, thereby, gene regulation (7, 12). In somatic cells, the histone variant H2A.Z is enriched in a narrow region near active gene promoters and enhancers and the enrichment is directly correlated with gene activation. In contrast, the ESC distribution of H2A.Z is rather atypical in that it occupies a large set of silent development genes and, paradoxically, almost always coexists with the active gene mark H3K4Me3 (7). Knockdown of H2A.Z compromises self-renewal and leads to aberrant differentiation (13). The histone variant H3.3 is also generally associated with gene activation in somatic cells, but in ESCs, H3.3 is present in both active and inactive gene promoters. Further studies showed that in active genes, H3.3 is also enriched in the transcribed region of the gene and correlated with transcribing RNA polymerase II (POL II). Deficiency of H3.3, and its chaperone HIRA, had no detectable effect on ESC self-renewal and gene expression, but there was a significant reduction in H3K27Me3 and the polycomb repressive complex 2 (PrC2) enrichment at bivalent promoters, suggesting a potential function in lineage specification (13, 14). The insights obtained to date underscore the intimate relation between epigenetics and developmental biology. The remainder of this review will discuss how AA metabolism affects these processes.
AA Metabolism in mESCs
mESC self-renewal and metabolism.
mESCs are extremely small cells with diameters of only a few micrometers compared with the 10- to 30-μm sizes of other mammalian cells and volumes approaching 1/1000th less (15, 16). The mESCs have a shortened G1 phase relative to fully differentiated cells, 1.5 h compared with 18 h (17), and as a consequence, their replication time is much shorter, ∼5 h compared with 24 h (15). This property does not apply to human ESCs (hESCs), which exhibit a doubling time of ∼35 h (18). The rapid growth observed for mESCs necessitates metabolic shifts that are compatible with the requirement for a high rate of cellular content duplication. mESCs have fewer and poorly developed mitochondria and consume less oxygen than somatic cells. Similar to cancer cells, the preferred energy source of mESCs is the oxidation of glucose via glycolysis, with minimal conversion of glucose-derived pyruvate to acetyl-CoA (17). During differentiation, the degree of glucose catabolism by oxidative phosphorylation (OX-PHOS) increases in proportion to mitochondrial production. Conversely, during reprogramming of induced pluripotent stem cells (iPSCs) from somatic cells, metabolism also reverts from glucose oxidation via OX-PHOS in the differentiated cells to primarily glycolysis. The reliance on glycolysis instead of OX-PHOS in ESCs/iPSCs is reminiscent of the Warburg effect in cancer cells. To maintain the citric acid cycle, anapleurotic metabolism from AAs and lipids generate acetyl-CoA (19). Genes encoding enzymes associated with AA metabolism are among the most altered during ESC differentiation and, conversely, much of this gene expression is reversed during reprogramming of somatic cells to iPSCs. That AA availability can modulate early embryonic development has been recognized for some time. For example, during in vitro embryo culture, the addition of glutamine and other “nonessential” AAs increases blastocyst formation, whereas removal of these same AAs causes embryo arrest at the 2-cell stage (20). Consequently, the somatic cell list of nonessential AAs, a designation derived from their effect on animal growth, does not apply to ESCs. This observation will be underscored by many of the studies described in this review. Beyond the 8-cell stage, AA utilization within the embryo appears to transition such that the embryo no longer requires the nonessential AAs for development in culture (20). As described below, recent research that used mESCs as a model system has led to surprising discoveries about the unique properties of AA metabolism in these cells. The research outlined in this review will underscore that it is now recognized that individual AAs affect the pluripotency, self-renewal, and differentiation properties of both hESCs and mESCs, as well as reprogramming of somatic cells to iPSCs.
Proline metabolism.
Washington et al. (21) discovered that elevated proline (Pro) in the culture medium, at concentrations of ≥100 μM, promotes proliferation and differentiation of mESCs into EpiSCs or primitive ectoderm (PrEc), even in the presence of the differentiation suppressor LIF. Although the mammalian target of rapamycin (mTOR) inhibitor rapamycin blocked the proline effect, treatment of mESCs with leucine to activate mTOR did not induce differentiation to EpiSCs/PrEc, suggesting that mTOR may be necessary but not sufficient. Furthermore, the addition of an excess of glycine to the medium blocked the proline effect. The glycine inhibition was later shown to result from competition for uptake by the sodium-coupled neutral AA transporter 2 (SNAT2) AA transporter (4). We have shown that expression of the SNAT2 transporter is induced by AA limitation (22). Proline also caused the expression of the mesendoderm marker brachyury 24 h earlier during the differentiation program of ESCs and suppressed the development of neuroectoderm. Casalino et al. (23) later confirmed that the proline-induced cells could differentiate into general mesendoderm [α-fetoprotein (Afp) as a marker] and cardiac mesoderm [(α-myosin heavy chain α-Mhc) and NK2 transcription factor related, locus 5 (Nkx2.5) as markers].
Casalino et al. (23) subsequently extended the studies by Washington et al. (21) to show that ornithine (Orn) can also induce the mESC to EpiSC transition, but the amount required is 20 times greater (100 μM Pro compared with 2 mM Orn). It was established that this effect requires the metabolism of proline or ornithine to Δ1-pyrroline-5-carboxylate, a common metabolite in their catabolism. The proline induction of EpiSCs/PrEc was complete (90% of colonies) after 18 h of treatment and, as shown by Washington et al. (21) and Tan et al. (4), other AAs that compete for proline transport could block the response. Several independent mESC cell lines were tested and the proliferation of each one was enhanced by proline, whereas growth rates for fully differentiated cells (embryonic fibroblasts and HEK293 and 3T3 cells) were unaffected. In proline-induced cells, the PrEc marker Fgf5 was increased by 100-fold, but conversely, not all mESC markers [stage-specific embryonic antigen (SSEA), Oct4] are completely lost. Proline-induced EpiSCs/PrEc cells appear to have all of the properties of embryo-derived EpiSCs/PrEc or those generated in culture by removal of LIF. The differentiation step triggered by proline was completely reversible; thus, the removal of proline from the medium resulted in a return to the mESC phenotype. Expression of Fgf5 was necessary for the mESC-to-EpiSC transition, and given that Fgf5 is a marker for epiblast formation in vivo, the authors suggested that the proline-triggered transition in culture might be analogous to the blastocyst-to-epiblast transition that occurs during implantation. Interestingly, embryo implantation requires degradation of the uterine extracellular matrix, which is 80% proline-rich collagen. Thus, localized proline release may serve to generate a signaling molecule that is critical to the implantation process. Casalino et al. (23) also made the discovery that ascorbic acid antagonized the proline effect, possibly through a mechanism independent of its antioxidant properties.
Comes et al. (24) noted that the morphologic changes for the proline-induced EpiSCs were reminiscent of a shift toward a mesenchymal phenotype. They showed that the EpiSCs migrate through Matrigel-coated trans-wells (Corning Inc.), whereas mESCs do not. Furthermore, when the proline-induced EpiSCs were injected into nude mice they formed lung teratomas. By analogy, Comes et al. (24) suggested that proline might act as a signal molecule to regulate tumor progression and invasiveness by modulating the epithelial-to-mesenchymal transition. It is known that proline metabolism is altered in cancer (25, 26), and Comes et al. (24) showed that proline resulted in a global increase in histone H3K9me2/H3K9me3 and H3K36me3 concentrations, preferentially at noncoding intergenic and in transcriptionally repressed constitutive heterochromatin regions. These increases in H3K9 and H3K36 methylation would lead to repression of gene expression. Interestingly, these changes in histone modification were reversed either by the removal of proline or treatment with ascorbic acid, the latter effect consistent with the antagonistic relation between proline and ascorbic acid established by Casalino et al. (23). Mechanistically, one can imagine this relation because ascorbic acid serves as a cofactor for several members of the Jumonji family of histone demethylases (27). Thus, proline may lead to suppression of gene expression by modulating the chromatin environment during an epithelial-to-mesenchymal transition. Conversely, reprogramming of somatic cells to iPSCs requires the reverse, a mesenchymal-to-epithelial–like transition (28, 29), that is characterized by an ascorbic acid–dependent decrease in H3K36 methylation concentrations (30). Thus, proline and ascorbic acid mediate opposite effects on H3K9 and H3K36 methylation status and promote the opposite directions of stem cell programming. Interestingly, we have identified jumonji domain-containing protein 3 (JMJD3) as an AA-responsive gene that is transcriptionally induced by AA limitation (31, 32). Whether or not the change in JMJD3 protein expression leads to altered histone modification in stem cells or during differentiation requires further study, because it has been proposed that the protein has important functions independent of its enzyme activity as an H3K27 demethylase (33).
Date et al. (34) showed that collagen hydrolysate, and hydroxyproline (Hyp) in particular, promoted EB differentiation to mesoendoderm and further toward cardiomyocytes. Although Hyp did not alter the EB growth rate or colony size, the expression of mesendoderm [Brachyury and forkhead box A2 (Foxa2)] and cardiomyocyte (α-Mhc and Nkx2.5) markers were increased, as was the number of contracting cells. For unknown reasons, these authors did not see an effect of proline itself, perhaps because of cell density (24). Glycine, another AA that is a major component of collagen, antagonized the Hyp effect on cell morphology, decreased the expression of PrEc marker genes [Fgf5 and orthodenticle homeobox 2 (Otx2)], and increased the mESC markers indicative of ground state pluripotency (Nanog, Rex1, and alkaline phosphatase). Consistent with the observations of Date et al., Sato et al. (35) showed that an epimer of Hyp, cis-4-hydroxy-d-proline, inhibited cardiac differentiation.
In summary, proline and its downstream metabolites appear to be critical for ESC maintenance and also affect some of the earliest steps of differentiation. Identification of the exact metabolites and their mechanism of action will provide interesting insight into the relation between AA metabolism and the biology of differentiation.
Threonine metabolism.
After a metabolomic study, the McKnight laboratory discovered that mESCs are uniquely sensitive to threonine (Thr) depletion because the conversion of threonine to glycine (for one-carbon metabolism) and to acetyl-CoA (for energy production) by threonine dehydrogenase (TDH) is essential for mouse stem cell survival (15). TDH activity is several thousand–fold higher in mESCs than in differentiated EBs or in adult mouse tissues. TDH activity is greatly reduced during the first few days of differentiation to EBs (15). Threonine-deficient medium inhibits mESC proliferation but not that of EBs or differentiated mouse cell lines, such as 3T3 or mouse embryonic fibroblast (MEF) cells (15). Given that growth of the latter cells, but not mESCs, was highly suppressed by cysteine-free medium, the authors speculated that a medium high in threonine and low in cysteine would be useful to select for mESCs. Conversely, a threonine-deficient medium could be useful to select against mESCs. Inhibition of TDH activity by selective inhibitors leads to selective mESC cell death by autophagy (36) and inhibition of the activity in vivo by the AA analog 3-hydroxy-norvaline (3HNV) resulted in suppression of development of the inner cell mass, the region of the blastocyst embryo (15). The latter observation shows that the threonine dependence of mESCs in culture is faithfully reproducing stem cell metabolism in vivo. Supplying additional glycine and pyruvate in the culture medium to maintain one-carbon metabolism and as a source of acetyl-CoA can reverse mESC death induced by threonine deprivation (37).
Ryu and Han (38) extended the initial studies by showing that if mESCs were threonine-deprived for 1 d, refeeding threonine led to increased cyclin D1 and E expression and increased DNA synthesis. Threonine also appears to prevent mESC differentiation. Even in the presence of LIF, when mESCs were incubated in DMEM lacking threonine for 1–4 d, most stem cell–specific markers were decreased, trophoectodermal and mesodermal markers were increased, and endodermal and ectodermal (except for Sox1) markers were unchanged (38). A signaling pathway that involved PI3K/Akt, all 3 primary MAPK pathways, and mammalian target of rapamycin complex 1 (mTORC1) activation mediated the threonine effect on mESC proliferation (38). Furthermore, small interfering RNA knockdown of TDH activity causes a decline in the expression of pluripotency markers and an increase in markers reflecting differentiation (37).
As discussed above, ESC genes associated with lineage specification often exist in a bivalent or poised epigenetic state as shown by the coexistence of H3K4me3 activating and H3K27me3 repressive marks (39). Methylation of histones is mediated by S-adenosylmethionine (SAM), for which production is dependent on one-carbon metabolism. Interestingly, threonine deprivation or knockdown of TDH activity in mESCs adversely affects SAM concentrations and, consequently, selectively suppresses di- and trimethylation of H3K4 (37). Given the relation between increased H3K4me3 and gene activation, suppression of this epigenetic mark could have a negative effect on gene expression in mESCs or during reprogramming of iPSCs. Indeed, Han et al. (40) confirmed previous reports that TDH activity is absent in MEF, but they went on to show that TDH activity was high in iPSCs prepared from those MEFs. Furthermore, overexpression of TDH in MEF enhanced reprogramming and TDH suppression hampered reprogramming. Those authors also showed that miR-9 inhibits TDH translation and therefore suppresses reprogramming and that protein arginine methyltransferase 5 (PRMT5) methylates R180, which leads to increased TDH activity. Consequently, miR-9 suppresses TDH-dependent reprogramming, whereas PRMT5 enhances it. These studies document that threonine metabolism is also critical for mESC function and iPSC formation and that the regulation of TDH activity can influence the efficiency of reprogramming.
Methionine Metabolism in hESCs
Human ESCs do not have TDH activity as the result of inactivating mutations within the coding region (41). Consequently, the relation of threonine to ESC function observed in the mouse is not likely to be the same, and indeed, hESCs or iPSCs deprived of threonine do not show a decline in cell growth (42). However, they do exhibit the same requirement for SAM-mediated methylation of H3K4me3, but the maintenance of SAM concentrations is more tightly linked to methionine (Met) metabolism. Deprivation of individual AAs showed that methionine, leucine (Leu), and lysine (Lys) were necessary for hESC and iPSC proliferation and their absence from the culture medium induced G1 cell cycle arrest and eventually apoptosis (42). However, significant differences between these 3 AAs were also noted. Whereas leucine- and lysine-depleted medium induced expression of the proapoptotic gene CCAAT/enhancer-binding protein (C/EBP) homologous protein (CHOP), methionine deprivation did not. Instead, methionine limitation induced apoptosis via a p53/p38-dependent mechanism that could be prevented by supplementation of methionine deficient medium with SAM. Furthermore, the inhibition of growth in methionine deficient medium could be rescued by medium supplementation with other metabolic intermediates of the methionine-SAM cycle. Extensive analysis led Shiraki et al. (42) to conclude that hESCs/iPSCs have a high rate of methionine metabolism, which serves an analogous role to threonine in mESCs/iPSCs by maintaining SAM concentrations for methylation. Interestingly, as in the case of threonine deprivation in mouse cells, limitation of methionine in human cells led to a decline in H3K4me3 but had no effect on H3K9me3, H3K27me3, or H3K36me3 (42). Using protocols to direct human iPSCs to each of 3 primary germ lines, the authors also showed that methionine deprivation actually enhanced differentiation in each case. Furthermore, short-term treatment with methionine-free medium of iPSCs before initiating their differentiation to hepatocyte-like cells resulted in the loss of the residual iPSCs by apoptosis and a significant enrichment of cells expressing the liver markers AFP and albumin (ALB). Collectively, these results show that, whereas hESCs do not rely on threonine-derived one-carbon metabolism for self-renewal and pluripotency, they utilize methionine metabolism to achieve the same result.
Impact of the AA Response on mESCs
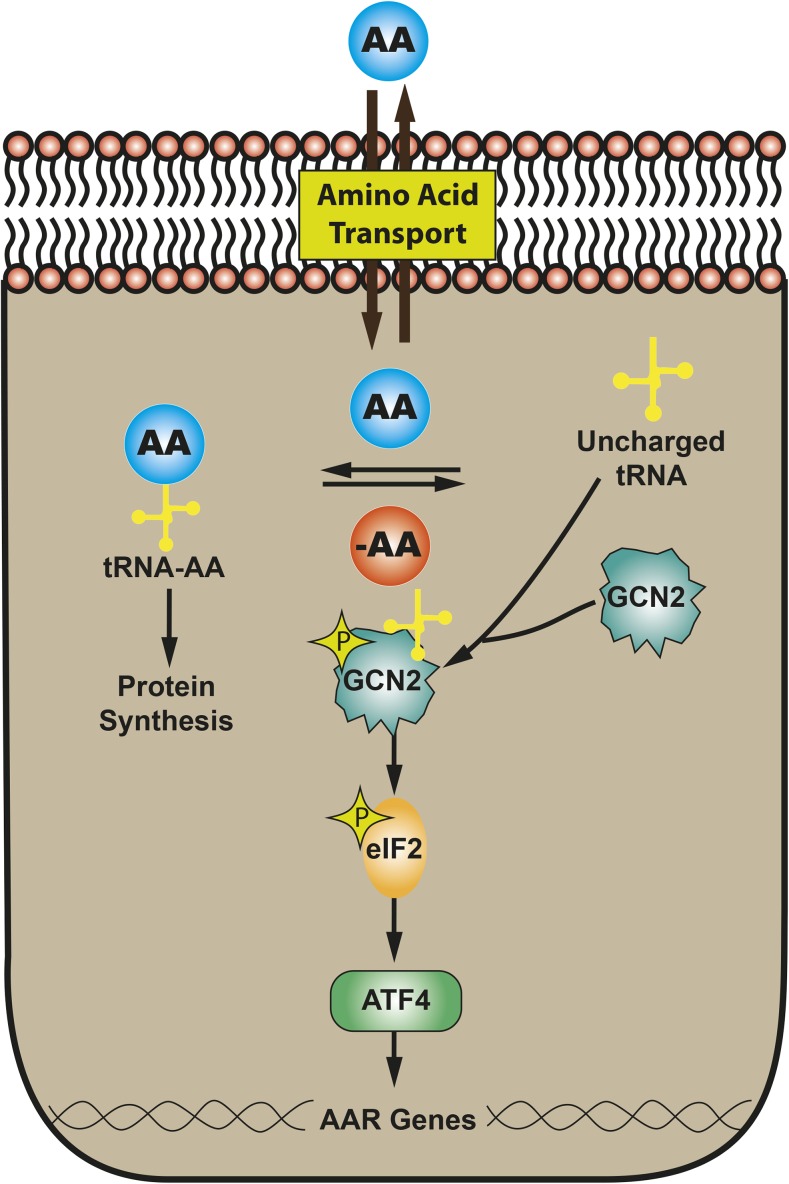
In vivo ESCs and the developing embryo may encounter a microenvironment in which nutrients are limited, which can have long-term effects on embryogenesis and growth. Even in highly developed countries, a significant number of infants are born small-for-gestational-age as the result of intrauterine growth retardation (IUGR) (reviewed in reference 43). IUGR can arise from many causes, including decreased placental function or poor maternal nutrition during pregnancy; in either of these cases, one possible outcome is a reduction in maternal-to-fetal transfer of AAs (44). Consistent with this hypothesis, placentas from human IUGR pregnancies have elevated eukaryotic initiation factor 2 (eIF2) phosphorylation (45), revealing activation of the general control non-derepressible 2 (Gcn2)–eIF2–activating transcription factor 4 (Atf4) pathway, a component of the AA response (AAR). Well documented in somatic cells and tissues, dietary protein or AA deprivation triggers a collection of signaling processes collectively referred to as the AAR (46, 47). Principal among these is the Gcn2-eIF2-Atf4 pathway (Figure 3). Atf4 mediates activation of hundreds of target genes (31, 48).
FIGURE 3.
The AAR pathway that mediates the detection and response to AA limitation. AA, amino acid; AAR, amino acid response; ATF4, activating transcription factor 4; eIF2, eukaryotic initiation factor 2; GCN2, general control non-derepressible 2; tRNA, transfer RNA.
It has long been recognized that in vivo AA availability can modulate early embryonic development (20). Likewise, during in vitro embryo culture, the addition of nonessential AAs, including glutamine, increases blastocyst formation, whereas limiting nonessential AAs causes embryo arrest at the 2-cell stage. These observations led Gardner (20) to conclude that “AAs appear to be fundamental regulators of cell function, including energy metabolism, in the preimplantation embryo.” Recent research that used cultured ESCs and iPSCs has provided mechanistic insight into the processes that link intermediary metabolism and stem cell function. In addition to the immediate effects of AA limitation on fetal development, it is now recognized that long-term consequences mediated by epigenetics are also a factor. For example, maternal dietary protein limitation during pregnancy leads to genomewide changes in DNA methylation of the fetus and, subsequently, altered gene expression during adulthood of the resulting offspring (49). Such epigenetic-mediated modulation of gene expression is the basis for the “fetal origins of adult disease” hypothesis (50). It is recognized that epigenetic mechanisms, such as DNA methylation and histone modification, help to establish correct gene expression patterns for cell lineage specification (51). Microarray results from somatic cells showed that AA limitation increased or decreased (≥2-fold, P ≤ 0.001) the expression of 190 genes encoding proteins associated with chromatin structure and histone modification (31), including 19 histone methylase/demethylase enzymes (32). For many of these AA-responsive histone-modifying enzymes, critical contributions to the regulation of ESC differentiation have been well documented (52). Thus, AAR-induced changes in chromatin structure, DNA methylation, and histone modifications may influence early differentiation of the primary germ layers, long-term organ development, and ultimately, organ function during adulthood.
Although investigation of AA sensing and regulatory mechanisms in cultured ESCs and EBs is limited, Shan et al. (53) published evidence to show that the AAR is functional in mESCs and that a low level of AAR activation influences the in vitro differentiation outcome in EBs. In somatic cells, activation of the AAR leads to a transient suppression of global protein synthesis via a decrease in translation initiation (54). mESCs respond in a similar manner, exhibiting a decrease of ∼20% in protein synthesis 8 h after activating the AAR (53). To investigate the impact of the AAR on the earliest stages of embryonic development, Shan et al. (53) activated a low level of the AAR pathway in differentiating EBs for 12 d. The AAR suppressed the overall differentiation process as evidenced by retention of ESCs, despite incubation in differentiating medium and the typical loss of ESCs in the control group. There was decreased formation of primitive ectoderm, mesoderm, and ectoderm in response to the AAR. In contrast, the relative abundance of primitive endoderm, visceral endoderm, and certain endoderm-derived lineages was greater after AAR activation (53). The relative increase in primitive and visceral endoderm is particularly interesting because these cells contribute to the formation of extraembryonic tissues (3, 55). Given the central role in maternal-to-fetal nutrient transport, these tissues may be particularly sensitive to nutrient limitation.
In addition to Gcn2 kinase, there are 3 other eIF2 kinases, one of which is protein kinase R-like endoplasmic reticulum kinase (Perk) (56). Perk signaling is one of the 3 arms of the unfolded protein response (UPR), a collection of pathways activated by endoplasmic reticulum (ER) stress resulting from misfolded proteins, calcium perturbation, and many other ER insults. Like Gcn2, Perk activation leads to increased synthesis of Atf4 and, as a consequence, the downstream transcriptional programs triggered by Perk and Gcn2 exhibit considerable overlap. However, for reasons unknown, the terminal gene targets are not completely identical (57). To determine the impact of the UPR on ESC differentiation, Xu et al. (58) treated mESCs with thapsigargin or tunicamycin, small molecule drugs used experimentally to cause/induce ER stress and activate the UPR pathways (59). Consistent with our observations for the AAR, Xu et al. showed that induction of the UPR in EBs resulted in increased abundance of markers for the endodermal lineage, a process that required Tgfβ/small for body size mothers against decapentaplegic (Smad) and wingless-related integration site (Wnt)/β-catenin signaling (58). Treatment of EBs with thapsigargin or tunicamycin for 2 d followed by 4 d of culture without the drugs caused an increase in both primitive endoderm [Sox7, GATA binding protein (Gata)4, and Gata6] and definitive endoderm (Sox17 and FoxA2) markers. Conversely, markers for ESCs (Oct4), trophectoderm [caudal type homeobox 2 (Cdx2)], epiblast/primitive ectoderm (Fgf5), ectoderm [paired box 6 (Pax6) and cytokeratin 18 (Ck18)], and mesoderm [T/brachyury and fetal liver kinase 1 (Flk1)] were all diminished. The authors also presented evidence that UPR activation within EBs leads to increased formation of hepatic and pancreatic lineages when differentiation for those cell types was targeted by specific culture treatments (58). Collectively, the AAR and the UPR studies suggest that nutrient-related cell stress can affect lineage specification to a significant degree.
Nutrient-Driven, Lineage-Specific Differentiation
Considerable evidence is emerging to indicate that the lineage specification path from either mESCs or hESCs and selection of specific differentiated cell types can be modulated by environmental nutritional queues. Nutrient-directed hESC differentiation has led to a potential method for mass production of human cardiomyocytes. It has been recognized for some time that fetal hearts prefer lactate rather than glucose to fuel energy production via OX-PHOS (60). On the basis of that observation, as well as gene expression and metabolic flux studies, Tohyama et al. (61) reasoned that a glucose-depleted medium containing lactate might favor cardiomyocyte survival. Culture of differentiating EBs from either hESCs or mESCs in the high-lactate medium resulted in an enrichment of cardiomyocytes to >95% purity and with relatively high yield. Similarly, Tomizawa et al. (62) also used tissue-specific differences in metabolic pathways to propose that a medium deficient in arginine, tyrosine, glucose, and pyruvate, but enriched in ornithine, phenylalanine, galactose, and glycerol, would enrich the population for hepatocytes. After 18 d of EB growth in the “hepatocyte-selection medium,” the number of cells was reduced by 60–70%. However, the remaining cells exhibited a morphology and gene expression pattern indicative of hepatoblast-like cells. Dietary leucine supplementation (2% leucine in the drinking water) of pregnant rats between days 13.5 and 20 of gestation led to reduced pancreatic β cell mass and impaired glucose tolerance tests in the 4-wk-old offspring (63). It was discovered that leucine activation of mTOR in the developing fetal pancreas leads to increased expression of hypoxia inducible factor 1, α subunit (Hif-1α), which functions as a repressor of the transcription factor neurogenin-3. Neurogenin-3–positive endocrine progenitor cells are a necessary step in the differentiation from endoderm to pancreatic endocrine tissue, including mature β cells. Hence, excess dietary leucine suppresses formation of this progenitor cell population. The observation that nutrients can modulate stem cell differentiation also applies to the hematopoietic system. Oburoglu et al. (64) showed that the commitment of hematopoietic stem cells to the erythroid rather than the myeloid lineage requires glutamine utilization for nucleotide biosynthesis. Blocking glutamine uptake or its metabolism to de novo nucleotide synthesis, in vitro or in vivo, diverts hematopoietic stem cell differentiation from the erythroid lineage to the myeloid lineage. Collectively, these investigations support the role of medium nutrient composition in modulating lineage specification, and begin to provide approaches for generating cell populations enriched in particular cell types.
Future Perspectives
The developing field of ESC and EB metabolism underscores the following insight from Shyh-Chang et al. (65): “Once thought to be a mere consequence of the state of the cell, metabolism is now known to play a pivotal role in dictating whether a cell proliferates, differentiates, or remains quiescent.” Emerging data show that the interplay between the extracellular nutrient supply and pluripotent cells regulates self-renewal and cell fate. The results from many laboratories show that the manipulation of cell culture nutrient composition can enrich specific cell lineages. Interestingly, this phenomenon does not apply to just one lineage, but may be universal across all 3 primary germ layers. It is highly likely that nutrient-driven specification not only applies to ESCs but to other stem cell populations as well. One of the goals of future studies in this area of investigation will be to identify the mechanisms by which nutrient supply directs specific lineage formation and then harness that knowledge for in vitro production of tissue-specific cell populations. It is clear from the early investigations that nutrient availability not only alters differentiation from ESCs but also the reprogramming of somatic cells to iPSC populations. Many of these in vitro stem cell manipulation systems will become even more important as the age of personalized medicine develops. Consequently, in the context of therapeutic uses of ESCs, iPSCs, or in vitro differentiated tissue, investigators will need to consider cell culture nutrient composition as a critical factor.
Acknowledgments
All authors read and approved the final manuscript.
Footnotes
Abbreviations used: AA, amino acid; AAR, AA response; Afp, α-fetoprotein; ALB, albumin; Atf4, activating transcription factor 4; Cdx2, caudal type homeobox 2; CHOP, CCAAT/enhancer-binding protein (C/EBP) homologous protein; Ck18, cytokeratin 18; CpG, cytosine-phospho-guanine; EB, embryoid body; eIF2, eukaryotic initiation factor 2; EpiSC, epiblast stem cell; ER, endoplasmic reticulum; ESC, embryonic stem cell; Fgf5, fibroblast growth factor 5; Flk1, fetal liver kinase 1; Foxa2, forkhead box A2; Gata, GATA binding protein; Gcn2, general control non-derepressible 2; hESC, human embryonic stem cell; Hif-1α, hypoxia inducible factor 1, α subunit; Hyp, hydroxyproline; ICM, inner cell mass; iPSC, induced pluripotent stem cell; IUGR, intrauterine growth retardation; JMJD3, jumonji domain-containing protein 3; LIF, leukemia inhibitory factor; MEF, mouse embryonic fibroblast; mESC, mouse embryonic stem cell; miR-9, microRNA-9; mTOR, mammalian target of rapamycin; mTORC1, mammalian target of rapamycin complex 1; Nkx2.5, NK2 transcription factor related, locus 5; Oct4, octamer-binding transcription factor 4; Otx2, orthodenticle homeobox 2; OX-PHOS, oxidative phosphorylation; Pax6, paired box 6; Perk, protein kinase R-like endoplasmic reticulum kinase; Pol II, RNA polymerase II; PrC2, polycomb repressive complex 2; PrEc, primitive ectoderm; PRMT5, protein arginine methyltransferase 5; Rex1, reduced expression protein 1; SAM, S-adenosylmethionine; Smad, small for body size mothers against decapentaplegic; SNAT2, sodium-coupled neutral amino acid transporter 2; Sox1, sex-determining region Y-box 1; Sox2, sex-determining region Y-box 2; Sox17, sex-determining region Y-box 17; SSEA, stage-specific embryonic antigen; TDH, threonine dehydrogenase; UPR, unfolded protein response; Wnt, wingless-related integration site; α-Mhc, α-myosin heavy chain; 3HNV, 3-hydroxy-norvaline.
References
- 1.Burton A, Torres-Padilla ME. Chromatin dynamics in the regulation of cell fate allocation during early embryogenesis. Nat Rev Mol Cell Biol 2014;15:723–34. [DOI] [PubMed] [Google Scholar]
- 2.Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev 2005;19:1129–55. [DOI] [PubMed] [Google Scholar]
- 3.Tam PP, Loebel DA. Gene function in mouse embryogenesis: get set for gastrulation. Nat Rev Genet 2007;8:368–81. [DOI] [PubMed] [Google Scholar]
- 4.Tan BS, Lonic A, Morris MB, Rathjen PD, Rathjen J. The amino acid transporter SNAT2 mediates L-proline-induced differentiation of ES cells. Am J Physiol Cell Physiol 2011;300:C1270–9. [DOI] [PubMed] [Google Scholar]
- 5.Hackett JA, Surani MA. DNA methylation dynamics during the mammalian life cycle. Philos Trans R Soc Lond B Biol Sci 2013;368:20110328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seisenberger S, Peat JR, Hore TA, Santos F, Dean W, Reik W. Reprogramming DNA methylation in the mammalian life cycle: building and breaking epigenetic barriers. Philos Trans R Soc Lond B Biol Sci 2013;368:20110330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kraushaar DC, Zhao K. The epigenomics of embryonic stem cell differentiation. Int J Biol Sci 2013;9:1134–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thiagarajan RD, Morey R, Laurent LC. The epigenome in pluripotency and differentiation. Epigenomics 2014;6:121–37. [DOI] [PubMed] [Google Scholar]
- 9.Boland MJ, Nazor KL, Loring JF. Epigenetic regulation of pluripotency and differentiation. Circ Res 2014;115:311–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X, Li L, Pandey R, Byun JS, Gardner K, Qin Z, Dou Y. The histone acetyltransferase MOF is a key regulator of the embryonic stem cell core transcriptional network. Cell Stem Cell 2012;11:163–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gifford CA, Ziller MJ, Gu H, Trapnell C, Donaghey J, Tsankov A, Shalek AK, Kelley DR, Shishkin AA, Issner R, et al. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell 2013;153:1149–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turinetto V, Giachino C. Histone variants as emerging regulators of embryonic stem cell identity. Epigenetics 2015;10:563–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu G, Cui K, Northrup D, Liu C, Wang C, Tang Q, Ge K, Levens D, Crane-Robinson C, Zhao K. H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell Stem Cell 2013;12:180–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banaszynski LA, Wen D, Dewell S, Whitcomb SJ, Lin M, Diaz N, Elsasser SJ, Chapgier A, Goldberg AD, Canaani E, et al. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell 2013;155:107–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Alexander P, Wu L, Hammer R, Cleaver O, McKnight SL. Dependence of mouse embryonic stem cells on threonine catabolism. Science 2009;325:435–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J, Alexander P, McKnight SL. Metabolic specialization of mouse embryonic stem cells. Cold Spring Harb Symp Quant Biol 2011;76:183–93. [DOI] [PubMed] [Google Scholar]
- 17.Chen G, Wang J. Threonine metabolism and embryonic stem cell self-renewal. Curr Opin Clin Nutr Metab Care 2014;17:80–5. [DOI] [PubMed] [Google Scholar]
- 18.Amit M, Carpenter MK, Inokuma MS, Chiu CP, Harris CP, Waknitz MA, Itskovitz-Eldor J, Thomson JA. Clonally derived human embryonic stem cell lines maintain pluripotency and proliferative potential for prolonged periods of culture. Dev Biol 2000;227:271–8. [DOI] [PubMed] [Google Scholar]
- 19.Gaspar JA, Doss MX, Hengstler JG, Cadenas C, Hescheler J, Sachinidis A. Unique metabolic features of stem cells, cardiomyocytes, and their progenitors. Circ Res 2014;114:1346–60. [DOI] [PubMed] [Google Scholar]
- 20.Gardner DK. Changes in requirements and utilization of nutrients during mammalian preimplantation embryo development and their significance in embryo culture. Theriogenology 1998;49:83–102. [DOI] [PubMed] [Google Scholar]
- 21.Washington JM, Rathjen J, Felquer F, Lonic A, Bettess MD, Hamra N, Semendric L, Tan BS, Lake JA, Keough RA, et al. L-Proline induces differentiation of ES cells: a novel role for an amino acid in the regulation of pluripotent cells in culture. Am J Physiol Cell Physiol 2010;298:C982–92. [DOI] [PubMed] [Google Scholar]
- 22.Palii SS, Thiaville MM, Pan YX, Zhong C, Kilberg MS. Characterization of the amino acid response element within the human sodium-coupled neutral amino acid transporter 2 (SNAT2) system A transporter gene. Biochem J 2006;395:517–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Casalino L, Comes S, Lambazzi G, De Stefano B, Filosa S, De Falco S, De Cesare D, Minchiotti G, Patriarca EJ. Control of embryonic stem cell metastability by L-proline catabolism. J Mol Cell Biol 2011;3:108–22. [DOI] [PubMed] [Google Scholar]
- 24.Comes S, Gagliardi M, Laprano N, Fico A, Cimmino A, Palamidessi A, De Cesare D, De Falco S, Angelini C, Scita G, et al. L-proline induces a mesenchymal-like invasive program in embryonic stem cells by remodeling H3K9 and H3K36 methylation. Stem Cell Reports 2013;1:307–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phang JM, Liu W, Hancock C, Christian KJ. The proline regulatory axis and cancer. Front Oncol 2012;2:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu W, Le A, Hancock C, Lane AN, Dang CV, Fan TW, Phang JM. Reprogramming of proline and glutamine metabolism contributes to the proliferative and metabolic responses regulated by oncogenic transcription factor c-MYC. Proc Natl Acad Sci USA 2012;109:8983–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simmons JM, Muller TA, Hausinger RP. Fe(II)/alpha-ketoglutarate hydroxylases involved in nucleobase, nucleoside, nucleotide, and chromatin metabolism. Dalton Trans 2008;5132–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 2010;7:64–77. [DOI] [PubMed] [Google Scholar]
- 29.Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 2010;7:51–63. [DOI] [PubMed] [Google Scholar]
- 30.Wang T, Chen K, Zeng X, Yang J, Wu Y, Shi X, Qin B, Zeng L, Esteban MA, Pan G, et al. The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell 2011;9:575–87. [DOI] [PubMed] [Google Scholar]
- 31.Shan J, Lopez MC, Baker HV, Kilberg MS. Expression profiling after activation of the amino acid deprivation response in HepG2 human hepatoma cells. Physiol Genomics 2010;41:315–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shan J, Fu L, Balasubramanian MN, Anthony T, Kilberg MS. ATF4-dependent regulation of the JMJD3 gene during amino acid deprivation can be rescued in Atf4-deficient cells by inhibition of deacetylation. J Biol Chem 2012;287:36393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller SA, Mohn SE, Weinmann AS. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol Cell 2010;40:594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Date Y, Hasegawa S, Yamada T, Inoue Y, Mizutani H, Nakata S, Akamatsu H. Major amino acids in collagen hydrolysate regulate the differentiation of mouse embryoid bodies. J Biosci Bioeng 2013;116:386–90. [DOI] [PubMed] [Google Scholar]
- 35.Sato H, Takahashi M, Ise H, Yamada A, Hirose S, Tagawa Y, Morimoto H, Izawa A, Ikeda U. Collagen synthesis is required for ascorbic acid-enhanced differentiation of mouse embryonic stem cells into cardiomyocytes. Biochem Biophys Res Commun 2006;342:107–12. [DOI] [PubMed] [Google Scholar]
- 36.Alexander PB, Wang J, McKnight SL. Targeted killing of a mammalian cell based upon its specialized metabolic state. Proc Natl Acad Sci USA 2011;108:15828–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shyh-Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Teo RY, Ratanasirintrawoot S, Zhang J, Onder T, Unternaehrer JJ, Zhu H, et al. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science 2013;339:222–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryu JM, Han HJ. L-threonine regulates G1/S phase transition of mouse embryonic stem cells via PI3K/Akt, MAPKs, and mTORC pathways. J Biol Chem 2011;286:23667–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 2006;125:315–26. [DOI] [PubMed] [Google Scholar]
- 40.Han C, Gu H, Wang J, Lu W, Mei Y, Wu M. Regulation of L-threonine dehydrogenase in somatic cell reprogramming. Stem Cells 2013;31:953–65. [DOI] [PubMed] [Google Scholar]
- 41.Edgar AJ. The human L-threonine 3-dehydrogenase gene is an expressed pseudogene. BMC Genet 2002;3:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shiraki N, Shiraki Y, Tsuyama T, Obata F, Miura M, Nagae G, Aburatani H, Kume K, Endo F, Kume S. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab 2014;19:780–94. [DOI] [PubMed] [Google Scholar]
- 43.Figueras F, Gardosi J. Intrauterine growth restriction: new concepts in antenatal surveillance, diagnosis, and management. Am J Obstet Gynecol 2011;204:288–300. [DOI] [PubMed] [Google Scholar]
- 44.Brown LD, Green AS, Limesand SW, Rozance PJ. Maternal amino acid supplementation for intrauterine growth restriction. Front Biosci (Schol Ed) 2011;3:428–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yung HW, Calabrese S, Hynx D, Hemmings BA, Cetin I, Charnock-Jones DS, Burton GJ. Evidence of placental translation inhibition and endoplasmic reticulum stress in the etiology of human intrauterine growth restriction. Am J Pathol 2008;173:451–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kilberg MS, Balasubramanian M, Fu L, Shan J. The transcription factor network associated with the amino acid response in mammalian cells. Adv Nutr 2012;3:295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kilberg MS, Shan J, Su N. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol Metab 2009;20:436–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 2000;6:1099–108. [DOI] [PubMed] [Google Scholar]
- 49.Lillycrop KA, Phillips ES, Jackson AA, Hanson MA, Burdge GC. Dietary protein restriction of pregnant rats induces and folic acid supplementation prevents epigenetic modification of hepatic gene expression in the offspring. J Nutr 2005;135:1382–6. [DOI] [PubMed] [Google Scholar]
- 50.Barker DJ. The origins of the developmental origins theory. J Intern Med 2007;261:412–7. [DOI] [PubMed] [Google Scholar]
- 51.Shi L, Wu J. Epigenetic regulation in mammalian preimplantation embryo development. Reprod Biol Endocrinol 2009;7:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Christophersen NS, Helin K. Epigenetic control of embryonic stem cell fate. J Exp Med 2010;207:2287–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shan J, Hamazaki T, Tang TA, Terada N, Kilberg MS. Activation of the amino acid response modulates lineage specification during differentiation of murine embryonic stem cells. Am J Physiol Endocrinol Metab 2013;305:E325–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kimball SR. Regulation of global and specific mRNA translation by amino acids. J Nutr 2002;132:883–6. [DOI] [PubMed] [Google Scholar]
- 55.Pfister S, Steiner KA, Tam PP. Gene expression pattern and progression of embryogenesis in the immediate post-implantation period of mouse development. Gene Expr Patterns 2007;7:558–73. [DOI] [PubMed] [Google Scholar]
- 56.Wek RC, Cavener DR. Translational control and the unfolded protein response. Antioxid Redox Signal 2007;9:2357–71. [DOI] [PubMed] [Google Scholar]
- 57.Dang Do AN, Kimball SR, Cavener DR, Jefferson LS. eIF2alpha kinases GCN2 and PERK modulate transcription and translation of distinct sets of mRNAs in mouse liver. Physiol Genomics 2009;38:328–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu H, Tsang KS, Wang Y, Chan JC, Xu G, Gao WQ. Unfolded protein response is required for the definitive endodermal specification of mouse embryonic stem cells via Smad2 and beta-catenin signaling. J Biol Chem 2014;289:26290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell 2000;5:897–904. [DOI] [PubMed] [Google Scholar]
- 60.Werner JC, Sicard RE. Lactate metabolism of isolated, perfused fetal, and newborn pig hearts. Pediatr Res 1987;22:552–6. [DOI] [PubMed] [Google Scholar]
- 61.Tohyama S, Hattori F, Sano M, Hishiki T, Nagahata Y, Matsuura T, Hashimoto H, Suzuki T, Yamashita H, Satoh Y, et al. Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell 2013;12:127–37. [DOI] [PubMed] [Google Scholar]
- 62.Tomizawa M, Toyama Y, Ito C, Toshimori K, Iwase K, Takiguchi M, Saisho H, Yokosuka O. Hepatoblast-like cells enriched from mouse embryonic stem cells in medium without glucose, pyruvate, arginine, and tyrosine. Cell Tissue Res 2008;333:17–27. [DOI] [PubMed] [Google Scholar]
- 63.Rachdi L, Aiello V, Duvillie B, Scharfmann R. L-leucine alters pancreatic beta-cell differentiation and function via the mTor signaling pathway. Diabetes 2012;61:409–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oburoglu L, Tardito S, Fritz V, de Barros SC, Merida P, Craveiro M, Mamede J, Cretenet G, Mongellaz C, An X, et al. Glucose and glutamine metabolism regulate human hematopoietic stem cell lineage specification. Cell Stem Cell 2014;15:169–84. [DOI] [PubMed] [Google Scholar]
- 65.Shyh-Chang N, Daley GQ, Cantley LC. Stem cell metabolism in tissue development and aging. Development 2013;140:2535–47. [DOI] [PMC free article] [PubMed] [Google Scholar]