ABSTRACT
Polyhydroxybutyrate (PHB) is a carbon and energy reserve polymer in various prokaryotic species. We determined that, when grown with mannitol as the sole carbon source, Bradyrhizobium diazoefficiens produces a homopolymer composed only of 3-hydroxybutyrate units (PHB). Conditions of oxygen limitation (such as microoxia, oxic stationary phase, and bacteroids inside legume nodules) were permissive for the synthesis of PHB, which was observed as cytoplasmic granules. To study the regulation of PHB synthesis, we generated mutations in the regulator gene phaR and the phasin genes phaP1 and phaP4. Under permissive conditions, mutation of phaR impaired PHB accumulation, and a phaP1 phaP4 double mutant produced more PHB than the wild type, which was accumulated in a single, large cytoplasmic granule. Moreover, PhaR negatively regulated the expression of phaP1 and phaP4 as well as the expression of phaA1 and phaA2 (encoding a 3-ketoacyl coenzyme A [CoA] thiolases), phaC1 and phaC2 (encoding PHB synthases), and fixK2 (encoding a cyclic AMP receptor protein [CRP]/fumarate and nitrate reductase regulator [FNR]-type transcription factor of genes for microoxic lifestyle). In addition to the depressed PHB cycling, phaR mutants accumulated more extracellular polysaccharides and promoted higher plant shoot dry weight and competitiveness for nodulation than the wild type, in contrast to the phaC1 mutant strain, which is defective in PHB synthesis. These results suggest that phaR not only regulates PHB granule formation by controlling the expression of phasins and biosynthetic enzymes but also acts as a global regulator of excess carbon allocation and symbiosis by controlling fixK2.
IMPORTANCE In this work, we investigated the regulation of polyhydroxybutyrate synthesis in the soybean-nodulating bacterium Bradyrhizobium diazoefficiens and its influence in bacterial free-living and symbiotic lifestyles. We uncovered a new interplay between the synthesis of this carbon reserve polymer and the network responsible for microoxic metabolism through the interaction between the gene regulators phaR and fixK2. These results contribute to the understanding of the physiological conditions required for polyhydroxybutyrate biosynthesis. The interaction between these two main metabolic pathways is also reflected in the symbiotic phenotypes of soybeans inoculated with phaR mutants, which were more competitive for nodulation and enhanced dry matter production by the plants. Therefore, this knowledge may be applied to the development of superior strains to be used as improved inoculants for soybean crops.
INTRODUCTION
Bradyrhizobium diazoefficiens is an important soil bacterium that fixes atmospheric N2 in symbiosis with soybean plants, a key crop for food production worldwide (1). In addition to its agricultural relevance, B. diazoefficiens may be of industrial interest because it produces significant quantities of polyhydroxybutyrate (PHB), a polymer that accumulates as granules in the cytoplasm and has potential use as biodegradable plastic (2–6). PHB granules are synthesized as sinks of excess carbon and reducing power and are used as carbon and energy reserves when bacteria face starvation conditions (7). However, this cycle of synthesis and degradation must be regulated because if the two pathways occur simultaneously (8), the net result would be consumption of energy and reducing power. The proteins involved in the different steps of the PHB cycle are well-characterized (9–12), and all of them are present in B. diazoefficiens (3, 13, 14); however, regulation of the cycle in this bacterium was not yet studied. In other rhizobium species, such as Rhizobium etli and Ensifer meliloti, the gene product of phaR (PHA [polyhydroxyalkanoate] regulator, previously known as aniA for anaerobically induced gene A) was reported to control, at least in part, PHB synthesis (15, 16).
The regulatory circuit in which PhaR takes part was best studied in Ralstonia eutropha (17–19). In this bacterial species, PhaR binds to the promoter of its own gene and to the promoter of phaP, which encodes a protein that binds to the surface of PHB granules called phasins (PhaP). Phasins contain amphipathic α-helices whereby these proteins may coat the hydrophobic granules during their growth, exposing a polar surface to the bulk cytoplasm (20, 21). It is believed that, in this way, the phasins may control the final size of the PHB granules (22). In addition, PhaR also binds to the surface of PHB granules, although with less affinity than phasins (18). Thus, the model proposed for the regulation of PHB granule growth through PhaR/PhaP activities suggests that, while the granules are growing, their surface area is enough to accommodate PhaR and PhaP, thus maintaining a low cytoplasmic concentration of free PhaR, which keeps the phaR and phaP promoters free. As the granules reach a critical volume, their surface areas become limiting for PhaR and PhaP, which are continuously synthesized. At this point, PhaP displaces PhaR from the granule surface, raising the PhaR cytoplasmic concentration. The free PhaR binds phaR and phaP promoters, and the expression of the two genes is inhibited, arresting PHB granule growth (18, 23). However, a recent study reported that the association constant of PhaR to its target DNA sequence in the phaR promoter is similar to that of an unspecific DNA sequence (24).
In E. meliloti, the symbiont of alfalfa, PhaP and PhaR (AniA) activities are necessary for PHB synthesis (16, 25). In addition, phaR (aniA) may play a more ample regulatory role in the carbon flow in R. etli. A phaR::Tn5 mutant synthesized around 40% of the PHB level of the wild type and had significantly increased extracellular polysaccharide (EPS) levels, with extensive changes in its proteome (15). However, the regulatory circuit through which phaR may control EPS synthesis is unknown.
The genome of B. diazoefficiens USDA 110 harbors one copy of phaR (blr0227), which is located adjacent to PHB-related genes phaA2 (bll0226) and phaB2 (bll0225) but is transcribed in the opposite direction (see Fig. S1A in the supplemental material). Meanwhile, at least four paralogs of phaP occur as isolated genes at locations elsewhere in the genome. The expression of these genes depends on the culture conditions. In particular, conditions permissive for PHB synthesis, such as microoxia (26) and growth in yeast extract-mannitol (YM) (14), increase the expression of phaP1 and phaP4, the last being the paralog with the highest expression in YM. PhaR and the four PhaP paralogs bind to PHB in vitro (14), with PhaP4 being the phasin with the highest affinity. Further, an increasing concentration of PhaP4 triggered a competitive displacement of PhaR bound to PHB fine powder in suspension (14). These results suggested that PhaP4 may be the main phasin responsible for PhaP biological function in B. diazoefficiens. However, mutations of these genes were not obtained, and thus their implication in the regulation of PHB synthesis could not be directly established in vivo.
To better understand the regulation of PHB in B. diazoefficiens USDA 110, we constructed mutant strains in phaR and in two phaP genes and evaluated their roles in PHB and EPS syntheses as well as in the symbiotic interaction with soybean plants.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Strains and plasmids are summarized in Table S1 in the supplemental material. B. diazoefficiens was grown oxically or microoxically in Götz minimal medium with mannitol as the sole carbon source (27). For oxic growth, 50-ml cultures were grown in 250-ml Erlenmeyer flasks at 30°C with rotary shaking at 180 rpm in contact with the air. For microoxic growth, cells were inoculated into 500-ml serum bottles containing 35 ml of Götz medium to an optical density at 500 nm (OD500) of 0.03. The bottles were closed with rubber septa and maintained at 30°C and 60 rpm under microoxic conditions by exchange of gas phase (0.5% O2 and 99.5% N2) every 8 to 16 h until the desired growth state (26). Total biomass was estimated by OD500 and by the number of viable bacteria by CFU counts on YM agar plates (28). Conjugation was performed in a modified peptone salts-yeast extract medium (26). Escherichia coli was grown in Luria-Bertani medium (29). Antibiotics were added to the media at the following concentrations: 400 (B. diazoefficiens) or 100 (E. coli) μg ml−1 streptomycin (Sm); 200 (B. diazoefficiens) or 100 (E. coli) μg ml−1 spectinomycin (Sp); 150 (B. diazoefficiens) or 25 (E. coli) μg ml−1 kanamycin (Km); 200 μg ml−1 (E. coli) ampicillin (Ap); 100 (B. diazoefficiens) or 10 (E. coli) μg ml−1 gentamicin (Gm); 20 and 100 (B. diazoefficiens liquid and solid cultures, respectively) or 10 (E. coli) μg ml−1 tetracycline (Tc); and 20 μg ml−1 (B. diazoefficiens) chloramphenicol (Cm).
Genetic techniques and DNA manipulation.
Cloning procedures, including DNA isolation, restriction digestion, ligation, and transformation, were performed as described previously (29). Tri- or biparental matings were performed with E. coli DH5α or S17-1 strains as described previously (3). Electroporation was performed with a Gene Pulser system (Bio-Rad, Hercules, CA) at 1.5 V, 25 μF, and 200 Ω in a 0.1-cm-gap-width electroporation cuvette.
Oligonucleotide primers were purchased from Life Technologies (Buenos Aires, Argentina) or Sigma-Aldrich (Madrid, Spain). DNA amplification was performed by PCR using Taq DNA polymerase (Life Technologies, Buenos Aires, Argentina) for routine PCR or Kapa HiFi HotStart (HS) DNA polymerase (Kapa Biosystems, Woburn, MA) or Pfx (Life Technologies, Buenos Aires, Argentina) for the amplification of blunt-ended amplicons and/or targets longer than 1,000 bp. DNA sequencing was performed at Macrogen Corp. (Seoul, South Korea).
To construct the B. diazoefficiens phaR mutant, primers specific for the blr0227 locus tag were designed (see Table S2 in the supplemental material). An internal 244-bp PCR fragment was amplified with primers Fw1 and Rv1 and cloned into pGEM-T Easy, creating pIQ35. This plasmid was digested with EcoRI and ligated into pG18mob2, generating pIQ36, which was conjugated into B. diazoefficiens LP 3004 by biparental mating using E. coli S17-1 as the donor. Transconjugants were selected in Gm, Sm, and Cm. We obtained two independent clones by single homologous recombination of pIQ36 in the middle of the phaR coding sequence, leading a predicted truncated polypeptide of 108 amino acids (58% of the full-length protein sequence). The results obtained regarding growth, PHB synthesis, and extracellular polysaccharide production were similar in the two clones, and, therefore, we selected one of them that was designated LP 0227 (here referred to as the phaR mutant) (see Fig. S1A in the supplemental material).
For complementation, the complete sequence of phaR (amplified with primers Fw1c/Rv1c) was integrated into a replicative vector. Briefly, a 1,245-bp target sequence and its own putative promoter (inferred by the BPROM program) were amplified from B. diazoefficiens LP 3004 and cloned into the SmaI site of pBBR1-MCS-5 to create pIQ37. Then, pIQ37 was digested with XbaI and PstI, and the phaR-containing fragment was inserted in the replicative broad-host-range promoter-probe vector pCB303 (30), generating plasmid pIQ38. The construction was confirmed by sequencing. The plasmid was transferred into the LP 0227 strain by biparental mating, selected by growth on Tc, Gm, and Cm. The LP 0227 strain carrying pIQ38 turned blue in YM agar plates supplemented with X-phosphate, thus confirming that the 1,245-bp fragment, cloned in the same orientation as the promoterless phoA gene in pIQ38, carried both the phaR promoter and the phaR coding sequence.
To construct the B. diazoefficiens phasin mutants, primers specific to the bll5155 (phaP1) or bll7395 (phaP4) locus tags were designed (see Table S2 in the supplemental material). Fragments of the B. diazoefficiens USDA 110 genomic DNA were generated from the two sides of the target coding sequences. For the two phasins, the strategy was similar. An upstream fragment of 249/253 bp and a downstream fragment of 235/222 bp were amplified using primer pair Fw2/Rv2 or Fw3/Rv3 (fragments phaP1-1 and phaP4-1, respectively) and primer pair Fw4/Rv4 or Fw5/Rv5 (fragments phaP1-2 and phaP4-2, respectively). The PCR fragments phaP1-2 and phaP4-2 were cloned into the SmaI site of pBBR1-MCS-4, creating pIQ39 and pIQ40. These plasmids were then digested with EcoRV, and the fragments phaP1-1 and phaP4-1 were introduced, creating pIQ41 and pIQ42, respectively. These plasmids were linearized with EcoRI. In parallel, pHP45ΩSmSp and pHP45ΩKm were EcoRI-digested, and the resulting fragments ΩSm-Sp and ΩKm were cloned into pIQ41 and pIQ42, creating pIQ43 and pIQ44, respectively. Finally, these two plasmids were digested with XbaI and KpnI, and the fragments of pIQ43 and pIQ44 were ligated into XbaI/KpnI-digested pK18mob and pG18mob2, creating pIQ45 and pIQ46, respectively.
For phaP1, gene replacement was performed by introducing pIQ45 into B. diazoefficiens USDA 110 by biparental mating, selecting by growth on Sm-Sp and Cm, and screening for Km sensitivity. The resulting strain was designated LP 5155 (ΔphaP1); this strain carries the Ω-Sm-Sp interposon in place of the genomic DNA from base 5711611 to base 5711972, thus removing 301 bp from the 339-bp open reading frame (ORF) bll5155 (see Fig. S1B in the supplemental material). For phaP4, gene replacement was performed by introducing pIQ46 into B. diazoefficiens USDA 110 by triparental mating using the pRK2013 helper plasmid, selecting by growth on Km and Cm, and screening for Gm sensitivity. The resulting strain was designated LP 7395 (ΔphaP4); this strain carries the Ω-Km interposon in place of the genomic DNA from base 8130454 to base 8130880, thus removing 426 bp from the 435-bp ORF bll7395 (Fig. S1B). The double mutant LP 5173 (ΔphaP1 ΔphaP4) was constructed as described above, introducing pIQ45 in the background of LP 7395.
All of the mutations were confirmed by PCR using external primers (see Table S2 and Fig. S1 in the supplemental material) as described previously (3) and by DNA sequencing.
Bioinformatic characterization of PhaR and PhaP.
PhaR (blr0227), PhaP1 (bll5155), and PhaP4 (bll7395) were aligned with their putative homologs in E. meliloti 1021, R. etli CFN42, Azorhizobium caulinodans ORS571, and Rhizobium leguminosarum bv. viciae 3841 using T-Coffee (31). The secondary structures of PhaP1 and PhaP4 were predicted using Phyre2 alpha helix prediction (http://www.sbg.bio.ic.ac.uk/phyre2) and the PSIPRED v 3.3 server (http://bioinf.cs.ucl.ac.uk/psipred/).
RNA isolation, cDNA synthesis, and qRT-PCR analysis.
B. diazoefficiens LP 3004 and phaR mutant cultures were grown microoxically as described above. The cultures were grown to mid-exponential phase at OD500 values of 0.25 to 0.33 (phaR mutant) or 0.50 to 0.62 (LP 3004), stopped by adding cool stop solution (equilibrated phenol and ethanol at a ratio of 1:9), collected by centrifugation at 4°C, and frozen at −80°C as previously described (26, 32). This material was used for RNA isolation and cDNA synthesis as described elsewhere (33). Primers for quantitative reverse transcription-PCRs (qRT-PCRs) were designed (see Table S2 in the supplemental material) to have a melting temperature of 57°C to 62°C and to generate PCR products of 87 to 134 bp. Each PCR mixture contained 9.5 μl of iQ SYBR green supermix (Bio-Rad, Hercules, CA), 2 μM individual primers, and appropriate dilutions of different cDNAs in a total volume of 19 μl. Reactions were run in triplicate in a Bio-Rad iCycler. Melting curves were generated with Bio-Rad iQ5 software to verify the specificity of the amplification. Relative changes in gene expression (fold changes) were calculated as described elsewhere (34). The fold changes observed were considered significant when their absolute values were higher than 2.0. Expression of the primary sigma factor gene sigA was used as a reference for normalization.
Quantification and analysis of PHB and extracellular polysaccharides.
For quantification of PHB, B. diazoefficiens cultures were centrifuged twice at 12,000 × g for 40 min. Then, the pellets were homogenized overnight with sodium hypochlorite at room temperature, washed with double-distilled water, precipitated with 1:1 alcohol-acetone, and resuspended in chloroform. Then, PHB was determined as crotonic acid in H2SO4 (35). For analysis of composition, PHB was extracted from lyophilized B. diazoefficiens cells as described previously (36), and monomer composition was analyzed by gas chromatography (GC) after conversion of PHB to 3-hydroxybutyryl-methyl esters via acid methanolysis (36). Benzoate-methyl ester was used as an internal standard.
To determine the PHB content of bacteroid cells, 10 fresh mature nodules (70 to 90 mg) were washed, crushed and homogenized, suspended in 0.5 ml of double-distilled water, and centrifuged for 5 min at 500 × g (3). The supernatants were then processed for PHB determination as above.
Extracellular polysaccharides (EPSs) include exopolysaccharide and capsular polysaccharide, which have the same sugar composition in B. diazoefficiens USDA 110 (37). Exopolysaccharides were precipitated from the culture supernatants as described previously (38), and capsular polysaccharides were released from bacterial cells and precipitated as described previously (37). Finally, the polysaccharides were hydrolyzed, and the reducing sugars were quantified with anthrone as described previously (38).
TEM.
Transmission electron microscopy (TEM) was performed essentially as described previously (3, 38). Briefly, bacteria from liquid cultures or nodules (each one transversally cut in half) were collected and fixed in 2% (vol/vol) glutaraldehyde. Next, the samples were dehydrated, infiltrated with epoxy resin, sectioned, and stained. Glutaraldehyde-fixed samples were placed in grids and viewed at 80 kV in a JEM 1200 EX II transmission electron microscope (JEOL Ltd., Tokyo, Japan).
Some TEM micrographs of free-living cells were used to determine the diameters of PHB granules, using ImageJ software (v 1.49). The largest diameter of every individual granule was registered from each individual cell (n = 132 to 411).
Plant experiments.
DM 3810 soybean seeds, kindly provided by DonMario SA, Argentina, were surface sterilized, germinated, and cultivated in vermiculite pots as described previously (3). The stability of plasmid pIQ38 was verified by bacteroid isolation and CFU determination on plates containing Cm or Cm and Tc. Competition for nodulation was studied by inoculating LP 3004 and phaR mutant strains in a 1:1 proportion (as CFU ml−1) to soybean plants as described previously (39). Nodule occupation was evaluated on the basis of the antibiotic resistance of the strains recovered from the nodules 21 days after inoculation. Statistical analysis was carried out with the χ2 test, considering 45% nodule occupation by each strain alone and 10% double occupation as the null hypothesis (38).
Plant growth parameters were assessed in 67-day-old plants. Shoots from each plant were cut and dried in an oven at 60°C to a constant weight, and the materials from each plant were weighed individually. The statistical analysis was performed by one-way analysis of variance (ANOVA), with an α of <0.05. All results shown are representative of duplicate experiments.
RESULTS
B. diazoefficiens produces PHB homopolymer.
The polyhydroxyalkanoates extracted from the lyophilized cells of B. diazoefficiens USDA 110 (wild type), LP 3004 (wild-type, spontaneous Sm-resistant derivative of USDA 110), and phaR mutant strains from exponential-phase (7 days) and late-stationary-phase (25 days) cultures under oxic conditions (Fig. 1A) were characterized for composition of their monomer units by GC analysis. GC chromatograms from the two culture ages were similar, and those corresponding to the stationary phase are shown in Fig. S2 in the supplemental material. As observed, polyhydroxyalkanoates of the three strains tested possess only 3-hydroxybutyrate units. This finding allowed us to quantify the polyhydroxyalkanoate contents by the detection of crotonic acid at 235 nm because this specific analysis method applies only for poly(3-hydroxybutyrate) [poly(3HB)] (35).
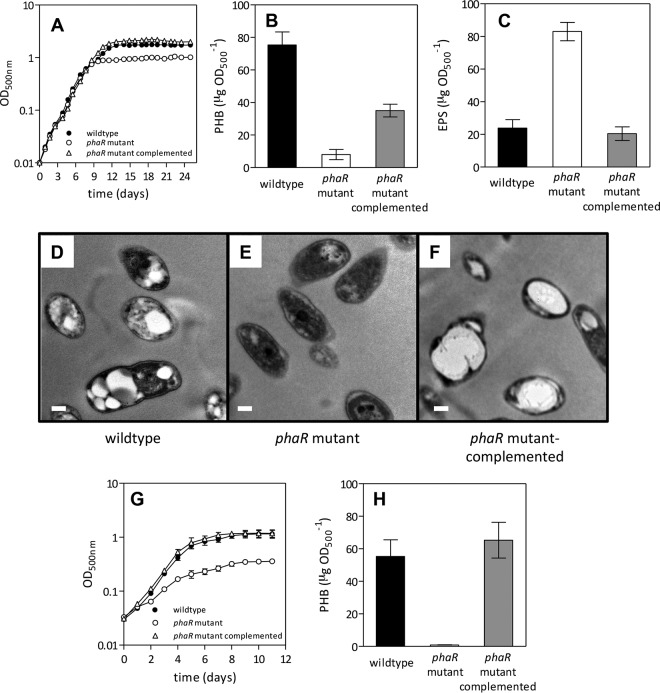
FIG 1.
Physiological characterization of the phaR mutant under oxic or microoxic conditions. (A to F) Oxic conditions, with rotary shaking at 180 rpm in contact with the air. (A) Growth kinetics. (B) PHB extracted from 25-day-old cells. (C) EPS obtained from 25-day-old cultures. (D to F) Transmission electron micrographs of cells from 25-day-old cultures showing bright PHB granules (scale bar, 0.2 μm). (G and H) Microoxic conditions, with rotary shaking at 60 rpm in contact with an atmosphere of 0.5% O2 and 99.5% N2. (G) Growth kinetics. (H) PHB extracted from 11-day-old cells. In all cases, quantitative results are the average ± standard error of the mean (SEM) from four independent cultures. When error bars are not visible, they are smaller than the symbol.
PHB and EPS syntheses are controlled by phaR under oxygen-limiting conditions.
We measured total biomass (as OD500) daily in oxic cultures for 25 days in the wild-type, phaR mutant, and mutant-complemented strains (Fig. 1A). There were no differences in exponential growth rates, but the stationary phase was reached earlier in the phaR mutant, resulting in reduced biomass with respect to the wild-type and mutant-complemented strains. Next, we measured PHB levels at 7 (logarithmic), 13 (early stationary phase), 17 (middle stationary phase), and 25 (late stationary phase) days of growth. The production of PHB at 7 days of growth was higher in the phaR mutant than in the wild type, but later, there was a strong increase in PHB production in the wild type at the early, middle, and late stationary phases that was not observed in the phaR mutant (see Fig. S3A in the supplemental material). At the late stationary phase, the mutant-complemented strain accumulated more PHB than the mutant but only half of the wild-type PHB levels (Fig. 1B). In addition, we measured EPS contents at the same growth states. While there were no differences between strains at the logarithmic and early and middle stationary phases (data not shown), at the late stationary phase the phaR mutant produced about 4-fold more EPS than the wild-type and phaR-complemented strains (Fig. 1C), which was also reflected macroscopically in the high mucoidy of the phaR mutant cell pellets after centrifugation (see Fig. S3B in the supplemental material). TEM of 25-day-old cells (Fig. 1D to F) showed that, while the phaR mutant cells were devoid of PHB granules, the wild-type and the phaR mutant-complemented strains had PHB granules (in the complemented strain, most cells had a single large granule).
In contrast, in microoxic cultures, the growth rate was lower and total biomass of the phaR mutant was smaller than those of the wild-type strain or the mutant-complemented strain from the beginning of the culture (Fig. 1G). Likewise, PHB production was affected in the mutant at the early stationary phase (11 days of growth) and was restored in the mutant-complemented strain (Fig. 1H). These results, in agreement with previous investigations (3, 27, 40), indicated that the microoxia and the stationary phase of oxic cultures were permissive conditions for PHB synthesis while the exponential phase of the oxic cultures was nonpermissive. In addition, the oxygen-limiting bacteroid state inside root nodules is also a permissive condition (3). Our results indicate that the influence of PhaR on PHB production is mainly exerted under these permissive conditions.
PhaR exerts a negative control on genes related to PHB and fixK2 expression.
The permissive conditions indicated above both involve oxygen limitation. Because oxic cultures of the phaR mutant at the late stationary phase produce too much EPS, which interferes with RNA extraction, we evaluated the role of PhaR as a transcriptional regulator in mid-exponential-phase microoxic cultures. We compared the levels of certain transcripts in the phaR mutant and the wild type by qRT-PCR as summarized in Fig. 2. The levels of most of the transcripts were lower in the wild type, indicating that phaR is a repressor, which is in agreement with previous reports (41). However, the mutation of phaR had no effect on its own transcription, which is in line with the low association constant of PhaR with respect to its target DNA sequence at the phaR promoter region described in R. eutropha H16 (24).
FIG 2.
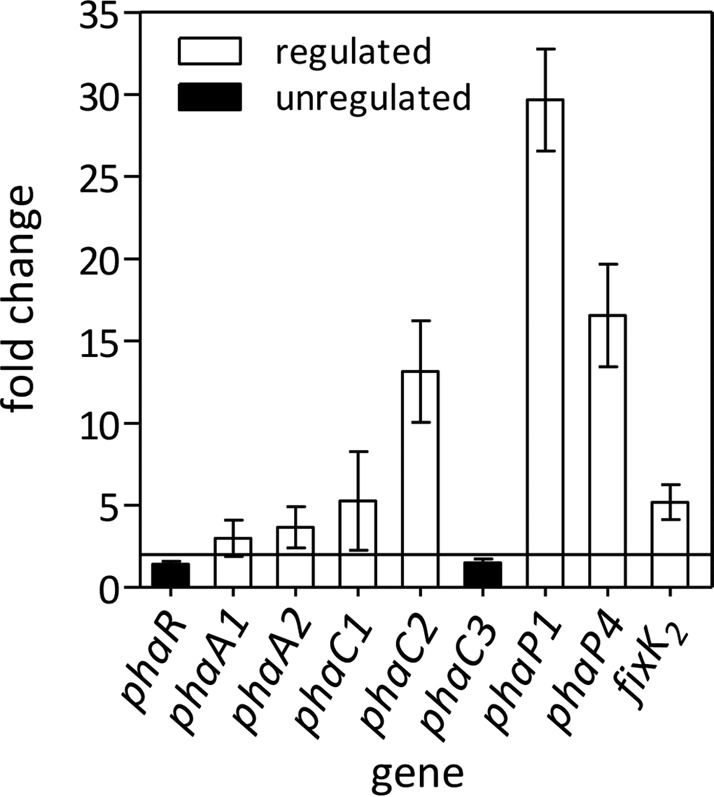
Differential expression of selected genes among the wild type and the phaR mutant, evaluated by qRT-PCR in cells grown microoxically. Fold change refers to the relative expression in the mutant compared to that in the wild type. An absolute value fold change of ≥2 (line of reference) was considered statistically significant. Results are the average from three runs ± SEM.
Phasin genes are known as direct targets of phaR. In B. diazoefficiens, there are at least four paralogs encoding phasins, of which phaP1 and phaP4 have the highest expression levels (14, 26) and PhaP4 has the highest affinity for PHB in vitro (14). Therefore, we assessed the expression of these two phasin genes and found that transcription of phaP1 and phaP4 was strongly derepressed in the phaR mutant, with phaP1 being the most affected.
In addition, we evaluated the transcript levels of genes encoding enzymes of the PHB biosynthetic pathway. Particularly, we assessed the 3-ketoacyl-coenzyme A (CoA) thiolase paralogs phaA1 and phaA2 and the PHB synthase paralogs phaC1, phaC2, and phaC3. Of these three PHB synthase-coding genes, only phaC1 is required (3). In turn, a deletion of phaC2 causes significant increases in PHB synthesis, while PhaC3 seems active only in a phaC2 mutant background (3). We observed that phaC2 was significantly (13-fold) upregulated, and transcription of phaC1 was increased 5-fold while transcription of phaC3 was not affected in the phaR mutant. The most upregulated gene in the mutant was phaC2, and interestingly, this gene was previously reported as positively controlled by the global regulator fixK2 (26). In the same line as the ΔphaC2 strain, a ΔfixK2 strain also accumulated more PHB than the wild type in the stationary phase (see Fig. S4 in the supplemental material). Therefore, we also measured the amounts of fixK2 transcripts and found that, as with the other genes, mutation of phaR resulted in a ∼6-fold upregulation of fixK2 transcription.
Phasin mutants showed altered patterns of PHB granule size and accumulation.
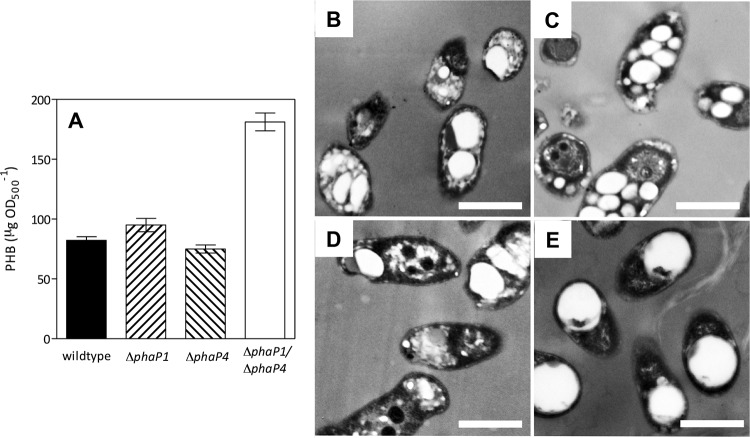
The secondary structure prediction analyses of PhaP1 and PhaP4 indicated high prevalences of alpha helix structures (91% and 86%, respectively, data not shown), which are compatible with known phasins. Moreover, growth kinetics of the ΔphaP1, ΔphaP4, and ΔphaP1 ΔphaP4 deletion mutants were similar to the wild type, indicating that none of the phasin genes are essential for growth in B. diazoefficiens. In addition, PHB accumulations in the single mutants were similar to that of the wild type at the middle stationary phase, but the ΔphaP1 ΔphaP4 strain accumulated ∼3 times more PHB (Fig. 3A). However, although the ΔphaP4 mutant produced PHB granules that did not differ significantly from those seen with the wild type (0.420 ± 0.134 μm and 0.521 ± 0.145 μm in diameter, respectively), the ΔphaP1 mutant produced smaller granules (0.304 ± 0.115 μm) but in greater quantity than the wild type (see Fig. S5 in the supplemental material). In addition, ΔphaP1 ΔphaP4 cells had large PHB granules (0.774 ± 0.140 μm) (Fig. 3B to D; see also Fig. S5 in the supplemental material) that occupied a considerable portion of the cytoplasm (Fig. 3E), similar to a phaP1 mutant in R. eutropha (23, 42). Thus, an influence of PhaP1 on granule surface/volume ratio as well as an interaction between PhaP1 and PhaP4 for controlling the growth of the granules may be appreciated.
FIG 3.
PHB accumulation in the wild type and three ΔphaP mutant derivatives, evaluated at 17 days under oxic conditions. (A) PHB extracted from cells, expressed as the average from four independent determinations ± SEM. Transmission electron micrographs of the wild-type (B), ΔphaP1 (C), ΔphaP4 (D), and ΔphaP1 ΔphaP4 (E) strains. Note the PHB granules as bright spots inside the cytoplasm. Scale bars, 0.5 μm.
The phaR mutant has a better symbiotic performance than the wild type.
Previous reports on the nodulation of phaR mutants are contradictory. On the one hand, the E. meliloti phaR mutant formed many pseudonodules in alfalfa that were negatively altered in N2 fixation (16); on the other hand, the R. etli phaR mutant produced macroscopically normal nodules in the common bean without significant differences in nitrogenase activity with respect to that of the wild type (15). However, nodulation differs among these host plants because alfalfa produces indeterminate nodules without PHB while the common bean produces determinate nodules, which have typical PHB granules in the bacteroidal cytoplasm. Since soybean plants produce determinate nodules, we wanted to know if our phaR mutant behaves similarly to R. etli for its symbiotic interaction with soybean roots.
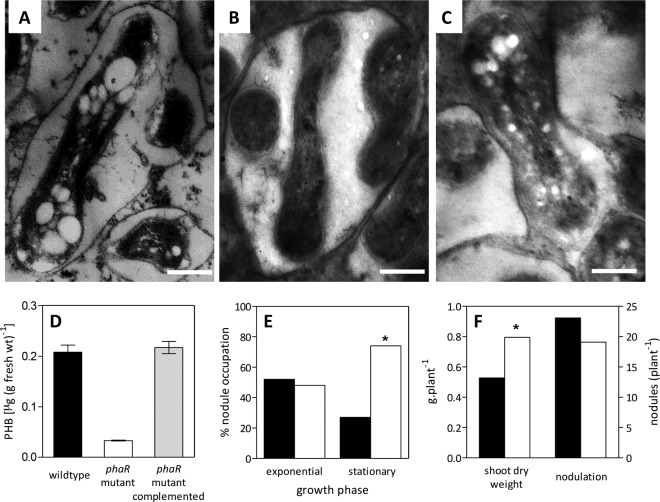
As in the common bean inoculated with R. etli, soybean plants inoculated with the B. diazoefficiens phaR mutant produced morphologically normal nodules. However, when nodules from 67-day-old plants were observed by TEM, it became evident that they were completely devoid of PHB granules (Fig. 4B). When this mutant was complemented with the wild-type phaR gene in trans, the production of PHB granules in soybean nodules was restored, indicating that the mutation in phaR was responsible for the lack of PHB granule production in the mutant bacteroids (Fig. 4C) as observed above under free-living conditions. Furthermore, the analytic determination of PHB levels from crushed nodules corroborated these observations (Fig. 4D). It is possible that the low PHB levels detected in nodules infected by the phaR strain (0.03 μg · mg nodule fresh weight−1) correspond to PHB granules from bacteria (not bacteroids) that remained in the infection threads.
FIG 4.
Effects of phaR on the symbiotic interaction with soybean plants. (A to C) Transmission electron micrographs of ultrathin cuts of 67-day-old soybean nodules showing bacteroids. (A) Wild type; (B) phaR mutant; (C) phaR mutant complemented. Note the PHB granules as bright spots in panels A and C. Scale bars, 0.5 μm. (D) Average PHB content (±SEM) from soybean nodules obtained from ten 67-day-old plants inoculated with the indicated strains. (E) Percentages of nodules occupied by the wild type (black bars) or the phaR mutant (white bars) inoculated in 1:1 mixtures on 10 soybean plants. Inoculants were prepared with cultures harvested at the physiological states indicated. (F) Shoot dry weight and nodulation of 10 soybean plants inoculated with the wild type (black bars) or the phaR mutant (white bars) after 67 days of growth. Values shown are the results determined by subtracting the value corresponding to the uninoculated control (shoot dry weight, 0.658 ± 0.09 g plant−1; nodules, 0). Asterisks in panels E and F indicate statistically significant differences (α < 0.05).
Competition for nodulation was evaluated by inoculating soybean plants with 1:1 mixtures of wild-type and phaR mutant strains, each one at a concentration on the order of 105 CFU ml−1. Because late-stationary-phase cultures are poor competitors (40) and we wanted to compare permissive and nonpermissive conditions, we prepared the inocula from oxic cultures either in the exponential phase (nonpermissive) or in the middle stationary phase (permissive). As a result, we observed in two independent experiments that under nonpermissive conditions the levels of competitiveness of the two strains were similar, while under permissive conditions the phaR mutant was more competitive than the wild type, occupying 74% of the nodules (Fig. 4E).
Despite differences in competitiveness, there were no differences in the numbers of nodules produced by the wild type and the phaR mutant (Fig. 4F). However, the shoot dry weights of 67-day-old plants cultured in nutrient solution without combined nitrogen were significantly higher in the plants inoculated with the phaR mutant (Fig. 4F). This result suggests that each nodule occupied by the phaR mutant was more active than those occupied by the wild type.
DISCUSSION
As observed in other bacterial species, phaR was necessary for the production of PHB in both microoxic and stationary-phase oxic cultures of B. diazoefficiens. In addition, mutation of phaR impaired biomass accumulation in stationary-phase oxic cultures and compromised the growth of microoxic cultures at all growth phases. These results indicate that PhaR exerts its activity under conditions of oxygen limitation, in which accumulation of the substrates for PHB synthesis, similarly to reducing power and acetyl-CoA levels, may take place. In addition, the lack of PHB granules in our phaR mutant was accompanied by a higher production of EPS, a correlation previously observed in E. meliloti (16), R. etli (15), and N-limited B. diazoefficiens wild-type cultures (27, 40). However, a plausible explanation of how phaR also controls EPS synthesis is lacking. Interestingly, PhaR also appeared as a repressor of fixK2, a global regulator which activates the expression of more than 200 genes needed for life under low-oxygen tension, including essential operons such as fixNOQP and fixGHIS (26, 43, 44). Because the expression of phaR is not affected in a ΔfixK2 strain grown under oxygen-limiting conditions (26), it seems that phaR is at the highest hierarchy of this regulatory circuit. Therefore, the regulation scope of PhaR may be wider than previously thought. In Fig. 5, we propose an extended model for the regulation of PHB synthesis, including the findings obtained in this work.
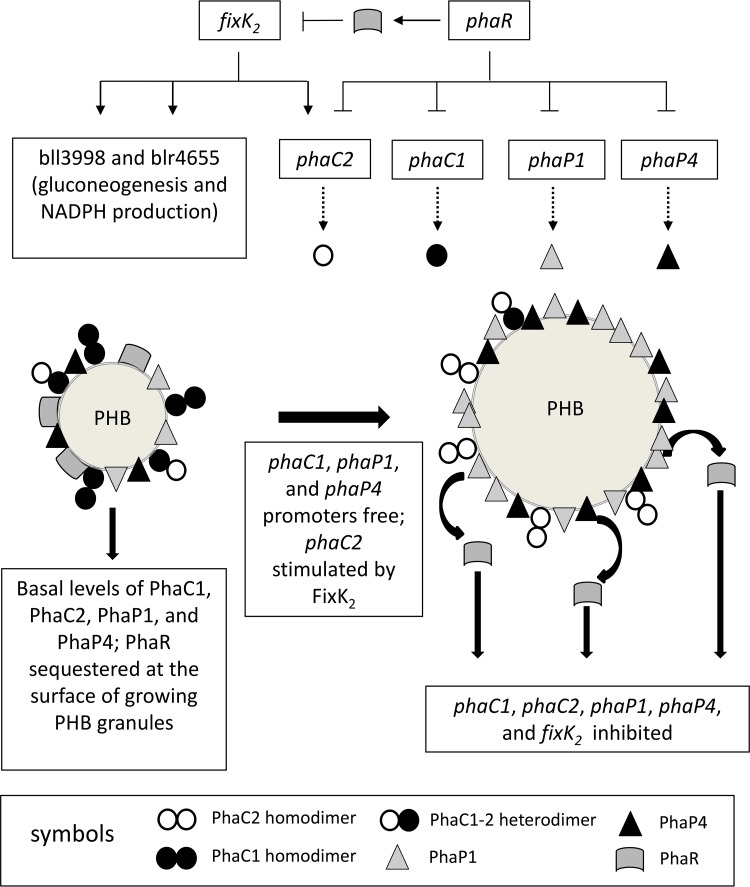
FIG 5.
Proposed model for the regulation of PHB synthesis in B. diazoefficiens, including the possible role of FixK2 (→, stimulation; ⫞, inhibition; solid lines indicate transcription; dashed lines depict translation). Permissive conditions, e.g., low O2, lead to an increase of NADPH and acetyl-CoA concentrations; thus, PHB biosynthesis starts using these metabolites as the substrates. This biosynthetic process is catalyzed initially by high levels of 3-ketoacyl-CoA thiolase, acetoacetyl-CoA reductase, and PHB synthase due to PhaR sequestration at the PHB granule surface. Meanwhile, phaC2 expression is stimulated by FixK2, increasing the relative PhaC2/PhaC1 concentration, which progressively decreases total PhaC activity during PHB granule growth. As granules grow, less PHB surface becomes available for PhaR and PhaP, which may provoke a competitive displacement of PhaR by PhaP from the surface that in turn may repress PhaR-controlled promoters. In the phaR mutant, these promoters are not repressed; therefore, PhaP1, PhaP4, and FixK2 levels are increased. Hence, FixK2 stimulates the expression of phaC2 and the biosynthesis of EPS. Altogether, these changes result in low levels of PHB in the phaR mutant. Moreover, in the ΔphaP1 ΔphaP4 strain, the very large PHB granule devoid of PhaP1 and PhaP4 may be able to allocate more PhaR than the wild type at the beginning of PHB biosynthesis. This PhaR sequestration may promote more PHB biosynthetic activity, leading to higher PHB accumulation before the PhaC2/PhaC1 ratio reaches its critical value and PhaR begins to accumulate in the cytoplasm. Protein symbols are indicated at the bottom of the figure. For more details, see the text.
At least three models may explain the initiation of PHB synthesis in R. eutropha (6), but it is unknown which of them is functional in B. diazoefficiens. Our results indicate that after initiation under permissive conditions, PHB synthesis would be catalyzed by basal levels of 3-ketoacyl-CoA thiolase, acetoacetyl-CoA reductase, and PHB synthase using microoxically accumulated reducing power and acetyl-CoA as the substrates. At this point, phaR may be expressed, but since the surface/volume ratio of the PHB granule is large, most of the PhaR produced should bind the PHB granule surface, leaving the phaC1, phaC2, phaP1, phaP4, and fixK2 promoters free. FixK2 stimulates the expression of phaC2, which encodes an inactive PHB synthase subunit that, by mass action, may impair the formation of PhaC1/PhaC1 fully active homodimers while increasing the concentration of PhaC1/PhaC2 less-active heterodimers and PhaC2/PhaC2 inactive homodimers (3, 14, 17, 45). Thus, the effect of the juxtaposition of phaR and fixK2 on phaC2 regulation may allow for a fine-tuning of the whole PHB synthase activity at the stage of PHB granule growth. Interestingly, fixK2 and phaC2 are also induced under conditions of oxidative burst, where an increase in EPS production was also observed (46). Therefore, we can speculate that PHB synthesis might be diminished under oxidative damage, perhaps by downregulation of phaR. In addition to PhaC2, the phasins PhaP1 and PhaP4 may play important roles in controlling the size of PHB granules. Although PhaP4 seems to be the main protein covering PHB granules (14), only the double mutant ΔphaP1 ΔphaP4 synthesized more PHB than the wild type. Moreover, when the two phasins were missing, the PHB granule surface/volume ratio decreased, and the bacteria possessed only one large granule per cell, resembling other bacterial phasin mutants (23, 47). Hence, in B. diazoefficiens, the combined function of PhaP1 and PhaP4 may prevent PHB granule coalescence, as reported in R. eutropha (42). This combined function suggests that in B. diazoefficiens, phasins may form trimers and/or tetramers as in other PHB-producing bacteria (48–50) as homo- and/or hetero-oligomers. In this way, the relative concentrations of PhaC1 and PhaC2 may modulate PHB synthase activity, while the production of PhaP1 and PhaP4 may control the number and size of PHB granules. As the granules reach their final sizes, PhaR would be released from their surface, exerting its inhibitory effect toward all of these PHB synthesis proteins. According to this model, the higher accumulation of PHB in the ΔphaP1 ΔphaP4 strain might be explained by a high PhaR sequestration at the PhaP1- and PhaP4-devoid PHB granule in formation, which might promote higher expression of PHB biosynthetic genes before the PhaC2/PhaC1 ratio reaches its critical value to inhibit PHB synthase activity and free PhaR begins to increase. The presence of one large granule per cell indicates that each granule grows rapidly within a single cell cycle.
In addition to its action upon phaC2 expression, FixK2 was reported to stimulate the expression of bll3998 (a succinate-semialdehyde dehydrogenase) and blr4655 (a phosphoenolpyruvate synthase) (26). The succinate-semialdehyde dehydrogenase, which catalyzes the oxidation of succinate-semialdehyde to succinate using NADP+ as an electron acceptor, was characterized in a sucA mutant of B. diazoefficiens as part of a bypass of the tricarboxylic acid (TCA) cycle (51). The phosphoenolpyruvate synthase is a key enzyme of gluconeogenesis and may participate in the secretion of excess carbon as EPS. Thus, induction of fixK2 under permissive conditions may increase the provision of NADPH to PHB synthesis while this pathway is active or may contribute to the switch of the carbon flow to gluconeogenesis when PHB synthesis is arrested by PhaP coverage of PHB granules and PhaC2 relative increased participation in the PHB synthase dimers (Fig. 5). This hypothesis might explain the inhibition of PHB synthesis and the increase in EPS content observed with the phaR mutants in E. meliloti (16), R. etli (15), and in this work. If phaR is knocked out, the expression of phaC2 and phaP becomes uncontrolled, and therefore, the accumulation of their products would lead to rapid inhibition of PHB synthesis and granule growth. In parallel, fixK2 expression is induced, which would stimulate the expression of the succinate-semialdehyde dehydrogenase and the phosphoenolpyruvate synthase. The precursors provided by the first enzyme could not be used for PHB synthesis because it is arrested and, as gluconeogenesis may be stimulated by the second enzyme, those precursors could be channeled to the synthesis and secretion of EPS.
In symbiosis, the phenotype observed here also indicated that phaR has a pleiotropic function. Previously, we observed that the lack of PHB in B. diazoefficiens ΔphaC1 strains led to decreased competitiveness for nodulation but had no effect on dry matter accumulation in plants cultured without combined nitrogen, indicating that N2 fixation was not affected (3). In contrast, here we observed an increase in competitiveness for nodulation of the phaR mutant grown under permissive conditions for PHB synthesis as well as an increase in dry matter accumulation in plants inoculated with the phaR mutant and cultured under N2-fixing conditions. Because the phaR mutant bacteroids also had no PHB granules in their cytoplasm, the comparison of the results obtained with phaC1 and phaR mutants indicated that the symbiotic phenotypes of the phaR mutants are not related only to PHB depletion. It is possible that in ΔphaC1 strains, the cytoplasmic levels of PhaR are high because of the absence of PHB granules, and in this way, the expression of the fixK2 regulon, which includes the regulators rpoN1, fixK1, and nnrR (26), might be repressed by PhaR. In contrast, derepression of fixK2 in the phaR mutants would favor the expression of these genes, and in particular, it might lead to higher N2 fixation via the expression of the alternative sigma factor RpoN1 (26). Nevertheless, we should not rule out an active role of EPS in the symbiotic performance of the phaR mutant due to its higher production in this strain.
Thus, the present results confirm the role of phaR as a global regulator in both free-living and symbiotic states, suggesting that this role may be exerted at two levels: directly on PHB synthesis and indirectly on other pathways through regulation of the transcription factor fixK2.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Paula Giménez, Silvana Tongiani, Abel Bortolameotti, and Jesús Chacón for technical assistance. We also thank Susana Jurado and Roxana Peralta (Servicio Central de Microscopía, Facultad de Ciencias Veterinarias, UNLP, La Plata, Argentina) for excellent assistance with electron microscopy.
This work was supported by ANPCyT, CONICET (Argentina), European Regional Development Fund (ERDF)/Ministerio de Economía y Competitividad (MINECO/Spain) grant AGL2011-23383), and the Deutsche Forschungsgemeinschaft (Germany) (grant Je 152/17-1).
Funding Statement
This work, including the efforts of Anibal Roberto Lodeiro, was funded by ANPCyT-CONICET (PICT-2013-2542). This work, including the efforts of Socorro Mesa, was funded by European Regional Development Fund (ERDF)/Ministerio de Economía y Competitividad (MINECO/Spain) (AGL2011-23383). This work, including the efforts of Dieter Jendrossek, was funded by Deutsche Forschungsgemeinschaft (DFG) (Je 152/17-1).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00757-16.
REFERENCES
- 1.Delamuta JR, Ribeiro RA, Ormeño-Orrillo E, Melo IS, Martínez-Romero E, Hungria M. 2013. Polyphasic evidence supporting the reclassification of Bradyrhizobium japonicum group Ia strains as Bradyrhizobium diazoefficiens sp. nov. Int J Syst Evol Microbiol 63:3342–3351. doi: 10.1099/ijs.0.049130-0. [DOI] [PubMed] [Google Scholar]
- 2.Madison LL, Huisman GW. 1999. Metabolic engineering of poly (3-hydroxyalkanoates): from DNA to plastic. Microbiol Mol Biol Rev 63:21–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quelas JI, Mongiardini EJ, Pérez Giménez J, Parisi G, Lodeiro AR. 2013. Analysis of two polyhydroxyalkanoate synthases in Bradyrhizobium japonicum USDA 110. J Bacteriol 195:3145–3155. doi: 10.1128/JB.02203-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khanna S, Srivastava AK. 2005. Recent advances in microbial polyhydroxyalkanoates. Process Biochem 40:607–619. doi: 10.1016/j.procbio.2004.01.053. [DOI] [Google Scholar]
- 5.Urtuvia V, Villegas P, González M, Seeger M. 2014. Bacterial production of the biodegradable plastics polyhydroxyalkanoates. Int J Biol Macromol 70:208–213. doi: 10.1016/j.ijbiomac.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 6.Jendrossek D, Pfeiffer D. 2014. New insights in the formation of polyhydroxyalkanoate granules (carbonosomes) and novel functions of poly(3-hydroxybutyrate). Environ Microbiol 16:2357–2373. doi: 10.1111/1462-2920.12356. [DOI] [PubMed] [Google Scholar]
- 7.Ratcliff WC, Kadam SV, Denison RF. 2008. Poly-3-hydroxybutyrate (PHB) supports survival and reproduction in starving rhizobia. FEMS Microbiol Ecol 65:391–399. doi: 10.1111/j.1574-6941.2008.00544.x. [DOI] [PubMed] [Google Scholar]
- 8.Taidi B, Mansfield D, Anderson AJ. 1995. Turnover of poly(3-hydroxybutyrate) (PHB) and its influence on the molecular mass of the polymer accumulated by Alcaligenes eutrophus during batch culture. FEMS Microbiol Lett 129:201–205. doi: 10.1016/0378-1097(95)00158-2. [DOI] [Google Scholar]
- 9.Pötter M, Steinbüchel A. 2005. Poly(3-hydroxybutyrate) granule-associated proteins: impacts on poly(3-hydroxybutyrate) synthesis and degradation. Biomacromolecules 6:552–560. doi: 10.1021/bm049401n. [DOI] [PubMed] [Google Scholar]
- 10.Uchino K, Saito T, Jendrossek D. 2008. Poly(3-hydroxybutyrate) (PHB) depolymerase PhaZa1 is involved in mobilization of accumulated PHB in Ralstonia eutropha H16. Appl Environ Microbiol 74:1058–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eggers J, Steinbüchel A. 2013. Poly(3-hydroxybutyrate) degradation in Ralstonia eutropha H16 is mediated stereoselectively to (S)-3-hydroxybutyryl coenzyme A (CoA) via crotonyl-CoA. J Bacteriol 195:3213–3223. doi: 10.1128/JB.00358-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sznajder A, Pfeiffer D, Jendrossek D. 2015. Comparative proteome analysis reveals four novel polyhydroxybutyrate (PHB) granule-associated proteins in Ralstonia eutropha H16. Appl Environ Microbiol 81:1847–1858. doi: 10.1128/AEM.03791-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aneja P, Dai M, Lacorre DA, Pillon B, Charles TC. 2004. Heterologous complementation of the exopolysaccharide synthesis and carbon utilization phenotypes of Sinorhizobium meliloti Rm1021 polyhydroxyalkanoate synthesis mutants. FEMS Microbiol Lett 239:277–283. doi: 10.1016/j.femsle.2004.08.045. [DOI] [PubMed] [Google Scholar]
- 14.Yoshida K, Takemoto Y, Sotsuka T, Tanaka K, Takenaka S. 2013. PhaP phasins play a principal role in poly-β-hydroxybutyrate accumulation in free-living Bradyrhizobium japonicum. BMC Microbiol 13:290. doi: 10.1186/1471-2180-13-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Encarnación S, del Carmen Vargas M, Dunn MF, Dávalos A, Mendoza G, Mora Y, Mora J. 2002. AniA regulates reserve polymer accumulation and global protein expression in Rhizobium etli. J Bacteriol 184:2287–2295. doi: 10.1128/JB.184.8.2287-2295.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Povolo S, Casella S. 2000. A critical role for aniA in energy-carbon flux and symbiotic nitrogen fixation in Sinorhizobium meliloti. Arch Microbiol 174:42–49. doi: 10.1007/s002030000171. [DOI] [PubMed] [Google Scholar]
- 17.Seo MC, Shin HD, Lee YH. 2004. Transcription level of granule-associated phaP and phaR genes and granular morphogenesis of poly-beta-hydroxyalkanoate granules in Ralstonia eutropha. Biotechnol Lett 26:617–622. doi: 10.1023/B:BILE.0000023018.00625.c4. [DOI] [PubMed] [Google Scholar]
- 18.Pötter M, Madkour MH, Mayer F, Steinbüchel A. 2002. Regulation of phasin expression and polyhydroxyalkanoate (PHA) granule formation in Ralstonia eutropha H16. Microbiology 148:2413–2426. doi: 10.1099/00221287-148-8-2413. [DOI] [PubMed] [Google Scholar]
- 19.York GM, Stubbe J, Sinskey AJ. 2002. The Ralstonia eutropha PhaR protein couples synthesis of the PhaP phasin to the presence of polyhydroxybutyrate in cells and promotes polyhydroxybutyrate production. J Bacteriol 184:59–66. doi: 10.1128/JB.184.1.59-66.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mezzina MP, Wetzler DE, Catone MV, Bucci H, Di Paola M, Pettinari MJ. 2014. A phasin with many faces: structural insights on PhaP from Azotobacter sp. FA8. PLoS One 9(7):e103012. doi: 10.1371/journal.pone.0103012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hauf W, Watzer B, Roos N, Klotz A, Forchhammer K. 2015. Photoautotrophic polyhydroxybutyrate granule formation is regulated by cyanobacterial phasin PhaP in Synechocystis sp. strain PCC 6803. Appl Environ Microbiol 81:4411–4422. doi: 10.1128/AEM.00604-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuchta K, Chi L, Fuchs H, Pötter M, Steinbüchel A. 2007. Studies on the influence of phasins on accumulation and degradation of PHB and nanostructure of PHB granules in Ralstonia eutropha H16. Biomacromolecules 8:657–662. doi: 10.1021/bm060912e. [DOI] [PubMed] [Google Scholar]
- 23.Pötter M, Müller H, Steinbüchel A. 2005. Influence of homologous phasins (PhaP) on PHB accumulation and regulation of their expression by the transcriptional repressor PhaR in Ralstonia eutropha H16. Microbiology 151:825–833. doi: 10.1099/mic.0.27613-0. [DOI] [PubMed] [Google Scholar]
- 24.Yamada M, Takahashi S, Okahata Y, Doi Y, Numata K. 2013. Monitoring and kinetic analysis of the molecular interactions by which a repressor protein, PhaR, binds to target DNAs and poly[(R)-3-hydroxybutyrate]. AMB Express 3:6. doi: 10.1186/2191-0855-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang C, Sheng X, Equi RC, Trainer MA, Charles TC, Sobral BW. 2007. Influence of the poly-3-hydroxybutyrate (PHB) granule-associated proteins (PhaP1 and PhaP2) on PHB accumulation and symbiotic nitrogen fixation in Sinorhizobium meliloti Rm1021. J Bacteriol 189:9050–9056. doi: 10.1128/JB.01190-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mesa S, Hauser F, Friberg M, Malaguti E, Fischer H-M, Hennecke H. 2008. Comprehensive assessment of the regulons controlled by the FixLJ-FixK2-FixK1 cascade in Bradyrhizobium japonicum. J Bacteriol 190:6568–6579. doi: 10.1128/JB.00748-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quelas JI, López-García SL, Casabuono A, Althabegoiti MJ, Mongiardini EJ, Pérez-Giménez J, Couto A, Lodeiro AR. 2006. Effects of N-starvation and C-source on Bradyrhizobium japonicum exopolysaccharide production and composition, and bacterial infectivity to soybean roots. Arch Microbiol 186:119–128. doi: 10.1007/s00203-006-0127-3. [DOI] [PubMed] [Google Scholar]
- 28.Vincent JM. 1970. A manual for the practical study of the root nodule bacteria. IBP handbook no. 15. Blackwell Scientific Publications, Oxford, United Kingdom. [Google Scholar]
- 29.Sambrook J, Russell D. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 30.Schneider K, Beck CF. 1987. New expression vectors for identifying and testing signal structures for initiation and termination of transcription. Methods Enzymol 153:452–461. doi: 10.1016/0076-6879(87)53071-3. [DOI] [PubMed] [Google Scholar]
- 31.Notredame C, Higgins DG, Heringa J. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 32.Hauser F, Pessi G, Friberg M, Weber C, Rusca N, Lindemann A, Fischer H-M, Hennecke H. 2007. Dissection of the Bradyrhizobium japonicum NifA+sigma54 regulon, and identification of a ferredoxin gene (fdxN) for symbiotic nitrogen fixation. Mol Genet Genomics 278:255–271. doi: 10.1007/s00438-007-0246-9. [DOI] [PubMed] [Google Scholar]
- 33.Torres MJ, Argandoña M, Vargas C, Bedmar EJ, Fischer H-M, Mesa S, Delgado MJ. 2014. The global response regulator RegR controls expression of denitrification genes in Bradyrhizobium japonicum. PLoS One 9(6):e99011. doi: 10.1371/journal.pone.0099011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nass/1.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Law JH, Slepecky RA. 1961. Assay of poly-hydroxybutyric acid. J Bacteriol 82:33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brandl H, Gross RA, Lenz RW, Fuller RC. 1988. Pseudomonas oleovorans as a source of poly(beta-hydroxyalkanoates) for potential applications as biodegradable polyesters. Appl Environ Microbiol 54:1977–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mort AJ, Bauer WD. 1980. Composition of the capsular and extracellular polysaccharides of Rhizobium japonicum. Changes with culture age and correlations with binding of soybean seed lectin to the bacteria. Plant Physiol 66:158–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quelas JI, Mongiardini EJ, Casabuono A, López-García SL, Althabegoiti MJ, Covelli JM, Pérez-Giménez J, Couto A, Lodeiro AR. 2010. Lack of galactose or galacturonic acid in Bradyrhizobium japonicum USDA 110 exopolysaccharide leads to different symbiotic responses in soybean. Mol Plant Microbe Interact 23:1592–1604. doi: 10.1094/MPMI-05-10-0122. [DOI] [PubMed] [Google Scholar]
- 39.Althabegoiti MJ, Covelli JM, Pérez-Giménez J, Quelas JI, Mongiardini EJ, López MF, López-García SL, Lodeiro AR. 2011. Analysis of the role of the two flagella of Bradyrhizobium japonicum in competition for nodulation of soybean. FEMS Microbiol Lett 319:133–139. doi: 10.1111/j.1574-6968.2011.02280.x. [DOI] [PubMed] [Google Scholar]
- 40.López-García S, Vázquez TEE, Favelukes G, Lodeiro A. 2001. Improved soybean root association of N-starved Bradyrhizobium japonicum. J Bacteriol 183:7241–7252. doi: 10.1128/JB.183.24.7241-7252.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maehara A, Taguchi S, Nishiyama T, Yamane T, Doi Y. 2002. A repressor protein, PhaR, regulates polyhydroxyalkanoate (PHA) synthesis via its direct interaction with PHA. J Bacteriol 184:3992–4002. doi: 10.1128/JB.184.14.3992-4002.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wieczorek R, Pries A, Steinbüchel A, Mayer F. 1995. Analysis of a 24-kilodalton protein associated with the polyhydroxyalkanoic acid granules in Alcaligenes eutrophus. J Bacteriol 177:2425–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mesa S, Reutimann L, Fischer H-M, Hennecke H. 2009. Posttranslational control of transcription factor FixK2, a key regulator for the Bradyrhizobium japonicum-soybean symbiosis. Proc Natl Acad Sci U S A 106:21860–21865. doi: 10.1073/pnas.0908097106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nellen-Anthamatten D, Rossi P, Preisig O, Kullik I, Babst M, Fischer H-M, Hennecke H. 1998. Bradyrhizobium japonicum FixK2, a crucial distributor in the FixLJ-dependent regulatory cascade for control of genes inducible by low oxygen levels. J Bacteriol 180:5251–5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jendrossek D. 2009. Polyhydroxyalkanoate granules are complex subcellular organelles (carbonosomes). J Bacteriol 191:3195–3202. doi: 10.1128/JB.01723-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Donati AJ, Jeon JM, Sangurdekar D, So JS, Chang WS. 2011. Genome-wide transcriptional and physiological responses of Bradyrhizobium japonicum to paraquat-mediated oxidative stress. Appl Environ Microbiol 77:3633–3643. doi: 10.1128/AEM.00047-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cai S, Cai L, Liu H, Liu X, Han J, Zhou J, Xiang H. 2012. Identification of the haloarchaeal phasin (PhaP) that functions in polyhydroxyalkanoate accumulation and granule formation in Haloferax mediterranei. Appl Environ Microbiol 78:1946–1952. doi: 10.1128/AEM.07114-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Neumann L, Spinozzi F, Sinibaldi R, Rustichelli F, Pötter M, Steinbüchel A. 2008. Binding of the major phasin, PhaP1, from Ralstonia eutropha H16 to poly(3-hydroxybutyrate) granules. J Bacteriol 190:2911–2919. doi: 10.1128/JB.01486-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao M, Li Z, Zheng W, Lou Z, Chen GQ. 2006. Crystallization and initial X-ray analysis of polyhydroxyalkanoate granule-associated protein from Aeromonas hydrophila. Acta Crystallogr Sect F Struct Biol Cryst Commun 62:814–819. doi: 10.1107/S1744309106025000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pfeiffer D, Jendrossek D. 2011. Interaction between poly(3-hydroxybutyrate) granule-associated proteins as revealed by two-hybrid analysis and identification of a new phasin in Ralstonia eutropha H16. Microbiology 157:2795–2807. doi: 10.1099/mic.0.051508-0. [DOI] [PubMed] [Google Scholar]
- 51.Green LS, Li Y, Emerich DW, Bergersen FJ, Day DA. 2000. Catabolism of alpha-ketoglutarate by a sucA mutant of Bradyrhizobium japonicum: evidence for an alternative tricarboxylic acid cycle. J Bacteriol 182:2838–2844. doi: 10.1128/JB.182.10.2838-2844.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.