Abstract
The consumption of amino acids by animals is controlled by both oral and postoral mechanisms. We used a genetic approach to investigate these mechanisms. Our studies have shown that inbred mouse strains differ in voluntary amino acid consumption, and these differences depend on sensory and nutritive properties of amino acids. Like humans, mice perceive some amino acids as having a sweet (sucrose-like) taste and others as having an umami (glutamate-like) taste. Mouse strain differences in the consumption of some sweet-tasting amino acids (d-phenylalanine, d-tryptophan, and l-proline) are associated with polymorphisms of a taste receptor, type 1, member 3 gene (Tas1r3), and involve differential peripheral taste responsiveness. Strain differences in the consumption of some other sweet-tasting amino acids (glycine, l-alanine, l-glutamine, and l-threonine) do not depend on Tas1r3 polymorphisms and so must be due to allelic variation in other, as yet unknown, genes involved in sweet taste. Strain differences in the consumption of l-glutamate may depend on postingestive rather than taste mechanisms. Thus, genes and physiologic mechanisms responsible for strain differences in the consumption of each amino acid depend on the nature of its taste and postingestive properties. Overall, mouse strain differences in amino acid taste and appetite have a complex genetic architecture. In addition to the Tas1r3 gene, these differences depend on other genes likely involved in determining the taste and postingestive effects of amino acids. The identification of these genes may lead to the discovery of novel mechanisms that regulate amino acid taste and appetite.
Keywords: mouse, inbred strain, behavior, consumption, intake, preference, gustatory nerves, sweet, umami
Introduction
Amino acids are essential components of organisms. They play multiple roles in their free form and as building blocks of proteins. Dietary amino acids and proteins constitute one of the macronutrients. Animals can detect macronutrients in food by using taste. For example, the carbohydrate content of food can be predicted on the basis of the sweet taste of sugars. Similarly, the protein content of food can be predicted on the basis of the taste of amino acids, which are often present in free form in protein-containing foods. Consistent with this, most amino acids have a taste, which makes some of them important as taste-active components in food. In addition to taste, postingestive mechanisms can also guide the choice and consumption of amino acids. Once ingested, amino acids and their metabolites may generate signals that affect appetite and satiety (1–6). Dietary amino acid deficiency may activate a specific hunger for the missing amino acid(s) and thereby prevent the pathological consequences of amino acid deficiency (7–11). A better understanding of the mechanisms involved in processing different amino acids by the organism may open new avenues for uses of these amino acids as flavor, nutritive, and therapeutic agents.
This review summarizes our studies of amino acid taste and appetite in mice. Genetic analyses were accompanied by physiologic experiments aimed at understanding the mechanisms responsible for genetic variation in amino acid consumption. We used amino acids that differ with respect to taste quality (e.g., sweet compared with bitter), chemical structure (e.g., d- compared with l-enantiomers), nutritional value (e.g., essential compared with nonessential), or metabolism (e.g., glucogenic compared with ketogenic) (Table 1). Humans perceive amino acids and their salts as having ≥1 basic taste qualities: sweet, umami, bitter, salty, and/or sour. Analyses of amino acid taste quality perception in laboratory animals with the use of behavioral and electrophysiologic approaches have shown that it has much in common with human taste perception. For example, if an amino acid tastes sweet to humans, rodents typically perceive it tasting similar to sucrose, show appetitive responses to it, and show activation of the same taste nerve fibers as those activated by sucrose. For brevity, we use in this review human psychophysical descriptors for taste qualities of amino acids, but it is more accurate to describe taste quality perception by nonhuman animals with the use of chemical names of taste stimuli (e.g., “sucrose-like taste" or “NaCl-like taste" for “sweet” or “salty,” respectively).
TABLE 1.
Amino acids used in the reviewed studies1
| Experiment5 |
|||||||||||
| B6 and 129 inbred mice (12–18) |
B6 × 129 F2 hybrid mice (19) |
129.B6-Tas1r3 congenic mice (20) |
|||||||||
| Amino acid | Predominant taste quality2 | Nutritional value3 | Metabolism4 | 2BT | CT | CTA | 2BT | CT | 2BT | BAT | CT |
| d-Histidine | Sweet | 8.5% | — | + | |||||||
| d-Phenylalanine | Sweet | 51.6% | — | + | + | + | + | + | + | + | |
| d-Tryptophan | Sweet | 24.7% | — | + | + | + | + | ||||
| Glycine | Sweet | Nonessential | Glucogenic | + | + | + | + | + | + | + | + |
| l-Alanine | Sweet | Nonessential | Glucogenic | + | + | + | + | + | |||
| l-Glutamate monoammonium | Umami | Nonessential | Glucogenic | + | + | ||||||
| l-Glutamate monosodium | Umami | Nonessential | Glucogenic | + | + | + | + | + | + | ||
| l-Glutamine | Sweet | Nonessential | Glucogenic | + | + | + | |||||
| l-Histidine | Bitter | Essential | Glucogenic | + | |||||||
| l-Proline | Sweet | Nonessential | Glucogenic | + | + | + | + | + | |||
| l-Serine | Sweet | Nonessential | Glucogenic | + | |||||||
| l-Threonine | Sweet | Essential | Glucogenic and ketogenic | + | + | + | |||||
| l-Tryptophan | Bitter | Essential | Glucogenic and ketogenic | + | |||||||
| l-Valine | Bitter | Essential | Glucogenic | + | |||||||
B6, C57BL/6ByJ inbred mouse strain; BAT, brief-access test; CT, chorda tympani nerve response; CTA; conditioned taste aversion; F2, hybrids of the second filial generation; Tas1r3, taste receptor, type 1, member 3 gene; 129, 129P3/J inbred mouse strain; 2BT, 2-bottle choice test.
Human sensory data are from references 21–27 and AA Bachmanov (unpublished data, 2015). Humans perceive the taste of l-glutamic acid and its salts as umami, but depending on its chemical form, l-glutamate may also have other taste components: for example, salty (for monosodium l-glutamate), bitter (for monoammonium l-glutamate), or sour (for free l-glutamic acid). Although l-histidine and l-valine are described as having a predominantly bitter taste, their bitterness is weak, and l-valine also has a weak sweet taste; consistent with the lack of strong taste to humans, mice did not strongly prefer or avoid these amino acids (Figure 1).
Nutritional values of l-amino acids are described as being essential or nonessential for mice (28). For d-amino acids, we provide nutritional values expressed relative to a corresponding l-form in mice (29). These values were calculated in 14-d growth assays with mice maintained on synthetic diets with variable amounts of l- and d-isomers of the same amino acid and constant amounts of the remaining amino acids (detailed methods are described in references 30 and 31).
Data were available only for l-amino acids and glycine.
The "+" symbols indicate that an amino acid was used in the study for the column; cells are blank for amino acids not used in the study for the column.
When amino acid–responsive taste bud cells are activated, the signal is transmitted via afferent gustatory nerves to the nucleus of the solitary tract in the brainstem; from this point, the gustatory neuraxis provides input to multiple processing centers in the brain, which evokes the taste perception and modulates ingestive behavior. In addition to taste, amino acid consumption can also be influenced by postoral and motivational factors. To reflect these aspects of processing taste information and ingestive behavior, we used multiple measures of responsiveness to amino acids: taste intensity (based on electrophysiologic recordings of activity in taste nerves), taste quality perception [based on conditioned taste aversion (CTA)12 generalization], palatability (based on initial licking responses in brief-access tests), and measures of voluntary consumption (intakes and preferences, based on long-term 2-bottle choice tests).
On the basis of our initial surveys of inbred mouse strains (32, 33), we selected 2 strains for detailed genetic and physiological analyses: C57BL/6ByJ (B6) and 129P3/J (129). In addition to exhibiting large differences in consumption of amino acids, these strains are well suited for genetic analyses because they have distant genealogy and because their genome sequence is available.
Inbred Strain Differences in Behavioral and Neural Taste Responses to Amino Acids
Two-bottle choice tests
To examine the contribution of taste and nutritive properties to the strain differences in amino acid consumption, we measured voluntary consumption of 10 amino acids in B6 and 129 mice with the use of 48-h 2-bottle tests (Figure 1). These amino acids differed with respect to taste quality, chemical structure, nutritive value, and metabolism (Table 1).
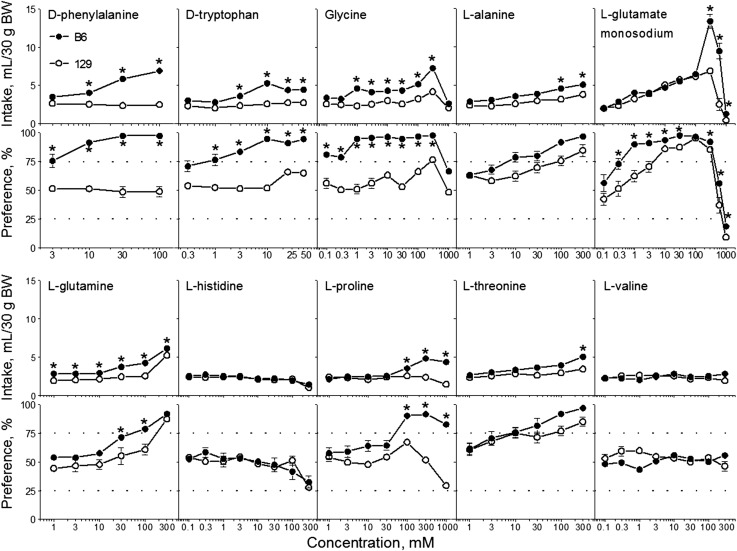
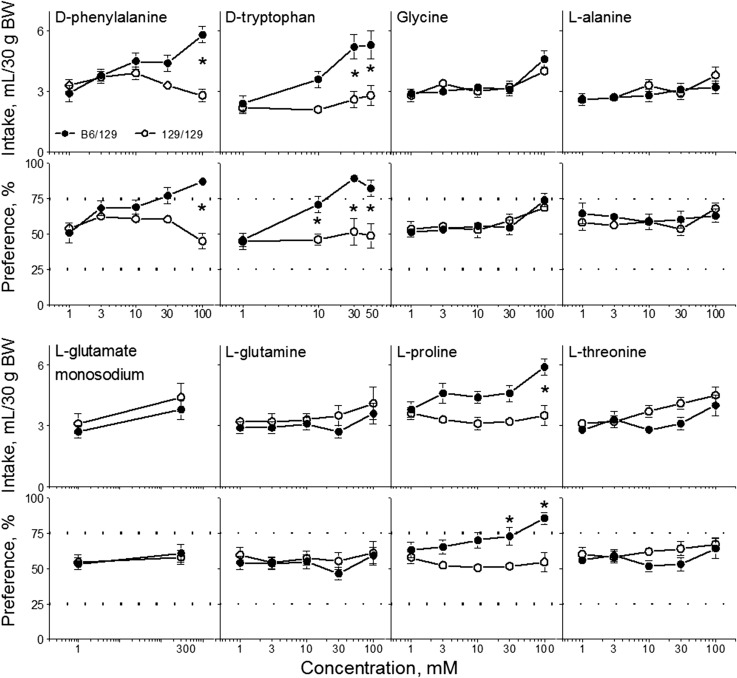
FIGURE 1.
Amino acid solution intakes (top) and preference scores (bottom) of B6 and 129 mice in 48-h 2-bottle choice tests with ascending concentrations of amino acid solutions. Values are means ± SEs, n = 7–18. The dotted horizontal lines show thresholds of preference (75%) and avoidance (25%). *Significant difference between B6 and 129 strains at a given concentration, P < 0.05 (post hoc or planned comparison tests). B6, C57BL/6ByJ inbred mouse strain; BW, body weight; 129, 129P3/J inbred mouse strain. Adapted from references 12–14 with permission.
Compared with the 129 mice, the B6 mice exhibited higher consumption of (intakes and preferences; Figure 1) and lower preference thresholds (Table 2) for sweet-tasting (d-phenylalanine, d-tryptophan, glycine, l-alanine, l-glutamine, l-proline, and l-threonine) and umami-tasting (l-glutamate) amino acids. Mice from the 2 strains did not differ in consumption of l-histidine or l-valine, which have tastes that are neither sweet nor umami.
TABLE 2.
Taste preference thresholds (in mM) of B6 and 129 inbred and 129.B6-Tas1r3 congenic mice in 48-h 2-bottle choice tests with ascending concentrations of amino acid solutions1
| Inbred mice |
129.B6-Tas1r3 congenic mice (20) |
|||
| Amino acid | B6 vs. 129 | Reference | B6 vs. 1292 | Tas1r3 effect on consumption3 |
| d-Phenylalanine | 3 < ND | (12) | 30 < ND | + |
| d-Tryptophan | 1 < ND | (12) | 30 < ND | + |
| Glycine | 0.1 < 300 | (12) | ND = ND | − |
| l-Alanine | 10 < 100 | (13) | ND = ND | − |
| l-Glutamate monosodium | 1 < 10 | (14) | ND = ND | − |
| l-Glutamine | 100 < 300 | (13) | ND = ND | − |
| l-Histidine | ND = ND | (13) | ||
| l-Proline | 100 < ND | (12) | 100 < ND | + |
| l-Threonine | 30 < 100 | (13) | ND = ND | − |
| l-Valine | ND = ND | (13) | ||
Taste preference threshold was defined as the lowest solution concentration for which animals display preference score ≥75%. Thresholds reported in the original publications (12, 14) were defined using a different criterion, and in some cases differ from threshold values shown in this table. B6, C57BL/6ByJ inbred mouse strain; ND, not determined (the highest concentration tested was below preference threshold); Tas1r3, taste receptor, type 1, member 3 gene; 129, 129P3/J inbred mouse strain; < and >, presence and direction of the difference in taste preference threshold between strains or genotypes; =, no difference in threshold; +, significant difference between mice with different Tas1r3 genotypes; −, lack of significant difference between mice with different Tas1r3 genotypes.
Tas1r3 genotypes.
Tas1r3 effect on consumption as reported in Table 3.
Both B6 and 129 mice preferred certain concentrations of glycine, l-alanine, monosodium l-glutamate, l-glutamine, and l-threonine, but preference thresholds were lower in B6 mice than in 129 mice. For glycine, preference scores of 129 mice barely reached the preference threshold (75%) and then only at a single, high concentration (300 mM; preference score: 76% ± 3%). Whereas B6 mice preferred certain concentrations of d-phenylalanine, d-tryptophan, and l-proline, 129 mice did not prefer these amino acids at any concentration tested. For l-histidine and l-valine, both B6 and 129 mice were indifferent to all concentrations tested.
These results show that the strain differences in amino acid consumption do not depend on chemical structure, nutritive value, or metabolism. This is because B6 mice had higher consumption of sweet- and umami-tasting amino acids regardless of their chiral, nutritive, or metabolic properties. These results also suggest that B6 and 129 mice do not differ in a generalized amino acid appetite, and that strain differences in amino acid consumption are restricted to only the sweet- and umami-tasting amino acids we tested. We therefore next examined peripheral taste responsiveness to, and taste quality perception of, sweet- and umami-tasting amino acids (described below in the sections entitled “Gustatory nerve electrophysiology" and “Taste quality perception").
The B6 mice consumed remarkably large amounts of 300-mM monosodium l-glutamate: for some B6 mice, daily consumption of monosodium l-glutamate exceeded half of their body weight. This consumption was higher than for any of the other amino acids tested and rivals the amounts of some highly preferred sweet substances ingested by this same strain (12). Although B6 and 129 mice differ in voluntary consumption of NaCl and sweeteners (12, 13, 34–39), the strain differences in monosodium l-glutamate consumption are independent of the strain differences in the consumption of NaCl or sweeteners, as explained below.
Monosodium l-glutamate contains sodium, which contributes a salty component to its taste and also has its own postingestive effects when consumed. Therefore, strain differences in monosodium l-glutamate consumption potentially could be influenced by differential responses to its sodium cation. However, this is unlikely because the direction of the strain differences in monosodium l-glutamate and NaCl consumption is opposite (i.e., B6 mice have lower NaCl intakes and preferences than do the 129 mice) (34–38). Thus, if anything, the sodium will diminish the B6-129 strain difference in monosodium l-glutamate consumption. The point here is that the strain differences in monosodium l-glutamate consumption are most likely due to specific effects of the l-glutamate moiety rather than to the effects of the sodium moiety present in monosodium l-glutamate.
We found that, compared with 129 mice, B6 mice consumed more monosodium l-glutamate and several sweeteners (12, 13, 39). This observation raises the possibility that the elevated consumption of monosodium l-glutamate and sweeteners by B6 mice is due to a common underlying mechanism. However, there was no correlation between monosodium l-glutamate and sweetener preferences in the hybrids of the second filial generation (F2) derived from the B6 and 129 strains (14), which shows that these 2 traits have an independent genetic determination. Thus, strain differences in the consumption of monosodium l-glutamate depend on mechanisms different from those underlying strain differences in the consumption of sweet-tasting amino acids.
Our observation of strain-specific effects of exposure to monosodium l-glutamate on its subsequent consumption (14) suggested that differences in monosodium l-glutamate consumption between B6 and 129 mice depend on postingestive effects of l-glutamate. During the 96-h 2-bottle tests of B6 and 129 mice with 300 mM monosodium l-glutamate and water, the strain differences in monosodium l-glutamate consumption were larger at the end of the test than they were in the beginning of the test. During this test, monosodium l-glutamate intakes and preferences increased in B6 mice and decreased in 129 mice. This suggests that the postingestive effects of monosodium l-glutamate are rewarding to B6 mice and aversive to 129 mice (14, 40). Similar effects of exposure to monosodium l-glutamate were observed in B6 and 129 mice in subsequent studies (41, 42). Consistent with this, B6 mice acquired flavor preferences conditioned by oral monosodium l-glutamate, whereas 129 mice did not (42). The direct evidence that glutamate can generate postingestive signals that influence appetite comes from reports of flavor preferences conditioned by intragastric infusions of monosodium l-glutamate in rats and mice (4–6, 43). This signal may involve postingestive metabolism of glutamate that differs between B6 and 129 mice (40, 44). Overall, these data support the possibility that the strain differences in glutamate consumption depend on postingestive mechanisms.
Gustatory nerve electrophysiology
In long-term 2-bottle choice tests, B6 mice consumed more sweet- and umami-tasting amino acids than did 129 mice (Figure 1). The consumption of taste solutions in long-term tests can be influenced not only by perception of their sensory attributes but also by their postingestive effects. Because some amino acids have nutritive value to mice (see Table 1), postingestive factors may contribute to the strain differences in their consumption, in addition to their taste properties. We assessed the possible role of afferent gustatory input in these strain differences by measuring integrated responses to lingual application of solutions of 9 different amino acids in the mouse chorda tympani nerve, one of the nerves that carry afferent gustatory signals (Figure 2).
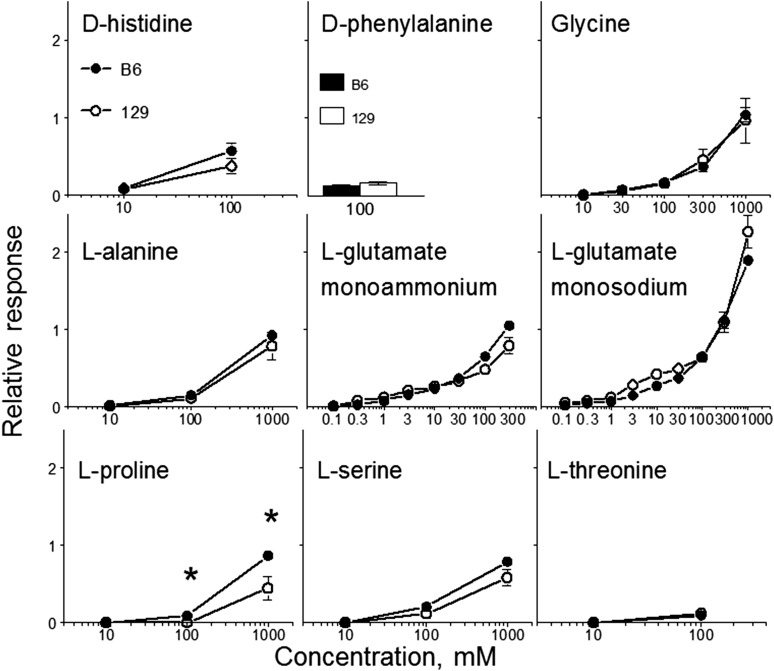
FIGURE 2.
Chorda tympani nerve responses to lingual stimulation with amino acids (relative to 100 mM NH4Cl) in B6 and 129 mice. Values are means ± SEs, n = 5–7. *Significant difference between B6 and 129 strains at a given concentration, P < 0.05 (t test). B6, C57BL/6ByJ inbred mouse strain; 129, 129P3/J inbred mouse strain. Adapted from references 15 and 16 with permission.
All amino acids tested evoked responses in the chorda tympani nerve at some concentrations. Compared with 129 mice, B6 mice had a greater response magnitude to, and lower response threshold for, l-proline. Responses to the other 8 amino acids did not differ significantly between the B6 and 129 strains. In tests with monosodium l-glutamate, possible strain differences in responsiveness to the l-glutamate moiety may be masked by differences in responsiveness to the associated sodium moiety. That is, if B6 mice have stronger responses to the l-glutamate moiety but weaker responses to the sodium moiety, the net response to monosodium l-glutamate could be similar in B6 and 129 mice. However, this was not the case because mice from both strains had similar responses to NaCl (15, 16). Thus, the lack of strain differences in responses to monosodium l-glutamate and NaCl implies that the 2 strains have similar responsiveness to the l-glutamate moiety of its sodium salt. This is further supported by similar chorda tympani responses of B6 and 129 mice to monoammonium l-glutamate.
Although B6 mice had a greater preference for d-phenylalanine, glycine, l-alanine, monosodium l-glutamate, l-proline, and l-threonine than did 129 mice (Figure 1), only l-proline evoked significantly larger whole-nerve chorda tympani responses in B6 mice. Thus, variation between the B6 and 129 strains in peripheral taste responsiveness may contribute to strain differences in the consumption of at least some sweet-tasting amino acids, exemplified by l-proline. It is possible, however, that the strains actually differ in peripheral taste responsiveness to these sweet-tasting amino acids, but these differences were not detected because of the following reasons: 1) whole-nerve rather than single-fiber recording was used, 2) the differences may reside in gustatory nerves other than the chorda tympani, or 3) low signal-to-noise ratio of responses to these amino acids could have led to a false-negative finding. The latter possibility is supported by genetic variation in chorda tympani responses to d-phenylalanine in B6 × 129 hybrids (see Table 3) and by a larger response to d-phenylalanine in the nucleus of the solitary tract of B6 mice than in 129 mice (45, 46), which shows that the strain differences in preferences for d-phenylalanine may also be related to gustatory mechanisms. Consistent with the chorda tympani results, however, B6 and 129 mice had similar responses in the nucleus of the solitary tract to glycine (45, 46). It is unlikely that strain differences in the consumption of sweet-tasting amino acids are influenced by their postingestive effects, because these amino acids differ in their nutritive properties (see Table 1). However, postingestive effects are likely involved in strain differences in the consumption of higher concentrations of monosodium l-glutamate (discussed in the previous section, “Two-bottle choice tests").
TABLE 3.
Strain differences and effects of allelic variations in the Tas1r3 gene on taste responsiveness to amino acids in mice1
| Inbred mice2 |
Tas1r3 effect3 |
||||||||
| 2BT4 |
CT5 |
B6 × 129 F2 hybrid mice (19) |
129.B6-Tas1r3 congenic mice (20) |
||||||
| Amino acid | Strain difference | Reference | Strain difference | Reference | 2BT4 | CT5 | 2BT4 | BAT6 | CT5 |
| d-Phenylalanine | + | (12) | − | (16) | + | + | + | + | − |
| d-Tryptophan | + | (12) | + | + | + | ||||
| Glycine | + | (12) | − | (16) | − | − | − | − | − |
| l-Alanine | + | (13) | − | (16) | − | − | − | ||
| l-Glutamate monosodium | + | (14) | − | (15) | − | − | − | ||
| l-Glutamine | + | (13) | − | − | |||||
| l-Proline | + | (12) | + | (16) | − | + | + | ||
| l-Threonine | + | (13) | − | (16) | − | ||||
Results for individual amino acids are generally consistent among measures and experimental populations. Minor discrepancies in effects of Tas1r3 genotype on responses to d-phenylalanine and l-proline are probably due to weak CTs to these stimuli, which lower signal-to-noise ratios and decrease statistical power to detect genetic differences. As a result, we might not have been able to detect the effect of Tas1r3 genotype on CTs to d-phenylalanine in inbred and 129.B6-Tas1r3 congenic mice and to l-proline in F2 hybrids. Blank cells indicate that there are no data for a particular test and taste stimulus. B6, C57BL/6ByJ inbred mouse strain; BAT, brief-access test; CT, chorda tympani nerve response; F2, hybrids of the second filial generation; Tas1r3, taste receptor, type 1, member 3 gene; 129, 129P3/J inbred mouse strain; 2BT, two-bottle choice tests.
The “+” and “−” symbols indicate a significant difference or lack of a significant difference between B6 and 129 strains. If there was significant strain difference, B6 mice always had higher taste responsiveness than did 129 mice.
The effect of Tas1r3 allelic variation is shown as a significant difference (+) or lack of significant difference (−) between mice with different Tas1r3 genotypes. If there was an effect of the Tas1r3 allelic variation, the B6 allele always increased taste responsiveness.
Long-term 2BTs; differences in taste solution intakes, preference scores, and/or preference thresholds.
Electrophysiologic recordings of integrated whole-nerve CTs to lingual application of taste stimuli; differences in response magnitude and/or response thresholds.
Initial licking responses in BATs; differences in standardized lick ratios.
Taste quality perception
The results of 2-bottle choice tests showed that B6 and 129 mice differed in their consumption of sweet- and umami-tasting amino acids. We examined whether these strain differences in taste preferences are due to genetic variation in taste quality perception by using the CTA generalization paradigm, a technique commonly used to assess taste quality perception in nonhuman animals (47). We used glycine and monosodium l-glutamate as representative sweet- and umami-tasting amino acids, respectively. We conditioned mice to avoid glycine or monosodium l-glutamate (conditioned stimuli) by pairing its ingestion with an injection of nausea-inducing lithium chloride (unconditioned stimulus), and then examined the suppression of licking responses in brief-access tests to prototypical taste stimuli representing the main taste qualities (sweet, salty, sour, and bitter).
Glycine.
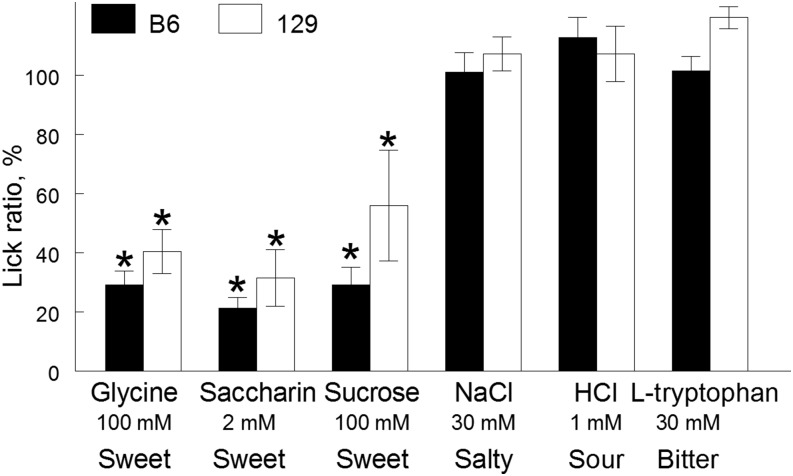
Although B6 mice strongly prefer glycine solutions at specific concentrations, 129 mice are indifferent to the same concentrations (Figure 1). We used the CTA generalization technique to examine whether this lack of robust glycine preference in 129 mice could be explained by their inability to perceive glycine sweetness. After conditioning B6 and 129 mice to avoid 100-mM glycine, both strains suppressed licking responses to sweet saccharin and sucrose but not to salty NaCl, sour hydrochloric acid, or bitter l-tryptophan (Figure 3). The fact that B6 and 129 mice both generalized the glycine CTA to sweet (saccharin and sucrose) but not other basic taste stimuli indicates that mice from these strains perceive glycine taste as predominantly sweet. This agrees with analyses of taste-evoked activity in the nucleus of the solitary tract, which showed that in both B6 and 129 mice 1) the across-neuron profile of responses to glycine was most highly correlated with that to sucrose and 2) glycine-evoked taste responses were most similar to those of other sweeteners on the basis of multidimensional scaling analyses (45).
FIGURE 3.
CTA generalization in B6 and 129 mice conditioned to avoid 100-mM glycine. Lick ratios were calculated by dividing lick numbers of individual conditioned (LiCl-treated) mice by the mean lick rate of the control (NaCl-treated) group. Values are means ± SEs, n = 7–10. *Significant decrease in lick rate in conditioned mice relative to control mice, P < 0.05 (post hoc test). B6, C57BL/6ByJ inbred mouse strain; CTA, conditioned taste aversion; LiCl, lithium chloride; 129, 129P3/J inbred mouse strain. Adapted from reference 17 with permission.
Our data are consistent with previous reports. CTA generalization between glycine and sucrose was also reported in previous studies in rodents (48–52). Perception of the sucrose-like taste of glycine by rodents is consistent with their appetitive responses to glycine ( (50, 53–56) and the perception of glycine as tasting sweet by humans (Table 1).
Thus, the lack of a robust glycine preference by 129 mice is unlikely to be explained by their inability to perceive the sweetness of glycine. Other aspects of taste perception likely underlie the strain differences in glycine preference. For example, it is possible that glycine is a more potent sweet-taste stimulus for B6 mice than for 129 mice, and that although 129 mice detect its sweet taste, the sweetness perception is not intense enough to drive behavior in 2-bottle choice tests.
l-Glutamate.
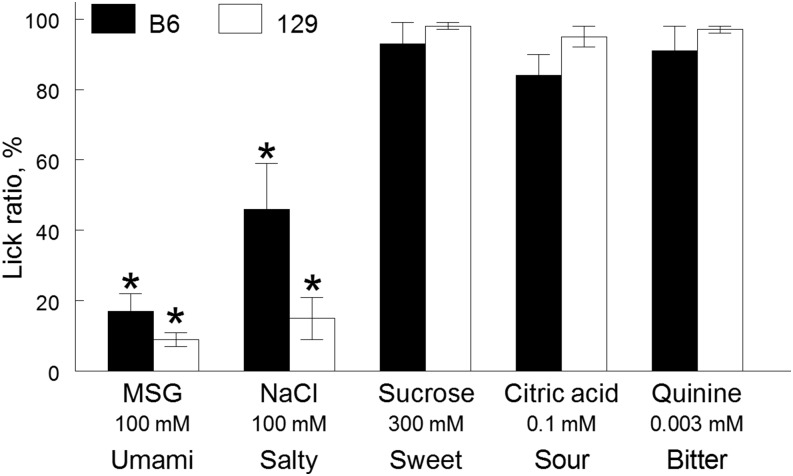
B6 mice consumed more monosodium l-glutamate than did 129 mice in 2-bottle choice tests (Figure 1). We examined whether this strain difference in intake is due to differences in the taste quality perception of monosodium l-glutamate. After conditioning mice to avoid 100-mM monosodium l-glutamate, both B6 and 129 mice suppressed licking responses to salty NaCl but not to sweet sucrose, sour citric acid, or bitter quinine (Figure 4). Therefore, both B6 and 129 mice generalized CTA from monosodium l-glutamate to salty but not to other basic taste stimuli, which indicates that mice from both strains perceive monosodium l-glutamate taste as predominantly umami and salty.
FIGURE 4.
CTA generalization in B6 and 129 mice conditioned to avoid 100-mM monosodium l-glutamate. Lick ratios were calculated by dividing lick numbers of individual conditioned (LiCl-treated) mice by the mean lick rate of the control (NaCl-treated) group. Values are means ± SEs, n = 6–10. *Significant decrease in lick rate in conditioned mice relative to control mice, P < 0.05 (post hoc test). B6, C57BL/6ByJ inbred mouse strain; CTA, conditioned taste aversion; LiCl, lithium chloride; MSG, monosodium l-glutamate; 129, 129P3/J inbred mouse strain. Adapted from reference 18 with permission.
These results show that both B6 and 129 mice perceive monosodium l-glutamate as a complex taste stimulus, which corresponds to results of other studies in humans and in other animals. These studies showed that mice perceive a unique taste of monosodium l-glutamate (57), which is probably equivalent to human perception of umami taste (Table 1). Humans and rodents also perceive a salty (NaCl-like) taste component of monosodium l-glutamate (Table 1) (21–23, 51, 57, 58), which is likely due to the presence of sodium. Consistent with this, monosodium l-glutamate and NaCl evoked similar patterns of responses in single fibers of the chorda tympani gustatory nerve in rodents (51, 59). Interestingly, rats conditioned to avoid a mixture of monosodium l-glutamate with amiloride (a blocker of the epithelial sodium channel, ENaC, which suppresses an amiloride-sensitive component of taste responses to sodium salts in rodents) generalized the CTA to sucrose (60–63). The lack of CTA generalization between monosodium l-glutamate and sucrose in our experiments may reflect the use of monosodium l-glutamate without amiloride as the conditioned stimulus in our study or species differences between mice and rats in taste quality perception of monosodium l-glutamate.
The similar patterns of monosodium l-glutamate CTA generalization in the B6 and 129 strains indicate that these 2 strains do not differ in their perception of the taste quality of monosodium l-glutamate (at least at the 100-mM concentration used as a conditioned stimulus). These CTA results are consistent with the similar gustatory nerve responses to glutamate salts in the B6 and 129 strains (Figure 2). Together, these data suggest that the strain differences in monosodium l-glutamate consumption are not due to differences in perception of the taste quality of monosodium l-glutamate or in peripheral taste responsiveness to monosodium l-glutamate.
Effect of the Polymorphisms in the taste receptor, type 1, member 3 gene (Tas1r3) Taste Receptor Gene on Behavioral and Neural Taste Responses to Amino Acids
The Tas1r3 gene encodes the taste receptor, type 1, member 3 protein (T1R3) that participates in forming both sweet [taste receptor, type 1, member 2 protein (T1R2)+T1R3] and umami [taste receptor, type 1, member 1 protein (T1R1)+T1R3] receptor dimers. In heterologous expression systems, the mouse T1R2+T1R3 receptor is activated by sweet-tasting d-amino acids, the mouse T1R1+T1R3 receptor is activated by l-amino acids, and both of these receptors are activated by the nonchiral glycine (see the section entitled “Interactions of the Proteins from the Taste Receptor, Type 1 Family (T1R) with Amino Acids").
Inbred mouse strains differ in sequences of the Tas1r3 gene. Some of these sequence variants, or polymorphisms, are associated with sweet-taste responsiveness. This genotype-phenotype association was a basis for positional cloning of the mouse saccharin preference locus (Sac) as the Tas1r3 gene (64). B6 mice carry a Tas1r3 allele that encodes a more sensitive receptor, and 129 mice carry a Tas1r3 allele for a less sensitive receptor (33, 64). These Tas1r3 polymorphisms may be responsible for strain differences in amino acid taste responses (see Figures 1 and 2). However, many other genes are also polymorphic between B6 and 129 mice, and allelic variations in these other genes may also influence strain differences in taste responses to amino acids. To assess the contribution of allelic variation in the Tas1r3 gene to strain differences in amino acid taste responses, we conducted experiments in hybrid and congenic mice.
Analyses of hybrid mice
We performed the following: 1) produced F2 hybrids between the B6 and 129 inbred mouse strains; 2) measured the consumption of amino acid solutions presented in the 2-bottle choice tests; 3) recorded integrated responses of the chorda tympani gustatory nerve to lingual application of amino acids; 4) determined the genotypes of markers on chromosome 4, where the Tas1r3 gene resides; and 5) conducted linkage analyses. For some, but not all, amino acids, we detected linkages to a distal (subtelomeric) region of chromosome 4, with linkage peaks in the vicinity of the Tas1r3 gene.
Two-bottle choice tests.
Three amino acids were tested in the 2-bottle choice tests: d-phenylalanine (30 mM), glycine (30 mM), and monosodium l-glutamate (1 and 300 mM). Significant linkages to Tas1r3 were found for d-phenylalanine but not for glycine or monosodium l-glutamate (Figure 5). Consistent with these interval mapping results, d-phenylalanine intakes and preferences were lower in mice homozygous for the 129 allele of Tas1r3 than in mice homozygous for the B6 allele of Tas1r3 or in 129/B6 Tas1r3 heterozygotes (Figure 6); this shows dominance of the B6 allele of the Tas1r3 gene over its 129 allele. F2 mice with different Tas1r3 genotypes did not differ significantly in their consumption of glycine or monosodium l-glutamate (Figure 6).
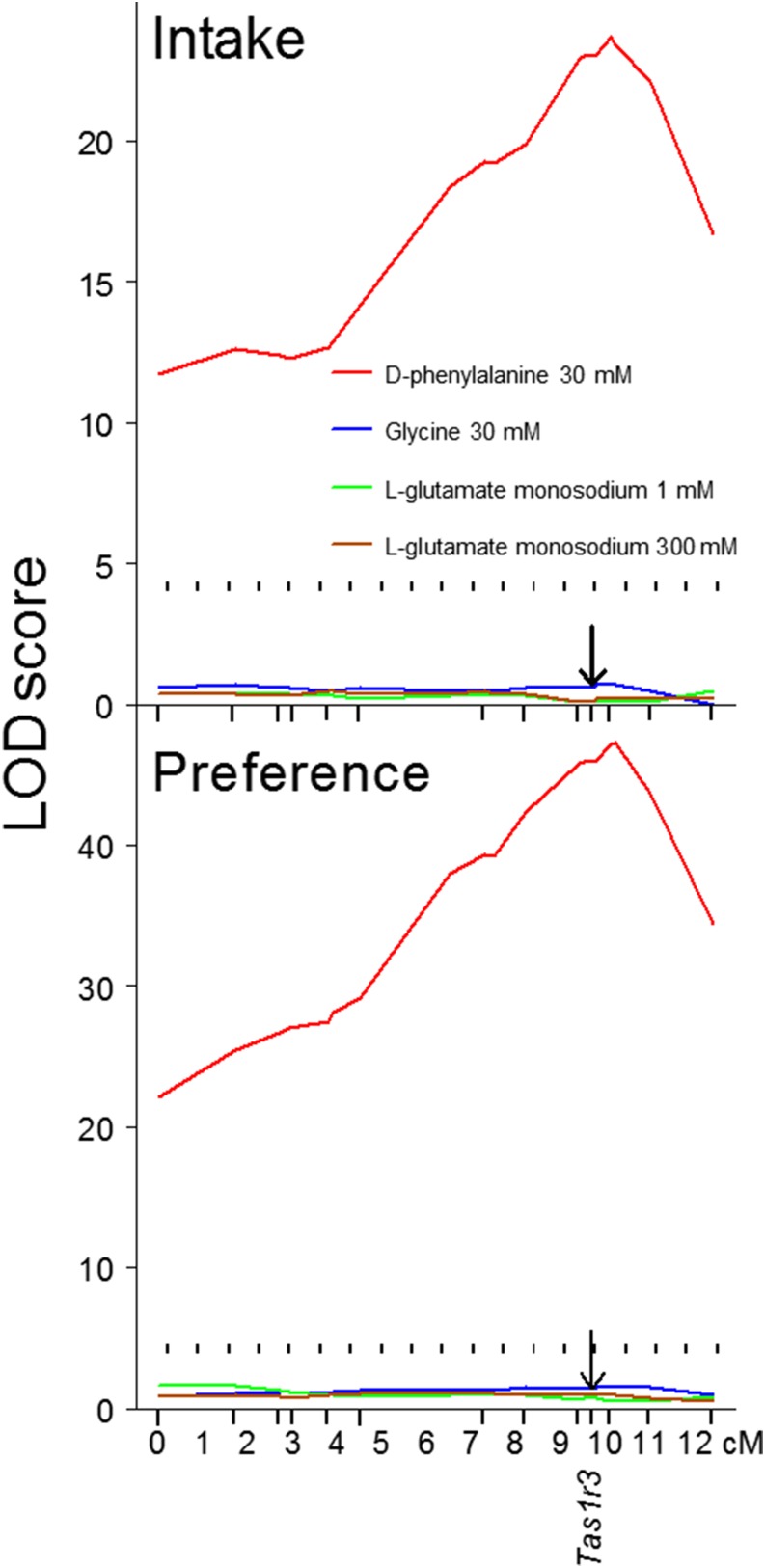
FIGURE 5.
Distal chromosome 4 interval mapping of amino acid intakes (in mL/mouse; top) and preference scores (in %; bottom) measured in 96-h 2-bottle choice tests, n = 450–455. The x axis shows distances between chromosomal markers in cM estimated by using MAPMAKER/EXP (http://www.broadinstitute.org/genome_software). Marks on the x axis show marker positions. The arrow indicates the position of the Tas1r3 gene. The curves trace the LOD scores calculated under an unconstrained (free) model by using MAPMAKER/QTL. The dotted horizontal lines show the threshold for significant (LOD: 4.3) linkage. cM, centimorgan; LOD, logarithm of the odds; Tas1r3, taste receptor, type 1, member 3 gene. Adapted from reference 19 with permission.
FIGURE 6.
Amino acid solution intakes (top) and preference scores (bottom) of F2 mice with different Tas1r3 genotypes in 96-h 2-bottle choice tests. Values are means ± SEs, n = 110–219. The dotted horizontal lines show thresholds of preference (75%) and avoidance (25%). The solid horizontal lines indicate significant differences between genotypes, P < 0.01 (planned comparison test). B6, genotype of the C57BL/6ByJ inbred mouse strain; F2, hybrids of the second filial generation; Tas1r3, taste receptor, type 1, member 3 gene; 129, genotype of the 129P3/J inbred mouse strain. Adapted from reference 19 with permission.
Gustatory nerve electrophysiology.
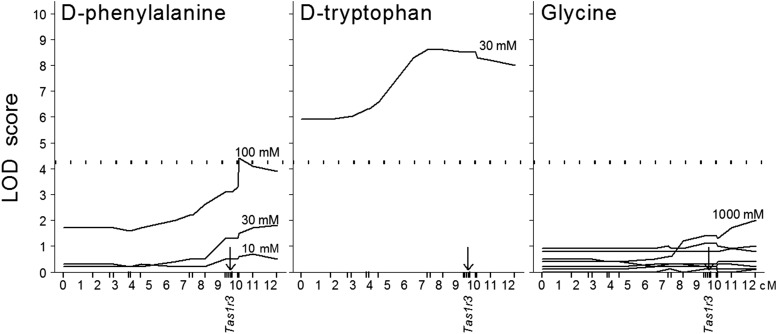
The chorda tympani nerve responses of the hybrid mice were measured for series of concentrations of 7 amino acids. Significant linkages to Tas1r3 were found for 100-mM d-phenylalanine and 30-mM d-tryptophan but not for glycine (Figure 7), l-alanine, monosodium l-glutamate, monoammonium l-glutamate, l-glutamine, or l-proline (data not shown). Consistent with these interval mapping results, chorda tympani responses to 100-mM d-phenylalanine and 30-mM d-tryptophan were lower in mice with the 129 alleles of Tas1r3 than in mice with the B6 alleles (Figure 8). F2 mice with different Tas1r3 genotypes did not differ significantly in chorda tympani responses to glycine, l-alanine, monoammonium l-glutamate, monosodium l-glutamate, l-glutamine, or l-proline (Figure 8). The results of these linkage analyses were consistent between behavioral and neural responses: both types of responses to d-phenylalanine were linked to Tas1r3, and both types of responses to glycine and monosodium l-glutamate were not linked to this region.
FIGURE 7.
Distal chromosome 4 interval mapping of chorda tympani responses to oral stimulation with amino acids, n = 42–58. Concentrations are shown next to the corresponding curves, with the exception of 1- to 300-mM glycine with no significant linkages to this chromosomal region. The x axis shows distances between chromosomal markers in cM estimated by using MAPMAKER/EXP. Marks on the x axis show marker positions. The arrows indicate the position of the Tas1r3 gene. The curves trace the LOD scores calculated under an unconstrained (free) model by using MAPMAKER/QTL. The dotted horizontal lines show the threshold for significant (LOD: 4.3) linkage. cM, centimorgan; LOD, logarithm of the odds; Tas1r3, taste receptor, type 1, member 3 gene. Adapted from reference 19 with permission.
FIGURE 8.
Chorda tympani nerve responses to lingual stimulation with amino acids (relative to 100-mM NH4Cl) in F2 mice with different Tas1r3 genotypes. Values are means ± SEs, n = 13–23. *Significant main effect of genotype at a given concentration, P < 0.01 (1-factor ANOVA). B6, genotype of the C57BL/6ByJ inbred mouse strain; F2, hybrids of the second filial generation; Tas1r3, taste receptor, type 1, member 3 gene; 129, genotype of the 129P3/J inbred mouse strain. Adapted from reference 19 with permission.
Analyses of congenic mice
To confirm and expand our finding that Tas1r3 polymorphisms affect taste responses to some but not all amino acids in B6 × 129 F2 hybrids ( ), we analyzed taste responses to amino acids in 129.B6-Tas1r3 congenic mice. We used 3 different measures: consumption in 48-h 2-bottle choice tests, initial licking responses, and responses of the chorda tympani nerve.
The 129.B6-Tas1r3 congenic strain was produced by serial backcrossing of offspring from the 129 × B6 intercross onto the 129 strain and selection of mice carrying a fragment of B6 chromosome 4 including the Tas1r3 gene (65). As a result, the congenic mice had the genetic background of the 129 strain and a small (<194 kb) donor chromosomal fragment containing the Tas1r3 gene from the B6 strain (64). We maintained 129.B6-Tas1r3 mice as a segregating congenic strain by mating congenic mice that had only one chromosome containing the B6 donor fragment (B6/129 genotype at the Tas1r3 locus) with 129 inbred mice. As a result, in each backcross generation we obtained mice with 2 different Tas1r3 genotypes: B6/129 heterozygotes and 129/129 homozygotes. Because the B6 allele of the Tas1r3 gene is dominant, B6/129 heterozygotes are phenotypically different from 129/129 homozygotes. Congenic littermates with B6/129 and 129/129 Tas1r3 genotypes were used in this study.
2-bottle choice tests.
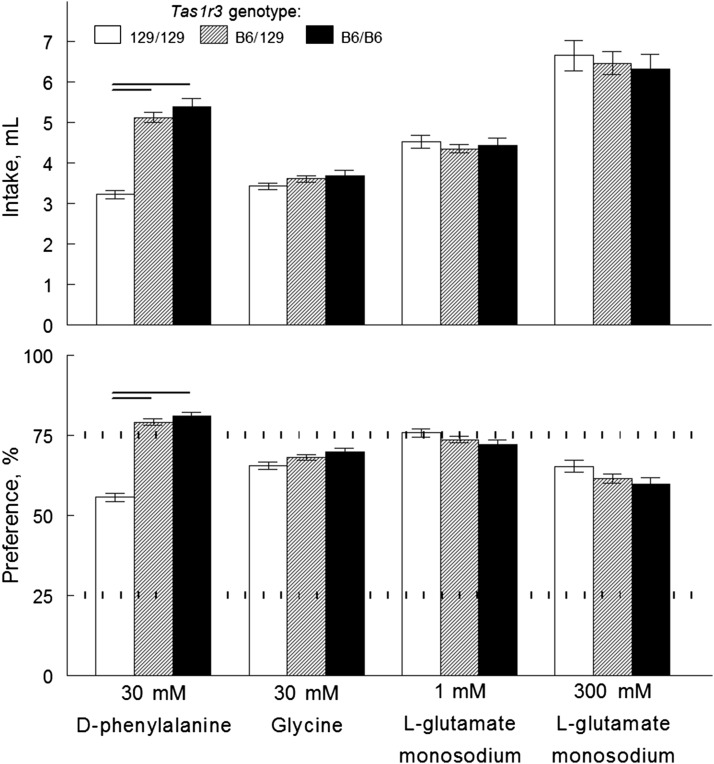
We tested concentration series of 8 amino acids. Compared with 129/129 mice, B6/129 congenic mice had higher intakes and preferences (Figure 9) and lower preference thresholds (Table 2) for d-phenylalanine, d-tryptophan, and l-proline. B6/129 and 129/129 mice had similar intakes and preferences for glycine, l-alanine, monosodium l-glutamate, l-glutamine, and l-threonine (Figure 9); preference scores for these 5 amino acids did not exceed preference thresholds in mice of both genotypes (Table 2).
FIGURE 9.
Amino acid solution intakes (top) and preference scores (bottom) of congenic mice with different Tas1r3 genotypes in 48-h 2-bottle choice tests with ascending concentrations of amino acid solutions. Values are means ± SEs, n = 10–14. The dotted horizontal lines show thresholds of preference (75%) and avoidance (25%). *Significant difference between mice with B6/129 and 129/129 Tas1r3 genotypes at a given concentration, P < 0.05 (planned comparison tests). B6, genotype of the C57BL/6ByJ inbred mouse strain; BW, body weight; Tas1r3, taste receptor, type 1, member 3 gene; 129, genotype of the 129P3/J inbred mouse strain. Adapted from reference 20 with permission.
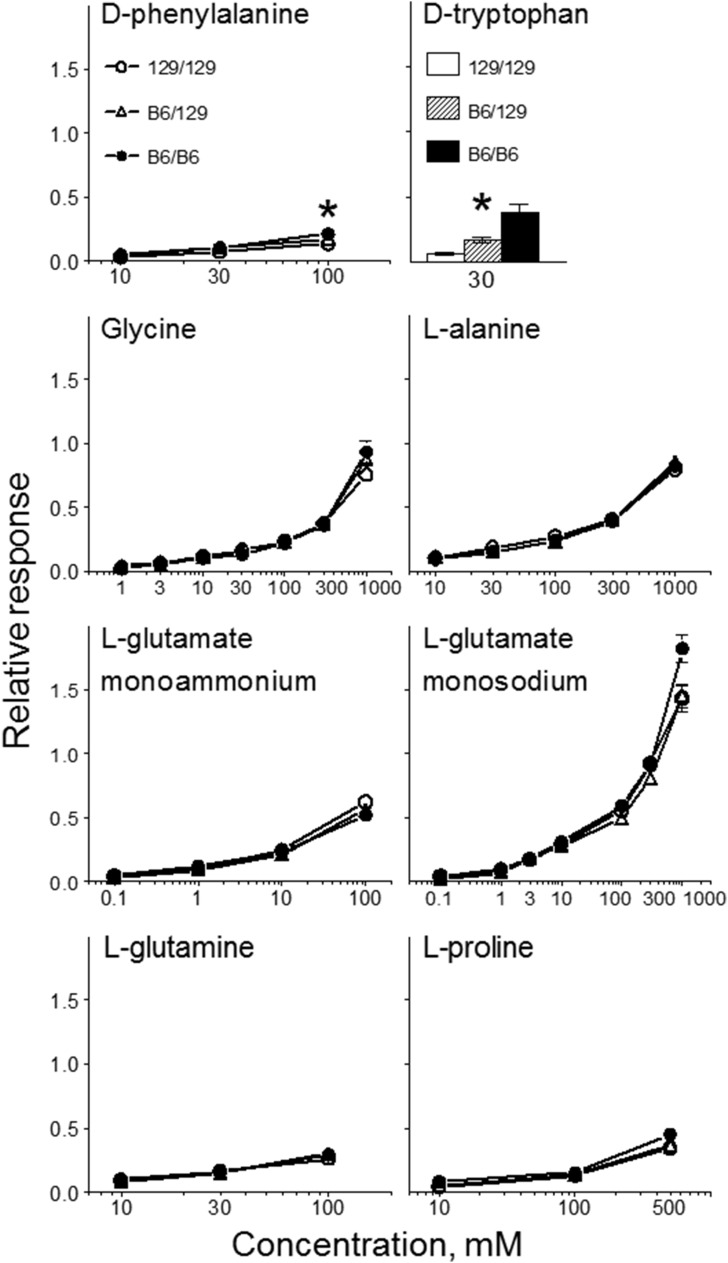
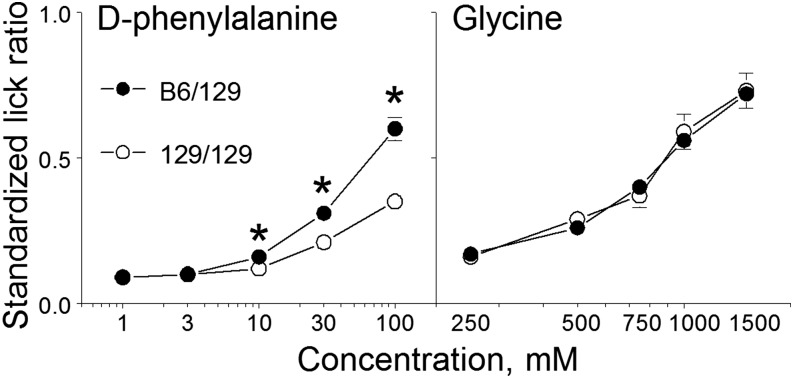
Brief-access tests.
Both B6/129 and 129/129 congenic mice showed concentration-dependent increases in licking responses to d-phenylalanine and glycine (Figure 10). B6/129 congenic mice had higher licking responses to 10–100-mM d-phenylalanine than did 129/129 mice. B6/129 and 129/129 congenic mice had similar licking responses for glycine at all concentrations tested. Thus, Tas1r3 genotype affected licking responses to d-phenylalanine but not glycine.
FIGURE 10.
Initial licking responses to a range of concentrations of amino acid solutions in brief-access tests of congenic mice with different Tas1r3 genotypes. The y axis shows the standardized lick ratios calculated by dividing the mean number of licks taken per trial by the maximum number of licks that the same mouse could potentially take across a 5-s trial. Values are means ± SEs, n = 17–20. *Significant difference between mice with B6/129 and 129/129 Tas1r3 genotypes at a given concentration, P < 0.05 (planned comparison test). B6, genotype of the C57BL/6ByJ inbred mouse strain; Tas1r3, taste receptor, type 1, member 3 gene; 129, genotype of the 129P3/J inbred mouse strain. Adapted from reference 20 with permission.
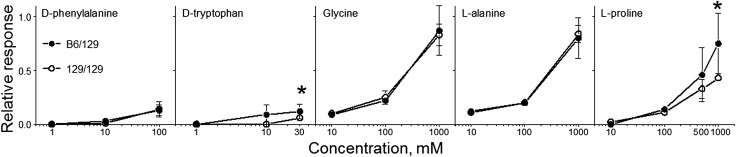
Gustatory nerve electrophysiology.
We tested a series of concentrations of 5 amino acids (Figure 11). Compared with 129/129 mice, B6/129 congenic mice had higher chorda tympani responses to d-tryptophan and l-proline. Responses to d-phenylalanine, glycine, and l-alanine were similar in B6/129 and 129/129 congenic mice.
FIGURE 11.
Chorda tympani nerve responses to lingual stimulation with amino acids (relative to 100-mM NH4Cl) in congenic mice with different Tas1r3 genotypes. Values are medians ± median absolute deviations, n = 10–14. *Significant difference between mice with B6/129 and 129/129 Tas1r3 genotypes at a given concentration, P < 0.05 (Mann-Whitney U test). B6, genotype of the C57BL/6ByJ inbred mouse strain; Tas1r3, taste receptor, type 1, member 3 gene; 129, genotype of the 129P3/J inbred mouse strain. Adapted from reference 20 with permission.
To summarize, taste responses of 129.B6-Tas1r3 congenic mice to amino acids were analyzed by using 3 different measures: responses of the chorda tympani nerve, initial licking responses, and sweetener consumption in the 48-h 2-bottle choice tests. The results were generally consistent across the 3 measures of taste responsiveness (summarized in Tables 2 and 3). That is, the Tas1r3 genotype influenced responses to d-phenylalanine, d-tryptophan, and l-proline. The B6 allele of the Tas1r3 gene was associated with higher sweet-taste responsiveness and sensitivity. The Tas1r3 genotype did not affect responses to glycine, l-alanine, monosodium l-glutamate, l-glutamine, or l-threonine. This pattern of results fits a model in which changes in properties of the T1R3 taste receptor protein affect afferent activity in sensory gustatory nerves evoked by some amino acids, which, in turn, influences consumption behavior toward these amino acids. The results are also generally consistent between experiments with hybrid and congenic mice (Table 3) and show that allelic variation of the Tas1r3 gene affects taste responses to some but not all amino acids.
Interactions of the Proteins from the Taste Receptor, Type 1 Family (T1R) with Amino Acids
Traditionally, T1R receptor-ligand interactions have been characterized in vitro by using heterologous expression experiments in which responses to taste stimuli in cells transfected with T1Rs were analyzed. Our data show that the in vivo approach can also be used to understand the function and specificity of taste receptors and to validate the findings of in vitro studies (66). The in vivo experiments examined the effects of the alleles of genes from the taste receptor, type 1 family (Tas1r) on taste responses in mice. Two types of gene variation were studied in vivo: targeted mutations disrupting a gene (67–69) and natural allelic variation (19, 20) (Table 4). The premise of analyses of receptor-ligand interactions based on the in vivo data is that if a taste response (either behavioral or neural) to an amino acid is affected by the Tas1r genotype, then that amino acid must interact with a taste receptor involving T1R.
TABLE 4.
Amino acid ligands of the T1R receptors1
| In vitro |
In vivo |
|||||||||||||||||||||
| T1R1+T1R3 |
T1R2+T1R3 |
Tas1r1 KO |
Tas1r2 KO |
Tas1r3 KO |
Tas1r3 polymorphisms2 |
|||||||||||||||||
| Human |
Mouse |
CT |
Behavior3 |
CT |
||||||||||||||||||
| Amino acids | Human (70) | Rat (70) | Mouse (71) | (70) | (72–75) | (76) | Rat (70) | (71) | (72–74) | Behavior3 (68) | (68) | (69) | GL (69) | Behavior3 (68) | CT (68) | (68) | (67) | (68) | (67) | GL (67) | Behavior3 (19, 20) | CT (19, 20) |
| d-Alanine | +* | – | + | − | − | + | + | |||||||||||||||
| d-Asparagine | −** | + | + | + | ||||||||||||||||||
| d-Aspartate | −** | |||||||||||||||||||||
| d-Glutamate | −** | −** | ||||||||||||||||||||
| d-Glutamine | −** | + | ||||||||||||||||||||
| d-Histidine | −** | + | + | |||||||||||||||||||
| d-Isoleucine | + | |||||||||||||||||||||
| d-Leucine | + | |||||||||||||||||||||
| d-Phenylalanine | −** | + | + | − | − | + | + | + | + | + | +/− | |||||||||||
| d-Threonine | − | |||||||||||||||||||||
| d-Tryptophan | −** | −** | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + | |||||
| d-Valine | + | |||||||||||||||||||||
| Glycine | −** | + | + | + | + | + | + | − | − | − | ||||||||||||
| l-Alanine | + | − | − | + | +* | + | − | + | +* | − | − | |||||||||||
| l-Arginine | + | +* | +* | |||||||||||||||||||
| l-Asparagine | + | − | +* | +* | ||||||||||||||||||
| l-Aspartate | +* | +* | +* | +* | +* | |||||||||||||||||
| l-Cysteine | + | |||||||||||||||||||||
| l-Glutamate | + | + | +* | − | − | + | +* | + | − | + | + | +* | + | −** | − | − | ||||||
| l-Glutamine | −** | + | − | − | ||||||||||||||||||
| l-Histidine | −** | + | − | |||||||||||||||||||
| l-Isoleucine | +* | − | ||||||||||||||||||||
| l-Leucine | −** | +* | − | |||||||||||||||||||
| l-Lysine | −** | +* | ||||||||||||||||||||
| l-Methionine | + | |||||||||||||||||||||
| l-Phenylalanine | +* | − | − | |||||||||||||||||||
| l-Proline | −** | +* | − | − | − | + | +/− | |||||||||||||||
| l-Serine | −** | + | − | + | +* | + | +* | |||||||||||||||
| l-Threonine | + | − | − | − | ||||||||||||||||||
| l-Tryptophan | −** | − | − | − | ||||||||||||||||||
| l-Tyrosine | −** | |||||||||||||||||||||
| l-Valine | + | − | ||||||||||||||||||||
References are shown in parentheses. "+" Under “In vitro” indicates a response to a taste stimulus and under “In vivo” indicates an effect on response to a taste stimulus; "−" under “In vitro” indicates a lack of response to a taste stimulus and under “In vivo” indicates a lack of effect on response to a taste stimulus; “*” under “In vitro” indicates a response only when the taste stimulus is applied with IMP but no response without IMP and under “In vivo” indicates an effect on a response to a taste stimulus mixed with IMP. **No response with or without IMP. B6, C57BL/6ByJ inbred mouse strain; CT, chorda tympani nerve response; F2, hybrids of the second filial generation; GL, glossopharyngeal nerve response; IMP, inosine-5′-monophosphate; KO, knockout; T1R1, taste receptor, type 1, member 1 protein; T1R2, taste receptor, type 1, member 2 protein; T1R3, taste receptor, type 1, member 3 protein; Tas1r1, taste receptor, type 1, member 1 gene; Tas1r2, taste receptor, type 1, member 2 gene; Tas1r3, taste receptor, type 1, member 3 gene; 129, 129P3/J inbred mouse strain.
Effect of Tas1r3 polymorphisms in B6 × 129 F2 hybrid and 129.B6-Tas1r3 congenic mice (see Table 3). The most likely functional Tas1r3 polymorphism encodes an amino acid substitution of isoleucine to threonine at position 60 (33, 77).
Behavioral responses in 2-bottle choice tests and/or brief-access tests.
When T1R3 is co-expressed in a heterologous system with T1R2, it functions as a broad-spectrum sweet-taste receptor and is activated by sweet-tasting d-amino acids and glycine (Table 4). In contrast, a heterodimer of T1R1 and T1R3 proteins functions as an umami taste receptor in humans and is more broadly tuned in rodents to respond to l-amino acids and glycine. Thus, the T1R3 protein is involved in transduction of both sweet and umami tastes, and correspondingly, a disruption of the Tas1r3 gene in knockout mice diminishes taste responses to both sweet- and umami-taste stimuli (67, 68). Consistent with the in vitro results, taste receptor, type 1, member 1 gene (Tas1r1) knockout mice have impaired taste responses to l-amino acids and umami stimuli, and taste receptor, type 1, member 2 gene (Tas1r2) knockout mice have impaired taste responses to sweeteners (including sweet-tasting d-amino acids) (68). A more recent study with a different strain of Tas1r1 knockout mice confirmed a reduction in responses to umami-tasting compounds in the chorda tympani nerve (69). Surprisingly, this study also reported that Tas1r1 knockout mice have reduced chorda tympani nerve responses to sweeteners; the mechanisms responsible for this effect are not clear (69).
The effects of naturally occurring Tas1r3 polymorphisms in inbred mouse strains (19, 20) in some, but not all, cases were similar to the effects of targeted mutations of the Tas1r3 gene. Tas1r3 genotype influenced taste responses to d-phenylalanine, d-tryptophan, and l-proline, but not to glycine, l-alanine, monosodium l-glutamate, l-glutamine, or l-threonine (Tables 3 and 4). For d-phenylalanine and d-tryptophan, effects of the Tas1r3 allelic variation and gene knockout were consistent: in both cases, Tas1r3 genotype influenced taste responses to these amino acids. However, the effects of Tas1r3 genotype on taste responses to l-alanine and l-glutamate were discrepant: whereas targeted mutations of Tas1r3 affected responses to these amino acids, Tas1r3 polymorphisms did not affect the responses to them. Taste responses to glycine, l-glutamine, l-proline, and l-threonine were not tested in Tas1r3 knockout mice; however, because these amino acids activate T1R1+T1R3 and/or T1R2+T1R3 in vitro, it is likely that Tas1r3 knockout mice are deficient in taste responsiveness to them.
Why are responses to some amino acids not affected by Tas1r3 polymorphisms, despite the fact that these amino acids interact with taste receptor(s) involving T1R3? This could be explained by the fact that these amino acids bind to the following: 1) the T1R3 protein at a site that is not affected by the Tas1r3 polymorphisms, 2) a partner protein (T1R1 or T1R2) of the heterodimeric T1R receptor, or 3) an additional, non-T1R, taste receptor. Several studies suggested that the taste of l-glutamate is transduced not only by the T1R-dependent mechanism but also by alternative transduction mechanisms (69, 78–83). Similarly, T1R-independent mechanisms may be involved in taste transduction of the other amino acids, for which taste responses are not affected by Tas1r3 allelic variation. This is consistent with residual sweet-taste responsiveness in Tas1r3 knockout mice (67), which also indicates the presence of T1R-independent taste transduction mechanisms. Such mechanisms may involve glucose transporters and metabolic sensors implicated in sugar taste (84) and an ability of amphiphilic sweeteners to permeate cell membrane and directly interact with intracellular targets (85–87).
Our data show that the in vivo approach can be used to understand the function and specificity of taste receptors and to validate the findings of in vitro studies. Experiments involving the expression of taste receptors in heterologous systems require substantial modification of the conditions that exist in vivo. This includes, for example, modification of receptors to traffic them to the cell membrane, use of variable components of intracellular transduction, and an absence of regulatory influences existing in vivo. Thus, the in vivo and in vitro approaches complement each other by revealing functional characteristics of taste receptors.
Genetic Architecture of Amino Acid Taste and Appetite: Complex Genetics and Multiple Underlying Physiologic Mechanisms
We have shown that B6 and 129 mice differ in behavioral and neural taste responsiveness to several amino acids (Figures 1 and 2, Table 2; summarized in Table 3). The strain differences in responses to d-phenylalanine, d-tryptophan, and l-proline depend on Tas1r3 polymorphisms. However, Tas1r3 polymorphisms do not affect responses to glycine, l-alanine, l-glutamate, l-glutamine, or l-threonine. Therefore, strain differences in responses to these other amino acids must depend on genes other than Tas1r3. What are these other genes and what are the physiologic mechanisms that mediate their effects on amino acid taste and appetite?
It is unlikely that the other 2 members of the Tas1r gene family, Tas1r1 and Tas1r2, are responsible for Tas1r3-independent variation in responses to amino acids. All 3 members of the Tas1r gene family cluster in distal chromosome 4 (64, 65, 88). The lack of linkages to this region for taste responses to some amino acids (see Figures 5 and 7) shows that none of the known T1R receptors has allelic variants associated with this strain variation.
One of the genetic loci affecting sweet-taste responses is d-phenylalanine aversion locus (dpa). This locus affects the ability of mice to generalize a CTA between d-phenylalanine and sucrose, which indicates that dpa affects perception of the sweetness of d-phenylalanine. The dpa locus also affects responses of sucrose-sensitive fibers of the chorda tympani nerve to d-phenylalanine. The dpa locus was mapped to proximal chromosome 4, a region distinct from the subtelomeric chromosome 4 harboring the Tas1r genes (89–92). However, it is unlikely that the dpa locus mediates differences in taste responses to amino acids between the B6 and 129 strains because we did not detect linkages to the dpa chromosomal region in B6 × 129 hybrids (data not shown).
There is evidence that Tas1r3-independent strain differences in voluntary consumption of sweet-tasting (glycine, l-alanine, l-glutamine, and l-threonine) and umami-tasting (l-glutamate) amino acids depend on gustatory and postingestive mechanisms, respectively. The fact that the 4 sweet-tasting amino acids all elicit sweet taste but differ in their nutritive properties (see Table 1) suggests that Tas1r3-independent genetic variation in sweet taste is a more likely mechanism underlying strain differences than postingestive effects of these amino acids [e.g., see (93)].
In contrast, the difference between B6 and 129 mice in their consumption of concentrated monosodium l-glutamate (Figure 1) is likely mediated by genetic variation in the postingestive effects of l-glutamate. Indeed, B6 and 129 mice have similar peripheral taste responses to l-glutamate salts (Figure 2) and similar perception of the taste quality of monosodium l-glutamate (Figure 4). Together, these findings suggest that gustatory mechanisms are not involved in differential intake of monosodium l-glutamate. In contrast, several lines of evidence indicate that strain-specific postingestive effects of glutamate can affect its intake (14, 40, 42, 44).
Conclusions
We used the mouse as a model organism to study genetics of amino acid taste and appetite. These studies show that the voluntary consumption of amino acids is a trait with complex genetics and multiple underlying physiologic mechanisms. Appetitive responses to some amino acids seem to be determined primarily by their sweet (sucrose-like) taste and are influenced by genetic variation in peripheral taste responsiveness. For some of these sweet-tasting amino acids, genetic differences in taste responsiveness depend on Tas1r3 polymorphisms; others are Tas1r3-independent. Appetitive responses to l-glutamate depend on genetic variation in its postoral rewarding properties.
Because taste responses to some sweet-tasting amino acids are affected by the Tas1r3 genotype, these amino acids must interact with a taste receptor involving T1R3. These data show the in vivo approach to characterize ligand specificity of the T1R3 taste receptor, which validates the findings of in vitro studies and shows that both the in vivo and in vitro approaches complement each other in the characterization of taste receptors.
Our results indicate that Tas1r3 is not the only gene underlying strain differences in taste responsiveness to sweet amino acids. The identification of these as yet unknown genes may lead to the discovery of new mechanisms of sweet-taste transduction.
Specific appetites—for example, sodium appetite—are known to be either need-induced or need-free (94, 95). There is evidence that amino acid appetite can also be need-induced or need-free. An example of the need-induced amino acid appetite is a response to deficiency of particular amino acids, which involves learning to reject a deficient diet and to choose a more adequate diet (8). Our studies show that voluntary l-glutamate consumption is an example of a need-free specialized amino acid appetite exhibited by replete animals. Genes responsible for this variation may unveil new mechanisms that regulate appetite and satiety.
Acknowledgments
All authors read and approved the final manuscript.
Footnotes
Abbreviations used: B6, C57BL/6ByJ inbred mouse strain; CTA, conditioned taste aversion; dpa, d-phenylalanine aversion locus; F2, hybrids of the second filial generation; LiCl, lithium chloride; Sac, saccharin preference locus; T1R, proteins from the taste receptor, type 1 family; T1R1, taste receptor, type 1, member 1 protein; T1R2, taste receptor, type 1, member 2 protein; T1R3, taste receptor, type 1, member 3 protein; Tas1r, genes from the taste receptor, type 1 family; Tas1r1, taste receptor, type 1, member 1 gene; Tas1r2, taste receptor, type 1, member 2 gene; Tas1r3, taste receptor, type 1, member 3 gene; 129, 129P3/J inbred mouse strain.
References
- 1.Sclafani A. Macronutrient-conditioned flavor preferences. In: Berthoud HR, Seeley RJ, editors. Neural and metabolic control of macronutrient intake. Boca Raton (FL): CRC Press; 1999. p. 93–107. [Google Scholar]
- 2.Fromentin G, Darcel N, Chaumontet C, Marsset-Baglieri A, Nadkarni N, Tome D. Peripheral and central mechanisms involved in the control of food intake by dietary amino acids and proteins. Nutr Res Rev 2012;25:29–39. [DOI] [PubMed] [Google Scholar]
- 3.Ackroff K, Kondoh T, Sclafani A. Dried bonito dashi: a preferred fish broth without postoral reward actions in mice. Chem Senses 2014;39:159–66. [DOI] [PubMed] [Google Scholar]
- 4.Ackroff K, Sclafani A. Flavor preferences conditioned by intragastric monosodium glutamate in mice. Chem Senses 2013;38:759–67. [DOI] [PubMed] [Google Scholar]
- 5.Uematsu A, Tsurugizawa T, Kondoh T, Torii K. Conditioned flavor preference learning by intragastric administration of L-glutamate in rats. Neurosci Lett 2009;451:190–3. [DOI] [PubMed] [Google Scholar]
- 6.Ackroff K, Sclafani A. Flavor preferences conditioned by post-oral infusion of monosodium glutamate in rats. Physiol Behav 2011;104:488–94. [DOI] [PubMed] [Google Scholar]
- 7.Torii K, Kondoh T, Mori M, Ono T. Hypothalamic control of amino acid appetite. Ann N Y Acad Sci 1998;855:417–25. [DOI] [PubMed] [Google Scholar]
- 8.Gietzen DW, Hao S, Anthony TG. Mechanisms of food intake repression in indispensable amino acid deficiency. Annu Rev Nutr 2007;27:63–78. [DOI] [PubMed] [Google Scholar]
- 9.Harper AE. Protein and amino acids in the regulation of food intake. In: Novin D, Wyrwicka W, Bray G, editors. Hunger: basic mechanisms and clinical implications. New York: Raven Press; 1976. p. 103–13. [Google Scholar]
- 10.Markison S, Gietzen DW, Spector AC. Essential amino acid deficiency enhances long-term intake but not short-term licking of the required nutrient. J Nutr 1999;129:1604–12. [DOI] [PubMed] [Google Scholar]
- 11.Markison S, Thompson BL, Smith JC, Spector AC. Time course and pattern of compensatory ingestive behavioral adjustments to lysine deficiency in rats. J Nutr 2000;130:1320–8. [DOI] [PubMed] [Google Scholar]
- 12.Bachmanov AA, Tordoff MG, Beauchamp GK. Sweetener preference of C57BL/6ByJ and 129P3/J mice. Chem Senses 2001;26:905–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bachmanov AA, Beauchamp GK. Amino acid and carbohydrate preferences in C57BL/6ByJ and 129P3/J mice. Physiol Behav 2008;93:37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bachmanov AA, Tordoff MG, Beauchamp GK. Intake of umami-tasting solutions by mice: a genetic analysis. J Nutr 2000;130(4S Suppl):935S–41S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inoue M, Beauchamp GK, Bachmanov AA. Gustatory neural responses to umami taste stimuli in C57BL/6ByJ and 129P3/J mice. Chem Senses 2004;29:789–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoue M, McCaughey SA, Bachmanov AA, Beauchamp GK. Whole nerve chorda tympani responses to sweeteners in C57BL/6ByJ and 129P3/J mice. Chem Senses 2001;26:915–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manita S, Bachmanov AA, Li X, Beauchamp GK, Inoue M. Is glycine “sweet” to mice? Mouse strain differences in perception of glycine taste. Chem Senses 2006;31:785–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murata Y, Beauchamp GK, Bachmanov AA. Taste perception of monosodium glutamate and inosine monophosphate by 129P3/J and C57BL/6ByJ mice. Physiol Behav 2009;98:481–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inoue M, Reed DR, Li X, Tordoff MG, Beauchamp GK, Bachmanov AA. Allelic variation of the Tas1r3 taste receptor gene selectively affects behavioral and neural taste responses to sweeteners in the F2 hybrids between C57BL/6ByJ and 129P3/J mice. J Neurosci 2004;24:2296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inoue M, Glendinning JI, Theodorides ML, Harkness S, Li X, Bosak N, Beauchamp GK, Bachmanov AA. Allelic variation of the Tas1r3 taste receptor gene selectively affects taste responses to sweeteners: evidence from 129.B6-Tas1r3 congenic mice. Physiol Genomics 2007;32:82–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ninomiya T, Ikeda S, Yamaguchi S, Yoshikawa T [Tastes of various amino acids. ] Stat Quality Control 1966;17:69–73. (in Japanese). [Google Scholar]
- 22.Yoshida M, Saito S. Multidimensional scaling of the taste of amino acids. Jpn Psychol Res 1969;11:149–66. [Google Scholar]
- 23.Bartoshuk LM, Cain WS, Cleveland CT, Grossman LS, Marks LE, Stevens JC, Stolwijk JA. Saltiness of monosodium glutamate and sodium intake. JAMA 1974;230:670. [DOI] [PubMed] [Google Scholar]
- 24.Kawai M, Sekine-Hayakawa Y, Okiyama A, Ninomiya Y. Gustatory sensation of (L)- and (D)-amino acids in humans. Amino Acids 2012;43:2349–58. [DOI] [PubMed] [Google Scholar]
- 25.Solms J, Vuataz L, Egli RH. The taste of L- and D-amino acids. Experientia 1965;21:692–4. [DOI] [PubMed] [Google Scholar]
- 26.Schiffman SS, Sennewald K, Gagnon J. Comparison of taste qualities and thresholds of D- and L-amino acids. Physiol Behav 1981;27:51–9. [DOI] [PubMed] [Google Scholar]
- 27.Shallenberger RS. Taste chemistry. London: Blackie Academic and Professional; 1993. [Google Scholar]
- 28.Tobin G, Stevens KA, Russell RJ. Nutrition. In: Fox JG, Davisson MT, Quimby FW, Barthold SW, Newcomer CE, Smith AL, editors. The mouse in biomedical research. Volume III: Normative Biology, Husbandry, and Models. 2nd ed. New York: Academic Press; 2007. p. 321–83. [Google Scholar]
- 29.Friedman M. Origin, microbiology, nutrition, and pharmacology of D-amino acids. Chem Biodivers 2010;7:1491–530. [DOI] [PubMed] [Google Scholar]
- 30.Friedman M, Levin CE. Nutritional value of D-amino acids, D-peptides, and amino acid derivatives in mice. Methods Mol Biol 2012;794:337–53. [DOI] [PubMed] [Google Scholar]
- 31.Friedman M, Levin CE. Nutritional and medicinal aspects of D-amino acids. Amino Acids 2012;42:1553–82. [DOI] [PubMed] [Google Scholar]
- 32.Bachmanov AA, Li S, Li X, Lu K, Tordoff MG, West DB, Ohmen JD, Reed DR, Beauchamp GK. Polymorphisms of the mouse Tas1r3 gene are related to sweetener preferences in 30 strains of mice. Chem Senses 2002;27:A95–6 (abstr). [Google Scholar]
- 33.Reed DR, Li S, Li X, Huang L, Tordoff MG, Starling-Roney R, Taniguchi K, West DB, Ohmen JD, Beauchamp GK, et al. Polymorphisms in the taste receptor gene (Tas1r3) region are associated with saccharin preference in 30 mouse strains. J Neurosci 2004;24:938–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bachmanov AA, Tordoff MG, Beauchamp GK. Voluntary sodium chloride consumption by mice: differences among five inbred strains. Behav Genet 1998;28:117–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beauchamp GK, Fisher AS. Strain differences in consumption of saline solutions by mice. Physiol Behav 1993;54:179–84. [DOI] [PubMed] [Google Scholar]
- 36.Gannon KS, Contreras RJ. Sodium intake linked to amiloride-sensitive gustatory transduction in C57BL/6J and 129/J mice. Physiol Behav 1995;57:231–9. [DOI] [PubMed] [Google Scholar]
- 37.Lush IE. The genetics of bitterness, sweetness, and saltiness in strains of mice. In: Wysocki CJ, Kare MR, editors. Genetics of perception and communication. New York: Marcel Dekker; 1991. p. 227–41. [Google Scholar]
- 38.Bachmanov AA, Beauchamp GK, Tordoff MG. Voluntary consumption of NaCl, KCl, CaCl2, and NH4Cl solutions by 28 mouse strains. Behav Genet 2002;32:445–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bachmanov AA, Tordoff MG, Beauchamp GK. Ethanol consumption and taste preferences in C57BL/6ByJ and 129/J mice. Alcohol Clin Exp Res 1996;20:201–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bachmanov AA, Inoue M, Ji H, Murata Y, Tordoff MG, Beauchamp GK. Glutamate taste and appetite in laboratory mice: physiologic and genetic analyses. Am J Clin Nutr 2009;90(Suppl):756S–63S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ackroff K, Weintraub R, Sclafani A. MSG intake and preference in mice are influenced by prior testing experience. Physiol Behav 2012;107:207–17. [DOI] [PubMed] [Google Scholar]
- 42.Ackroff K, Sclafani A. Flavor preferences conditioned by oral monosodium glutamate in mice. Chem Senses 2013;38:745–58. [DOI] [PubMed] [Google Scholar]
- 43.Uematsu A, Tsurugizawa T, Uneyama H, Torii K. Brain-gut communication via vagus nerve modulates conditioned flavor preference. Eur J Neurosci 2010;31:1136–43. [DOI] [PubMed] [Google Scholar]
- 44.Ji H, Bachmanov AA. Differences in postingestive metabolism of glutamate and glycine between C57BL/6ByJ and 129P3/J mice. Physiol Genomics 2007;31:475–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCaughey SA. Taste-evoked responses to sweeteners in the nucleus of the solitary tract differ between C57BL/6ByJ and 129P3/J mice. J Neurosci 2007;27:35–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCaughey SA, Glendinning JI. Experience with sugar modifies behavioral but not taste-evoked medullary responses to sweeteners in mice. Chem Senses 2013;38:793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spector AC. Psychophysical evaluation of taste function in nonhuman mammals. In: Doty RL, editor. Handbook of olfaction and gustation. New York: Marcel Dekker; 2003. p. 861–79. [Google Scholar]
- 48.Nowlis GH, Frank ME, Pfaffmann C. Specificity of acquired aversions to taste qualities in hamsters and rats. J Comp Physiol Psychol 1980;94:932–42. [DOI] [PubMed] [Google Scholar]
- 49.Pritchard TC, Scott TR. Amino acids as taste stimuli. II. Quality coding. Brain Res 1982;253:93–104. [DOI] [PubMed] [Google Scholar]
- 50.Kasahara T, Iwasaki K, Sato M. Taste effectiveness of some D- and L-amino acids in mice. Physiol Behav 1987;39:619–24. [DOI] [PubMed] [Google Scholar]
- 51.Yamamoto T, Matsuo R, Kiyomitsu Y, Kitamura R. Taste effects of “umami” substances in hamsters as studied by electrophysiological and conditioned taste aversion techniques. Brain Res 1988;451:147–62. [DOI] [PubMed] [Google Scholar]
- 52.Danilova V, Hellekant G, Tinti JM, Nofre C. Gustatory responses of the hamster Mesocricetus auratus to various compounds considered sweet by humans. J Neurophysiol 1998;80:2102–12. [DOI] [PubMed] [Google Scholar]
- 53.Iwasaki K, Kasahara T, Sato M. Gustatory effectiveness of amino acids in mice: behavioral and neurophysiological studies. Physiol Behav 1985;34:531–42. [DOI] [PubMed] [Google Scholar]
- 54.Lush IE, Hornigold N, King P, Stoye JP. The genetics of tasting in mice. VII. Glycine revisited, and the chromosomal location of Sac and Soa. Genet Res 1995;66:167–74. [DOI] [PubMed] [Google Scholar]
- 55.Eylam S, Spector AC. Stimulus processing of glycine is dissociable from that of sucrose and glucose based on behaviorally measured taste signal detection in Sac ‘taster’ and ‘non-taster’ mice. Chem Senses 2004;29:639–49. [DOI] [PubMed] [Google Scholar]
- 56.Dotson CD, Spector AC. The relative affective potency of glycine, L-serine and sucrose as assessed by a brief-access taste test in inbred strains of mice. Chem Senses 2004;29:489–98. [DOI] [PubMed] [Google Scholar]
- 57.Ninomiya Y, Funakoshi M. Behavioural discrimination between glutamate and the four basic taste substances in mice. Comp Biochem Physiol A Comp Physiol 1989;92:371–6. [DOI] [PubMed] [Google Scholar]
- 58.Yamamoto T, Yuyama N, Kato T, Kawamura Y. Gustatory responses of cortical neurons in rats. III. Neural and behavioral measures compared. J Neurophysiol 1985;53:1370–86. [DOI] [PubMed] [Google Scholar]
- 59.Ninomiya Y, Funakoshi M. Peripheral neural basis for behavioural discrimination between glutamate and the four basic taste substances in mice. Comp Biochem Physiol A Comp Physiol 1989;92:371–6. [DOI] [PubMed] [Google Scholar]
- 60.Yamamoto T, Matsuo R, Fujimoto Y, Fukanaga I, Miyasaka A, Imoto T. Electrophysiological and behavioral studies on the taste of umami substances in the rat. Physiol Behav 1991;49:919–25. [DOI] [PubMed] [Google Scholar]
- 61.Chaudhari N, Yang H, Lamp C, Delay E, Cartford C, Than T, Roper S. The taste of monosodium glutamate: membrane receptors in taste buds. J Neurosci 1996;16:3817–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stapleton JR, Roper SD, Delay ER. The taste of monosodium glutamate (MSG), L-aspartic acid, and N-methyl-D-aspartate (NMDA) in rats: are NMDA receptors involved in MSG taste? Chem Senses 1999;24:449–57. [DOI] [PubMed] [Google Scholar]
- 63.Heyer BR, Taylor-Burds CC, Tran LH, Delay ER. Monosodium glutamate and sweet taste: generalization of conditioned taste aversion between glutamate and sweet stimuli in rats. Chem Senses 2003;28:631–41. [DOI] [PubMed] [Google Scholar]
- 64.Bachmanov AA, Li X, Reed DR, Ohmen JD, Li S, Chen Z, Tordoff MG, de Jong PJ, Wu C, West DB, et al. Positional cloning of the mouse saccharin preference (Sac) locus. Chem Senses 2001;26:925–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li X, Inoue M, Reed DR, Huque T, Puchalski RB, Tordoff MG, Ninomiya Y, Beauchamp GK, Bachmanov AA. High-resolution genetic mapping of the saccharin preference locus (Sac) and the putative sweet taste receptor (T1R1) gene (Gpr70) to mouse distal chromosome 4. Mamm Genome 2001;12:13–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bachmanov AA. Genetic approach to characterize interaction of sweeteners with sweet taste receptors in vivo. Chem Senses 2005;30(Suppl 1):i82–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Damak S, Rong M, Yasumatsu K, Kokrashvili Z, Varadarajan V, Zou S, Jiang P, Ninomiya Y, Margolskee RF. Detection of sweet and umami taste in the absence of taste receptor T1r3. Science 2003;301:850–3. [DOI] [PubMed] [Google Scholar]
- 68.Zhao GQ, Zhang Y, Hoon MA, Chandrashekar J, Erlenbach I, Ryba NJ, Zuker CS. The receptors for mammalian sweet and umami taste. Cell 2003;115:255–66. [DOI] [PubMed] [Google Scholar]
- 69.Kusuhara Y, Yoshida R, Ohkuri T, Yasumatsu K, Voigt A, Hubner S, Maeda K, Boehm U, Meyerhof W, Ninomiya Y. Taste responses in mice lacking taste receptor subunit T1R1. J Physiol 2013;591:1967–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li X, Staszewski L, Xu H, Durick K, Zoller M, Adler E. Human receptors for sweet and umami taste. Proc Natl Acad Sci USA 2002;99:4692–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nelson G, Chandrashekar J, Hoon MA, Feng L, Zhao G, Ryba NJ, Zuker CS. An amino-acid taste receptor. Nature 2002;416:199–202. [DOI] [PubMed] [Google Scholar]
- 72.Jiang P, Ji Q, Liu Z, Snyder LA, Benard LM, Margolskee RF, Max M. The cysteine-rich region of T1R3 determines responses to intensely sweet proteins. J Biol Chem 2004;279:45068–75. [DOI] [PubMed] [Google Scholar]
- 73.Jiang P, Cui M, Zhao B, Snyder LA, Benard LM, Osman R, Max M, Margolskee RF. Identification of the cyclamate interaction site within the transmembrane domain of the human sweet taste receptor subunit T1R3. J Biol Chem 2005;280:34296–305. [DOI] [PubMed] [Google Scholar]
- 74.Jiang P, Cui M, Zhao B, Liu Z, Snyder LA, Benard LM, Osman R, Margolskee RF, Max M. Lactisole interacts with the transmembrane domains of human T1R3 to inhibit sweet taste. J Biol Chem 2005;280:15238–46. [DOI] [PubMed] [Google Scholar]
- 75.Jiang P, Cui M, Ji Q, Snyder L, Liu Z, Benard L, Margolskee RF, Osman R, Max M. Molecular mechanisms of sweet receptor function. Chem Senses 2005;30(Suppl 1):i17–8. [DOI] [PubMed] [Google Scholar]
- 76.Bassoli A, Borgonovo G, Caremoli F, Mancuso G. The taste of D- and L-amino acids: In vitro binding assays with cloned human bitter (TAS2Rs) and sweet (TAS1R2/TAS1R3) receptors. Food Chem 2014;150:27–33. [DOI] [PubMed] [Google Scholar]
- 77.Nie Y, Vigues S, Hobbs JR, Conn GL, Munger SD. Distinct contributions of T1R2 and T1R3 taste receptor subunits to the detection of sweet stimuli. Curr Biol 2005;15:1948–52. [DOI] [PubMed] [Google Scholar]
- 78.Brand JG. Receptor and transduction processes for umami taste. J Nutr 2000;130(4S Suppl):942S–5S. [DOI] [PubMed] [Google Scholar]
- 79.Chaudhari N, Landin AM, Roper SD. A metabotropic glutamate receptor variant functions as a taste receptor. Nat Neurosci 2000;3:113–9. [DOI] [PubMed] [Google Scholar]
- 80.San Gabriel A, Uneyama H, Yoshie S, Torii K. Cloning and characterization of a novel mGluR1 variant from vallate papillae that functions as a receptor for L-glutamate stimuli. Chem Senses 2005;30(Suppl 1):i25–6. [DOI] [PubMed] [Google Scholar]
- 81.Maruyama Y, Pereira E, Margolskee RF, Chaudhari N, Roper SD. Umami responses in mouse taste cells indicate more than one receptor. J Neurosci 2006;26:2227–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yasuo T, Kusuhara Y, Yasumatsu K, Ninomiya Y. Multiple receptor systems for glutamate detection in the taste organ. Biol Pharm Bull 2008;31:1833–7. [DOI] [PubMed] [Google Scholar]
- 83.Chaudhari N, Maruyama Y, Roper S, Trubey K. Multiple pathways for signaling glutamate taste in rodents. Chem Senses 2005;30(Suppl 1):i29–30. [DOI] [PubMed] [Google Scholar]
- 84.Yee KK, Sukumaran SK, Kotha R, Gilbertson TA, Margolskee RF. Glucose transporters and ATP-gated K+ (KATP) metabolic sensors a re present in type 1 taste receptor 3 (T1r3)-expressing taste cells. Proc Natl Acad Sci USA 2011;108:5431–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Naim M, Nir S, Spielman AI, Noble AC, Peri I, Rodin S, Samuelov-Zubare M. Hypothesis of receptor-dependent and receptor-independent mechanisms for bitter and sweet taste transduction: implications for slow taste onset and lingering aftertaste. In: Given P, Parades D, editors. Chemistry of taste: mechanisms, behaviors, and mimics. ACS symposium series 825. Washington (DC): American Chemical Society; 2002. p. 2–17. [Google Scholar]
- 86.Peri I, Mamrud-Brains H, Rodin S, Krizhanovsky V, Shai Y, Nir S, Naim M. Rapid entry of bitter and sweet tastants into liposomes and taste cells: implications for signal transduction. Am J Physiol Cell Physiol 2000;278:C17–25. [DOI] [PubMed] [Google Scholar]
- 87.Zubare-Samuelov M, Shaul ME, Peri I, Aliluiko A, Tirosh O, Naim M. Inhibition of signal termination-related kinases by membrane-permeant bitter and sweet tastants: potential role in taste signal termination. Am J Physiol Cell Physiol 2005;289:C483–92. [DOI] [PubMed] [Google Scholar]
- 88.Li X, Bachmanov AA, Li S, Chen Z, Tordoff MG, Beauchamp GK, de Jong PJ, Wu C, Chen L, West DB, et al. Genetic, physical, and comparative map of the subtelomeric region of mouse chromosome 4. Mamm Genome 2002;13:5–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ninomiya Y, Sako N, Katsukawa H, Funakoshi M. Taste receptor mechanisms influenced by a gene on chromosome 4 in mice. In: Wysocki CJ, Kare MR, editors. Genetics of perception and communication. New York: Marcel Dekker; 1991. p. 267–78. [Google Scholar]
- 90.Ninomiya Y, Higashi T, Mizukoshi T, Funakoshi M. Genetics of the ability to perceive sweetness of d-phenylalanine in mice. Ann N Y Acad Sci 1987;510:527–9. [Google Scholar]
- 91.Ninomiya Y, Nomura T, Katsukawa H. Genetically variable taste sensitivity to D-amino acids in mice. Brain Res 1992;596:349–52. [DOI] [PubMed] [Google Scholar]
- 92.Shigemura N, Yasumatsu K, Yoshida R, Sako N, Katsukawa H, Nakashima K, Imoto T, Ninomiya Y. The role of the dpa locus in mice. Chem Senses 2005;30(Suppl 1):i84–5. [DOI] [PubMed] [Google Scholar]
- 93.Katsumori S, Inoue M, Kitamura A, Bosak N, Theodorides M, Golden GJ, Ohmoto M, Matsumoto I, Bachmanov AA. T1R3-independent sweet receptor mechanism involved in glycine preference. Japan J Taste Smell Res 2013;20:215–8. [Google Scholar]
- 94.Flynn FW, Ramos R. Bombesin suppresses need-free and need-induced salt intake in rats. Behav Neurosci 1994;108:780–8. [DOI] [PubMed] [Google Scholar]
- 95.Stellar E. Salt appetite: its neuroendocrine basis. Acta Neurobiol Exp (Warsz) 1993;53:475–84. [PubMed] [Google Scholar]